Abstract
Chloroplasts evolved from an ancient cyanobacterial endosymbiont more than 1.5 billion years ago. During subsequent coevolution with the nuclear genome, the chloroplast genome has remained independent, albeit strongly reduced, with its own transcriptional machinery and distinct features, such as chloroplast-specific innovations in gene expression and complicated post-transcriptional processing. Light activates the expression of chloroplast genes via mechanisms that optimize photosynthesis, minimize photodamage, and prioritize energy investments. Over the past few years, studies have moved from describing phases of chloroplast gene expression to exploring the underlying mechanisms. In this review, we focus on recent advances and emerging principles that govern chloroplast gene expression in land plants. We discuss engineering of pentatricopeptide repeat proteins and its biotechnological effects on chloroplast RNA research; new techniques for characterizing the molecular mechanisms of chloroplast gene expression; and important aspects of chloroplast gene expression for improving crop yield and stress tolerance. We also discuss biological and mechanistic questions that remain to be answered in the future.
Key words: chloroplast, chloroplast gene expression, transcription, post-transcriptional processing, translation
The chloroplast remains independent, with its own transcriptional machinery and distinct features. This review highlights recent advances in chloroplast gene expression in land plants and discusses open questions for future research. The application of new techniques will enable further characterization of mechanisms that underlie chloroplast gene expression, providing useful insights for improvement of crop yield and stress tolerance.
Introduction
Chloroplasts are a type of double membrane–bound cytoplasmic organelle found in plants, algae, and some protists. The “energy factories” of the cell, they are the site where photosynthesis takes place, converting light energy into chemical energy to sustain life on Earth. Chloroplasts are likely derived from an ancient photosynthetic cyanobacterium that was engulfed by a eukaryotic host cell, giving rise to an ancestral eukaryotic plant cell more than 1.5 billion years ago (Archibald, 2015). However, during subsequent endosymbiotic evolution, the chloroplast genome shrank markedly, as most genes of cyanobacterial origin were transferred into the nucleus or lost altogether (Martin et al., 2002; Daniell et al., 2016). In land plants, the chloroplast genome generally ranges in size from 120 to 160 kb and exhibits a highly conserved structure. It comprises a single circular DNA molecule with a quadripartite structure in which two copies of a 26-kb inverted repeat region separate it into one large and one small single-copy region (Daniell et al., 2016; de Vries and Archibald, 2018).
On average, the chloroplast genomes of land plants have retained about 120 genes with conserved content. Most of these genes are particularly important for plant viability because they encode core subunits of the photosynthetic apparatus and the gene expression system of chloroplasts (i.e., RNA polymerase subunits, ribosomal proteins, ribosomal RNAs [rRNAs], and transfer RNAs [tRNAs]). Substantial alteration of chloroplast gene expression can kill plants, underscoring the importance of these genes even in a reduced complement (Zhang et al., 2020).
Similar to those of bacteria, most chloroplast genes are organized as operons transcribed from single promoters and are separated by relatively small non-coding intergenic regions. However, coevolution of the chloroplast and nuclear genomes also allowed many chloroplast-specific innovations in the gene expression machinery to arise. Primary chloroplast transcripts must undergo complicated post-transcriptional processing steps to form mature mRNA, such as RNA editing, RNA splicing, 5′ and 3′ trimming, and intercistronic cleavage (Barkan, 2011). These post-transcriptional processing events ensure that transcripts in chloroplasts have proper stability and secondary structures more suitable for translation by chloroplast 70S ribosomes (Zoschke and Bock, 2018; Zhang et al., 2020). However, chloroplast genes are not constitutively expressed, and light is one of the most important environmental cues that switches on their expression through at least two layers of regulation, transcriptional and translational.
Numerous discoveries have substantially expanded our understanding of chloroplast gene expression over the last several years. However, systematic summary and discussion of chloroplast gene expression are generally lacking. Here, we highlight recent advances and emerging principles that govern chloroplast gene expression in land plants, especially at the levels of transcription, post-transcriptional processing, and translation. Because the activation of chloroplast gene expression is tightly controlled by light, we also discuss current proposed mechanisms for light-induced transcription and translation of these genes. Finally, we discuss a number of biological and mechanistic questions about chloroplast gene expression that remain to be answered.
