Abstract
JUJUNCAO (Cenchrus fungigraminus; 2n = 4x = 28) is a Cenchrus grass with the highest biomass production among cultivated plants, and it can be used for mushroom cultivation, animal feed, and biofuel production. Here, we report a nearly complete genome assembly of JUJUNCAO and reveal that JUJUNCAO is an allopolyploid that originated ∼2.7 million years ago (mya). Its genome consists of two subgenomes, and subgenome A shares high collinear synteny with pearl millet. We also investigated the genome evolution of JUJUNCAO and suggest that the ancestral karyotype of Cenchrus split into the A and B ancestral karyotypes of JUJUNCAO. Comparative transcriptome and DNA methylome analyses revealed functional divergence of homeologous gene pairs between the two subgenomes, which was a further indication of asymmetric DNA methylation. The three types of centromeric repeat in the JUJUNCAO genome (CEN137, CEN148, and CEN156) may have evolved independently within each subgenome, with some introgressions of CEN156 from the B to the A subgenome. We investigated the photosynthetic characteristics of JUJUNCAO, revealing its typical C4 Kranz anatomy and high photosynthetic efficiency. NADP-ME and PEPCK appear to cooperate in the major C4 decarboxylation reaction of JUJUNCAO, which is different from other C4 photosynthetic subtypes and may contribute to its high photosynthetic efficiency and biomass yield. Taken together, our results provide insights into the highly efficient photosynthetic mechanism of JUJUNCAO and provide a valuable reference genome for future genetic and evolutionary studies, as well as genetic improvement of Cenchrus grasses.
Key words: genome assembly, allotetrapolyploid, centromere architecture, evolutionary trajectory, photosynthetic efficiency, Cenchrus grass
This study reports the assembly of a gapless, T2T genome of the allotetrapolyploid Cenchrus fungigraminus (JUJUNCAO). Using the near-complete genome, the authors present a full picture of its centromere architecture, reveal the evolutionary trajectory of its genome, establish a model of the ancestral grass karyotype of its two subgenomes, and provide insights into the mechanisms that underlie its high photosynthetic efficiency.
Introduction
Cenchrus is a widespread genus of the Poaceae family. Cenchrus grasses are C4 plants characterized by high photosynthetic efficiency, massive biomass production, and high stress tolerance. Most species are important for animal forage, cultivation material for edible and medicinal mushrooms (so called JUNCAO 菌草), and biofuel production. JUJUCNAO (Cenchrus fungigraminus; named JUJUNCAO for giant JUNCAO, 巨菌草; Supplemental Figure 1) is the most productive Cenchrus species, with an average height of ∼4.0 m and a green-matter yield of ∼400 ton/ha annually (Lin, 2013; Lin et al., 2022), which is about twice that of the two elephant grasses Cenchrus purpureus (Yan et al., 2020) and Pennisetum purpureum (Zhang et al., 2022b) and other high-biomass C4 grasses (Mullet, 2017). JUJUNCAO is able to grow in arid and semi-arid regions as well as ecologically fragile areas (Lin, 2013; Xu et al., 2014; He et al., 2017; Hayat et al., 2020; Jia et al., 2020). Cultivation of Cenchrus grasses such as JUJUNCAO is one of the most efficient strategies for using marginal land that is not optimal for traditional crops and satisfying increasing demands for food, including meat, milk, and mushrooms (Samson et al., 2005; Lin, 2013; Eisler et al., 2014; Gu et al., 2019; Xie and Xu, 2019; Kuang et al., 2022).
JUJUNCAO is morphologically similar to another Cenchrus grass cultivar (P. purpureum × Pennisetum americanum cv. Reyan No. 4), which is a triploid offspring of elephant grass (Luo et al., 2016). JUJUNCAO can be distinguished from other Cenchrus grasses by its stem, leaf, flower, or color phenotypes (Lin et al., 2022). These other species include pearl millet (Cenchrus americanus; syn. Pennisetum glaucum) (Varshney et al., 2017), C. purpureus (Yan et al., 2020) and P. purpureum (Zhang et al., 2022b), three grasses with assembled genome sequences. Although many efforts have been made to decipher the molecular biology of JUJUNCAO (Lin et al., 2015; Ye et al., 2015; Zhu et al., 2015; Chen et al., 2016; Li et al., 2020; Zhou et al., 2021), little is known about the molecular basis of its agronomic traits and evolutionary trajectory, owing to the lack of a high-quality reference genome.
Polyploidization is an important evolutionary force in angiosperm plants and provides diverse genetic resources to benefit the domestication of crops (Adams and Wendel, 2005; Soltis et al., 2009). Polyploidization may also result in high sequence similarity between homologous/homeologous chromosomes and thus in frequent rearrangements between these chromosomes (Wendel et al., 2016), a feature that poses major challenges in genomic biology, such as deciphering the architecture of telomeres and centromeres. Centromeres are essential for genome integrity because they mediate the junction between sister chromatids and the proper separation of chromosomes during mitosis and meiosis (Liu et al., 2020b). Plant centromeres often consist of satellite arrays interrupted by long terminal repeat (LTR) retrotransposons (Zhong et al., 2002; Hall et al., 2004; Comai et al., 2017), making it challenging to understand centromere structure. Assembly of a highly contiguous genome is therefore essential for resolving centromere structure and mechanisms of chromosome fusion and fission in the allotetraploid JUJUNCAO.
Recent advances in DNA sequencing technology and assembly methodology have enabled gapless, telomere-to-telomere (T2T) chromosome/genome assemblies for human cells (Nurk et al., 2022), barley (Navrátilová et al., 2022), banana (Belser et al., 2021), and maize (Liu et al., 2020a), which enable in-depth investigation of the structure and evolutionary trajectory of chromosomes, especially in regions with complex repetitive sequences such as telomeres and centromeres. Here, we generated a T2T gapless genome of the allotetraploid JUJUNCAO, performed bisulfite whole-genome sequencing, and analyzed its genome evolution and the genomic basis of its C4 characteristics. These results provide insights into the evolution of JUJUNCAO, particularly its centromere architecture, and the molecular b asis of its high photosynthetic efficiency.
Results
Assembly of a T2T genome of JUJUNCAO
To decipher the genome structure of JUJUNCAO, we first estimated its genome size to be 2.05–2.09 Gb (Supplemental Table 1) using flow cytometry analysis (Dolezel and Bartos, 2005). A total of 120 Gb (∼60× coverage of the estimated genome size) of PacBio long high-fidelity (HiFi) circular consensus sequencing (CCS) data were generated and assembled into 2.13 Gb of contigs using the Hifiasm assembler (Cheng et al., 2021). A final total of 116 contigs were obtained after removal of 145 Mb of heterozygous sequences, resulting in a total assembly size of 1.99 Gb, with 1.97 Gb (∼99.0%) anchored into 14 pseudo-chromosomes based on Hi-C data (Figure 1; Supplemental Figure 2; Table 1 and Supplemental Table 2). The assembled JUJUNCAO genome contains 24 annotated telomeres and 13 T2T chromosomes, eight of which are gap-free (Chr2B, 3A, 3B, 4B, 5A, 5B, 6A, and 7A), four of which have only one gap (Chr1B, 2A, 6B, and 7B), and one of which (Chr4A) has three gaps at the telomeric region of the short arm (Supplemental Figure 2C).
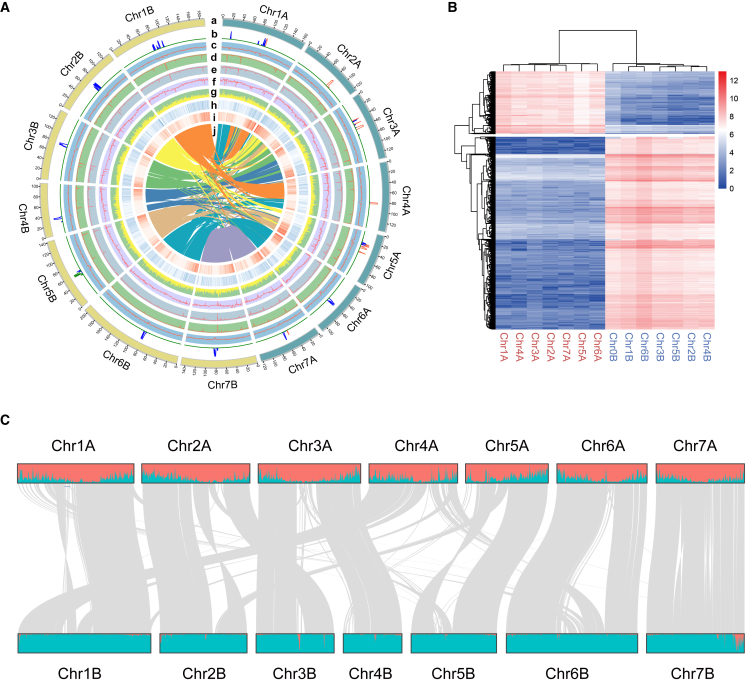
Figure 1.
Allotetraploid genome of JUJUNCAO (C. fungigraminus).
(A) Circos plot of the JUJUNCAO genome assembly. Quantitative tracks (b to j) are aggregated in 500-kbp bins, and independent y axis labels are as follows: (a) chromosomes; (b) centromeric repeat sequences; (c) GC content; (d) CG methylation; (e) CHG methylation; (f) CHH methylation; (g) LTRs; (H) SNPs; (h) gene expression; (i) gene density; (j) syntenic relationships.
(B) Clustering of counts of 13-mers that differentiate homeologous chromosomes enabled consistent partitioning of the genome into two subgenomes. Red and blue chromosome names correspond to the A and B subgenomes, respectively.
(C) Distribution of subgenome-specific 13-mer sequences (red for subgenome A and blue for subgenome B) for 14 chromosomes and synteny between the two subgenomes.
We used benchmarking universal single-copy orthologs (BUSCO; Simao et al., 2015) and the LTR retrotransposon assembly index (LAI; Ou et al., 2018) to assess the completeness and contiguity of the assembly. The total complete BUSCO score was 98.4%, and the LAI of the JUJUNCAO assembly was 15.9 (Table 1 and Supplemental Tables 3 and 4).
Table 1.
Genome assembly and annotation statistics of JUJUNCAO
| Assembly | Initial assembly | Purge haplotigs | Hi-C |
|---|---|---|---|
| Total assembly size of contigs (bp) | 2 131 895 497 | 1 986 757 443 | – |
| Number of contigs | 815 | 116 | – |
| Contig N50 (bp) | 131 346 082 | 134 074 806 | – |
| Contig N90 (bp) | 94 754 406 | 99 768 082 | – |
| Longest contig (bp) | 168 262 481 | 162 376 048 | – |
| GC content (%) | 47.11 | 47.11 | 47.11 |
| BUSCO completeness of assembly (%) | – | 98.40 | – |
| LAIa | – | 15.90 | – |
| Length of chromosomes (bp) | – | – | 1 966 989 277 |
| Number of chromosomes | – | – | 14 |
| Number of gaps | – | – | 13 |
| Anchored rate (%) | – | – | 99.00 |
| Annotation | – | – | – |
| Percentage of repeat elements | 73.38 | – | – |
| Total number of genes | 68 526 | – | – |
| BUSCO completeness of annotation (%) | 93.00 | – | – |
| Subgenome | A subgenome | B subgenome | – |
| Length of chromosomes (bp) | 899 693 551 | 1 067 295 726 | – |
| Number of chromosomes | 7 | 7 | – |
| Number of genes | 34 630 | 33 779 | – |
LAI was used to assess genome assembly quality.
