Abstract
Correct measurement of environmental parameters is fundamental for plant fitness and survival, as well as for timing developmental transitions, including the switch from vegetative to reproductive growth. Important parameters that affect flowering time include day length (photoperiod) and temperature. Their response pathways have been best described in Arabidopsis, which currently offers a detailed conceptual framework and serves as a comparison for other species. Rice, the focus of this review, also possesses a photoperiodic flowering pathway, but 150 million years of divergent evolution in very different environments have diversified its molecular architecture. The ambient temperature perception pathway is strongly intertwined with the photoperiod pathway and essentially converges on the same genes to modify flowering time. When observing network topologies, it is evident that the rice flowering network is centered on EARLY HEADING DATE 1, a rice-specific transcriptional regulator. Here, we summarize the most important features of the rice photoperiodic flowering network, with an emphasis on its uniqueness, and discuss its connections with hormonal, temperature perception, and stress pathways.
Key words: rice, photoperiod, temperature, flowering, stress, florigens
Rice flowering time depends on external environmental parameters, among which photoperiod is the most important. Temperature variations, hormonal balance, and occasional stress conditions also modify normal flowering patterns by integrating into the molecular network of regulatory genes.
Distinctive features of the rice photoperiodic flowering pathway
Flowering time is a key adaptive trait that allows plants to synchronize reproduction with the most favorable environmental conditions. Seasonal changes in day length (photoperiod) follow a sinusoidal curve whose amplitude varies with latitude but, at any given location, is invariant from one year to another. Thus, photoperiod variations offer very stable and measurable parameters to anchor plant reproduction to a specific time of the year, and plant species can be categorized according to the photoperiodic regime required to promote flowering. Short-day (SD) plants flower when day length falls below a critical threshold, long-day (LD) plants flower when day length exceeds a critical threshold, and day-neutral plants do not use photoperiodic cues to time reproduction.
Rice is a facultative SD plant that flowers faster if exposed to day lengths shorter than 13.5 h; it can also flower under LD conditions, although this takes more time (Itoh et al., 2010). Genetic mapping enabled the isolation of several flowering-time genes, starting with HEADING DATE 1 (Hd1) which belongs to the CCT family of transcriptional regulators (Yano et al., 2000). Hd1 shows high sequence similarity to CONSTANS (CO), a flowering promoter central to the photoperiodic pathway of Arabidopsis. This feature suggested the existence of an evolutionarily shared flowering network common to monocots and dicots. In Arabidopsis, CO transcription is controlled by the circadian clock through GIGANTEA (GI), and CO is required to activate transcription of FLOWERING LOCUS T (FT), which encodes a mobile florigenic protein (Andrés and Coupland, 2012). A similar network arrangement was also demonstrated in rice, where OsGI promotes Hd1 expression, which in turn promotes transcription of the FT homolog Hd3a under inductive photoperiodic conditions (Hayama et al., 2003). The strong homology between genes and their similar arrangement in gene regulatory networks (GRNs) further supported the idea of a conserved architecture. However, as more genes were cloned, it became evident not only that rice-specific regulators existed but also that genes homologous to Arabidopsis flowering regulators were arranged differently within the flowering network. Therefore, we wish to revisit the concept of a shared network and suggest that a strict comparison to Arabidopsis is misleading.
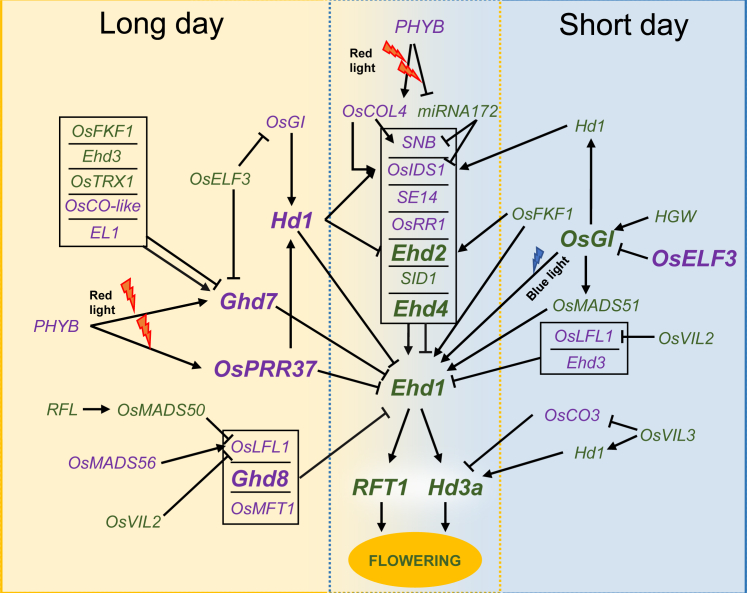
The EARLY HEADING DATE 1 (Ehd1) B-type response regulator was the first rice-specific promoter of flowering to be isolated. Ehd1 induces expression of Hd3a and RICE FT1 (RFT1) florigens under both LDs and SDs (Doi et al., 2004; Zhao et al., 2015). This gene occupies a central position in the network, operating as a hub that integrates signals mediated by several genes (Figure 1). All major flowering-time regulators cloned after Hd1, including GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (Ghd7, also known as Hd4), Ghd8 (also known as Hd5 or DTH8), PSEUDO RESPONSE REGULATOR 37 (PRR37, also known as Hd2, DTH7, or Ghd7.1), and RICE INDETERMINATE 1 (RID1, also known as Ehd2 or OsID1), encode strong repressors of Ehd1 that reduce its transcription under LDs (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008, 2013; Xue et al., 2008; Wei et al., 2010; Koo et al., 2013). As a result of this arrangement, and unlike flowering-time regulation in Arabidopsis, LD regulation in rice is characterized by active repression of florigen expression, with induction of flowering taking place under SD only when transcriptional blocks are released. Hd1 itself is an LD repressor of Ehd1, suggesting that the OsGI–Hd1 module evolved in connection with, and not in parallel to, Ehd1-mediated regulation (Gómez-Ariza et al., 2015; Nemoto et al., 2016). Almost all regulators of flowering cloned to date either activate or repress Ehd1 (Figure 1). An additional list of genes not discussed in the main text is provided in Supplemental Table 1.
Figure 1.
Gene regulatory networks controlling rice photoperiodic flowering.
The networks represent the transcriptional relationships that take place under LDs and SDs. Regulatory signals ultimately converge on Ehd1 and florigen transcription. Genes indicated in purple act as flowering inhibitors, and those in green act as promoters. Genes indicated in bold have a stronger impact on flowering time, as inferred from the effects of their corresponding loss-of-function mutants. Some positive and negative regulators of Ehd1 and Ghd7 have been grouped in boxes to simplify graphical representation. Arrows and flat-ended arrows indicate transcriptional activation and repression, respectively. The interaction of light with gene expression is indicated by lightning symbols.
A second aspect that distinguishes rice from Arabidopsis stems from interpretation of the connections between photoperiod measurement and flowering-time control. This relationship is summarized by the external coincidence model of photoperiodism, which postulates that flowering is induced when a sensitive phase of expression of a circadian-regulated factor coincides with a favorable environmental input (Thomas and Vince-Prue, 1997).
In Arabidopsis, CO is central to this model. Its expression is controlled by the circadian clock that induces a peak of transcription at the end of the light period only under LDs. The presence of light during this phase of the cycle leads to CO protein stabilization and accumulation, FT induction, and flowering (Valverde et al., 2004; Song et al., 2012). Under SDs, peak expression occurs during the night, preventing accumulation of the CO protein. By contrast, Hd1 is not as central to external coincidence because Hd1 protein abundance follows gene transcription and is not modified by changes in day length or the presence of light (Yang et al., 2015). Therefore, Hd1 protein accumulation does not predict LD and SD flowering behaviors, although it remains possible that post-translational modifications affect protein activity, but not abundance, in a day-length-dependent manner (Ishikawa et al., 2011).
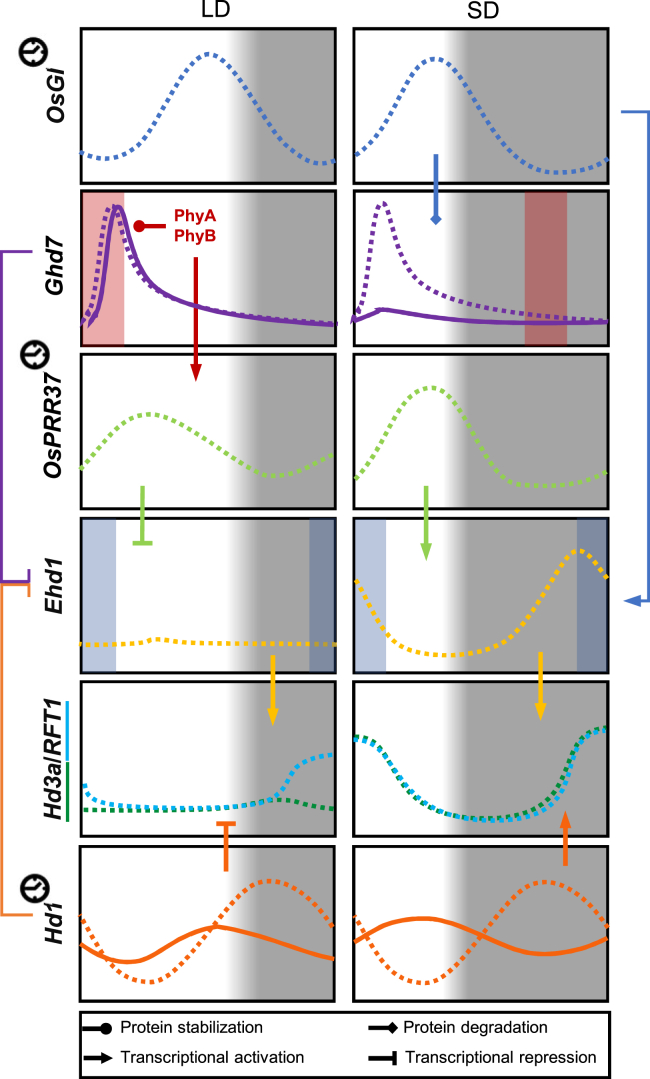
The mRNA and protein accumulation profiles of several flowering regulators, depending on the photoperiod, suggest that Ghd7 might be key to interpreting external coincidence in rice (Figure 2). Transcription of Ghd7 is promoted by red light and gated in the morning under LDs. Its cognate protein accumulates to reduce Ehd1 expression and delay flowering (Itoh et al., 2010; Zheng et al., 2019). Under SDs, Ghd7 transcription is reduced, and its gate of inducibility shifts toward the night. With the reduction in Ghd7 expression, Ehd1 repression is relaxed, and a gate for its induction opens during the morning in response to blue light signals mediated by OsGI (Itoh et al., 2010). Most importantly, stability of the Ghd7 protein depends on the photoperiod, and Ghd7 does not accumulate under SDs, even if overexpressed (Zheng et al., 2019). Ghd7 stability is influenced by direct interaction with OsGI that promotes its degradation in a proteasome-dependent manner. Conversely, phytochromes have a positive effect on Ghd7 stability, and mutants of PHYTOCHROME B (PhyB) or PHOTOPERIODIC SENSITIVITY 5, which encodes a plastid heme oxygenase essential for biosynthesis of the chromophore of phytochromes, never accumulate Ghd7 (Izawa et al., 2000; Andrés et al., 2009; Osugi et al., 2011; Weng et al., 2014; Zheng et al., 2019). Therefore, the antagonistic activities of OsGI and phytochromes shape the diurnal accumulation pattern of Ghd7, both transcriptionally and post-transcriptionally, and Ghd7 accumulation patterns discriminate between LD and SD. Thus, a plausible interpretation of external coincidence in rice suggests that it releases LD repression by preventing accumulation of Ghd7. Red and blue light signals have antagonistic effects on the flowering network, both of them converging on Ehd1 transcription, a behavior substantially different from that of LD species such as Arabidopsis (Figure 2). Post-translational regulation of other important components of the flowering network remains to be evaluated before a final model can be defined.
Figure 2.
Diurnal accumulation patterns of major flowering regulators under LD (boxes on the left) and SD (boxes on the right) show the central position of Ghd7 in the external coincidence model of rice flowering.
The peak of GI transcription tracks dusk under LDs and SDs. The GI protein interacts with Ghd7 and contributes to its degradation in a 26S proteasome–dependent manner. Transcription of Ghd7 is sensitive to red light, with a gate of inducibility (red shading) that occurs during the morning under LDs. The gate shifts to the night under SDs, and although few publications have reported reduced transcription under SDs, a larger consensus indicates a transcriptional peak in the morning, not different from the one detected under LDs. Irrespective of transcription, the Ghd7 protein does not accumulate under SDs or in phyB mutants, but it does show reduced accumulation in GI overexpressors. Thus, light- and photoperiod-dependent regulatory layers determine Ghd7 abundance. Ehd1 expression is gated in the morning by blue light signals (blue shading). OsGI can induce Ehd1 transcription under SDs when not antagonized by the Ghd7 protein. The diurnal profile of Ehd1 transcription is also determined by Hd1 and PRR37, which promote its expression under SDs and repress it under LDs. Finally, Hd3a and RFT1 are transcribed under SDs by a combination of Hd1- and Ehd1-mediated induction. Under LDs, florigen expression is repressed by Hd1, and induction by Ehd1 is limited. Eventually, RFT1 escapes repression under LDs and is transcribed to promote flowering. Solid and dashed lines indicate protein and mRNA accumulation patterns, respectively. A clock symbol indicates that the gene is under circadian clock control.