Transcription
Chloroplast genes are transcribed by two distinct polymerases: nucleus-encoded polymerase (NEP) and plastid-encoded polymerase (PEP). NEP is a phage-type RNA polymerase composed of a single catalytic subunit, whereas PEP is a bacterial-type, multi-subunit RNA polymerase (Yagi and Shiina, 2014; Borner et al., 2015). Three typical NEPs have been identified in land plants, including plastid-targeted RpoTp, mitochondrion-targeted RpoTm, and dual-targeted RpoTmp that is localized to both chloroplasts and mitochondria (Borner et al., 2015). These NEPs share a strong evolutionary relationship with T3/T7 phage-type polymerase. Unlike chloroplasts that have both NEP and PEP, mitochondria contain only NEP for transcription, and the unique PEP of chloroplasts may be associated with their photosynthetic function.
The NEP promoter is characterized by a conserved YRTA motif upstream from the transcription start site. A subfamily of NEP promoters has an additional GAA box about 18- to 20-nt upstream of the YRTA motif (Weihe and Börner, 1999; Ortelt and Link, 2021). The PEP promoter is distinct from the NEP promoter and retains the –35 (TTGACA) and –10 (TATAAT) consensus sequence motifs, resembling bacterial σ70 promoters (Ortelt and Link, 2021). Except for the rpoB operon and the accD gene, which are exclusively transcribed by NEP, most genes in chloroplasts are transcribed by both NEP and PEP (Ortelt and Link, 2014). Currently, at least 215 transcription start sites of PEP and NEP have been determined in mature chloroplasts of Arabidopsis using Terminome-seq (Castandet et al., 2019).
The activities of both NEP and PEP increase considerably with chloroplast development. However, PEP activity undergoes a more pronounced increase than NEP activity. In mature chloroplasts, PEP serves as the major transcriptional machinery and produces more than 80% of all primary plastid transcripts (Cahoon et al., 2004; Zhelyazkova et al., 2012; Kindgren and Strand, 2015). The activities of NEP and PEP are much higher in young leaves than in old leaves, although old leaves still maintain substantial chloroplast transcription activity (Zoschke et al., 2007). Although NEP and PEP are still active in plastids of non-photosynthetic tissues, their activities are much lower compared with that in mature chloroplasts of photosynthetic tissues (Puthiyaveetil et al., 2021).
During evolution, chloroplasts of land plants inherited the PEP catalytic core from the ancestral cyanobacterium but lost all genes encoding transcription factors, several σ factors, and nucleoid-associated proteins (e.g., the HU protein that was first named the U factor in Escherichia coli strain U93 and later renamed HU because the U factor shared many characteristics with eukaryotic histones) (Drlica and Rouviere-Yaniv, 1987; Yagi and Shiina, 2014). With the acquisition of novel host cell–derived polymerase-associated proteins (PAPs), the PEP transcription apparatus, or PEP complex, of land plants became more complicated. In addition to transcriptional initiation, transcriptional pausing by the PEP complex has an essential role in transcriptional output. Transcription by the PEP complex is also highly induced by light, which helps chloroplasts synchronize their gene expression program to support the needs of photosynthesis. We focus below on our current understanding of the PEP complex, highlighting PAP functions, transcriptional pausing, and light-induced transcription.
The PEP complex is a hybrid system with a prokaryotic core and peripheral PAPs of eukaryotic origin
The PEP catalytic core is composed of α, β, β′, and β′′ subunits that are encoded by the plastid genes rpoA, rpoB, rpoC1, and rpoC2, respectively. Similar to bacterial RNA polymerase, a σ factor is required for PEP to recognize its target promoters harboring the –35 and –10 motifs. Plant σ factors belong to the bacterial σ70 family and contain all four conserved σ domains also found in bacterial σ factors. In land plants, genes encoding σ factors were transferred to the nucleus and duplicated several times, yielding many nuclear copies (Fu et al., 2021). For example, there are as many as six σ factors in Arabidopsis (Arabidopsis thaliana), named SIG1 to SIG6 (Macadlo et al., 2020; Puthiyaveetil et al., 2021).
However, distinct from bacterial RNA polymerase, PEP has acquired many PAPs for executing RNA biosynthesis. Twelve PAPs have been identified in Arabidopsis chloroplasts, PAP1 to PAP12 (Steiner et al., 2011; Pfannschmidt et al., 2015). These PAPs bind specifically and tightly to the Rpo catalytic core, forming well-conserved protrusions as in other RNA polymerases, although the 3D map resolution of the PEP complex is currently insufficient to determine the relative positions of these PAPs within the PEP complex (Ruedas et al., 2022). All these PAPs are necessary for chloroplast transcription and show a low level of functional redundancy. Loss of function of each PAP results in similar phenotypes: a severe block of chloroplast transcription and an albino (cells devoid of chlorophyll) phenotype, resembling that seen in knockout mutants of chloroplast rpo genes (Pfalz et al., 2006; Garcia et al., 2008; Myouga et al., 2008; Arsova et al., 2010; Schroter et al., 2010; Steiner et al., 2011; Gao et al., 2011; Yagi et al., 2012; Chang et al., 2017).