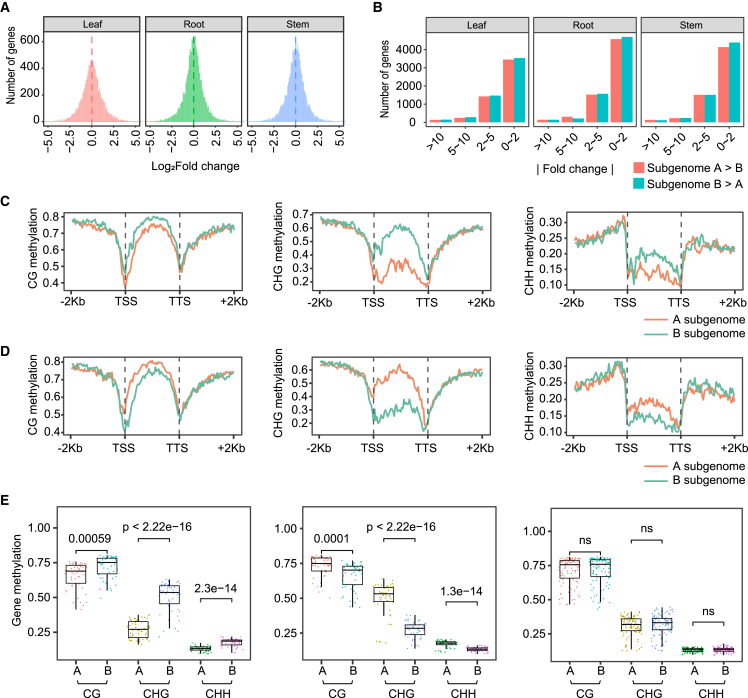
Figure 4.
Expression and DNA methylation divergence in the allotetraploid genome of JUJUNCAO.
(A) The overall distribution of the log2 (fold change) of expression between homeologous gene pairs in leaves, stems, and roots.
(B) The number of homeologous gene pairs between the A and B subgenomes with different degrees of expression bias. The expression bias (absolute value of fold change) was divided into four bins: 0–2, 2–5, 5–10, >10.
(C) The methylation profiles of homeologous gene pairs with subgenome A–biased expression in the stem (fold change >2).
(D) The methylation profiles of homeologous gene pairs with subgenome B–biased expression in the stem (fold change >2).
(E) Comparison of three types of methylation level in the gene body of A and B subgenomes in the stem. From left to right panel: the methylation levels of subgenome A–biased homeologous gene pairs (fold change >2), subgenome B–biased homeologous gene pairs (fold change >2), and differentially expressed gene pairs (with 0–2 fold change). All pairwise comparisons were performed by t test.
Genome annotation
We predicted 68 562 gene models using ab initio gene prediction and homology-based methods. Subgenomes A and B harbor 34 630 and 33 779 genes, respectively. BUSCO (v5.4.1) analysis was performed to evaluate the gene annotations (Simao et al., 2015), and 93.0% of the conserved genes from the BUSCO database were present in the JUJUNCAO genome (Table 1 and Supplemental Tables 4 and 5).
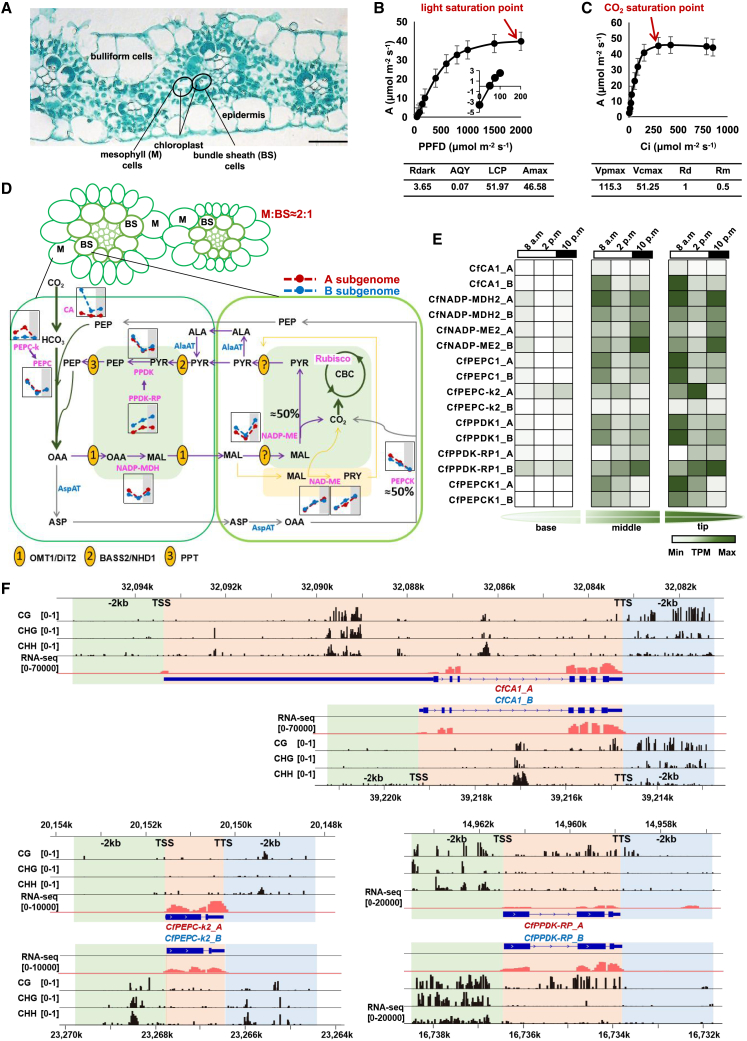
Figure 5.
C4 pathway in JUJUNCAO.
(A) Cross-sections of JUJUNCAO leaves. Horizontal bars indicate 20 μm.
(B) The light response of net CO2 assimilation rate (n = 5) (mean ± SD). PPFD, photosynthetic photon flux density; A, net photosynthetic rate; Rdark, dark respiration; AQY, apparent quantum yield; LCP, light compensation point; Amax, maximum CO2 net assimilation rate.
(C) The response of net CO2 assimilation rate to intercellular CO2 concentration (Ci) (n = 5) (mean ± SD). Vpmax, maximal PEP carboxylation activity; Vcmax, maximal Rubisco activity; Rd, leaf mitochondrial respiration; Rm, mesophyll mitochondrial respiration.
(D) Schematic of the C4 pathway in JUJUNCAO. The NADP-ME (purple) and PEPCK (grey) pathway predominates, whereas the NAD-ME (orange) pathways function as subsidiary pathways. CA, carbonic anhydrase; PEPC, phosphoenolpyruvate carboxylase; PEPC-k, PEPC kinase; NADP-MDH, NADP-malate dehydrogenase; NADP-ME, NADP-malic enzyme; NAD-ME, NAD-malic enzyme; PEPCK, phosphoenolpyruvate carboxykinase; PPDK, pyruvate orthophosphate dikinase; PPDK-RP, PPDK regulatory protein. Transporters of C4 metabolites, including BASS2 (probable sodium/metabolite cotransporter 2), NHD1 (sodium/proton antiporter 1), OMT1 (2-oxoglutarate/malate transporter 1), and DiT2 (dicarboxylate transporter 2), and accessory proteins, including AspAT (aspartate transaminase) and AlaAT (alanine transaminase), were also identified (Supplemental Table 11). The figure shows the expression of genes at different time points (08:00 h, 14:00 h, and 22:00 h). Shaded boxes indicate the time points for the night period (22:00 h). Red indicates genes in the A subgenome, and blue indicates genes in the B subgenome.
(E) Expression patterns of JUJUNCAO carbon fixation genes across the leaf gradient at different time points.
(F) IGV tracks showing methylation and RNA-seq levels of CfCA1, CfPEPC-K2, and CfPPDK-RP1. The patterns of CG, CHG, and CHH methylation in C4 genes are shown from top to bottom. From left to right, the green background represents the 2-kb region upstream of the gene, the red background represents the gene region, and the blue background represents the 2-kb region downstream of the gene.
A total of 1.16 Gb of repeat sequences were found in the JUJUNCAO genome, accounting for 68.87% of the assembly. The total proportion of transposable elements (TEs) in JUJUCAO was comparable to that in Sorghum bicolor, higher than that in Setaria italica and Panicum miliaceum, but lower than that in Zea mays (Supplemental Figure 3; Supplemental Table 6). We also investigated the LTR insertion times and found a smaller wave of LTR bursts in JUJUNCAO at ∼1.2 mya, similar to that in Z. mays. By contrast, the other species investigated, such as C. americanus, P. miliaceum, S. bicolor, and S. italica, had a stronger LTR burst within 1 mya (Supplemental Figure 3B). LTR retrotransposons were the most abundant TEs in the JUJUNCAO genome, accounting for 35.69% of the genome, followed by unknown repeats (28.67%), and DNA transposons occupied only 4.51% of the genome. The Ty3/Gypsy superfamily was the most abundant type of LTR retrotransposon, making up more than half of the retrotransposons and 22.40% of the JUJUNCAO genome (Supplemental Figure 3C; Supplemental Table 6).
Disentangling the subgenomes
To further confirm the chromosome phasing results, we identified 32 629 chromosome-specific 13-bp sequences (13-mers), which clustered into two distinct groups, indicative of the two subgenomes in JUJUNCAO (Figure 1B and 1C). These results are consistent with the allopolyploidy features revealed by our karyotyping results (Supplemental Figure 2A; Zhu et al., 2015). The average nucleotide identity of homologous regions in the two subgenomes was 90.1%, and chromosome rearrangements occurred frequently after the split of the common ancestor of the two subgenomes (Figure 1C). Except for Chr07A/Chr07B, in which large segment arrangements were observed in the other six chromosome pairs, subgenome A of JUJUNCAO exhibited highly conserved collinear synteny with the genome of pearl millet, with 90.7% average nucleotide identity. By contrast, subgenome B of JUJUNCAO and the genome of pearl millet showed more genome rearrangements and had 86.4% average nucleotide identity (Supplemental Figure 4).