A different perspective relates to the evolutionary interpretation of CO/Hd1 functions, arguing in favor of their different origins (Ballerini and Kramer, 2011). The CO gene originated from a tandem duplication of COL1 and evolved a transcriptional pattern and protein features that made it a key photoperiod sensor. Its appearance can be traced to the common ancestor of the Brassicaceae, in which it connected transcriptionally to FT and LD flowering induction arose (Simon et al., 2015). Thus, the CO function is a recent acquisition that occurred long after the split between monocots and dicots. Hd1 was likely recruited independently to regulate photoperiodic flowering in rice, and the similar network arrangement is most probably the result of convergent evolution. In fact, it is possible that CCT domain proteins are particularly suited to controlling florigen expression and flowering. Not surprisingly, Ghd7 and PRR37 encode CCT domain proteins that are as central as Hd1 to rice flowering regulation (Xue et al., 2008; Koo et al., 2013; Gao et al., 2014; Nemoto et al., 2016).
Finally, a distinctive feature of rice is its evolution of a double florigen system that is essential for flowering under any photoperiod (Komiya et al., 2008, 2009). Florigen induction is not dependent only upon SD, and RFT1 expression can also be promoted under LD (Komiya et al., 2009). This flexibility in florigen expression allows rice to use both Hd3a and RFT1 in different environments and latitudes (Wang et al., 2021a). A major example of such flexibility is the relaxation of day-length dependency that occurs at higher latitudes and is enhanced by artificial selection, enabling expansion of the species and its cultivation area (Takahashi et al., 2009; Goretti et al., 2017). Thus, photoperiodic induction of florigens is fundamental to both SD and LD flowering, and, in contrast to Arabidopsis, no other florigen-independent flowering-time pathway has been described in rice to date.
Post-translational aspects of flowering-time control
Formation of higher-order complexes
Recent studies have shed light on higher levels of coordination among components of the photoperiod pathway, dependent upon combinations of protein–protein interactions, higher-order complex formation, and post-translational modifications. This level tunes network outputs by interacting with light quantity and quality signals from the environment.
From this perspective, the Hd1 protein has been most widely studied, owing to its strong effects on flowering and because of its homology to CO, which is subject to several levels of post-transcriptional and post-translational regulation (Jang et al., 2008; Song et al., 2012, 2014; Sarid-Krebs et al., 2015; Graeff et al., 2016).
The CCT domain of Hd1 is localized at the C terminus and is necessary for DNA binding and protein–protein interactions, whereas the N terminus contains two B-boxes required for protein–protein interactions and transcriptional regulation (Gangappa and Botto, 2014).
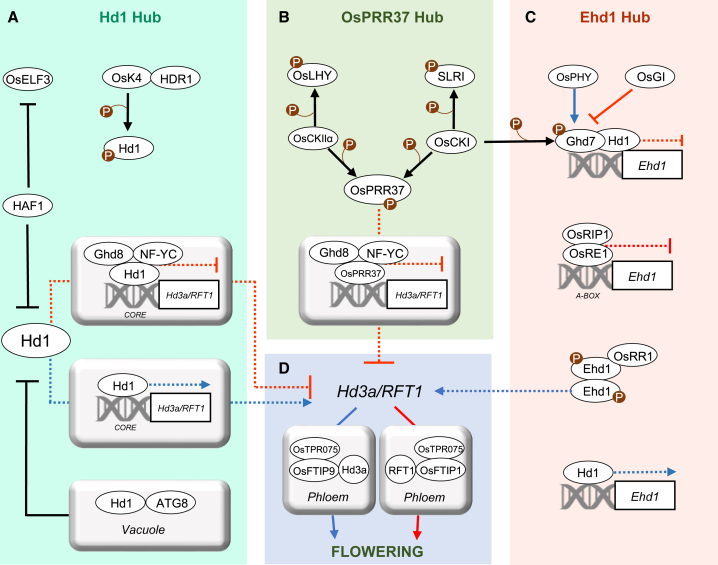
The molecular activity of Hd1 can be explained by its ability to form complexes with other nuclear proteins. Transcriptional repression activity under LDs is dependent upon assembly of NUCLEAR TRANSCRIPTION FACTOR Y (NF-Y) heterotrimeric complexes formed by Hd1, NF-YB, and NF-YC. The latter subunits encode histone-like proteins that, upon dimerization, construct a histone-fold domain scaffold that exhibits affinity for DNA in a non-sequence-specific manner. The third element of the trimer confers sequence specificity to DNA binding. The Hd1/NF-Y complex directly binds the Hd3a promoter, recognizing TGTGG sequences called CO-responsive elements (COREs) because they were identified in Arabidopsis as being recognized by CO and present in the FT promoter (Adrian et al., 2010; Tiwari et al., 2010; Gnesutta et al., 2017; Goretti et al., 2017; Shen et al., 2020; Lv et al., 2021). Structural studies have since determined the precise conformation of the Hd1/NF-Y and CO/NF-Y heterotrimers bound to DNA, corroborating previous observations (Shen et al., 2020; Lv et al., 2021). The CO/NF-Y structure suggests a certain degree of flexibility in DNA binding. Specifically, only a TGTG sequence is strictly necessary for protein binding in the TGTGG CORE of Arabidopsis, whereas the last base does not affect DNA recognition (Lv et al., 2021). If the same feature were demonstrated for the Hd1/NF-Y heterotrimer, its potential DNA-binding sites would expand. However, all DNA interaction studies have been performed at florigen loci. The full repertoire of Hd1 or CO binding sites on a genome-wide scale in vivo would help to better define DNA-binding properties and possibly identify novel target genes (Figure 3).
Figure 3.
Post-transcriptional levels of regulation in the flowering-time network.
We identified four hubs corresponding to Hd1, PRR37, Ehd1, and the florigens.
(A) Hd1 hub. Hd1 forms Hd1/NF-Y complexes that directly repress expression of florigens under LDs. Repression is released in SDs, and Hd1 becomes an activator. Hd1 stability depends on HAF1 and on components of the autophagy pathway, including ATG proteins, in the vacuole. Hd1 can be phosphorylated by OsK4, and this modification might impact Hd1 stability.
(B) OsPRR37 hub. OsPRR37 can replace Hd1 in an NF-Y complex and repress florigen expression under LDs. It can be phosphorylated by CKI and CKIIα. Phosphorylation might affect PRR37 stability or activity.
(C) Ehd1 hub. Ehd1 is repressed under LDs by the Ghd7/Hd1 and OsRE1/OsRIP1 complexes. Phosphorylation is essential for Ehd1 dimerization and activity. OsRR1 interacts with Ehd1 to form an inactive complex and inhibit its capacity to induce expression of the florigens. Phosphorylation of Ghd7 by CKI enhances its repressor activity.
(D) Florigen hub. Activity of the florigens depends on their transport in the phloem, which takes place by physical interaction with OsFTIP proteins and OsTPR075. Proteins are indicated by ovals and genes by rectangles. Names of DNA motifs bound by proteins or protein complexes are indicated below the double helix. Red and blue arrows indicate LD and SD regulation, respectively. Dashed arrows/flat-ended arrows indicate transcriptional activation/repression. Continuous arrows +P indicate phosphorylation. Continuous flat-ended arrows indicate protein degradation.
The NF-YB/C dimer can also accommodate NF-YA subunits, as well as other CCT domain proteins, including PRR37, PRR73, and Ghd7, thus expanding DNA accessibility through variation in motif recognition. Although NF-YA/B/C heterotrimers invariably recognize CCAAT box elements, how DNA-binding specificity would change with incorporation of PRR37, PRR73, and Ghd7 remains to be experimentally assessed (Gnesutta et al., 2018; Shen et al., 2020; Liang et al., 2021).
An additional element of complexity involves the expansion of gene families. The rice genome encodes 10 NF-YA, 11 NF-YB, and seven NF-YC genes (Petroni et al., 2012). The combinatorial assembly of their cognate proteins and their tissue specificity confer substantial transcriptional plasticity to the putative complexes, a feature shared with Arabidopsis (Kumimoto et al., 2008, 2010; Thirumurugan et al., 2008).
The Hd1 protein can heterodimerize with Ghd7 to repress Ehd1 expression, indicating the possibility of interaction between CCT domain proteins (Nemoto et al., 2016; Zhang et al., 2017). Whether these interactions take place in vivo between Hd1/NF-Y and Ghd7/NF-Y complexes, or between individual Hd1 and Ghd7, is unclear. However, biochemical characterization of CO/NF-Y suggests the possibility of multimerization between ternary complexes. Chromatographic studies indicate multiple oligomeric states for CO in vitro, with the most probable being trimeric or tetrameric assemblies (Lv et al., 2021). When the FT promoter region containing the COREs was incubated with CO/NF-Y in electrophoretic mobility shift assays, multivalent binding was observed, and three out of four COREs could be occupied simultaneously. These data raise the very interesting possibility that multiple (up to four) heterotrimers assemble on the DNA, recognizing several COREs possibly brought into proximity by the multimers. A consequence of this mode of action is that spacing between COREs might create a specific syntax read by the multimers, a long-range interaction model that we have already discussed elsewhere (Gnesutta et al., 2018).
Expanding on this concept, we can speculate that the substitution of Hd1 with PRR37, PRR73, or Ghd7 could lead to a variety of heteromultimers with distinct DNA-reading possibilities. Indeed, protein–protein interaction data support the idea that the Hd1–Ghd7 complex contains Ghd8 as well (Cai et al., 2019). Multimerization patterns could soon be demonstrated in rice also. One caveat of this idea is that CO (and possibly Hd1) multimerization takes place via the B-boxes, which are absent in PRR37, PRR73, and Ghd7. Nonetheless, other regions of the proteins might be able to mediate these interactions. For instance, Hd1 and Ghd7 contact each other through the CCT domain of Hd1 and the zinc finger plus central region (but not CCT domain) of Ghd7 (Zhang et al., 2017).
Hd1 promotes flowering and florigen expression under SDs but inhibits them under LDs (Zong et al., 2021). When considering protein–protein interactions, this photoperiodic conversion finds a relatively simple explanation because it clearly depends upon the presence of Ghd7 or Ghd8 (Du et al., 2017b; Sun et al., 2022). Under LDs, fully assembled complexes repress Ehd1, Hd3a, and RFT1 transcription. Under SDs, reduced expression of Ghd8 and instability of the Ghd7 protein deprive the complexes of these components, converting Hd1 into a transcriptional activator. Genetic data support this model because Hd1 ghd7 ghd8 mutants flower earlier than hd1 ghd7 ghd8 under any photoperiod (Zong et al., 2021). These data also indicate that Hd1 is intrinsically a constitutive activator of flowering, regardless of day length. Such a model might also imply changes in DNA accessibility (Zheng et al., 2019).
Interestingly, CO also has a dual function, acting as both an LD promoter and SD repressor of flowering (Luccioni et al., 2019). However, unlike that of rice florigens, expression of FT is not increased in co mutants under non-inductive conditions. Promotion of flowering by the co mutation under SDs depends upon reducing expression of TERMINAL FLOWER 1 (TFL1) at the apex, which in turn enhances sensitivity to FT. Thus, despite an apparent similarity, the effects of Hd1 and CO under non-inductive photoperiods depend on very different mechanisms (Luccioni et al., 2019).
Protein stability and phosphorylation
Differential protein stability has a central role in the regulation of photoperiodic flowering and the definition of external coincidence. Seasonal and diurnal windows of CO protein accumulation define the timing of FT expression. The abundance of rice Hd1 protein cycles, with peak accumulation occurring mostly during the day, and it is antiphasic relative to mRNA accumulation (Yang et al., 2015). This pattern is not a consequence of increased stability during the light phase but may result from cycling of Hd1 mRNA (Figure 2) (Ishikawa et al., 2011). That differential stability is not light dependent is also corroborated by the similar accumulation patterns observed under SDs and LDs (Yang et al., 2015; Hu et al., 2022).
Hd1 is targeted for degradation by HEADING DATE ASSOCIATED FACTOR 1, a RING-finger E3 ubiquitin ligase, via the 26S proteasome and by components of the autophagy pathway, including OsATG5, -7, and -8 (Yang et al., 2015; Hu et al., 2022). There is no clear time-of-day effect on Hd1 protein accumulation in haf1 or osatg5 mutants, as Hd1 levels are higher at any time point tested and in any photoperiod. However, autophagic degradation of Hd1 seems to be more effective in the dark. These data indicate that diurnal accumulation of the Hd1 protein is not shaped by degradation mechanisms or changes in day length.
In addition to protein turnover, phosphorylation is another important step in the post-translational control of Hd1 regulatory activity. HEADING DATE REPRESSOR 1 (HDR1) is a transcription factor that delays flowering by increasing transcription of Hd1 and reducing that of Ehd1 and the florigens (Sun et al., 2016). At the post-translational level, HDR1 can bind to the kinase OsK4, which phosphorylates Hd1. These three proteins form a complex in vivo, suggesting that Hd1, possibly in its phosphorylated form, may be involved in a positive loop of self-regulation that involves HDR1 and OsK4 (Figure 3). Additionally, either the phosphorylated or unphosphorylated forms might be preferentially subjected to degradation or incorporation into higher-order complexes.
Florigens as final outputs of leaf regulatory networks
Florigens are small globular proteins that belong to the phosphatidyl ethanolamine binding protein (PEBP) family and are present in all taxa from bacteria to mammals. They are responsible for triggering the flowering process in higher plants but also have roles in tuberization, nodulation, and seed development and as modifiers of plant architecture (Navarro et al., 2011; Chen et al., 2014; Wang et al., 2021b). They are produced in specialized companion cells of the leaves from which they enter sieve elements through plasmodesmata and reach distant plant tissues (Chen et al., 2018a).