Limited functional information is available on each PAP based on experimentation. PAP5, also named PLASTID TRANSCRIPTIONALLY ACTIVE 12 (pTAC12) and HEMERA, can bind to single-stranded RNA and single-stranded DNA, suggesting that PAP5 may localize close to the active sites of the PEP complex and facilitate the positioning of tethered DNA and/or RNA (Pfalz et al., 2015). PAP5 and PAP3/pTAC10 are also required for formation and/or stability of the entire PEP complex (Pfalz et al., 2015; Chang et al., 2017). PAP8/pTAC6 contains an RNA-binding motif homologous to a rhinoviral RNA-dependent RNA polymerase and shows RNA-binding activity in vitro, suggesting a role in handling nascent RNA from the PEP catalytic core (Chambon et al., 2022). PAP10/THIOREDOXIN Z contains a thioredoxin domain; together with its interacting protein PAP6/FRUCTOKINASE-LIKE 1, PAP10 is thought to confer redox regulation of transcriptional activity (Arsova et al., 2010; Schroter et al., 2010; Diaz et al., 2018; Yang et al., 2021). Two other paralogous proteins with superoxide dismutase domains, PAP4/FE SUPEROXIDE DISMUTASE 3 and PAP9/FE SUPEROXIDE DISMUTASE 2, are implicated in protecting the PEP complex from reactive oxygen species produced by the photosynthetic apparatus (Myouga et al., 2008; Favier et al., 2021). Crystal structure analysis of recombinant PAP9 showed a fold similar to that of iron or manganese superoxide dismutases, although its catalytic center is coordinated with a zinc ion (Favier et al., 2021).
Transcriptional pausing exists in chloroplasts
In bacteria and metazoans, RNA synthesis by RNA polymerase is discontinuous. RNA polymerase tends to pause (from several seconds to minutes or even longer) at specific DNA regions, a phenomenon termed “transcriptional pausing” (Core and Adelman, 2019; Wissink et al., 2019). Transcriptional pausing is a major regulatory mechanism for transcription, as it allows time for paused RNA polymerases to make regulatory decisions at specific genomic locations and thereby modulate transcriptional output (Landick, 2021; Noe Gonzalez et al., 2021).
In metazoans, pausing of RNA polymerase II (Pol II) at promoter-proximal positions is a key rate-limiting step in transcriptional output and an essential point at which opposing or reinforcing signals are integrated (Core and Adelman, 2019). Promoter-proximal pausing by RNA Pol II also controls gene expression by opening chromatin architecture in the vicinity of promoters (Core and Adelman, 2019). Transcriptional pausing is also implicated in diverse gene regulatory mechanisms such as transcription–translation coupling in bacteria (Larson et al., 2014), splicing (Akhtar et al., 2019), RNA folding (Muniz et al., 2021), and genome stability (Saponaro et al., 2014; Lavigne et al., 2017). In Arabidopsis, RNA Pol II in the nucleus pauses at approximately 20%–40% of protein-coding genes, and pausing both over promoter-proximal regions and after the polyadenylation site influences transcriptional efficiency (Hetzel et al., 2016; Zhu et al., 2018; Kindgren et al., 2020).