Genome evolution
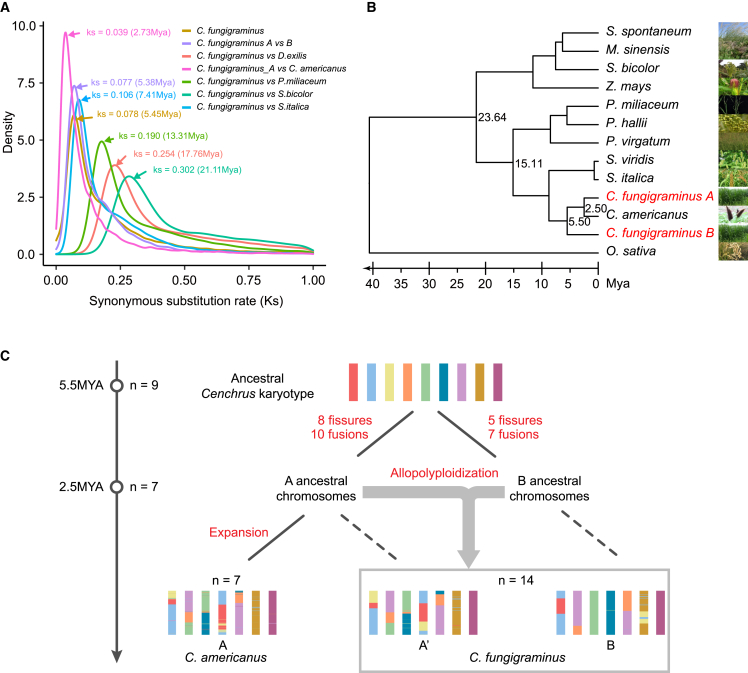
The near-T2T complete assembly enabled us to investigate the evolution of the JUJUNCAO genome. To establish the timeline of paleo-tetraploidy in JUJUNCAO, we performed inter- and intra-subgenomic comparisons between JUJUNCAO and other Panicoideae species using protein sequences of the primary transcripts from 10 representative Panicoideae species (Saccharum spontaneum, Miscanthus sinensis, S. bicolor, Z. mays, Panicum hallii, P. miliaceum, Panicum virgatum, C. americanus, S. italica, and Setaria viridis) and the outgroup species Oryza sativa (Figure 2A). Our results showed that JUJUNCAO diverged from S. italica, P. miliaceum, and S. bicolor approximately 7.4 mya, 13.3 mya, and 21.1 mya, respectively (Figure 2A). The allotetraploid origin of JUJUNCAO was estimated at ∼2.7 mya based on the Ks value of 0.039.
Figure 2.
Phylogenetic relationships between JUJUNCAO and species of Panicoideae.
(A) Distributions of synonymous substitution rate (Ks) between C. fungigraminus and seven representative species of Panicoideae. The Ks peaks for pairwise comparisons are indicated by arrows.
(B) Phylogenetic tree of 11 species and the A/B subgenomes of C. fungigraminus.
(C) Chromosome evolution of JUJUNCAO and C. americanus from ancestral chromosomes. The nine chromosomes of the ancestral Cenchrus karyotype (ACK) were reconstructed from syntenic blocks of related species, and the origin of the two JUJUNCAO subgenomes was simulated. Nine colors indicate the composition of the ACK from two subgenomes with distinct origins. These chromosomes are for illustrative purposes of fissure and fusion only and do not represent the actual dimensions.
We then traced the evolutionary trajectory of JUJUNCAO and representative species from different genera of Panicoideae, including Z. mays, S. bicolor, Echinochloa haploclada, Digitaria exilis, P. miliaceum, P. hallii, and S. italica (Supplemental Figure 5). According to the number and structure of chromosomes in these species, we inferred that the Paniceae ancestor originated at ∼15.1 mya and had nine chromosomes. Most of the Paniceae species evolved from the ancestral grass karyotype (AGK) through 10–18 chromosome rearrangement events, including fusions and fissures. For example, E. haploclada evolved with the fewest chromosomal rearrangement events (four fusions and six fissures) and most closely resembled the AGK. Ancestors of JUJUNCAO subgenomes A and B are estimated to have diverged ∼5.5 mya from Cenchrus ancestors, following the divergence of Setaria and Cenchrus at ∼8.1 mya (Figure 2B and Supplemental Figure 5). Our results showed that each of the two JUJUNCAO subgenomes contained one chromosome (Chr6A and Chr4B) that resembled the AGK, suggesting that the two subgenomes of JUJUNCAO may have evolved independently from the same ancestral Cenchrus karyotype. Based on the chromosome synteny of Paniceae species, we propose the ancestral Cenchrus karyotype (ACK) (Figure 2C; Supplemental Figure 6). Compared with the ACK, 18 large rearrangements (eight fissures and 10 fusions) were found in subgenome A and 12 large rearrangements (five fissures and seven fusions) in subgenome B. We also found that JUJUNCAO and P. purpureum diverged ∼0.3 mya and displayed similar karyotypes compared with the AGK, but the sequence similarity between homologous genes differed among different chromosome pairs (Supplemental Figure 7).
Centromere architecture in JUJUNCAO
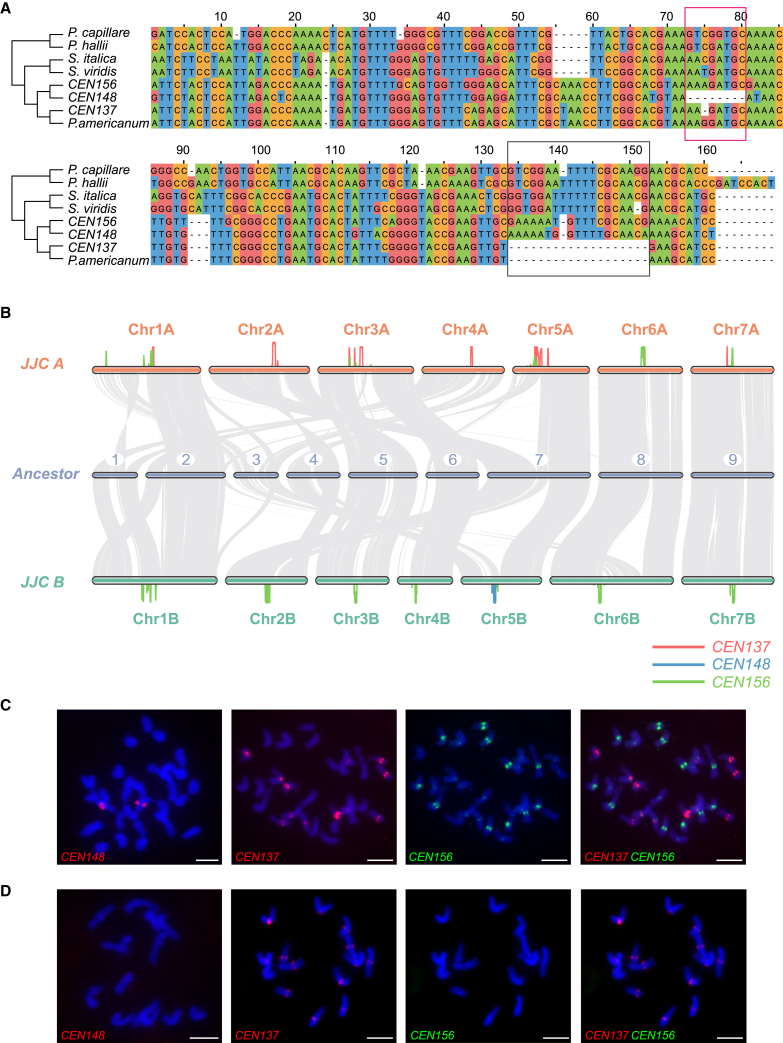
Achieving high contiguity of centromeres is one of the major challenges in T2T genome assembly. Here, the complete genome enabled us to annotate the centromeres of all 14 chromosomes. We identified three types of centromeric repeats with high sequence similarity in the JUJUNCAO genome, CEN137, CEN148, and CEN156, each composed of satellite repeats with lengths of ∼137 bp, ∼148 bp, and ∼156 bp, respectively (Supplemental Table 7). These satellite repeats shared high sequence similarity and were relatively conserved among Paniceae species (identity: 59%–98%) such as S. viridis, S. italica, P. hallii, and Panicum capillare (Figure 3A). Among them, CEN156 showed the highest sequence similarity to centromeric monomers from other species, whereas CEN137 and CEN148 showed 19-bp and 8-bp deletions, respectively (Figure 3A).
Figure 3.
Characteristics of centromere sequences in JUJUNCAO.
(A) Multiple sequence alignment of the three centromeric satellite sequences from JUJUNCAO and their homologous sequences from S. viridis, S. italica, P. hallii, P. capillare, and P. americanum. Green, red, orange, and blue represent A, G, C, and T bases, respectively. The red and blue boxes represent variant loci within the three centromeric satellites (CEN137, CEN156, and CEN148).
(B) Syntenic blocks between the A and B subgenomes of JUJUNCAO (JJC A and B) and the expected Paniceae ancestral genome (ACK). Red, blue, and green lines show the distributions of different centromeric sequences in the JUJUNCAO genome.
(C and D) FISH assays of JUJUNCAO (C) and the C. americanum cultivar MZL (D) using repeat-specific probes generated from the three centromeric satellites (CEN137, CEN148, and CEN156). Scale bars, 10 μm.
We used a k-mer method to accurately locate the three similar satellite repeats and found that they were distributed in distinct patterns. CEN156 is the most abundant centromeric array in the JUJUNCAO genome and is enriched on almost all chromosomes, with the exception of Chr2A and Chr4A. CEN137 is enriched mainly on chromosomes from subgenome A, and CEN148 is found specifically on Chr5B (Figure 3B). We performed fluorescence in situ hybridization (FISH) to confirm these results in JUJUNCAO and the diploid P. americanum cultivar MZL using probes generated from repeat-specific oligomers of the three centromeric repeats. There were two strong signals representing the Chr5B-specific localization of CEN148 in JUJUNCAO, and these were absent in MZL. CEN137 signals were present on 12 of the chromosomes of JUJUNCAO and MZL, with the strongest signal on Chr5A. For CEN156, there were 11 signals representing six of the chromosomes of JUJUNCAO, and these were absent in MZL. There were also weak signals for CEN156 in the JUJUNCAO genome, some of which co-localized with that of CEN137 (Figure 3C). Taken together, our results revealed high sequence similarity and a distinct distribution pattern of the three centromeric satellites identified in the JUJUNCAO genome. The similarity in centromeric structure between the JUJUNCAO A subgenome and the diploid P. americanum MZL genome suggest that they are derived from common A ancestral chromosomes (Figure 2C).