PEBPs with a particularly strong influence on flowering can be divided into two major functional classes, FT-LIKE and TFL1-LIKE. In Arabidopsis, FT and TFL1, the founding members of each class, share an amino acid identity of over 98% but have antagonistic functions. FT promotes flowering by mediating both photoperiod and temperature signals, whereas TFL1 represses flowering (Wickland and Hanzawa, 2015; Susila et al., 2021).
There are 13 rice genes in the FT-like gene family (Chardon and Damerval, 2005). Hd3a and RFT1 are paralogs separated by only 11.5 kb that arose from a local duplication event that occurred after the divergence of monocots from dicots (Komiya et al., 2008). They share a high degree of identity, but their expression patterns have diverged, resulting in partly distinct functions. Both genes are transcribed in response to SDs, and their cognate proteins can move to the meristem and trigger flowering (Tamaki et al., 2007; Komiya et al., 2009). Thus, under inductive conditions, they are redundant and compensate each other’s function. Only Hd3a single mutants show a mild delay of flowering. However, the Hd3a–RFT1 double RNAi plant never flowers under SD, indicating that, in contrast to Arabidopsis, the switch to inflorescence development is fully dependent upon florigens in rice (Komiya et al., 2008; Tamaki et al., 2015). Under LD conditions, expression of RFT1, but not Hd3a, is induced in leaves. This is sufficient to trigger flowering, albeit later than in SD, and shows how rice facultative photoperiodic behavior is always mediated by florigens. No florigen-independent pathway of flowering induction has been described to date.
The closest homolog of Hd3a and RFT1, FT LIKE 1 (FT-L1), has florigenic activity and can induce flower formation in seedlings grown in vitro when overexpressed (Izawa et al., 2002). FT-L1 expression is directly induced by Hd3a and RFT1 (Giaume et al., 2023). Its transcripts and protein can be detected at all stages of inflorescence development in the same tissues, indicating a meristematic cell-autonomous activity (Furutani et al., 2006; Zong et al., 2022; Giaume et al., 2023). Loss-of-function mutations delay flowering and enhance the lateness of hd3a and rft1 single mutants. Interestingly, the mutants develop panicles with a larger number of secondary branches, indicating reduced determinacy, and this effect is genetically separable from the control of flowering time. Thus, rice evolved a unique triple florigenic system that times the transition to reproductive growth as well as shaping panicle architecture (Giaume et al., 2023).
Four homologs of TFL1, RICE CENTRORADIALIS (RCN) 1 to 4, have been described in rice (Kaneko-Suzuki et al., 2018). Overexpression of RCN1 and RCN2 delays flowering and increases the number of panicle branches (Nakagawa et al., 2002), whereas rcn knockout plants possess small panicles with a reduced number of branches (Liu et al., 2013). RCNs are transcribed in the vasculature but not in the shoot apical meristem (SAM), in contrast to TFL1. However, the proteins are translocated to the SAM to repress flowering. This mode of action resembles that of the florigens and suggests competition between RCNs and Hd3a and RFT1 proteins at the SAM. It remains unclear how the two opposing activities are balanced when both flowering-activating and -repressing PEBPs are present at the SAM (Kaneko-Suzuki et al., 2018).
Florigens move through plasmodesmata to reach distant compartments of the plant. FT is loaded into the phloem by FT INTERACTING PROTEIN 1 (FTIP1) (Liu et al., 2012). It has been demonstrated that rice FTIP1 (OsFTIP1), the closest homolog of Arabidopsis FTIP1, is necessary to promote rice flowering under LDs via its specific modulation of RFT1 transport from companion cells to sieve elements. OsFTIP1 interacts with RFT1, and in osftip1 mutants, RFT1 accumulates to high levels in companion cells but decreases in sieve elements, suggesting that OsFTIP1 promotes RFT1 export from companion cells to sieve elements in the phloem (Song et al., 2017). Although this mechanism is limited to RFT1 transport under LDs, a parallel mechanism determines Hd3a transport under SDs. OsFTIP9 encodes a homolog of OsFTIP1, and its protein product interacts with Hd3a to mediate its loading into sieve elements (Zhang et al., 2022). Consistent with this function, osftip9 mutants flower late under SDs but not LDs. Thus, the OsFTIP1-RFT1 and OsFTIP9-Hd3a dimers mirror each other’s functions under LDs and SDs, respectively (Figure 3). Whether dimerization could also take place by swapping the interactors between dimers remains to be determined. Nonetheless, the interaction of both dimers is strengthened by OsTPR075, a tetratricopeptide repeat protein active under both SDs and LDs (Zhang et al., 2022). When mutated, it decreases the amount of Hd3a and RFT1 reaching the apex, leading to late flowering under any photoperiod.
Such mechanisms of transport might require endosomal trafficking mediated by SNARE proteins within intracellular membranes. In Arabidopsis, SYNTAXIN OF PLANTS121 (SYP121) encodes a SNARE protein that interacts with QUIRKY (QKY). The SYP121–QKY complex regulates endosomal transport of FT in vesicles directed to the plasma membrane of companion cells. FT export from companion cells to sieve elements is impaired in SYP121 or QKY loss-of-function mutants, delaying flowering under LDs (Liu et al., 2019a). Endosomal trafficking could also be implicated in florigen transport in rice, as OsFTIP1 and OsFTIP9 are localized in the endoplasmic reticulum (Song et al., 2017; Zhang et al., 2022). However, homologs of SYP121 and QKY in rice have not yet been studied.
The regulation of florigen loading into the phloem stream is likely subject to several layers of control. The phosphatidylinositol 3-/4-kinase family protein OsUbDKγ4 reduces OsFTIP1 protein abundance by proteasome-mediated degradation and accelerates flowering if mutated (Song et al., 2017). How this post-translational mechanism interacts with day length and whether it also targets OsFTIP9 under SDs should be assessed.
Florigens, including FT, Hd3a, and RFT1, bind to phosphatidylcholine (PC), a phospholipid that is more abundant in the outer membrane layer of the SAM, facing the apoplast (Nakamura et al., 2014, 2019; Qu et al., 2021). Artificial manipulation of PC levels at the Arabidopsis SAM modifies flowering, consistent with PC promoting the floral transition in an FT-dependent manner (Nakamura et al., 2014). In rice, a phospholipase D (spPLD) hydrolyzes PC, and the corresponding loss-of-function mutants flower earlier than the wild type, promoting expression of Hd3a and RFT1 targets at the SAM (Qu et al., 2021). Interestingly, the activity of spPLD in delaying flowering depends upon its secretion in the apoplast, suggesting that the PC–florigen interaction takes place outside of the cell, mediating aspects of florigen activity that might involve their transport at the apex. Whatever the mechanism, this evidence indicates that interaction with PC potentiates the activity of the florigens.
The response of the SAM to flowering inductive signals
Variability of florigen complexes
Once translocated to the SAM, florigens induce its conversion from vegetative to reproductive growth.
The meristem is the ultimate recipient of flowering signals, where integration of several environmental inputs takes place. Commitment to a flowering fate is irreversible for most species and must be precisely timed and executed, particularly in annuals whose life cycles end after a single flowering episode. A proper threshold of inductive signals should be reached before the reproductive switch takes place.
It is still unclear how florigenic proteins move from conductive tissues, i.e., mature phloem or protophloem, into meristematic cells at the apex and how they move within it. However, research in rice, Arabidopsis, and several other model and non-model species indicates a common mode of action for florigens. Central to their activity is the florigen activation/repressor complex (FAC/FRC) (Taoka et al., 2011; Park et al., 2014; Li et al., 2015; Tylewicz et al., 2015; Abe et al., 2019; Collani et al., 2019; Sun et al., 2020; Cerise et al., 2021; Liu et al., 2021). The FAC is a heterohexamer assembled around a dimer of 14-3-3 proteins, which form a W-shaped structure. Upon entering meristematic cells, Hd3a and RFT1 bind to the 14-3-3 dimer in the cytoplasm (Taoka et al., 2011; Zhao et al., 2015). The florigen/14-3-3 complex enters the nucleus, where it binds to a transcription factor from the bZIP family that confers DNA-binding properties to the complex. Two florigen molecules rest on the C-terminal regions of each of the 14-3-3 proteins, and the two angles at the base of the W form pockets to which the C-terminal portion of the bZIP binds.
The structure of the FAC was first shown to contain the OsFD1 transcription factor, but it was later demonstrated that several bZIPs can replace OsFD1 (Tsuji et al., 2013; Brambilla et al., 2017; Jang et al., 2017; Cerise et al., 2021; Kaur et al., 2021). bZIPs act as dimers, and the complex orients their DNA-binding domain towards the DNA. However, whereas the florigen/14-3-3 dimer was resolved using full-length proteins, only nine amino acids of OsFD1 were crystalized, with the structure of the remainder of the protein being inferred by modeling. Thus, the exact conformation of the bZIP dimer within the FAC still needs to be resolved in more detail.
Similar to the diversity of bZIPs that take part in the formation of FACs/FRCs, a variety of 14-3-3 homo- or heterodimers can form the core of these complexes (Cerise et al., 2021). It is still unclear how this plasticity impacts gene expression. The binding motifs of bZIPs are almost identical within and between species, as well as for promoters and repressors of flowering (Taoka et al., 2011; Collani et al., 2019; Cerise et al., 2021). Therefore, selectivity of the complexes might depend on additional interacting partners or on the binding syntax (the number of and spacing between motifs) typical of each promoter (Cerise et al., 2021). Upon binding to the DNA, FAC targets, which promote inflorescence development, are activated. The most relevant include members of the MADS-box family of transcription factors. In rice, OsMADS14, -15, -18, and -34/PANICLE PHYTOMER 2 (PAP2) redundantly control panicle formation (Kobayashi et al., 2012). A quadruple mutant of these genes replaces inflorescence branches with vegetative shoots, and no flowers are formed. However, the inflorescence meristem is initiated normally, as indicated by the change from alternate to spiral phyllotaxis that can be observed in both the mutant and wild type as they switch from vegetative to reproductive growth. These observations indicate that OsMADS14, -15, -18, and -34 may not be the very first factors responsible for conversion of the vegetative meristem into inflorescence meristem, and other targets, activated earlier or in parallel, are likely present.
The FRCs share the same heterotrimeric architecture as the FACs but incorporate elements that repress the floral transition. Most notably, RCNs can replace Hd3a and RFT1, binding to 14-3-3s and delaying transition (Kaneko-Suzuki et al., 2018). Also, bZIPs with a floral repression function, such as Hd3a BINDING FACTORS 1 and 2, can form FRCs even if their activity occurs mostly in leaves and their precise role at the SAM needs to be more thoroughly defined (Brambilla et al., 2017).
Phosphorylation of the C-terminal SAP/TAP motif of bZIPs that form FACs/FRCs is necessary for their interaction with 14-3-3 proteins, and mutations in this region reduce the functionality of the complex. Conversely, mutations that mimic constitutive phosphorylation confer stronger flowering promotion activities to FD proteins (Taoka et al., 2011; Collani et al., 2019). In rice, several protein kinases that affect both the functional and interaction properties of OsFDs have been isolated. Calcineurin B-like-interacting protein kinase 3 (OsCIPK3) interacts with and phosphorylates OsFD1 (Peng et al., 2021). Interestingly, oscipk3 mutants show a late-flowering phenotype and accumulate less phosphorylated OsFD1 only under LDs, whereas plants grown in SD conditions have a wild-type phenotype. Thus, OsCIPK3 specifically affects the assembly of an RFT1/14-3-3/OsFD1 complex under LDs, suggesting that another unknown kinase operates under SDs. The calcium-dependent protein kinases OsCDPK41 and OsCDPK49 interact with and phosphorylate OsFD7, which forms FACs with Hd3a and RFT1 as well as with FT-L1 (Kaur et al., 2021). The phenotypic consequences of their mutation have not yet been determined, but they could be good candidates contributing to bZIP phosphorylation under SDs. Finally, a high-throughput study that interrogated more than 100 interactions between stress-activated protein kinases (SAPKs) and bZIPs identified SAPK4, -9, and -10 as interactors of OsFD1 (Liu et al., 2019b). Among these, at least SAPK10 can phosphorylate OsFD1, probably targeting the RXXS/T at the SAP domain, although this has not been directly demonstrated (Liu et al., 2019b). Overexpression of SAPK10 under a constitutive promoter accelerates flowering under both LDs and SDs and elevates transcription levels of OsFD1 and OsMADS15. Collectively, these studies suggest that the kinase–bZIP modules share a common mode of action.