Expression bias of homeologous gene pairs
Polyploidization may cause expression bias between homeologous gene pairs (Yang et al., 2016; Liang and Schnable, 2018). To investigate homeolog expression bias in allotetraploid JUJUNCAO, we defined 22 260 homoeologous gene pairs between the A and B subgenomes. Comparative transcriptome analysis using RNA sequencing (RNA-seq) data generated from leaves, stems, and roots showed that A-subgenome genes are expressed at a slightly lower level than those of the B subgenome, but there is no major overall bias for one subgenome (Figure 4A and Supplemental Figure 8). We identified 5668 and 5708 expression-biased genes in subgenomes A and B, respectively (Figure 4B). Subgenome B–biased homeologs were enriched in Chr1B, the subtelomeric region of the long arm of Chr4B, and the subtelomeric region of the short arms of Chr5B and Chr6B. Subgenome A–biased homeologs were enriched in Chr1A, the centromere proximal region, and the short arm of Chr7A (Supplemental Figure 9). In total, 1124 differentially expressed homeologous gene pairs (DEHGs) were identified: 557 showed subgenome-A bias and 567 showed subgenome-B bias. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional classification showed that DEHGs from the two subgenomes were enriched in different metabolism, biosynthesis, and signaling pathways in roots, stems, and leaves. Carboxylic acid biosynthesis, RNA metabolic process, and mitogen-activated protein kinase (MAPK)-signaling pathways were specifically enriched in subgenome A–biased DEHGs, whereas hydrogen peroxide metabolic process, response to oxidative stress, and AMPK-signaling pathways were enriched in subgenome B–biased DEHGs (Supplemental Figures 10 and 11). These results revealed functional divergence between the two subgenomes of JUJUNCAO.
DNA methylation of homeologous gene pairs
Allele-specific DNA methylation is one of the mechanisms associated with expression bias of homeologs in allopolyploid plants (Edger et al., 2017; Bird et al., 2021). To investigate whether the expression bias of homeologous gene pairs in JUJUNCAO was determined by DNA methylation, we performed high-throughput bisulfite sequencing using libraries generated from leaves, stems, and roots, with a genome coverage of about 30× and a cytosine depth ranging from 15.1× to 19.1× in the three tissues (Supplemental Table 8). On average, about 67% of the reads generated from JUJUNCAO tissues could be mapped to the assembled genome, and the de-repetitive retention rate was ∼75% (Supplemental Table 9). The percentage methylation of CG, CHG, and CHH contexts was highest in roots (73.3%, 53%, and 2.4%, respectively), followed by stems (63.5%, 47.2%, and 2.3%) and leaves (63.0%, 44.3%, and 2.1%; Supplemental Table 10). Cytosine methylation was mainly enriched in regions around centromeres, although the methylation patterns of centromeres differed from each other. These results are consistent with the hypermethylation of repetitive sequences (Supplemental Figure 12).
We next compared overall DNA methylation patterns between the two subgenomes in the examined tissues. In upstream and downstream regions, methylation of all CG, CHG, and CHH contexts was significantly higher in subgenome B than in subgenome A for leaves (p < 1.9e−05) and stems (p < 0.037) (Supplemental Figure 13). In the gene body, only CHH methylation was significantly higher in subgenome B than in subgenome A for all tissues (p <8.5e−05). To further explore the association between DNA methylation and gene expression, we compared DNA methylation patterns of subgenome-biased genes and found that CG and CHG methylation rates in the gene body were significantly lower in subgenome-biased genes (whose expression was more than twice that of their homeologs) than in their subgenome homeologs. By contrast, CHH methylation showed the opposite pattern in the three tissues. We found no significant differences in methylation of gene pairs that showed a lower level of differential expression (0–2 fold change), with the exception of CHH methylation in leaves (p = 0.0084) (Figure 4C–4E, Supplemental Figures 14 and 15).
C4 photosynthetic characteristics of JUJUNCAO
To explore the C4 photosynthesis features of JUJUNCAO, we first performed a histological analysis and demonstrated that JUJUNCAO has typical Kranz anatomy, with two mesophyll (M) cells separated by consecutive enlarged bundle sheaths (BS) (BS-M-M-BS) (Figure 5A), similar to other C4 plants such as maize and sugarcane (Langdale, 2011; Sage et al., 2014). We next investigated the photosynthetic response of JUJUNCAO to light and CO2 concentration. At 400 μmol mol−1 CO2, JUJUNCAO had a light saturation point (LSP) near 2000 μmol mol−1 (Figure 5B) and a light-saturated net CO2 assimilation rate (A) of 40 μmol m−2 s−1. The apparent quantum yield (AQY) was 0.07, the light compensation point (LCP) was 51.97 μmol photons m−2 s−1, and the Amax was 46.58 μmol m−2 s−1 (Figure 5B). Investigation of the primary method of carbon fixation in JUJUNCAO showed that at a light intensity of 2000 μmol photons m−2 s−1, the CO2 response curve with a low CO2 compensation point (154 μmol mol−1) presents a steep slope typical of C4 species (Figure 5C) (Sage et al., 2014). After fitting, the maximal PEP carboxylation activity (Vpmax) was 115.30 μmol m−2 s−1, and the maximal Rubisco activity (Vcmax) was 51.25 μmol m−2 s−1 (Figure 5C). These results confirmed that JUJUNCAO is a C4 grass.
Genes related to the C4 photosynthetic pathway of JUJUNCAO
We identified C4 pathway genes in JUJUNCAO based on their C3 and C4 orthologs from sugarcane, maize, sorghum, and rice (Supplemental Figure 16; Supplemental Table 11). The JUJUNCAO genome contained 44 putative genes involved in the C4 carbon fixation module, including those encoding the key enzymes carbonic anhydrase (CA), NAD-malic enzyme (NAD-ME), NADP-malate dehydrogenase (NADP-MDH), NADP-malic enzyme (NADP-ME), phosphoenolpyruvate carboxylase (PEPC), phosphoenolpyruvate carboxykinase (PEPCK), PEPC kinase (PEPC-k), pyruvate orthophosphate dikinase (PPDK), and PPDK regulatory protein (PPDK-RP). These genes were evenly distributed in the two subgenomes (Supplemental Table 11). To further investigate the photosynthetic and diel expression patterns of C4 pathway genes, we collected RNA-seq samples at 08:00 h, 14:00 h, and 22:00 h from the immature zone (base), transition-zone (middle), and mature zone (tip) of field-grown JUJUNCAO leaves (Figure 5D and 5E). In the gradient leaf tissues, we were able to distinguish gene family members involved in carbon fixation from non-C4-related genes involved in other processes. Eight of the putative C4 pathway genes, namely CfCA1, CfPEPC1, CfPEPC-k2, CfNADP-MDH2, CfNADP-ME2, CfPEPCK, CfPPDK1, and CfPPDK-RP1, displayed the expected expression pattern of genes involved in photosynthesis: their expression increased from the leaf base to the tip (Li et al., 2010), and they had a diurnal expression pattern in the mature zone, with little or no expression in the immature zone (Figure 5D and 5E). In addition, expression of these genes was higher in leaves than in stems and roots (Supplemental Figure 17). These results suggested that these eight genes are related to C4 photosynthesis in JUJUNCAO. Notably, CfPEPC-k2 displayed a C4 expression pattern in JUJUNCAO leaves but was clustered with non-C4 genes of sugarcane, maize, and sorghum (Figure 5D and 5E; Supplemental Figure 17). In the daytime, expression of CfPEPC-k2 and CfPPDK-RP1 increased from 08:00 h to 14:00 h (Figure 5E), a pattern opposite to that of the other C4 genes. These results indicated that a neofunctionalization of these two genes occurred in JUJUNCAO after its divergence from other Poaceae species.
The genes CfPEPC-k2, CfPPDK-RP1, and CfCA1 in the A and B subgenomes were differentially expressed at different time points (Figure 5D). CfCA1 and CfPPDK-RP1 in subgenome B (CfCA1_B and CfPPDK-RP1_B) displayed enhanced expression (>2.75-fold) in leaves compared with those of subgenome A (CfCA1_A and CfPPDK-RP1_A), and expression of CfPEPC-k2 was 2.88-fold higher in subgenome A (CfPEPC-k2_A) than in subgenome B (CfPEPC-k2_B) (Supplemental Figure 17). To investigate the potential influence of gene methylation on gene expression, we examined differences in CG, CHG, and CHH methylation in the 2-kb upstream region, 5′ untranslated region (UTR), exon region, intron region, 3′ UTR, and 2-kb downstream region of these three genes. CfPEPC-k2, CfPPDK-RP1, and CfCA1 in subgenomes A and B had different methylation levels in the three methylation contexts (Figure 5F). CfCA1_B and CfPPDK-RP1_B had higher CG methylation levels in the intron region than CfCA1_A and CfPPDK-RP1_A. In addition, CfCA1_A, with a significantly longer 5′ UTR, contained high CG, CHG, and CHH methylation levels (Figure 5F). By contrast, CfPEPC-k2_A had the lowest CG methylation levels in the intron region. CG, CHG, and CHH methylation levels were lower in CfPEPC-k2_A than in CfPEPC-k2_B.
C4 decarboxylation is mainly dependent on NADP-ME with the cooperation of PEPCK
There are three subtypes of C4 plants, each of which utilizes one of the major decarboxylating enzymes: NADP-ME, NAD-ME, and PEPCK (Maier et al., 2011). Comparative transcriptome analysis revealed that genes encoding NADP-ME (CfNADP-ME2) and PEPCK (CfPEPCK) showed comparable expression levels during the daytime (Figure 5D and 5E); the TPM of CfNADP-ME2_A and CfNADP-ME2_B was 648 and 965, respectively, and that of CfPEPCK1_A and CfPEPCK1_B was 1060 and 799, indicative of a consistent rhythmic trend between the two enzymes. However, the NAD-ME genes CfNAD-ME1 and CfNAD-ME2 showed a rhythmic trend opposite to that of CfNADP-ME2 and CfPEPCK (Figure 5E). Therefore, we speculated that NADP-ME and PEPCK are the two major decarboxylating enzymes and contribute equally to the C4 decarboxylation reaction of JUJUNCAO, with NAD-ME acting in a complementary manner to the two major enzymes.