When the SAM is reprogrammed to become a panicle, plant architecture changes to facilitate reproduction. The uppermost internodes, compressed below the SAM during vegetative growth, start to elongate when flowering signals reach the apex. This arrangement ensures coordination between flowering and stem elongation such that, when both are complete, a mature panicle can open its flowers on top of a long stem, above the leaves, releasing pollen to the wind. The Hd3a and RFT1 florigens induce internode elongation by reducing the expression of PREMATURE INTERNODE ELONGATION 1 (PINE1), a C2H2 zinc finger transcription factor that represses growth during the vegetative phase (Gómez-Ariza et al., 2019). PINE1 is expressed at the SAM and very strongly in the basal nodes, where IMs are located. Elevated transcription of PINE1 maintains IMs in an inactive state, and their reactivation is thus florigen dependent. PINE1 represses growth by reducing stem responsiveness to gibberellins, although the exact molecular mechanism involved remains unclear. Equally unclear is how florigenic proteins that reach the SAM create a growth gradient along the stem whereby the 4th or 5th internode from the apex elongates first, followed sequentially by the uppermost internodes (Hoshikawa, 1989). A plausible hypothesis is that the florigens induce secondary signals that form a gradient along the stem. The gradient could depend on auxin, which is produced at the shoot tip and transported toward the root (Wolbang et al., 2004). Experiments in which the inflorescence of barley was removed and the decapitated tip was treated with auxin indicated that this hormone is necessary for stem elongation (Wolbang and Ross, 2001; Wolbang et al., 2004; Yin et al., 2007). Thus, the crosstalk between gibberellins and auxin might be key to interpreting PINE1 activity.
Independent work isolated PINE1 as the gene underlying a quantitative trait locus that represses internode elongation in deepwater rice varieties (Nagai et al., 2020). The gene was named DECELERATOR OF INTERNODE ELONGATION 1 (DEC1), and its reduced expression in deepwater rice upon submergence is responsible for rapid internode elongation. This excellent study showed how PINE1/DEC1 activity is central to pathways that lead to internode elongation, independent of the environmental triggers. Also, because variation in expression levels, rather than coding sequence diversity, is responsible for distinct growth behaviors, the regulatory sequences of PINE1/DEC1 could be targeted for breeding efforts aimed at controlling plant growth.
Antagonistic signaling pathways balance the switch to reproductive growth and panicle development
Commitment of the SAM to reproductive growth by FACs is necessary but not sufficient to correctly initiate reproductive growth and complete inflorescence development, and several pathways must balance their antagonistic forces to reach proper developmental equilibrium.
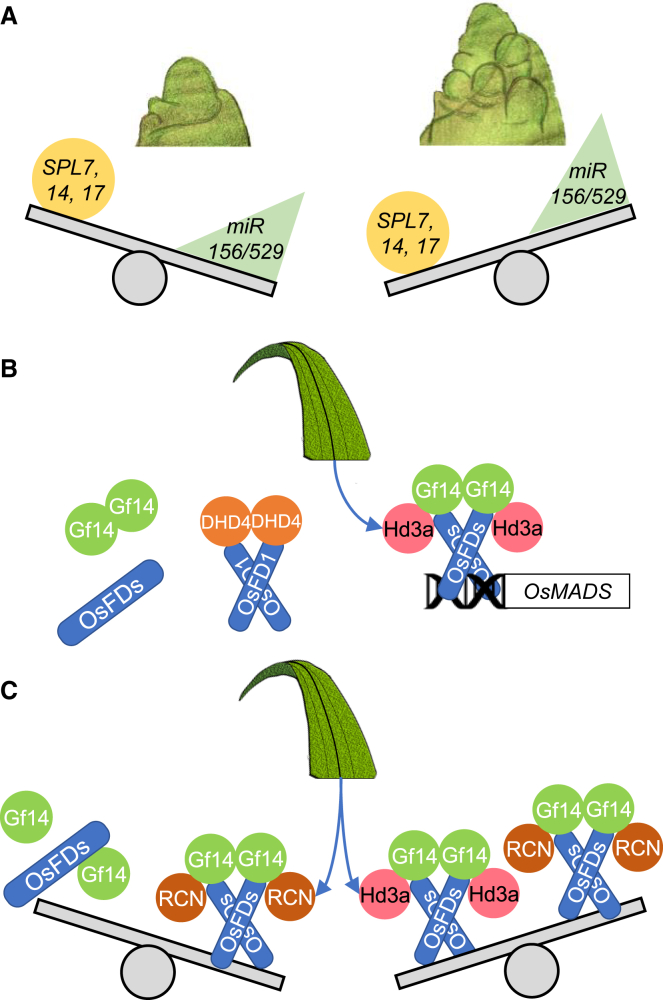
During or shortly before specification of the inflorescence, the vegetative program must be actively suppressed. Three SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) transcription factors, SPL7, -14, and -17, are necessary for suppression of bract outgrowth and promotion of inflorescence branching (Wang et al., 2021c). In a triple spl7 spl14 spl17 mutant, several vegetative shoots develop at positions normally occupied by bracts, replacing the primary branch meristems and indicating that vegetative development extends into the reproductive stage if not properly blocked. Expression of SPLs is regulated by microRNAs miR156 and miR529 at the post-transcriptional level, and their ectopic expression mimics the phenotypes of the spl7 spl14 spl17 mutant (Jiao et al., 2010; Miura et al., 2010; Wang et al., 2021c). Because miR156/529 act as intrinsic time rulers, creating a spatiotemporal gradient that controls developmental shifts during both vegetative and reproductive growth, the module has a central position in the network controlling the transition to inflorescence development. However, how miR/SPL-mediated suppression of vegetative development interacts with FAC-dependent promotion of reproductive development is unclear. One possibility is that the two pathways act independently. It is more likely that florigen signaling might interact with the miR/SPL module to block vegetative growth while reproductive meristems are being established (Figure 4). This perspective is supported by the finding that spl9 mutants show a marked reduction in RCN1 expression at the apex (Hu et al., 2021). Nonetheless, whether other SPLs have a similar effect remains to be tested.
Figure 4.
Balancing signals during the meristematic switch to reproductive growth.
Meristems on top represent the approximate stages during which the molecular events represented below occur.
(A) The balance between SPLs and miR156/529 determines the branching pattern and vegetative features of the inflorescence.
(B) Florigens transported from the leaves form FACs that induce transcription of MADS-box genes and switch the developmental fate of the meristem. DHD4 competes with OsFD1 to bind Gf14 under LDs.
(C) The reproductive switch is antagonized by FRCs, and RCNs transported from the leaves compete with the florigens for binding to Gf14s.
Another point of connection between the SPL and florigen pathways occurs at the level of regulation of OsMADS34/PAP2 expression. The florigens and SPL14 promote OsMADS34 transcription, whereas miR156 reduces it (Kobayashi et al., 2012; Wang et al., 2015). The balance between the two activities certainly impacts panicle development. In fact, mutations in OsMADS34 convert panicle branches into shoots (in combination with osmads14/15 and -18 mutants, as described above), increase the number of rachis branches, and fail to specify spikelets, which retain vegetative characters (Gao et al., 2010; Kobayashi et al., 2010). Overall, the mutant is unable to establish spikelet meristem identity and shows prolonged indeterminate growth of the panicle. These phenotypes are partly shared with those of spl or florigen mutants and miR156 overexpressors, suggesting that despite individual differences, these pathways balance vegetative vs. reproductive development and determinate vs. indeterminate growth.
An additional balancing mechanism involves the antagonism between florigens and RCNs. When RCNs reach the SAM, their structural identity with the florigens leads to competition for formation of FRCs at the expense of FACs (Kaneko-Suzuki et al., 2018). Mutations in RCNs reduce panicle branching and spikelet number, whereas their overexpression causes hyper-ramification (Nakagawa et al., 2002; Kaneko-Suzuki et al., 2018). These phenotypes are the opposite of those produced by florigen mutations or overexpression (Tamaki et al., 2015; Giaume et al., 2023). Thus, although florigens control development toward spikelet differentiation, whose direct effect is a reduction of branching, RCNs antagonize this trajectory, and the resulting equilibrium shapes inflorescence architecture. Artificial modulation of the two opposing forces, by means of genetics, might be of interest for yield increase if hyper-ramification, increased spikelet number, and floret fertility could be associated on the same varieties.
Among RCNs, at least RCN4 is a downstream direct target of OsMADS34/PAP2 and its paralog OsMADS5 (Zhu et al., 2022). Single rcn4 mutants do not show altered branching, likely because of redundancy with RCN1–3, but rcn4 mutation partially rescues the hyper-ramification of osmads34 mutants (Nakagawa et al., 2002; Zhang et al., 2005; Zhu et al., 2022). Thus, RCNs might act as negative regulators at both the VM-to-inflorescence meristem transition and the PBM-to-SM transition, downstream of OsMADS34/PAP2 and SPLs.
Finally, one last level of balance is provided by Delayed Heading Date 4 (DHD4), which encodes a CO-like transcription factor (Cai et al., 2021). The DHD4 protein can interact with OsFD1 and competes with 14-3-3s to limit formation of the Hd3a/14-3-3/OsFD1 complex. Mutations in DHD4 mildly accelerate flowering by inducing the expression of OsMADS14 and -15 transcription factors. This competition provides a totally new perspective on contrasting forces at the apex because it involves a novel class of proteins not previously implicated as balancing signals (Figure 4).
Selection of flowering-time genes during rice domestication and breeding
Flowering time is a trait of major applied interest because it affects two major aspects of rice cultivation: expansion to higher latitudes and adaptation to local environments. The LD regulatory pathway delays flowering (Figure 1), and mutations in major LD repressors accelerate the crop cycle, allowing rice to be cultivated at latitudes with shorter growing seasons (Shrestha et al., 2014). Mutations in Hd1, Ghd7, Ghd8, and PRR37 are widespread in both Asian and European germplasm and have been instrumental in bringing rice up to 55°N in China and 45°N in Europe (Gao et al., 2014; Gómez-Ariza et al., 2015; Goretti et al., 2017). Most European varieties share a high degree of genetic similarity with varieties from northern China, and mutant alleles of LD repressor genes are largely shared by both germplasms (Cai et al., 2013). It is likely that expansion to northern China followed domestication and preceded spread of the crop to Mediterranean Europe. Thus, a common pool of flowering-time alleles are under continuous selection by breeders in different areas of the globe (Zhao et al., 2011). Among them, hd1 mutant alleles are particularly abundant, probably because they also confer an adaptive advantage in cultivation under SDs. In tropical regions, functional Hd1 promotes flowering, shortening the cycle to the extent that varieties do not take advantage of the entire growing season, leading to severe yield penalties (Kim et al., 2018). An exception to this general rule involves varieties that harbor functional Hd1 but non-functional RFT1, which are found only in indica germplasm cultivated at lower latitudes (Ogiso-Tanaka et al., 2013).
Despite the major effect of single mutations on flowering, loss-of-function alleles of LD repressors are rarely found alone, and in modern varieties, combinations of multiple mutant alleles are common. This feature could be a consequence of the breeding history of each variety, which has been selected to have a flowering time whose cycle length perfectly matches the length of the local cropping season. Additive or epistatic effects depend on the molecular interactions described above and contribute to fine adaptation of cycle length (Figure 2), e.g., pyramiding of ghd7 and prr37 produces the strongest acceleration under LDs because it removes complexes that independently repress florigen expression, allowing access to the highest latitudes.
An additional element of variability that is important for breeding is represented by genes whose mutations have a minor effect on phenotype. These are instrumental in fine-tuning photoperiodic responses and adjusting flowering locally in addition to major-effect genes (Wu et al., 2013; Cai et al., 2021).
Sequencing of wild and cultivated accessions from all rice subgroups has revealed the existence of large natural allelic variation at flowering-time loci, which can also account for latitudinal expansion (Zhao et al., 2011; Huang et al., 2012). The contribution of several allelic variants to phenotypic diversity has been defined through the use of chromosome segment substitution lines, in which the effect of each allele can be unequivocally measured in an almost isogenic background, indicating that alleles do not necessarily fall in the extreme categories of fully functional or loss of function (Itoh et al., 2018). Rather, distinct haplotypes can confer various degrees of photoperiod sensitivity, reflecting adaptation to several geographic areas. These reconstructions of the history of selection give insights into the trajectories of domestication and rice subgroup differentiation. Several haplotypes are common to all subgroups and represent standing variation. This has occurred for major LD repressors, including Hd1, PRR37, Ghd7, and Ghd8. Other haplotypes arose after subgroup differentiation, also taking advantage of introgression events and local genomic rearrangements (Fujino et al., 2010; Itoh et al., 2018). Thus, these studies can also reconstruct gene flow among subgroups and reveal the history of human selection during the spread of rice to new environments. Further mining of natural variation will be key to advancing flowering-time research in the future.
Response of flowering time to variations in ambient temperature
Expansion of cultivation to higher latitudes has exposed rice to lower ambient temperatures during the cropping period. Phenotypic plasticity and artificial selection have adjusted the flowering response and adapted rice to new environments.
Lower ambient temperatures delay flowering under both LDs and SDs (Luan et al., 2009). In an excellent field study performed across nine LD environments, Guo et al. showed that an environmental index derived from temperatures at the early growth stage of rice had a perfect negative correlation with flowering time of a biparental mapping population. Genetic mapping of loci responsible for adaptation of flowering time demonstrated that variation at Hd1, PRR37, Ghd8, and Hd6 accounted for phenotypic variation (Takahashi et al., 2001; Guo et al., 2020). Extending the statistical treatment of environmental data to the 3000 Genomes collection enabled to distinguish accessions based on sensitivity of flowering time to temperature change. Accessions with higher sensitivity tended to be distributed at higher, colder latitudes, whereas accessions with lower sensitivity predominated in equatorial regions. This study showed that temperature can be used as an effective predictor of rice flowering time and that genes of the photoperiod pathway mediate between flowering induction and ambient temperature perception (Guo et al., 2020). Thus, the LD photoperiod pathway also operates as an ambient temperature flowering pathway.
The effect of Hd1 as an LD repressor is enhanced at lower ambient temperature, and the sensitivity of flowering to changes in temperature is strongly reduced in the hd1 mutant (Luan et al., 2009; Nagalla et al., 2021). A similar effect has been observed for Ghd7 (Nagalla et al., 2021). PRR37 has opposite effects across a temperature range. When mean ambient temperatures fall below a critical threshold, PRR37 represses flowering, but it reverts to a flowering promoter at higher temperatures (Guo et al., 2020).
Phytochromes act as thermosensors and integrate temperature information into developmental mechanisms (Jung et al., 2016). The reversion of the active Pfr form into its ground Pr state occurs more slowly at night, when temperatures are lower. In rice, PhyB enhances the repressor activity of Ghd7 at lower ambient temperatures, consistent with the idea that temperature perception mediated by PhyB is integrated into the flowering network via Ghd7 (Nagalla et al., 2021). Because PhyB interacts with Ghd7 to promote its degradation (Zheng et al., 2019), one could speculate that this mechanism is impaired at lower temperatures and that Ghd7 persists in the plant to delay flowering.
Hormonal control of flowering
Of the several pathways that control flowering in plants, hormonal ones are very important only in some species (Blazquez and Weigel, 2000; Trusov and Botella, 2006; Galvão and Schmid, 2014). The role of hormones in rice flowering time has not been studied extensively, and most evidence indicates that the photoperiodic pathway might be the only relevant one. Nonetheless, some hormones can affect flowering and, most importantly, shape panicle architecture upon reproductive commitment.
Auxin
The only link between auxin signaling and flowering time occurs at the level of OsmiR393, which targets the auxin receptor homologs OsAFB2 and OsTIR1 (Xia et al., 2012). Overexpression of OsmiR393 causes early flowering, although it is not clear which genes in the flowering network are responsible for the phenotype and how they exert their effects.
Upon floral commitment, activity of the DR5:VENUS auxin reporter has been observed in all panicle meristems and in the developing vasculature of the inflorescence. Moreover, auxin polar transporters colocalize with the reporter during flower formation and supposedly provide positional information for flower primordia initiation (Yang et al., 2017). Mutants with abnormal auxin content display panicle phenotypes, including anomalous panicle size, branching defects, and spikelets with altered organ identity (Yoshikawa et al., 2014).
Gibberellins
The role of gibberellins (GAs) as promoters of flowering is well established in Arabidopsis and other species, in which they act at the SAM to induce expression of floral integrator genes (Thomas and Vince-Prue, 1997; Reeves and Coupland, 2001; Eriksson et al., 2006). However, there is little evidence for the influence of GAs on flowering in rice. Treatments with GAs do not modify flowering time, although this does not exclude a role for GAs in this process. Also, it is unclear whether endogenous (or exogenous) GAs can reach the SAM, because just underneath the apical dome there is a ring-shaped area of GIBBERELLIN 2-OXIDASE 1 (GA2OX1) expression that is responsible for inactivation of bioactive GAs. The expression of GA2OX1 decreases drastically upon floral induction, indicating that the SAM may become accessible to GAs during reproductive development (Sakamoto et al., 2001). Overexpression of GA2OX1 delays flowering in transgenic rice, but this phenotype might be part of a more general and pleiotropic “GA deficiency syndrome” unrelated to flowering-time control (Sakamoto et al., 2003).
Another indirect link between flowering and GA signaling is offered by Hd16/Early Flowering 1/Casein Kinase I (hereafter CKI). Allelic variants with reduced activity and knockdown mutants cause early flowering. CKI encodes a kinase that phosphorylates the rice DELLA protein SLENDER RICE 1, thus stabilizing it. Unstable SLENDER RICE 1 could be the cause of the early flowering phenotype (Dai and Xue, 2010). However, CKI also phosphorylates the floral repressors Ghd7 and PRR37, and this modification might be essential for their activity, thus explaining the earliness of ckI mutants (Figure 3) (Hori et al., 2013; Kwon et al., 2015).
Cytokinins
Cytokinins affect both panicle formation and floral induction. A lack of cytokinin has been associated with a small SAM and abortive inflorescence meristems, leading to smaller panicles with reduced branches (Kurakawa et al., 2007; Ding et al., 2014; Du et al., 2017a; Wu et al., 2017; Song et al., 2018). Conversely, increasing cytokinin content in the inflorescence meristem leads to formation of a highly branched panicle and increases yield (Ashikari et al., 2005).
An elegant model that links cytokinin dynamics to flowering-time control has recently been proposed (Cho et al., 2022). Cytokinin signaling is mediated by type A and B response regulators (RRs), and Ehd1 is a type B RR. Ehd1 works as a homodimer; however, its homodimerization is inhibited by the type A RRs OsRR1 and OsRR2 (Cho et al., 2016). Transcription of OsRR1 and OsRR2 increases in response to cytokinin during the vegetative phase. Their cognate proteins can then bind and inactivate Ehd1, reducing transcription of Hd3a and RFT1 and delaying flowering (Cho et al., 2022). During floral commitment, a reduction in cytokinin levels reduces transcription of type A RRs, releasing Ehd1 inhibition and florigen expression.
Abscisic acid
The effect of abscisic acid (ABA) on flowering mostly relates to its role as an environmental stress hormone. The perception of ABA depends upon a group of proteins from the PYRABACTIN RESISTANCE 1/PYRABACTIN RESISTANCE 1-like/REGULATORY COMPONENTS OF THE ABA RECEPTOR family (hereafter called PYLs) that are essential for transmission of the ABA signal (Ma et al., 2009; Park et al., 2009). The rice genome encodes 13 PYLs that belong to two distinct groups. In a landmark study, Miao et al. showed that different combinations of pyl1, -2, -3, -4, -5, -6, and -12 mutations, belonging to group I, delay flowering to various extents (Miao et al., 2018). Because the same mutations also decrease sensitivity to ABA, a possible interpretation is that ABA can promote flowering. This concept is supported by studies with ABA biosynthetic mutants. Disturbing endogenous ABA levels with both knockout and overexpressors of the ABA biosynthetic gene MAO HUZI 4 (MHZ4) causes lateness (Ma et al., 2014). This effect might also depend upon interactions with the ethylene pathway, because mhz4 mutations abolish ABA biosynthesis but enhance ethylene emission. Delayed flowering has also been observed in ABA biosynthetic mutants of Arabidopsis, in which the relationship between ABA signaling and flowering regulation has been more thoroughly explored (Martignago et al., 2020).
ABA is antagonistic to GAs in several physiological processes, and connections between the two pathways determine the proper hormonal balance. OsAP2-39 encodes a transcription factor of the APETALA2 family that can directly activate expression of the 9-cis-epoxycarotenoid dioxygenase gene OsNCED-1 (an ABA biosynthetic gene) and increase the level of enzymes responsible for GA inactivation/degradation. Overexpression of OsAP2-39 causes a late-flowering phenotype that can be recovered by exogenous GAs, supporting the idea that GAs can act as promoters of flowering (Yaish et al., 2010).
Brassinosteroids (BRs)
The first evidence for a connection between BRs and flowering came from the finding that SDG725, an H3K36 methyltransferase essential for expression of genes involved in BR biosynthesis and signaling, can also affect flowering. In fact, its knockdown leads to a typical BR deficiency phenotype and late flowering. SDG725 promotes flowering by methylating several genes, including Ehd3, Ehd2, OsMADS50, Hd3a, and RFT1 (Sui et al., 2013).
More recently, BRASSINAZOLE-RESISTANT 1 (OsBZR1), a positive regulator of BR signaling, has emerged as an integrator of flowering-time control. The interaction of OsBZR1 with OsMED25, which mediates recruitment of the RNA polymerase to promote transcription, is essential for OsBZR1 to properly carry out its role in regulating the expression of BR-responsive genes (Ren et al., 2020). Knockdown of OsMED25 reduces Ehd1, Hd3a, and RFT1 expression and causes late flowering. Histone deacetylase HDA703 was also identified as an interactor of OsBZR1 and a promoter of flowering. OsBZR1 binding motifs present in the Ghd7 promoter recruit the dimer, and activity of HDA703 represses its transcription by histone deacetylation, leading to flowering promotion (Wang et al., 2020b).
Ethylene
The OsETR2 gene encodes an ethylene receptor expressed in the SAM and panicle. When overexpressed, it reduces ethylene sensitivity and causes late flowering, and its knockdown produces the opposite phenotypes. The authors proposed that OsETR2 can delay floral transition by increasing the transcription of OsGI and RCN1 (Wuriyanghan et al., 2009). Given the interactions of the ethylene pathway with the GA and ABA pathways, the effect on flowering time might be due to more complex interactions between hormones rather than single ones (Kuroha et al., 2018).
OsCTR2 is suggested to be a negative regulator of ethylene signaling, but its effects on flowering time are difficult to interpret because both overexpression and knockdown lines showed delayed flowering (Wang et al., 2013). These observations are emblematic of the difficulty in studying the dependency of flowering upon hormonal pathways, given their numerous and complex interconnections.
Flowering time under stress conditions
Although transition to flowering is mostly determined by interactions between the photoperiod and the allelic composition at flowering-time loci, other environmental parameters, including abiotic stresses, can modify the transition. All external stressors eventually converge upon transcriptional regulation of Ehd1, Hd3a, and RFT1, which thus act as integrators of multiple signals.
Drought stress
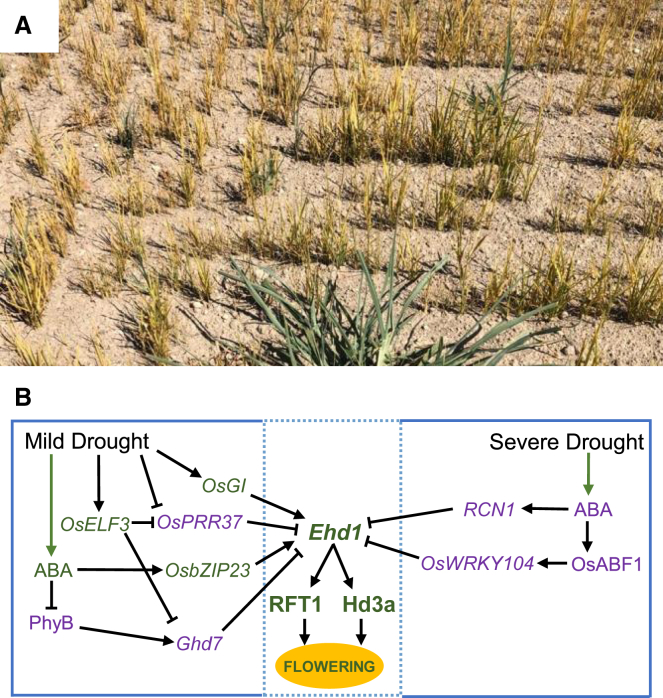
A considerable fraction of rice cultivation depends upon rainwater and is therefore subject to fluctuations in water availability. Even when rice is grown in paddy fields, extreme weather events linked to climate change can compromise water supply, imposing drought stress (Figure 5).
Figure 5.
Gene regulatory networks controlling flowering under drought stress.
(A) A rice paddy field experiencing severe drought during summer 2022 in northern Italy. Droughts hit several countries in 2022.
(B) Molecular network controlling Ehd1 expression in response to mild and severe drought stress. Arrows and flat-ended arrows indicate transcriptional activation and repression, respectively. Genes indicated in purple act as flowering inhibitors, and those in green act as promoters. Green arrows indicate increased biosynthesis.
Time to flowering can respond to drought in two opposite ways, either decreasing, a response known as drought escape (DE), or increasing. The final effect depends upon the severity of the drought. A mild water deficit triggers DE, and earlier flowering enables the plant to complete its life cycle before the stress becomes too severe (Weng et al., 2014; Du et al., 2018; Groen et al., 2020). Conversely, severe drought threatens plant survival, and a delay in flowering prevents entry into the delicate and energy-consuming reproductive phase (Galbiati et al., 2016; Zhang et al., 2016; Wang et al., 2020a).
In DE, ABA levels increase and induce expression of bZIP23, which acts as a positive regulator of the DE response. bZIP23 feeds back on the regulation of flowering-time genes, inducing transcription of OsTOC1, Ehd1, Hd3a, and RFT1 and reducing that of Ghd7 (Du et al., 2018). Genetic analyses indicate that mutations in PRR37, GI, and EARLY FLOWERING 3 (ELF3) delay flowering under mild water deficit compared with wild-type controls, producing an impaired DE response, and this occurs independently of ABA. The expression of Ehd1, Hd3a, and RFT1 correlates with the flowering time of the mutants. Thus, components of the photoperiod pathway are integrated with DE responses in a complex manner, only partly dependent upon ABA (Weng et al., 2014; Du et al., 2018) (Figure 5). Downstream of florigens, the OsMADS18 transcription factor has been identified as strongly induced during drought as an additional integrator of DE, consistent with the flowering-promotive role of MADS-box genes at the end of the photoperiodic cascade (Fornara et al., 2004; Kobayashi et al., 2012; Groen et al., 2020).
The flowering delay caused by severe drought is also ABA dependent but proceeds through a different molecular mechanism. High ABA levels induce expression of the OsABF1 bZIP transcription factor, a flowering repressor (Zhang et al., 2016). Reducing its expression by RNAi also accelerates flowering under drought stress and induces Ehd1. The activity of OsABF1 depends upon OsWRKY104 creating an ABA-dependent floral repressive module. Drought stress and ABA also induce expression of RCN1, with rcn1 attenuating the flowering delay caused by stress (Wang et al., 2020a). It remains to be determined whether and how the OsABF1- and RCN1-dependent mechanisms are integrated and in which tissue. Given that RCNs can form floral repressor complexes with bZIPs, an intriguing possibility is that OsABF1 and RCN1 interact to delay flowering when plants experience severe drought (Figure 5).