Phylogenetic analysis of Cenchrus grasses
Although many Cenchrus grasses have been identified to date, only a few genetic resources are available for them. To understand the phylogenetic relationships of other Cenchrus grasses, we recently performed genome sequencing of 19 Cenchrus grasses, including the Cenchrus cultivar Bajra, the P. americanum cultivar MZL, elephant grass (P. purpureum (Schumach.), ZXC), RY4HWC (P. purpureum × P. americanum cv. Reyan No. 4), ZJLWC (P. americanum × P. purpureum), G1Z (P. americanum × P. purpureum cv. Guimu 1), AXC (P. purpureum cv. dwarf), TXC (TAIXUCAO), DSXC (DUOSUIXIANGCAO), HNXC (Huanan XIANGCAO), and HNDS (Hainan DUOSUI) (Lin et al., 2022). Three S. italica varieties (SRR13414290, SRR13414328, and SRR13414313) were used as outgroups. These data comprise about 3.842 billion clean reads (∼0.5 Tbp) (Supplemental Table S12). We aligned the filtered reads to the JUJUNCAO genome and used the GATK pipeline for variant calling. A total of 149 319 SNPs shared by all the tested grasses were used for principal-component analysis (PCA) and phylogenetic analysis. Both results clustered the grasses into three clades: the three S. italica varieties were an outgroup, Bajra and MZL comprised group II, and JUJUNCAO (C. fungigraminus) and the other grasses comprised group I. This classification also coincides with their matching rates (Supplemental Figure 18; Supplemental Table S12). Our results also demonstrated that the two species closely related to JUJUNCAO are Bajra and MZL. We next estimated the population dynamics of Cenchrus using the A and B subgenomes of JUJUNCAO as ancestor genomes. The effective population of the Cenchrus genus was estimated to be ∼10 000 at 0.1 kya and more than 10 000 earlier. Our results demonstrated an abrupt decrease in the population size of Cenchrus grasses between 200 and 400 kya in both simulations. An additional decrease in population size at 20–50 kya was also observed using the B subgenome of JUJUNCAO as the ancestor genome. These results suggested the elimination of large numbers of Cenchrus individuals during the two stages, probably caused by extremely low temperature.
Discussion
A high-quality genome is very important for evolutionary and genetic studies. Although genomes have been assembled for many plant species, particularly economically important crops, only recently have a few genomes with T2T chromosomes been published. Here, we report a gapless, near-complete genome of the high-biomass grass JUJUNCAO. The contig N50 is 134.1 Mb, which is about 7000 times, 73 times, and 46 times that of pearl millet (Varshney et al., 2017), C. purpureus (Yan et al., 2020), and P. purpureum (Zhang et al., 2022b), respectively. To the best of our knowledge, this assembly is the highest-quality published Panicoideae genome. Because of the high biomass, stress tolerance, and broad adaptation of JUJUNCAO, its highly complete allopolyploid genome not only provides a high-quality reference genome for a Cenchrus grass but also serves as a valuable genetic resource for studying the mechanisms that underlie its robust performance and improving high-biomass grasses.
Genome evolution
Using the near-complete assembly, we discriminated the two subgenomes of JUJUNCAO and traced the evolutionary trajectory of this paleo-tetraploid. Comparison of genome structures revealed that the two subgenomes of JUJUCNAO were united and doubled ∼2.7 mya, much more recently than those of the two allotetraploid elephant grasses C. purpureus (6.61 mya; Yan et al., 2020) and P. purpureum Schum (15 mya; Zhang et al., 2022b). We proposed the ACK and suggested that the two subgenomes of JUJUNCAO evolved independently from the same ACK. This conclusion differs from that of the previous study by Zhang et al. (2022b), which suggested that after its divergence from pearl millet, the ancestral A genome split into the ancestors of the A′ and B subgenomes of P. purpureum. Frequent chromosomal rearrangements may have occurred between the two subgenomes after the allotetraploidization of JUJUNCAO. We found that subgenome A of JUJUNCAO is much more closely related to the two Cenchrus cultivars Bajra and MZL than to pearl millet (see also Lin et al., 2022). However, our results also suggested that these two Cenchrus cultivars are unlikely to be the parental species of JUJUNCAO. The relatives of JUJUNCAO subgenome B and the two elephant grasses remain unknown (Yan et al., 2020; Zhang et al., 2022b), and more Cenchrus species should be collected to resolve the phylogenetic relationships within this genus.
Polyploids generally benefit from whole-genome duplication that combines complementary pathways to confer adaptive advantages and robust agronomic performance. This has been attributed in part to subgenome-biased expression of homeologous genes in polyploids, particularly allopolyploids (Wang et al., 2011; Bertioli et al., 2019). Consistent with previous studies, subgenome-biased genes in JUJUNCAO demonstrated biological functional divergence between the two subgenomes, which was to some extent negatively associated with DNA methylation. We inferred from these results that DNA methylation may lead to divergent expression in the two subgenomes of JUJUNCAO, which is thought to have occurred after the divergence of the two subgenomes.
Centromere evolution
Centromeres in many plants and animals are often composed of a single satellite repeat, typically with a length ranging from 150 bp to 180 bp (Jiang et al., 2003; Yang et al., 2018). Although the function of centromeres is highly conserved among different species, the centromeric satellites are highly variable, and changes in sequences can be detected within and among species and even among different chromosomes (Cheng et al., 2002; Hall et al., 2003; Zhang et al., 2014; Presting, 2018; Yang et al., 2018; Ahmad et al., 2020). In this study, we revealed the dynamic architecture and evolution of JUJUNCAO centromeres and suggested that centromeric satellite repeats were conserved in Paniceae. Among the three centromeric satellite repeats identified in the JUJUNCAO genome, CEN156 shared the highest similarity with homologs from closely related species and was thought to be the founding sequence of ancestral Cenchrus chromosomes. It diverged into CEN137 in the common ancestral genome of subgenome A and P. americanum MZL and into CEN148 in the ancestral subgenome B after the divergence of the ancestral A and B subgenomes of JUJUNCAO. The subgenome A–specific CEN137 and subgenome B–specific CEN148 suggest that the three types of satellite repeats evolved independently within the two subgenomes. However, introgressions of CEN156 from subgenome B into subgenome A may have occurred after the paleo-polyploidization of the JUJUNCAO genome. This would be a reasonable explanation for the presence of CEN156 in subgenome A of JUJUNCAO and its absence from the genome of the diploid P. americanum MZL, which is phylogenetically close to subgenome A of JUJUNCAO. We also examined overall DNA methylation patterns in the centromeric regions and showed enrichment of cytosine methylation in the peri/centromeric regions, with different segments in different centromeres or the same centromere being either hypomethylated or hypermethylated. These results were consistent with those reported for other plants such as rice (Yan et al., 2010), Arabidopsis, and maize (Zhang et al., 2008; Koo and Jiang, 2011).
Mechanisms underlying high photosynthetic efficiency
C4 plants are among the most productive and efficient biomass producers. The Pennisetum grasses were identified as C4 plants decades ago on the basis of their low compensation point and Kranz anatomy (Coombs and Baldry, 1972; Coombs et al., 1973a, 1973c, 1973b; Huber and Sankhla, 1973). C4 plants are classically grouped into three biochemical subtypes according to whether they contain high levels of NADP-ME, PEPCK, or NAD-ME for decarboxylation of C4 acids (malate and/or aspartate) (Dengler and Nelson, 1999; Edwards and Voznesenskaya, 2011). Like other Pennisetum grasses, JUJUNCAO is also a C4 plant, with anatomical and physiological adaptations that optimize CO2 fixation. The centrifugal chloroplast arrangement of bundle sheath cells observed in JUJUNCAO was consistent with that of classic NADP-ME species such as sugarcane and maize (Sage et al., 2014) but different from the evenly distributed pattern of P. purpureum, which is typical of the PEPCK subtype. The anatomical variation between P. purpureum and JUJUNCAO could be influenced by environmental factors, such as light (Sales et al., 2018). Currently, Pennisetum grasses such as P. purpureum, P. americanum, and Pennisetum setaceum are classified as the NADP-ME subtype (Neto and Guerra, 2019). Many studies have suggested that more than one decarboxylation enzyme may co-exist in a single C4 plant; that the PEPCK pathway could be complementary in NADP-ME subtype species such as sugarcane, maize, and sorghum; and that the contribution of PEPCK to total decarboxylase activity was lower than that of NADP-ME and differed among species (Wang et al., 2014). Both NADP-ME and PEPCK enzymes may also be present in Pennisetum grasses (P. purpureum), owing to their use of malate and/or aspartate as the C4 translocated acid (Coombs et al., 1973c). Interestingly, JUJUNCAO CfPEPCK reached a transcript level comparable to that of CfNADP-ME. This is similar to results from sugarcane, which is categorized as an NADP-ME species, although PEPCK transcript abundance was higher than that of NADP-ME when measured by the serial analysis of gene expression (SAGE) technique (Calsa and Figueira, 2007). The rise in PEPCK activity can be modulated by various environmental factors (Furbank, 2011; Sales et al., 2018) such as shading and limited water supply (Sales et al., 2018; Cacefo et al., 2019). Therefore, we propose that high CfPEPCK transcription may potentially enable the maintenance of greater photosynthetic efficiency in regions where JUJUNCAO thrives.
Allopolyploid-derived subgenomes begin with distinct, global differences that would be expected to lead to global differences in gene expression levels. We observed no expression bias in genes encoding decarboxylases between the A and B subgenomes. However, the C4 genes CfCA1, CfPPDK-RP1, and CfPEPC-k2 showed some evidence of subgenome dominance, with CfCA1 and CfPPDK-RP1 exhibiting subgenome B dominance and CfPEPC-k2 exhibiting subgenome A dominance. Subgenome dominance led to divergence with respect to expression levels. The accurate regulation of gene expression in the A and B subgenomes might be indispensable for allopolyploid plant development, and DNA methylation may serve as a regulator of C4 gene expression (Reeves et al., 2017). Higher CG methylation levels in the intron region may confer higher expression of CfCA1 and CfPPDK-RP1 from subgenome B, given that gene-body methylation is positively correlated with gene expression (Jones, 2012; Li et al., 2012). However, the transcript level of CfPEPC-k2 was higher in subgenome A than in subgenome B. DNA methylation can repress gene expression (Chan et al., 2005). Thus, gene expression of CfPEPC-k2 in subgenome B might be repressed by DNA methylation of the downstream 2-kb region. In this study, we found that the relationship between gene expression and DNA methylation was complicated by genic regions, methylation contexts, and subgenomes in the allopolyploid.
Methods
Plant materials and sequencing
The JUJUNCAO (C. fungigraminus, 2n = 28) and C. americanus plants used in this study were grown in the greenhouse at Fujian Agriculture and Forestry University. For Illumina short-read sequencing, genomic DNA extracted from leaf tissue was used for construction of a 280-bp paired-end library with the NEBNext Ultra DNA Library Prep Kit. The library was sequenced using the Illumina NovaSeq 6000 platform. For PacBio HiFi library construction and sequencing, the g-TUBE was used to shear gDNA to ∼20-kb fragments for construction of SMRTbell libraries using the SMRTbell Express Template Prep Kit 2.0. The libraries were sequenced using the PacBio Sequel system at Novogene Company. Hi-C libraries were generated and sequenced from young leaves of JUJUCAO at Novogene as described previously (Xie et al., 2015). In brief, after fixation with formaldehyde, the fresh leaves were lysed for DNA extraction. The cross-linked DNA was digested overnight with MboI to generate sticky ends for biotinylation and then proximity ligated to form chimeric junctions. The enriched DNA was then physically sheared to a size of 200–600 bp. Chimeric fragments representing the original cross-linked long-distance physical interactions were used to construct paired-end sequencing libraries, which were sequenced using the Illumina HiSeq 2500 platform (PE 125 bp).