Salt stress
Rice cultivation in river deltas is threatened by soil salinization, which occurs when natural events return seawater to the fields. A well-described suite of protective mechanisms is activated in response to increasing salinity. However, the connections between salt stress and flowering-time control are just starting to be explored and suggest that circadian clock components are preferential integrators of these pathways. The evening complex (EC) is a central feature of the circadian clock that is assembled by LUX ARRHYTHMO, ELF3, and ELF4 and binds to DNA to repress gene expression (Silva et al., 2020). The rice genome encodes two orthologs of ELF3 and three of ELF4. The oself4a, oself3-1, and oslux single mutants are hyper-sensitive to salt stress, showing reduced survival rates when grown at high NaCl concentrations (Wang et al., 2021d). Additionally, under SDs, oself4a mutants flower late, whereas single oslux and double oself3-1 oself3-2 mutants never flower (Wang et al., 2021d; Andrade et al., 2022). These observations point to the EC as an integrator of flowering and salt stress signals. Direct targets of the EC include several PRRs as well as OsGI. OsELF4a, OsLUX, and OsELF3-1 can bind the OsGI promoter to repress its expression. Mutations in OsGI increase rice survival rates upon salt or osmotic stress treatments, increase the leaf concentrations of osmoprotectants such as proline and sucrose, and induce earlier flowering under LDs (Li et al., 2016; Wang et al., 2021d). Thus, the EC–OsGI module fine-tunes salt tolerance and promotes flowering, representing an interesting target for breeding efforts.
Temperature stress
Rice plants are sensitive to temperature variations, particularly during flowering and grain filling. A 1°C increase in the minimum night temperature is correlated with yield reductions of 10% (Peng et al., 2004). High temperatures can induce early flowering and reduce yield, whereas low temperatures delay flowering, indicating that temperature and day length measurements coordinately control the reproductive transition. In both cases, temperature perception converges on transcriptional regulation of Ehd1 and the florigens (Luan et al., 2009; Chen et al., 2018b). The qHd1 quantitative trait locus is a plausible candidate for part of a still-unexplored rice thermosensory pathway. Genetic variation at qHd1 partly explains the phenotypic variation in heading date at high ambient temperatures. The Zhenshan 97 allele of qHd1 maintains stable heading dates even when mean temperature increases. Heading date stabilization is observed when plants are grown at different temperatures but under the same day length, indicating that photoperiod and thermosensory pathways are genetically separable (Chen et al., 2018b). The causal gene underlying qHd1 has not yet been precisely mapped, but the OsMADS51 transcription factor is a strong candidate. An insertion in the first intron in Zhenshan 97 represents a functional polymorphism, reducing transcription of OsMADS51 compared with varieties that lack the insertion. The transcription of the OsMADS51 downstream targets Ehd1, Hd3a, and RFT1 is also reduced, explaining the flowering delay, particularly at high temperatures. Functional validation of temperature responses using osmads51 mutants is still missing. However, syntenic relationships and functional data from temperate grasses suggest that the monocot OsMADS51 clade includes orthologs of FLOWERING LOCUS C, a major controller of vernalization responses (Ruelens et al., 2013). It is thus tempting to speculate that OsMADS51-like genes regulate temperature-dependent flowering and that in rice, which lacks a vernalization pathway, they have subfunctionalized to control a high-ambient-temperature flowering pathway.
Nutrient availability
Maximizing yields requires optimal fertilization. Different nutrients have been shown to influence flowering. The supply of K and P accelerates flowering, whereas low or high N fertilization delays it (Ye et al., 2019; Zhang et al., 2021). The N-mediated heading date 1 (Nhd1) gene encodes an MYB transcription factor whose expression is induced upon N fertilization (Zhang et al., 2021). In the nhd1 mutant, flowering is delayed under both SDs and LDs, and transcription of Hd3a is reduced. Because NHD1 directly binds to the promoter of Hd3a, it lies at the interface between N perception and flowering.
Concluding remarks and future perspectives
In this section, we briefly indicate trajectories that we believe should be pursued to advance flowering-time research in its basic and applied facets.
Gene cloning and further refinement of GRNs
More flowering-time genes remain to be cloned in the future and placed in GRNs. For these, as well as for many known regulators, precise positioning needs to be thoroughly determined. Although expression analyses provide a first means of placement in the network, more refined genetic analyses can be laborious and time consuming yet are necessary to define complex relationships.
Understanding protein abundance and activity
Transcriptional data are relatively straightforward to produce and sufficient to build GRNs. However, full understanding of network activity will come only after studying regulation at the post-transcriptional level. Protein abundance, modifications, and interaction patterns can depend upon day length and may be largely independent of transcription. Studies of gene × environment interactions are needed to determine these features and would provide many benefits for breeding.
Quantitative integration of information
The complex interconnection of genes in GRNs makes it difficult to predict how perturbation of gene activity will impact the phenotype. This is particularly evident when trying to make quantitative predictions. To this end, in silico models can be a useful tool for both scientists and breeders. Initial models made use of quantitative analyses related to a few major regulators to assess latitudinal adaptation, predicting florigen expression and flowering responses (Qiu et al., 2021). The power of these models can be increased by integrating more genes, including minor controllers, and refining algorithms with expression data collected in more environments.
Exploitation of basic understanding for applied purposes
Finally, all the above is useful for guiding better breeding, driving selection with molecular rather than phenotypic data, and quickly tailoring new varieties to specific cultivation environments, possibly also with the use of gene-editing technologies. This will be the most daunting task, requiring close and constructive interactions between scientists and breeders.
Funding
This work was supported by funding from the Italian Ministry of Foreign Affairs and International Cooperation, Italy-Japan bilateral collaboration on Agrifood #PGR10097.
Acknowledgments
No conflict of interest is declared.
Published: May 4, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Abe M., Kosaka S., Shibuta M., Nagata K., Uemura T., Nakano A., Kaya H. Transient activity of the florigen complex during the floral transition in arabidopsis thaliana. Devenir. 2019;146 doi: 10.1242/dev.171504. [DOI] [PubMed] [Google Scholar]
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade L., Lu Y., Cordeiro A., Costa J.M.F., Wigge P.A., Saibo N.J.M., Jaeger K.E. The evening complex integrates photoperiod signals to control flowering in rice. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2122582119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Andrés F., Galbraith D.W., Talón M., Domingo C. Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol. (Wash. D C) 2009;151:681–690. doi: 10.1104/pp.109.139097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Ballerini E.S., Kramer E.M. In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2011;2:81. doi: 10.3389/fpls.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- Brambilla V., Martignago D., Goretti D., Cerise M., Somssich M., de Rosa M., Galbiati F., Shrestha R., Lazzaro F., Simon R., Fornara F. Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell. 2017;29:2801–2816. doi: 10.1105/tpc.17.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Fan J., Jiang Z., Basso B., Sala F., Spada A., Grassi F., Lu B.-R. The puzzle of Italian rice origin and evolution: determining genetic divergence and affinity of rice germplasm from Italy and Asia. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Chen S., Wu M., Zheng T., Zhou L., Li C., Zhang H., Wang J., Xu X., Chai J., et al. Early heading 7 interacts with DTH8, and regulates flowering time in rice. Plant Cell Rep. 2019;38:521–532. doi: 10.1007/s00299-019-02380-7. [DOI] [PubMed] [Google Scholar]
- Cai M., Zhu S., Wu M., Zheng X., Wang J., Zhou L., Zheng T., Cui S., Zhou S., Li C., et al. DHD4, a CONSTANS-like family transcription factor, delays heading date by affecting the formation of the FAC complex in rice. Mol. Plant. 2021;14:330–343. doi: 10.1016/j.molp.2020.11.013. [DOI] [PubMed] [Google Scholar]
- Cerise M., Giaume F., Galli M., Khahani B., Lucas J., Podico F., Tavakol E., Parcy F., Gallavotti A., Brambilla V., Fornara F. OsFD4 promotes the rice floral transition via florigen activation complex formation in the shoot apical meristem. New Phytol. 2021;229:429–443. doi: 10.1111/nph.16834. [DOI] [PubMed] [Google Scholar]
- Chardon F., Damerval C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005;61:579–590. doi: 10.1007/s00239-004-0179-4. [DOI] [PubMed] [Google Scholar]
- Chen M., MacGregor D.R., Dave A., Florance H., Moore K., Paszkiewicz K., Smirnoff N., Graham I.A., Penfield S. Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year. Proc. Natl. Acad. Sci. USA. 2014;111:18787–18792. doi: 10.1073/pnas.1412274111. –18792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Payyavula R.S., Chen L., Zhang J., Zhang C., Turgeon R. FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc. Natl. Acad. Sci. USA. 2018;115:2830–2835. doi: 10.1073/pnas.1719455115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., Zhang H.W., Zhang H.L., Ying J.Z., Ma L.Y., Zhuang J.Y. Natural variation at qHd1 affects heading date acceleration at high temperatures with pleiotropism for yield traits in rice. BMC Plant Biol. 2018;18:112. doi: 10.1186/s12870-018-1330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho L.H., Yoon J., Pasriga R., An G. Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. (Wash. D C) 2016;170:2159–2171. doi: 10.1104/pp.15.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho L.H., Yoon J., Tun W., Baek G., Peng X., Hong W.J., Mori I.C., Hojo Y., Matsuura T., Kim S.R., et al. Cytokinin increases vegetative growth period by suppressing florigen expression in rice and maize. Plant J. 2022;110:1619–1635. doi: 10.1111/tpj.15760. [DOI] [PubMed] [Google Scholar]
- Collani S., Neumann M., Yant L., Schmid M. FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol. (Wash. D C) 2019;180:367–380. doi: 10.1104/pp.18.01505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Xue H.-W. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010;29:1916–1927. doi: 10.1038/emboj.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., You J., Chen L., Wang S., Ding Y. Nitrogen fertilizer increases spikelet number per panicle by enhancing cytokinin synthesis in rice. Plant Cell Rep. 2014;33:363–371. doi: 10.1007/s00299-013-1536-9. [DOI] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Liu L., Li M., Fang S., Shen X., Chu J., Zhang Z. UNBRANCHED3 regulates branching by modulating cytokinin biosynthesis and signaling in maize and rice. New Phytol. 2017;214:721–733. doi: 10.1111/nph.14391. [DOI] [PubMed] [Google Scholar]
- Du A., Tian W., Wei M., Yan W., He H., Zhou D., Huang X., Li S., Ouyang X. The DTH8-hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant. 2017;10:948–961. doi: 10.1016/j.molp.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Du H., Huang F., Wu N., Li X., Hu H., Xiong L. Integrative regulation of drought escape through ABA dependent and independent pathways in rice. Mol. Plant. 2018;11:584–597. doi: 10.1016/j.molp.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. GA4 is the active gibberellin in the regulation of leafy transcription and arabidopsis floral initiation. Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Parenicová L., Falasca G., Pelucchi N., Masiero S., Ciannamea S., Lopez-Dee Z., Altamura M.M., Colombo L., Kater M.M. Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. (Wash. D C) 2004;135:2207–2219. doi: 10.1104/pp.104.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K., Wu J., Sekiguchi H., Ito T., Izawa T., Matsumoto T. Multiple introgression events surrounding the Hd1 flowering-time gene in cultivated rice, Oryza sativa L. Mol. Genet. Genom. 2010;284:137–146. doi: 10.1007/s00438-010-0555-2. [DOI] [PubMed] [Google Scholar]
- Furutani I., Sukegawa S., Kyozuka J. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J. 2006;46:503–511. doi: 10.1111/j.1365-313X.2006.02703.x. [DOI] [PubMed] [Google Scholar]
- Galbiati F., Chiozzotto R., Locatelli F., Spada A., Genga A., Fornara F. Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ. 2016;39:1982–1993. doi: 10.1111/pce.12760. –1993. [DOI] [PubMed] [Google Scholar]
- Galvão V.C., Schmid M. Advances in Botanical Research - the Molecular Genetics of Floral Transition and Flower Development. 2014. Regulation of flowering by endogenous signals; pp. 63–102. [Google Scholar]
- Gangappa S.N., Botto J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19:460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., Guo C., Kong H., Xue H., Zhang D. The SEPARATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. (Wash. D C) 2010;153:728–740. doi: 10.1104/pp.110.156711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Jin M., Zheng X.-M., Chen J., Yuan D., Xin Y., Wang M., Huang D., Zhang Z., Zhou K., et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA. 2014;111:16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaume F., Bono G.A., Martignago D., Miao Y., Vicentini G., Toriba T., Wang R., Kong D., Cerise M., Chirivì D., et al. Two florigens and a florigen-like protein form a triple regulatory module at the shoot apical meristem to promote reproductive transitions in rice. Native Plants. 2023;9:525–534. doi: 10.1038/s41477-023-01383-3. [DOI] [PubMed] [Google Scholar]
- Gnesutta N., Kumimoto R.W., Swain S., Chiara M., Siriwardana C., Horner D.S., Holt B.F., Mantovani R. CONSTANS imparts DNA sequence-specificity to the histone-fold NF-YB/NF-YC dimer. Plant Cell. 2017;29:1516–1532. doi: 10.1105/tpc.16.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N., Mantovani R., Fornara F. Plant flowering: imposing DNA specificity on histone-fold subunits. Trends Plant Sci. 2018;23:293–301. doi: 10.1016/j.tplants.2017.12.005. [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J., Galbiati F., Goretti D., Brambilla V., Shrestha R., Pappolla A., Courtois B., Fornara F. Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 2015;66:2027–2039. doi: 10.1093/jxb/erv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Ariza J., Brambilla V., Vicentini G., Landini M., Cerise M., Carrera E., Shrestha R., Chiozzotto R., Galbiati F., Caporali E., et al. A transcription factor coordinating internode elongation and photoperiodic signals in rice. Native Plants. 2019;5:358–362. doi: 10.1038/s41477-019-0401-4. [DOI] [PubMed] [Google Scholar]
- Goretti D., Martignago D., Landini M., Brambilla V., Gómez-Ariza J., Gnesutta N., Galbiati F., Collani S., Takagi H., Terauchi R., et al. Transcriptional and post-transcriptional mechanisms limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff M., Straub D., Eguen T., Dolde U., Rodrigues V., Brandt R., Wenkel S. MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in arabidopsis. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen S.C., Ćalić I., Joly-Lopez Z., Platts A.E., Choi J.Y., Natividad M., Dorph K., Mauck W.M., Bracken B., Cabral C.L.U., et al. The strength and pattern of natural selection on gene expression in rice. Nature. 2020;578:572–576. doi: 10.1038/s41586-020-1997-2. [DOI] [PubMed] [Google Scholar]
- Guo T., Mu Q., Wang J., Vanous A.E., Onogi A., Iwata H., Li X., Yu J. Dynamic effects of interacting genes underlying rice flowering-time phenotypic plasticity and global adaptation. Genome Res. 2020;30:673–683. doi: 10.1101/gr.255703.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hori K., Ogiso-Tanaka E., Matsubara K., Yamanouchi U., Ebana K., Yano M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013;76:36–46. doi: 10.1111/tpj.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa K. Nobunkyo; Tokyo, Japan: 1989. The Growing Rice Plant. [Google Scholar]
- Hu L., Chen W., Yang W., Li X., Zhang C., Zhang X., Zheng L., Zhu X., Yin J., Qin P., et al. OsSPL9 regulates grain number and grain yield in rice. Front. Plant Sci. 2021;12:682018. doi: 10.3389/fpls.2021.682018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yang Z., Zhang Y., Zhang A., Lu Q., Fang Y., Lu C. Autophagy targets Hd1 for vacuolar degradation to regulate rice flowering. Mol. Plant. 2022;15:1137–1156. doi: 10.1016/j.molp.2022.05.006. [DOI] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z.-X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W., et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Aoki M., Kurotani K.I., Yokoi S., Shinomura T., Takano M., Shimamoto K. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genom. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Itoh H., Wada K.C., Sakai H., Shibasaki K., Fukuoka S., Wu J., Yonemaru J.I., Yano M., Izawa T. Genomic adaptation of flowering-time genes during the expansion of rice cultivation area. Plant J. 2018;94:895–909. doi: 10.1111/tpj.13906. [DOI] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Tokutomi S., Okuno K., Shimamoto K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant) Plant J. 2000;22:391–399. doi: 10.1046/j.1365-313x.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Sugiyama N., Tanisaka T., Yano M., Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C.S., Wenkel S., Soppe W., Deng X.-W., Valverde F., Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Li H.-Y., Kuo M.-L. Ectopic expression of Arabidopsis FD and FD PARALOGUE in rice results in dwarfism with size reduction of spikelets. Sci. Rep. 2017;7 doi: 10.1038/srep44477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A.K., Box M.S., Charoensawan V., Cortijo S., et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- Kaneko-Suzuki M., Kurihara-Ishikawa R., Okushita-Terakawa C., Kojima C., Nagano-Fujiwara M., Ohki I., Tsuji H., Shimamoto K., Taoka K.I. TFL1-Like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 2018;59:458–468. doi: 10.1093/pcp/pcy021. [DOI] [PubMed] [Google Scholar]
- Kaur A., Nijhawan A., Yadav M., Khurana J.P. OsbZIP62/OsFD7, a functional ortholog of FLOWERING LOCUS D, regulates floral transition and panicle development in rice. J. Exp. Bot. 2021;72:7826–7845. doi: 10.1093/jxb/erab396. [DOI] [PubMed] [Google Scholar]
- Kim S.R., Torollo G., Yoon M.R., Kwak J., Lee C.K., Prahalada G.D., Choi I.R., Yeo U.S., Jeong O.Y., Jena K.K., Lee J.S. Loss-of-function alleles of heading date 1 (Hd1) are associated with adaptation of temperate japonica rice plants to the tropical region. Front. Plant Sci. 2018;9 doi: 10.3389/fpls.2018.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. PANICLE PHYTOMETRA2 (PAP2), encoding a SEPARATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2010;51:47–57. doi: 10.1093/pcp/pcp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., Yoshida H., Nagamura Y., Kyozuka J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPARATA MADS box gene. Plant Cell. 2012;24:1848–1859. doi: 10.1105/tpc.112.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Koo B.-H., Yoo S.-C., Park J.-W., Kwon C.-T., Lee B.-D., An G., Zhang Z., Li J., Li Z., Paek N.-C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant. 2013;6:1877–1888. doi: 10.1093/mp/sst088. [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Adam L., Hymus G.J., Repetti P.P., Reuber T.L., Marion C.M., Hempel F.D., Ratcliffe O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228:709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Zhang Y., Siefers N., Holt B.F. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010;63:379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Kuroha T., Nagai K., Gamuyao R., Wang D.R., Furuta T., Nakamori M., Kitaoka T., Adachi K., Minami A., Mori Y., et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science. 2018;361:181–186. doi: 10.1126/science.aat1577. [DOI] [PubMed] [Google Scholar]
- Kwon C.T., Koo B.H., Kim D., Yoo S.C., Paek N.C. Casein kinases I and 2α phosphorylate oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol. Cells. 2015;38:81–88. doi: 10.14348/molcells.2015.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lin H., Dubcovsky J. Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 2015;84:70–82. doi: 10.1111/tpj.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yue W., Wang M., Qiu W., Zhou L., Shou H. Mutation of OsGIGANTEA leads to enhanced tolerance to polyethylene glycol-generated osmotic stress in rice. Front. Plant Sci. 2016;7:465. doi: 10.3389/fpls.2016.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Zhang Z., Cheng N., Liu H., Song S., Hu Y., Zhou X., Zhang J., Xing Y. The transcriptional repressor OsPRR73 links circadian clock and photoperiod pathway to control heading date in rice. Plant Cell Environ. 2021;44:842–855. doi: 10.1111/pce.13987. [DOI] [PubMed] [Google Scholar]
- Liu L., Liu C., Hou X., Xi W., Shen L., Tao Z., Wang Y., Yu H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012;10:e1001313. doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Teo Z.W.N., Bi Y., Song S., Xi W., Yang X., Yin Z., Yu H. A conserved genetic pathway determines inflorescence architecture in arabidopsis and rice. Dev. Cell. 2013;24:612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Liu L., Li C., Teo Z.W.N., Zhang B., Yu H. The MCTP-SNARE complex regulates florigen transport in arabidopsis. Plant Cell. 2019;31:2475–2490. doi: 10.1105/tpc.18.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li Z., Hou Y., Wang Y., Wang H., Tong X., Ao H., Zhang J. Protein interactomic analysis of SAPKs and ABA-inducible bZIPs revealed key roles of SAPK10 in rice flowering. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20061427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Huang X., Ma B., Zhang T., Sang N., Zhuo L., Zhu J. Components and functional diversification of florigen activation complexes in cotton. Plant Cell Physiol. 2021;62:1542–1555. doi: 10.1093/pcp/pcab107. [DOI] [PubMed] [Google Scholar]
- Luan W., Chen H., Fu Y., Si H., Peng W., Song S., Liu W., Hu G., Sun Z., Xie D., Sun C. The effect of the crosstalk between photoperiod and temperature on the heading-date in rice. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luccioni L., Krzymuski M., Sánchez-Lamas M., Karayekov E., Cerdán P.D., Casal J.J. CONSTANS delays Arabidopsis flowering under short days. Plant J. 2019;97:923–932. doi: 10.1111/tpj.14171. [DOI] [PubMed] [Google Scholar]
- Lv X., Zeng X., Hu H., Chen L., Zhang F., Liu R., Liu Y., Zhou X., Wang C., Wu Z., et al. Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant Cell. 2021;33:1182–1195. doi: 10.1093/plcell/koab016. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Ma B., Yin C.-C., He S.-J., Lu X., Zhang W.-K., Lu T.-G., Chen S.-Y., Zhang J.-S. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (oryza sativa L.) seedlings. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignago D., Siemiatkowska B., Lombardi A., Conti L. Abscisic acid and flowering regulation: many targets, different places. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21249700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Yamanouchi U., Wang Z.-X., Minobe Y., Izawa T., Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. (Wash. D C) 2008;148:1425–1435. doi: 10.1104/pp.108.125542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C., Xiao L., Hua K., Zou C., Zhao Y., Bressan R.A., Zhu J.K. Mutations in a subfamily of abscisic acid recepto genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA. 2018;115:6058–6063. doi: 10.1073/pnas.1804774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.-J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Nagai K., Mori Y., Ishikawa S., Furuta T., Gamuyao R., Niimi Y., Hobo T., Fukuda M., Kojima M., Takebayashi Y., et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature. 2020;584:109–114. doi: 10.1038/s41586-020-2501-8. [DOI] [PubMed] [Google Scholar]
- Nagalla A.D., Nishide N., Hibara K.I., Izawa T. High ambient temperatures inhibit ghd7-mediated flowering repression in rice. Plant Cell Physiol. 2021;62:1745–1759. doi: 10.1093/pcp/pcab129. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Shimamoto K., Kyozuka J. Overexpression of RCN1 and RCN2, rice Terminal Flower 1/Centroradialis homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Andrés F., Kanehara K., Liu Y.C., Dörmann P., Coupland G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 2014;5:3553. doi: 10.1038/ncomms4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Lin Y.C., Watanabe S., Liu Y. chi, Katsuyama K., Kanehara K., Inaba K. High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates its Phospholipid-Binding Site in Flowering. iScience. 2019;21 doi: 10.1016/j.isci.2019.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Abelenda J.A., Cruz-Oró E., Cuéllar C.A., Tamaki S., Silva J., Shimamoto K., Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478:119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Nonoue Y., Yano M., Izawa T. Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016;86:221–233. doi: 10.1111/tpj.13168. [DOI] [PubMed] [Google Scholar]
- Ogiso-Tanaka E., Matsubara K., Yamamoto S.i., Nonoue Y., Wu J., Fujisawa H., Ishikubo H., Tanaka T., Ando T., Matsumoto T., Yano M. Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osugi A., Itoh H., Ikeda-Kawakatsu K., Takano M., Izawa T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. (Wash. D C) 2011;157:1128–1137. doi: 10.1104/pp.111.181792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim S.L., Lee S., Je B.I., Piao H.L., Park S.H., Kim C.M., Ryu C.H., Park S.H., Xuan Y.H., et al. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 2008;56:1018–1029. doi: 10.1111/j.1365-313X.2008.03667.x. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F.F., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 2014;46:1337–1342. doi: 10.1038/ng.3131. [DOI] [PubMed] [Google Scholar]
- Peng S., Huang J., Sheehy J.E., Laza R.C., Visperas R.M., Zhong X., Centeno G.S., Khush G.S., Cassman K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA. 2004;101:9971–9975. doi: 10.1073/pnas.0403720101. –5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Zhu C., Liu T., Zhang S., Feng S., Wu C. Phosphorylation of OsFD1 by OsCIPK3 promotes the formation of RFT1-containing florigen activation complex for long-day flowering in rice. Mol. Plant. 2021;14:1135–1148. doi: 10.1016/j.molp.2021.04.003. [DOI] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R.W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., Holt B.F., Mantovani R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012;24:4777–4792. doi: 10.1105/tpc.112.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L., Wu Q., Wang X., Han J., Zhuang G., Wang H., Shang Z., Tian W., Chen Z., Lin Z., et al. Forecasting rice latitude adaptation through a daylength-sensing-based environment adaptation simulator. Nat. Food. 2021;2:348–362. doi: 10.1038/s43016-021-00280-2. [DOI] [PubMed] [Google Scholar]
- Qu L., Chu Y.J., Lin W.H., Xue H.W. A secretory phospholipase D hydrolyzes phosphatidylcholine to suppress rice heading time. PLoS Genet. 2021;17:e1009905. doi: 10.1371/journal.pgen.1009905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.H., Coupland G. Analysis of flowering time control in arabidopsis by comparison of double and triple mutants. Plant Physiol. (Wash. D C) 2001;126:1085–1091. doi: 10.1104/pp.126.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Tian X., Li S., Mei E., He M., Tang J., Xu M., Li X., Wang Z., Li C., Bu Q. Oryza sativa mediator subunit OsMED25 interacts with OsBZR1 to regulate brassinosteroid signaling and plant architecture in rice. J. Integr. Plant Biol. 2020;62:793–811. doi: 10.1111/jipb.12914. [DOI] [PubMed] [Google Scholar]