Genome size estimation
Flow cytometry analysis was performed to estimate the C-value of JUJUNCAO using fresh leaves, and tomato was used as a reference genome control (Michaelson et al., 1991; Dolezel and Bartos, 2005).
Genome assembly and scaffolding
About 117 Gb of clean PacBio CCS reads were used for de novo assembly of the JUJUNCAO genome with default settings of Hifiasm v0.16.1-r375 (Cheng et al., 2021), and the primary output contigs with long stretches of phased blocks were sent to NextPolish v1.3.1 (Hu et al., 2020) for polishing using Illumina short reads. Purge_haplotigs v1.1.1 (Roach et al., 2018) was used to remove heterozygous redundant sequences with the parameters “purge_haplotigs contigcov -l 10 -m 80 -h 190.” The total span of the final filtered assembly was 1 986 757 443 bp in 116 contigs with a contig N50 size of 134 074 806 bp. To build a chromosome-level assembly, a Hi-C library was constructed and sequenced as described previously (Zhang et al., 2022a). A total of 233 Gb of Hi-C reads were mapped to the contig-level assembly using BWA v0.7.17 with default parameters (Li, 2013). We corrected the contig-level assembly based on the chromatin contact signals. All of the corrected contigs were remapped with Hi-C reads and reordered and scaffolded using the ALLHiC pipeline (https://github.com/tangerzhang/ALLHiC), and the resulting assembly was manually corrected according to the visualization of chromatin contact patterns. Finally, we generated a pseudo-chromosome assembled genome that included 14 chromosomes.
Gene annotation
In brief, we integrated sequence homology, de novo prediction, and transcriptome data to build consensus gene models using the GETA pipeline (https://github.com/chenlianfu/geta). Protein sequences from closely related grass species (C. americanus, S. bicolor, and O. sativa) were used to perform homology-based prediction. The GETA pipeline randomly selected 1000 homology-based genes to train AUGUSTUS v3.4.0 (Stanke et al., 2004) for de novo prediction on the pre-masked genome sequences. We provided the pipeline with RNAs isolated from root, stem, and leaf tissues of JUJUNCAO. To assemble the transcriptomes, trimmomatic v0.39 (Bolger et al., 2014) was used with default parameters to filter the RNA-seq reads. The filtered reads were aligned to the reference genome with HISAT2 v2.1.0 (Kim et al., 2015). The reliable intron and optimal transcript information were identified based on RNA-seq alignment. We next assembled and filtered the transcripts with the “sam2transfrag” function in the GETA pipeline, which retains the isoform of each gene with the highest RNA-seq read depth as the representative transcript. Finally, gene models from these three methods were integrated into a non-redundant high-confidence set of gene models.
Repeat sequence annotation
A repeat library of the JUJUNCAO genome was constructed ab initio using RepeatModeler v2.0.2 with a combination of de novo and homology strategies, including two de novo repeat-finding programs, RECON and RepeatScout, which we imported into RepeatMasker v4.1.2 (http://www.repeatmasker.org/) for identification and clustering of repetitive elements. We also integrated results from LTR_FINDER (Xu and Wang, 2007) and LTRharvest (Ellinghaus et al., 2008) and removed false positives from the initial predictions using the LTR_retriever pipeline (Ou and Jiang, 2018) to obtain the final full-length LTR-RTs.
Telomere and centromere detection
Tandem repeats were identified in the JUJUNCAO genome with Tandem Repeat Finder v4.09 (TRF; parameters: 2 7 7 80 10 50 500 -m -f -d) (Benson, 1999), and the telomere and three types of centromere sequences were found. To precisely investigate the distributions of the three centromere repeat monomers (CEN137, CEN148, CEN156), we divided the genome into adjacent 5-kbp windows, defined 12-mers, and exactly matched these sequences to the window. For each region, we calculated peak values of the distances between 13-mers to test for periodic repeats. For example, if a window contained CEN137 repeats (monomer size of 137 bp), the most frequent 13-mer distance would be 137. We merged the same kind of windows, and CEN137, CEN148, and CEN156 sequences were matched back to corresponding windows using matchPattern (max.mismatch = 3/peakvalues, with.indels = T) (https://github.com/Bioconductor/Biostrings). Finally, we identified 159 310 bp of CEN137 repeats, 24 075 bp of CEN148 repeats, and 216 886 bp of CEN156 repeats in all 14 chromosomes, and several were aligned by mafft.v 7.4 (Katoh et al., 2009) to generate the consensus sequence (Supplemental table 7).
Subgenome identification
We resolved the JUJUNCAO genome into subgenomes A and B using previously described methods (Session et al., 2016; Mitros et al., 2020). First, homeologous chromosomes were determined based on their conserved synteny to each other and to C. americanus. Then, Jellyfish v2.2.10 (Marçais and Kingsford, 2011) was used to scan 13-bp sequences (13-mers), and 32 629 subgenome-specific 13-mers were identified using the following criteria: (1) occurred >1000 times across the genome, and (2) were more than two-fold enriched in one member of each homeologous chromosome pair (setting aside the fusion-related chromosomes). These 13-mers could be clearly divided into two groupings (A 13-mers and B 13-mers) by hierarchical clustering. Similarly, chromosome clustering partitioned the genome into subgenomes A and B.
Phylogenetic analysis, genomic comparison, and divergence time estimation
Protein sequences of S. spontaneum, M. sinensis, S. bicolor, Z. mays, P. hallii, P. miliaceum, P. virgatum, C. americanus, C. fungigraminus, S. italica, S. viridis, and O. sativa were used to identify gene family clusters with Orthofinder v2.5.4. We used the option “-M msa” to obtain maximum likelihood trees from multiple sequence alignments. Divergence times were estimated using r8s v1.81. Calibration times were obtained from the TimeTree database (http://www.timetree.org/). Expansion and contraction of Orthofinder-derived gene clusters were determined using CAFÉ v4.2 (https://github.com/hahnlab/CAFE) and were based on changes in gene family size in the inferred phylogenetic history. Collinearity between genomes was analyzed using JCVI (https://github.com/tanghaibao/jcvi) with default parameters, and all orthologous and paralogous gene pairs were identified on the basis of syntenic blocks. Paired synonymous substitution rates (Ks) were calculated using the Nei-Gojobori method (https://github.com/tanghaibao/bio-pipeline/tree/master/synonymous_calculation).
Karyotype evolution analysis of Paniceae
The distribution of seven ancestral eudicot chromosomal lineages for each chromosome in each species was depicted by syntenic blocks between the ancestral chromosomes of rice (Murat et al., 2017) and those of the detected species.
SNP identification and population genomics analysis
Paired-end raw reads were filtered by removing reads with adapters, reads with >10% N content, and reads with more than 50% low-quality bases (Q ≤ 5). Filtered paired-end reads were mapped to the assembled JUJUNCAO genome using Bowtie2 v2.4.4 (Langdon, 2015). We then used Picard (v1.129) (https://github.com/broadinstitute/picard) to mark the duplicated reads. SNPs were identified using the GATK toolkit v.3.5 (https://github.com/broadinstitute/gatk) with the HaplotypeCaller module. Next, we merged the GCVF files using the GenotypeGVCFs tool in GATK to generate raw SNP files. The SNP filter application VCFtools v0.1.13 (Danecek et al., 2011) was used with “--max-missing 0.8 --maf 0.05 --max-alleles 2 --min-meanDP 4 --minQ 200.” Vcf2phylip software (https://github.com/edgardomortiz/vcf2phylip) was used to convert the format, and RaxML v8.12 (Stamatakis, 2014) was used to construct a maximum likelihood tree. PCA was performed using PLINK v1.90 (Purcell et al., 2007) to transform the VCF file into a PLINK file for input.
Demographic analysis
To calculate the site-frequency spectrum (SFS) of Pennisetum populations, we used the “-doSaf” parameter of ANGSD (Korneliussen et al., 2014) to calculate the site allele frequency likelihood based on individual genotype likelihoods assuming HWE from filtered BAM files. Then the “-realSFS” parameter was used to obtain a maximum likelihood estimate of the SFS by an expectation maximization (EM) algorithm. The folded SFS was used to estimate population demographic history by stairway plots (Liu and Fu, 2015) with 200 bootstrap iterations. We used 6.5e−9 as the mutation rate and a generation time of 1 year for the analysis.
Bisulfite whole-genome sequencing and bioinformatic analysis
The JUJUNCAO reference genome and the clean reads generated from bisulfite sequencing were transformed into bisulfite-converted sequences (C-to-T and G-to-A converted). The clean reads were then aligned to the reference genome using BISMARK with default parameters, and methylation information was extracted using BISMARK_METHYLATION_EXTRACTOR with the following options: --comprehensive --bedGraph –counts --CX_context --cytosine_report. The methylation level of an individual cytosine was calculated from the number of methylated cytosines divided by the total cytosine depth (i.e., mC/(mC + non-mC)). Only sites that covered more than three mapped reads were used for subsequent analysis. Sites with more than one methylated cytosine mapping were considered methylation sites.
FISH
FISH experiments were performed as described previously (Huang et al., 2021). Oligomers specific for each centromeric satellite repeat (Supplemental Table 13) were designed according to their sequence alignment results (Figure 3). The oligomers were labeled via nick-translation using digoxigenin-11-dUTP or biotin-16-dUTP (Roche Diagnostics). Digoxigenin-labeled probes and biotin-labeled probes were detected using rhodamine-conjugated anti-digoxigenin (Roche Diagnostics) and Alexa Fluor 488 streptavidin (Thermo Fisher Scientific), respectively. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the chromosomes. Images of the samples were captured with an Olympus BX63 fluorescence microscope using an Olympus DP80 CCD camera (Olympus).
Histological analysis
One-centimeter strips were cut crosswise to the leaf and fixed for 12 h in a solution of 50% ethanol, 5% acetic acid, and 37% formaldehyde at 4°C. The strips were then processed in a series of ethanol solutions for 1 h each (50%, 60%, 70%, 85%, 95%, 100%), followed by mixtures of xylene and ethanol for 30 min each (25% xylene + 75% ethanol, 50% xylene + 50% ethanol, 25% ethanol + 75% xylene, 100% xylene) and finally paraffin for 12 h. The tissue was then oriented in paraffin and sectioned at 8 μm using a rotary microtome (Leica, RM2255). Tissue was stained with 1% safranine and fast green solution and viewed under a microscope (Leica, BX3-CBH).
Identification of genes related to key characteristics of JUJUNCAO
C4 pathway genes from sorghum and S. spontaneum were used as query sequences for Basic Local Alignment Search Tool (BLASTP) searches against the predicted sequences of JUJUNCAO. Sequences with E-value <1e−5 were selected for further analysis.