- Ruelens P., De Maagd R.A., Proost S., Theißen G., Geuten K., Kaufmann K. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 2013;4:2280. doi: 10.1038/ncomms3280. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kobayashi M., Itoh H., Tagiri A., Kayano T., Tanaka H., Iwahori S., Matsuoka M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. (Wash. D C) 2001;125:1508–1516. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Morinaka Y., Ishiyama K., Kobayashi M., Itoh H., Kayano T., Iwahori S., Matsuoka M., Tanaka H. Genetic manipulation of gibberellin metabolism in transgenic rice. Nat. Biotechnol. 2003;21:909–913. doi: 10.1038/nbt847. [DOI] [PubMed] [Google Scholar]
- Sarid-Krebs L., Panigrahi K.C.S., Fornara F., Takahashi Y., Hayama R., Jang S., Tilmes V., Valverde F., Coupland G. Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 2015;84:451–463. doi: 10.1111/tpj.13022. [DOI] [PubMed] [Google Scholar]
- Shen C., Liu H., Guan Z., Yan J., Zheng T., Yan W., Wu C., Zhang Q., Yin P., Xing Y. Structural insight into DNA recognition by CCT/NF-YB/YC complexes in plant photoperiodic flowering. Plant Cell. 2020;32:3469–3484. doi: 10.1105/tpc.20.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R., Gómez-Ariza J., Brambilla V., Fornara F. Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 2014;114:1445–1458. doi: 10.1093/aob/mcu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C.S., Nayak A., Lai X., Hutin S., Hugouvieux V., Jung J.H., López-Vidriero I., Franco-Zorrilla J.M., Panigrahi K.C.S., Nanao M.H., et al. Molecular mechanisms of evening complex activity in arabidopsis. Proc. Natl. Acad. Sci. USA. 2020;117:6901–6909. doi: 10.1073/pnas.1920972117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Rühl M., de Montaigu A., Wötzel S., Coupland G. Evolution of CONSTANS regulation and function after gene duplication produced a photoperiodic flowering switch in the Brassicaceae. Mol. Biol. Evol. 2015;32:2284–2301. doi: 10.1093/molbev/msv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Estrada D.A., Johnson R.S., Kim S.K., Lee S.Y., MacCoss M.J., Imaizumi T. Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA. 2014;111:17672–17677. doi: 10.1073/pnas.1415375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Chen Y., Liu L., Wang Y., Bao S., Zhou X., Teo Z.W.N., Mao C., Gan Y., Yu H. OsFTIP1-Mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKγ4. Plant Cell. 2017;29:491–507. doi: 10.1105/tpc.16.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Li L., Li Y., Wang T., Fu X. Overexpression of gene OsSUI1 affects floral organ development in rice (Oryza sativa L.) Mol. Breed. 2018;38 [Google Scholar]
- Sui P., Shi J., Gao X., Shen W.-H., Dong A. H3K36 methylation is involved in promoting rice flowering. Mol. Plant. 2013;6:975–977. doi: 10.1093/mp/sss152. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang Z., Wu J., Cui X., Feng D., Wang K., Xu M., Zhou L., Han X., Gu X., Lu T. The oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering. PLoS Genet. 2016;12:e1005927. doi: 10.1371/journal.pgen.1005927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Wang C., Chen X., Liu H., Huang Y., Li S., Dong Z., Zhao X., Tian F., Jin W. dlf1 promotes floral transition by directly activating ZmMADS4 and ZmMADS67 in the maize shoot apex. New Phytol. 2020;228:1386–1400. doi: 10.1111/nph.16772. [DOI] [PubMed] [Google Scholar]
- Sun K., Huang M., Zong W., Xiao D., Lei C., Luo Y., Song Y., Li S., Hao Y., Luo W., et al. Hd1, Ghd7, and DTH8 synergistically determine the rice heading date and yield-related agronomic traits. J. Genet. Genomics. 2022;49:437–447. doi: 10.1016/j.jgg.2022.02.018. [DOI] [PubMed] [Google Scholar]
- Susila H., Jurić S., Liu L., Gawarecka K., Chung K.S., Jin S., Kim S.J., Nasim Z., Youn G., Suh M.C., et al. Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science. 2021;373:1137–1142. doi: 10.1126/science.abh4054. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Shomura A., Sasaki T., Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Teshima K.M., Yokoi S., Innan H., Shimamoto K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA. 2009;106:4555–4560. doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tamaki S., Tsuji H., Matsumoto A., Fujita A., Shimatani Z., Terada R., Sakamoto T., Kurata T., Shimamoto K. FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc. Natl. Acad. Sci. USA. 2015;112:E901–E910. doi: 10.1073/pnas.1417623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K.i., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., Yamaguchi M., Nakashima C., Purwestri Y.A., Tamaki S., et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Thirumurugan T., Ito Y., Kubo T., Serizawa A., Kurata N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- Thomas B., Vince-Prue D. Academic Press; 1997. Photoperiodism in Plants. [Google Scholar]
- Tiwari S.B., Shen Y., Chang H.C., Hou Y., Harris A., Ma S.F., Mcpartland M., Hymus G.J., Adam L., Marion C., et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis -element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- Trusov Y., Botella J.R. Silencing of the ACC synthase gene ACACS2 causes delayed flowering in pineapple [Ananas comosus (L.) Merr.] J. Exp. Bot. 2006;57:3953–3960. doi: 10.1093/jxb/erl167. [DOI] [PubMed] [Google Scholar]
- Tsuji H., Nakamura H., Taoka K.i., Shimamoto K. Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 2013;54:385–397. doi: 10.1093/pcp/pct005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylewicz S., Tsuji H., Miskolczi P., Petterle A., Azeez A., Jonsson K., Shimamoto K., Bhalerao R.P. Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. Proc. Natl. Acad. Sci. USA. 2015;112:3140–3145. doi: 10.1073/pnas.1423440112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang W., Yin Z., Wen C.-K. Rice CONSTITUTIVE TRIPLE-RESPONSE2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J. Exp. Bot. 2013;64:4863–4875. doi: 10.1093/jxb/ert272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sun S., Jin J., Fu D., Yang X., Weng X., Xu C., Li X., Xiao J., Zhang Q. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA. 2015;112:15504–15509. doi: 10.1073/pnas.1521949112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lu Y., Guo Z., Ding Y., Ding C. 2020. RICE CENTRORADIALIS 1, a TFL1-like Gene, Responses to Drought Stress and Regulates Rice Flowering Transition. Rice13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jiao X., Kong X., Liu Y., Chen X., Fang R., Yan Y. The histone deacetylase HDA703 interacts with OsBZR1 to regulate rice brassinosteroid signaling, growth and heading date through repression of Ghd7 expression. Plant J. 2020;104:447–459. doi: 10.1111/tpj.14936. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhou P., Huang R., Zhang J., Ouyang X. A daylength recognition model of photoperiodic flowering. Front. Plant Sci. 2021;12:778515. doi: 10.3389/fpls.2021.778515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Guo J., Peng Y., Lyu X., Liu B., Sun S., Wang X. Light-induced mobile factors from shoots regulate rhizobium-triggered soybean root nodulation. Science. 2021;374:65–71. doi: 10.1126/science.abh2890. [DOI] [PubMed] [Google Scholar]
- Wang L., Ming L., Liao K., Xia C., Sun S., Chang Y., Wang H., Fu D., Xu C., Wang Z., et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol. Plant. 2021;14:1168–1184. doi: 10.1016/j.molp.2021.04.013. [DOI] [PubMed] [Google Scholar]
- Wang X., He Y., Wei H., Wang L. A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ. 2021;44:3283–3301. doi: 10.1111/pce.14167. [DOI] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. (Wash. D C) 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X., Wang L., Wang J., Hu Y., Du H., Xu C., Xing Y., Li X., Xiao J., Zhang Q. Grain Number, Plant Height, and Heading Date7 is a central regulator of growth, development, and stress response. Plant Physiol. (Wash. D C) 2014;164:735–747. doi: 10.1104/pp.113.231308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickland D.P., Hanzawa Y. The FLOWERING LOCUS T/TERMINAL FLOWER1 gene family: functional evolution and molecular mechanisms. Mol. Plant. 2015;8:983–997. doi: 10.1016/j.molp.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Wolbang C.M., Ross J.J. Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta. 2001;214:153–157. doi: 10.1007/s004250100663. [DOI] [PubMed] [Google Scholar]
- Wolbang C.M., Chandler P.M., Smith J.J., Ross J.J. Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiol. (Wash. D C) 2004;134:769–776. doi: 10.1104/pp.103.030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., You C., Li C., Long T., Chen G., Byrne M.E., Zhang Q. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. USA. 2008;105:12915–12920. doi: 10.1073/pnas.0806019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Zheng X.-M., Lu G., Zhong Z., Gao H., Chen L., Wu C., Wang H.-J., Wang Q., Zhou K., et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA. 2013;110:2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Cui K., Wang W., Li Q., Fahad S., Hu Q., Huang J., Nie L., Mohapatra P.K., Peng S. Heat-induced cytokinin transportation and degradation are associated with reduced panicle cytokinin expression and fewer spikelets per panicle in rice. Front. Plant Sci. 2017;8:371. doi: 10.3389/fpls.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuriyanghan H., Zhang B., Cao W.-H., Ma B., Lei G., Liu Y.-F., Wei W., Wu H.-J., Chen L.-J., Chen H.-W., et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell. 2009;21:1473–1494. doi: 10.1105/tpc.108.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K., Wang R., Ou X., Fang Z., Tian C., Duan J., Wang Y., Zhang M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yaish M.W., El-kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.-M., Rothstein S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fu D., Zhu C., He Y., Zhang H., Liu T., Li X., Wu C. The RING-finger ubiquitin ligase HAF1 mediates heading date 1 degradation during photoperiodic flowering in rice. Plant Cell. 2015;27:2455–2468. doi: 10.1105/tpc.15.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yuan Z., Meng Q., Huang G., Périn C., Bureau C., Meunier A.-C., Ingouff M., Bennett M.J., Liang W., Zhang D. Dynamic regulation of auxin response during rice development revealed by newly established hormone biosensor markers. Front. Plant Sci. 2017;8:256. doi: 10.3389/fpls.2017.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., Sasaki T. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T., Li Y., Zhang J., Hou W., Zhou W., Lu J., Xing Y., Li X. Nitrogen, phosphorus, and potassium fertilization affects the flowering time of rice (Oryza sativa L.) Glob. Ecol. Conserv. 2019;20 [Google Scholar]
- Yin C., Gan L., Ng D., Zhou X., Xia K. Decreased panicle-derived indole-3-acetic acid reduces gibberellin A1 level in the uppermost internode, causing panicle enclosure in male sterile rice Zhenshan 97A. J. Exp. Bot. 2007;58:2441–2449. doi: 10.1093/jxb/erm077. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T., Ito M., Sumikura T., Nakayama A., Nishimura T., Kitano H., Yamaguchi I., Koshiba T., Hibara K.-I., Nagato Y., Itoh J.I. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014;78:927–936. doi: 10.1111/tpj.12517. [DOI] [PubMed] [Google Scholar]
- Zhang S., Hu W., Wang L., Lin C., Cong B., Sun C., Luo D. TFL1/CEN-like genes control intercalary meristem activity and phase transition in rice. Plant Sci. 2005;168:1393–1408. [Google Scholar]
- Zhang C., Liu J., Zhao T., Gomez A., Li C., Yu C., Li H., Lin J., Yang Y., Liu B., Lin C. A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol. (Wash. D C) 2016;171:334–343. doi: 10.1104/pp.16.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hu W., Shen G., Liu H., Hu Y., Zhou X., Liu T., Xing Y. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day condition. Sci. Rep. 2017;7:5388. doi: 10.1038/s41598-017-05873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhang Y., Li K., Yan M., Zhang J., Yu M., Tang S., Wang L., Qu H., Luo L., et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr. Biol. 2021;31:671–683.e5. doi: 10.1016/j.cub.2020.10.095. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang F., Zhou X., Poh T.X., Xie L., Shen J., Yang L., Song S., Yu H., Chen Y. The tetratricopeptide repeat protein OsTPR075 promotes heading by regulating florigen transport in rice. Plant Cell. 2022;34:3632–3646. doi: 10.1093/plcell/koac190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C.-W., Eizenga G.C., Wright M.H., Ali M.L., Price A.H., Norton G.J., Islam M.R., Reynolds A., Mezey J., et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen H., Ren D., Tang H., Qiu R., Feng J., Long Y., Niu B., Chen D., Zhong T., et al. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa) New Phytol. 2015;208:936–948. doi: 10.1111/nph.13503. [DOI] [PubMed] [Google Scholar]
- Zheng T., Sun J., Zhou S., Chen S., Lu J., Cui S., Tian Y., Zhang H., Cai M., Zhu S., et al. Post-transcriptional regulation of Ghd7 protein stability by phytochrome and OsGI in photoperiodic control of flowering in rice. New Phytol. 2019;224:306–320. doi: 10.1111/nph.16010. [DOI] [PubMed] [Google Scholar]
- Zhu W., Yang L., Wu D., Meng Q., Deng X., Huang G., Zhang J., Chen X., Ferrándiz C., Liang W., et al. Rice SEPARATA genes OsMADS5 and OsMADS34 cooperate to limit inflorescence branching by repressing the TERMINAL FLOWER1-like gene RCN4. New Phytol. 2022;233:1682–1700. doi: 10.1111/nph.17855. [DOI] [PubMed] [Google Scholar]
- Zong W., Ren D., Huang M., Sun K., Feng J., Zhao J., Xiao D., Xie W., Liu S., Zhang H., et al. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading. New Phytol. 2021;229:1635–1649. doi: 10.1111/nph.16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong J., Wang L., Zhu L., Bian L., Zhang B., Chen X., Huang G., Zhang X., Fan J., Cao L., et al. A rice single cell transcriptomic atlas defines the developmental trajectories of rice floret and inflorescence meristems. New Phytol. 2022;234:494–512. doi: 10.1111/nph.18008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.