Phylogenetic analysis of C4 pathway genes
Based on protein sequence alignment, a phylogenetic tree of the C4 pathway gene family was constructed by the neighbor-joining (NJ) method. The tree was constructed using MEGAX (Kumar et al., 2018) with the “pairwise deletion” option and the “Poisson correction” model, and the reliability of internal branches was assessed using 1000 bootstrap replicates. The results were imported into the interactive Tree of Life (iTOL) (Letunic and Bork, 2016) to create the phylogenetic tree.
Gas exchange measurements
Leaf gas exchange was measured in the +1 leaf (the first fully expanded leaf from the top to bottom) using a portable infrared gas analyzer (LI-COR 6800XT, LI-COR, Lincoln, NE, USA). Measurements were made at a leaf temperature of 30°C, humidity of 65%, and flow rate of 500 μmol s⁻1. The light-response curves were made at a CO2 concentration of 400 μmol mol⁻1 and photosynthetic photon flux densities (PPFDs) of 2000, 1500, 1000, 800, 600, 400, 200, 150, 100, 75, 50, and 0 μmol m⁻2 s⁻1. The Excel sheet used for curve fitting was downloaded from http://landflux.org/tools. CO2 response curves were measured at a PPFD of 2000 μmol m⁻2 s⁻1 and CO2 concentrations of 400, 300, 200, 120, 70, 40, 20, 0, 400, 400, 400, 600, 800, 1200, and 1500 μmol mol⁻1. The CO2 response curves were fitted according to von Caemmerer.
Funding
This work was supported by grants from the Major Special Project of Fujian Province (2021NZ029009) and the Natural Science foundation of Fujian Province (2019J01665).
Author contributions
J.Z., Z.L., and Z.W. conceived the project. X.H., H.Z., Y.H., Z.Z., and H.L. collected the samples and performed the experiments. B.W., X.H., R.G., Y.W., Y.Z., J.M., and Y.H. contributed to genome assembly and data analysis. J.Z., H.Z., X.H., and B.W. wrote the manuscript. J.Z. and R.M. revised the manuscript. All authors reviewed the manuscript.
Acknowledgments
We would like to thank Prof. Xinlong Liu at Yunnan Key Laboratory of Sugarcane Genetic Improvement, Sugarcane Research Institute, Yunnan Academy of Agricultural Sciences, Kaiyuan, China for kindly providing the Cenchrus cultivars. No conflict of interest is declared.
Published: June 3, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Zonghua Wang, Email: zonghuaw@163.com.
Ray Ming, Email: rayming@illinois.edu.
Jisen Zhang, Email: zjisen@126.com.
Zhanxi Lin, Email: lzxjuncao@163.com.
Supplemental information
Data availability
The whole-genome sequencing data (including Illumina short reads, HiFi reads, and Hi-C interaction reads), bisulfite whole-genome sequencing data, and transcriptomes of different tissues used in this study have been deposited at the National Genomics Data Center under accession number PRJCA013426. The genome assembly and gene annotation data for C. fungigraminus have been deposited at the Genome Warehouse (GWH) under accession number GWHBWDX00000000.1 and are publicly accessible at https://ngdc.cncb.ac.cn/gwh.
References
- Adams K.L., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ahmad S.F., Singchat W., Jehangir M., Suntronpong A., Panthum T., Malaivijitnond S., Srikulnath K. Dark matter of primate genomes: satellite DNA repeats and their evolutionary dynamics. Cells. 2020;9:e122714. doi: 10.3390/cells9122714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser C., Baurens F.C., Noel B., Martin G., Cruaud C., Istace B., Yahiaoui N., Labadie K., Hřibová E., Doležel J., et al. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun. Biol. 2021;4:1047. doi: 10.1038/s42003-021-02559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli D.J., Jenkins J., Clevenger J., Dudchenko O., Gao D., Seijo G., Leal-Bertioli S.C.M., Ren L., Farmer A.D., Pandey M.K., et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019;51:877–884. doi: 10.1038/s41588-019-0405-z. [DOI] [PubMed] [Google Scholar]
- Bird K.A., Niederhuth C.E., Ou S., Gehan M., Pires J.C., Xiong Z., VanBuren R., Edger P.P. Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. New Phytol. 2021;230:354–371. doi: 10.1111/nph.17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacefo V., Ribas A.F., Zilliani R.R., Neris D.M., Domingues D.S., Moro A.L., Vieira L.G.E. Decarboxylation mechanisms of C4 photosynthesis in Saccharum spp.: increased PEPCK activity under water-limiting conditions. BMC Plant Biol. 2019;19:144. doi: 10.1186/s12870-019-1745-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calsa T., Jr., Figueira A. Serial analysis of gene expression in sugarcane (Saccharum spp.) leaves revealed alternative C4 metabolism and putative antisense transcripts. Plant Mol. Biol. 2007;63:745–762. doi: 10.1007/s11103-006-9121-z. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Chen B., Ye J., Luo Z., Lin J. ISSR Analysis of genetic diversity of Pennisetum germplasm resource. Heilongjiang Anim. Sci. Vet. Med. 2016:129–133. [Google Scholar]
- Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Dong F., Langdon T., Ouyang S., Buell C.R., Gu M., Blattner F.R., Jiang J. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell. 2002;14:1691–1704. doi: 10.1105/tpc.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Maheshwari S., Marimuthu M.P.A. Plant centromeres. Curr. Opin. Plant Biol. 2017;36:158–167. doi: 10.1016/j.pbi.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Coombs J., Baldry C.W. C-4 pathway in Pennisetum purpureum. Nat. New Biol. 1972;238:268–270. doi: 10.1038/newbio238268a0. [DOI] [PubMed] [Google Scholar]
- Coombs J., Baldry C.W., Bucke C. The C-4 pathway in Pennisetum purpureum : I. The allosteric nature of PEP carboxylase. Planta. 1973;110:95–107. doi: 10.1007/bf00384832. [DOI] [PubMed] [Google Scholar]
- Coombs J., Baldry C.W., Bucke C. The C-4 pathway in Pennisetum purpureum : II. Malate dehydrogenase and malic enzyme. Planta. 1973;110:109–120. doi: 10.1007/bf00384833. [DOI] [PubMed] [Google Scholar]
- Coombs J., Baldry C.W., Brown J.E. The C-4 pathway in Pennisetum purpureum : III. Structure and photosynthesis. Planta. 1973;110:121–129. doi: 10.1007/bf00384834. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler N.G., Nelson T. In: C4 Plant Biology. Sage R.F., Monson R.K., editors. Academic Press; 1999. 5 - leaf structure and development in C4 Plants; pp. 133–172. [DOI] [Google Scholar]
- Dolezel J., Bartos J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P.P., Smith R., McKain M.R., Cooley A.M., Vallejo-Marin M., Yuan Y., Bewick A.J., Ji L., Platts A.E., Bowman M.J., et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017;29:2150–2167. doi: 10.1105/tpc.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G.E., Voznesenskaya E.V. In: C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Raghavendra A.S., Sage R.F., editors. Springer Netherlands; 2011. Chapter 4 C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants; pp. 29–61. [DOI] [Google Scholar]
- Eisler M.C., Lee M.R.F., Tarlton J.F., Martin G.B., Beddington J., Dungait J.A.J., Greathead H., Liu J., Mathew S., Miller H., et al. Agriculture: steps to sustainable livestock. Nature. 2014;507:32–34. doi: 10.1038/507032a. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinf. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank R.T. Evolution of the C(4) photosynthetic mechanism: are there really three C(4) acid decarboxylation types? J. Exp. Bot. 2011;62:3103–3108. doi: 10.1093/jxb/err080. [DOI] [PubMed] [Google Scholar]
- Gu B., Zhang X., Bai X., Fu B., Chen D. Four steps to food security for swelling cities. Nature. 2019;566:31–33. doi: 10.1038/d41586-019-00407-3. [DOI] [PubMed] [Google Scholar]
- Hall A.E., Keith K.C., Hall S.E., Copenhaver G.P., Preuss D. The rapidly evolving field of plant centromeres. Curr. Opin. Plant Biol. 2004;7:108–114. doi: 10.1016/j.pbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Hall S.E., Kettler G., Preuss D. Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res. 2003;13:195–205. doi: 10.1101/gr.593403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat K., Zhou Y., Menhas S., Bundschuh J., Hayat S., Ullah A., Wang J., Chen X., Zhang D., Zhou P. Pennisetum giganteum: an emerging salt accumulating/tolerant non-conventional crop for sustainable saline agriculture and simultaneous phytoremediation. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.114876. [DOI] [PubMed] [Google Scholar]
- He K., Huang Y., Jiang F., Lin J., Cheng Z., Zhang K., Xie W., Huang Z. Effects of two types of herb plants' roots on soil moisture in the alluvial soil in Changing County. Science of Soil and Water Conservation. 2017;15:25–34. [Google Scholar]
- Hu J., Fan J., Sun Z., Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36:2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- Huang Y., Ding W., Zhang M., Han J., Jing Y., Yao W., Hasterok R., Wang Z., Wang K. The formation and evolution of centromeric satellite repeats in Saccharum species. Plant J. 2021;106:616–629. doi: 10.1111/tpj.15186. [DOI] [PubMed] [Google Scholar]
- Huber W., Sankhla N. Eco-physiological studies on Indian arid zone plants : II. Effect of salinity and gibberellin on the activity of the enzymes of amino-acid metabolism in leaves of Pennisetum typhoides. Oecologia. 1973;13:271–277. doi: 10.1007/bf00360516. [DOI] [PubMed] [Google Scholar]
- Jia Y., Liao Z., Chew H., Wang L., Lin B., Chen C., Lu G., Lin Z. Effect of Pennisetum giganteum z.x.lin mixed nitrogen-fixing bacterial fertilizer on the growth, quality, soil fertility and bacterial community of pakchoi (Brassica chinensis L.) PLoS One. 2020;15 doi: 10.1371/journal.pone.0228709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Birchler J.A., Parrott W.A., Dawe R.K. A molecular view of plant centromeres. Trends Plant Sci. 2003;8:570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Katoh K., Asimenos G., Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.H., Jiang J. Super-stretched pachytene chromosomes for plant molecular cytogenetic mapping. Methods Mol. Biol. 2011;701:239–245. doi: 10.1007/978-1-61737-957-4_13. [DOI] [PubMed] [Google Scholar]
- Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinf. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang W., Liu J., Tian H., Shi H., Dong J., Song C., Li X., Du G., Hou Y., Lu D., et al. Cropland redistribution to marginal lands undermines environmental sustainability. Natl. Sci. Rev. 2022;9:nwab091. doi: 10.1093/nsr/nwab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale J.A. C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell. 2011;23:3879–3892. doi: 10.1105/tpc.111.092098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015;8:1. doi: 10.1186/s13040-014-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- Li P., Ponnala L., Gandotra N., Wang L., Si Y., Tausta S.L., Kebrom T.H., Provart N., Patel R., Myers C.R., et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- Li Q., Xiang C., Xu L., Cui J., Fu S., Chen B., Yang S., Wang P., Xie Y., Wei M., Wang Z. SMRT sequencing of a full-length transcriptome reveals transcript variants involved in C18 unsaturated fatty acid biosynthesis and metabolism pathways at chilling temperature in Pennisetum giganteum. BMC Genom. 2020;21:52. doi: 10.1186/s12864-019-6441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu J., Hu F., Ge S., Ye M., Xiang H., Zhang G., Zheng X., Zhang H., Zhang S., et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genom. 2012;13:300. doi: 10.1186/1471-2164-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Schnable J.C. Functional divergence between subgenomes and gene pairs after whole genome duplications. Mol. Plant. 2018;11:388–397. doi: 10.1016/j.molp.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Lin X., Mao Y., Qi Q., Zhang C., Tian Y., Chen Y., Lin Z. Sequence analysis on ITS and chloroplast matK gene of six Pennisetum JUNCAO. Diagn. Pathol. 2015;10:174–180. [Google Scholar]
- Lin Z. National School of Administration Press; Beijing: 2013. Juncao Science. [Google Scholar]
- Lin Z., Lin D., Liu Z., Siren L. Cenchrus fungigraminus Z. X, Lin & D. M, Lin & S. R. Lan sp. nov., a new species of Panicoideae ( Poaceae) : evidence from morphological, nuclear and plastid genome data. J. For. Environ. 2022;42:514–520. [Google Scholar]
- Liu J., Seetharam A.S., Chougule K., Ou S., Swentowsky K.W., Gent J.I., Llaca V., Woodhouse M.R., Manchanda N., Presting G.G., et al. Gapless assembly of maize chromosomes using long-read technologies. Genome Biol. 2020;21:121. doi: 10.1186/s13059-020-02029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fu Y.X. Exploring population size changes using SNP frequency spectra. Nat. Genet. 2015;47:555–559. doi: 10.1038/ng.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Su H., Zhang J., Shi L., Liu Y., Zhang B., Bai H., Liang S., Gao Z., Birchler J.A., Han F. Rapid birth or death of centromeres on fragmented chromosomes in maize. Plant Cell. 2020;32:3113–3123. doi: 10.1105/tpc.20.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z.-z., Lin J.-r., Lin Z.-k., Chen B.-c., Mei L., Lin Z.-x. Karyotype analysis of Pennisetum purpureum×P.americanum cv.Reyan No.4 and P .purpureum cv.Guiminyin. Pratacult. Sci. 2016;33:1711–1717. [Google Scholar]
- Maier A., Zell M.B., Maurino V.G. Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C(4) and C(3) photosynthesis. J. Exp. Bot. 2011;62:3061–3069. doi: 10.1093/jxb/err024. [DOI] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson M.J., Price H.J., Ellison J.R., Johnston J.S. Comparison of plant DNA contents determined by Feulgen microspectrophotometry and laser flow cytometry. Am. J. Bot. 1991;78:183–188. [Google Scholar]
- Mitros T., Session A.M., James B.T., Wu G.A., Belaffif M.B., Clark L.V., Shu S., Dong H., Barling A., Holmes J.R., et al. Genome biology of the paleotetraploid perennial biomass crop Miscanthus. Nat. Commun. 2020;11:5442. doi: 10.1038/s41467-020-18923-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J.E. High-biomass C(4) grasses-Filling the yield gap. Plant Sci. 2017;261:10–17. doi: 10.1016/j.plantsci.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Murat F., Armero A., Pont C., Klopp C., Salse J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 2017;49:490–496. doi: 10.1038/ng.3813. [DOI] [PubMed] [Google Scholar]
- Navrátilová P., Toegelová H., Tulpová Z., Kuo Y.T., Stein N., Doležel J., Houben A., Šimková H., Mascher M. Prospects of telomere-to-telomere assembly in barley: analysis of sequence gaps in the MorexV3 reference genome. Plant Biotechnol. J. 2022;20:1373–1386. doi: 10.1111/pbi.13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto M.A.M., Guerra M.P. A new method for determination of the photosynthetic pathway in grasses. Photosynth. Res. 2019;142:51–56. doi: 10.1007/s11120-019-00646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A., et al. The complete sequence of a human genome. Science (New York, N.Y.) 2022;376:44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Chen J., Jiang N. Assessing genome assembly quality using the LTR Assembly Index (LAI) Nucleic Acids Res. 2018;46:e126. doi: 10.1093/nar/gky730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presting G.G. Centromeric retrotransposons and centromere function. Curr. Opin. Genet. Dev. 2018;49:79–84. doi: 10.1016/j.gde.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves G., Grangé-Guermente M.J., Hibberd J.M. Regulatory gateways for cell-specific gene expression in C4 leaves with Kranz anatomy. J. Exp. Bot. 2017;68:107–116. doi: 10.1093/jxb/erw438. [DOI] [PubMed] [Google Scholar]
- Roach M.J., Schmidt S.A., Borneman A.R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinf. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R.F., Peixoto M.M., Sage T.L. 1st ed. Wiley; New York: 2014. Photosynthesis in Sugarcane. Sugarcane: Physiology, Biochemistry and Functional Biology; p. 121. [Google Scholar]
- Sales C.R., Ribeiro R.V., Hayashi A.H., Marchiori P.E., Silva K.I., Martins M.O., Silveira J.A., Silveira N.M., Machado E.C. Flexibility of C4 decarboxylation and photosynthetic plasticity in sugarcane plants under shading. Environ. Exp. Bot. 2018;149:34–42. doi: 10.1016/j.envexpbot.2017.10.027. [DOI] [Google Scholar]
- Samson R., Mani S., Boddey R., Sokhansanj S., Quesada D., Urquiaga S., Reis V., Ho Lem C. The potential of C4 perennial grasses for developing a global. BIOHEAT industry. 2005;24:461–495. [Google Scholar]
- Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., Depamphilis C.W., Wall P.K., Soltis P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Steinkamp R., Waack S., Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Shi C., Thudi M., Mariac C., Wallace J., Qi P., Zhang H., Zhao Y., Wang X., Rathore A., et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 2017;35:969–976. doi: 10.1038/nbt.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tang H., Paterson A.H. Seventy million years of concerted evolution of a homoeologous chromosome pair, in parallel, in major Poaceae lineages. Plant Cell. 2011;23:27–37. doi: 10.1105/tpc.110.080622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bräutigam A., Weber A.P.M., Zhu X.G. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 2014;65:3567–3578. doi: 10.1093/jxb/eru058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F., Jackson S.A., Meyers B.C., Wing R.A. Evolution of plant genome architecture. Genome Biol. 2016;17:37. doi: 10.1186/s13059-016-0908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Xu Z. Sustainable agriculture: from sweet sorghum planting and ensiling to ruminant feeding. Mol. Plant. 2019;12:603–606. doi: 10.1016/j.molp.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Xie T., Zheng J.F., Liu S., Peng C., Zhou Y.M., Yang Q.Y., Zhang H.Y. De novo plant genome assembly based on chromatin interactions: a case study of Arabidopsis thaliana. Mol. Plant. 2015;8:489–492. doi: 10.1016/j.molp.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Xu L., Zhou J., Liang J., Cui H., Tao M., Zhihui T., Zhu Z., Lin H. The remediation potential of Pennisetum sp on Cu、Cd contaminated soil. Acta Ecol. Sin. 2014;34:5342–5348. [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Kikuchi S., Neumann P., Zhang W., Wu Y., Chen F., Jiang J. Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 2010;63:353–365. doi: 10.1111/j.1365-313X.2010.04246.x. [DOI] [PubMed] [Google Scholar]
- Yan Q., Wu F., Xu P., Sun Z., Li J., Gao L., Lu L., Chen D., Muktar M., Jones C., et al. The elephant grass (Cenchrus purpureus) genome provides insights into anthocyanidin accumulation and fast growth. Mol. Ecol. Resour. 2021;21:526–542. doi: 10.1111/1755-0998.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Liu D., Wang X., Ji C., Cheng F., Liu B., Hu Z., Chen S., Pental D., Ju Y., et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- Yang X., Zhao H., Zhang T., Zeng Z., Zhang P., Zhu B., Han Y., Braz G.T., Casler M.D., Schmutz J., Jiang J. Amplification and adaptation of centromeric repeats in polyploid switchgrass species. New Phytol. 2018;218:1645–1657. doi: 10.1111/nph.15098. [DOI] [PubMed] [Google Scholar]
- Ye J., Lin J., Jiao W., Chen B. Guangdong Agricultural Sciences; 2015. RAPD Analysis of Genetic Diversity of Penniseum Germplasm Resources in Fujian; pp. 128–132. [Google Scholar]
- Zhang H., Koblížková A., Wang K., Gong Z., Oliveira L., Torres G.A., Wu Y., Zhang W., Novák P., Buell C.R., et al. Boom-bust turnovers of megabase-sized centromeric DNA in Solanum species: rapid evolution of DNA sequences associated with centromeres. Plant Cell. 2014;26:1436–1447. doi: 10.1105/tpc.114.123877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Qi Y., Pan H., Tang H., Wang G., Hua X., Wang Y., Lin L., Li Z., Li Y., et al. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 2022;54:885–896. doi: 10.1038/s41588-022-01084-1. [DOI] [PubMed] [Google Scholar]
- Zhang S., Xia Z., Li C., Wang X., Lu X., Zhang W., Ma H., Zhou X., Zhang W., Zhu T., et al. Chromosome-scale genome assembly provides insights into speciation of allotetraploid and massive biomass accumulation of elephant grass (Pennisetum purpureum Schum.) Mol. Ecol. Resour. 2022;22:2363–2378. doi: 10.1111/1755-0998.13612. [DOI] [PubMed] [Google Scholar]
- Zhang W., Lee H.R., Koo D.H., Jiang J. Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C.X., Marshall J.B., Topp C., Mroczek R., Kato A., Nagaki K., Birchler J.A., Jiang J., Dawe R.K. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chen S., Shi W., David-Schwartz R., Li S., Yang F., Lin Z. Transcriptome profiling reveals the effects of drought tolerance in Giant Juncao. BMC Plant Biol. 2021;21:2. doi: 10.1186/s12870-020-02785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Wang P., Lin X., Lin H., Su D., Lin Z. Karyotype analysis of Pennisetum purpureum and Pennisetum giganteum. Guizhou Agricultural Sciences. 2015;43:14–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-genome sequencing data (including Illumina short reads, HiFi reads, and Hi-C interaction reads), bisulfite whole-genome sequencing data, and transcriptomes of different tissues used in this study have been deposited at the National Genomics Data Center under accession number PRJCA013426. The genome assembly and gene annotation data for C. fungigraminus have been deposited at the Genome Warehouse (GWH) under accession number GWHBWDX00000000.1 and are publicly accessible at https://ngdc.cncb.ac.cn/gwh.