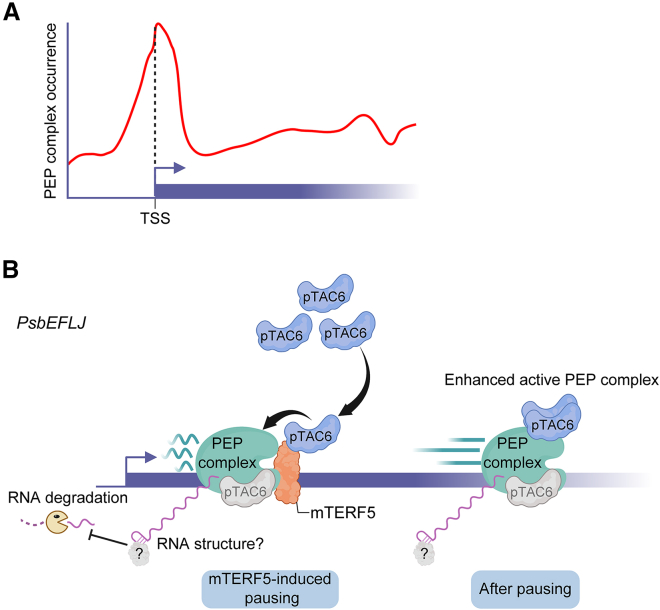
Transcriptional pausing has also been examined in Arabidopsis chloroplasts. A global run-on analysis suggested that transcriptional pausing is a widespread phenomenon that occurs at about 30% of all chloroplast genes (Zhu et al., 2018; Ding et al., 2019). This conclusion was supported by a high-resolution map of PEP binding patterns along chloroplast genes, which showed that PEP is frequently non-uniformly distributed, with a tendency to accumulate in close proximity to and downstream of transcription start sites (Palomar et al., 2022). This result indicated that the early elongation of the PEP complex is more prone to pause over promoter-proximal regions than to move forward further downstream (Figure 1A). However, the mechanism that establishes this widespread promoter-proximal pausing is unclear. Moreover, decreasing transcriptional initiation of the PEP complex by knocking out SIG2 or SIG6 results in an overall reduction in PEP occurrence both over promoter-proximal regions and further downstream. This observation indicates that PEP pausing is tightly coupled with transcriptional elongation (Chi et al., 2015; Palomar et al., 2022) and suggests that it might generally regulate transcriptional output in chloroplasts.
Figure 1.
Proposed model for the function of mTERF5-induced pausing and preferential binding of the PEP complex to chloroplast promoters.
(A) PEP complex distribution as a function of position along chloroplast genes. The model for PEP occurrence was redrawn according to Palomar et al. (2022). TSS, transcription start site.
(B) A working model for mTERF5-induced pausing in regulating psbEFLJ transcription is proposed based on Ding et al. (2019) and Meteignier et al. (2020). The PEP complex represents the major transcriptional machinery in chloroplasts and is composed of the core enzyme and PAPs. PAP8/pTAC6 is one of 12 PAPs. mTERF5 serves as a transcriptional pausing factor and induces PEP pausing at the psbEFLJ polycistron by binding to the +30 to +51 region relative to the transcription start site of this gene. mTERF5 interacts with pTAC6 outside of the PEP complex and helps recruit additional pTAC6 proteins into the PEP complex to enhance its activity, thereby positively regulating psbEFLJ transcription. In addition, transcriptional pausing caused by mTERF5 may provide time and location for the action of unknown RNA-binding proteins or the folding of nascent RNA, thus improving RNA stability.
Transcriptional pausing in chloroplasts can be regulated by proteinaceous trans-factors in a gene-specific manner. MITOCHONDRIAL TRANSCRIPTION TERMINATION FACTORs (mTERFs) were originally identified in mitochondria of metazoans. However, the mTERF family has expanded during the evolution of land plants, and its members are localized to mitochondria and/or chloroplasts (Kleine, 2012). mTERF5/MTERF DEFECTIVE IN ARABIDOPSIS1 is a member of the mTERF family that induces the PEP complex to pause at the psbEFLJ locus by binding to the +30 to +51 region relative to the transcription start site. mTERF5 also helps to recruit PAP8/pTAC6, an essential subunit of the PEP complex, into the paused PEP complex, yielding an enhanced PEP complex that may promote transcriptional elongation and thus positively regulate psbEFLJ transcription (Figure 1B) (Ding et al., 2019). In addition, stabilization of the 5′ ends of processed psbE RNA is clearly lower in mterf5 mutants, suggesting that transcriptional pausing caused by mTERF5 also regulates RNA stability (Meteignier et al., 2020). Considering that RNA stability is often maintained by RNA-binding proteins, mTERF5-induced pausing may facilitate the action of unknown RNA-binding proteins to improve the stability of psbE RNA. Overall, transcriptional pausing in chloroplasts represents an important point for the recruitment of regulatory proteins and for coupling transcription and post-transcriptional processes, such as RNA stability.
The PEP complex is activated by light-induced redox changes
Light is the major driver initiating transcription in chloroplasts of land plants. In the nucleus, light promotes the expression of genes encoding PAPs through histone H4 acetylation by the nucleosome acetyltransferase of the H4 complex at the early stage of light signaling (Barrero-Gil et al., 2022). In addition, light activates the formation of the PEP complex by eliminating PHYTOCHROME-INTERACTING TRANSCRIPTION FACTORs, which in turn synchronously generate a nucleus-to-plastid signal involved in σ factors (Yang et al., 2019; Yoo et al., 2019; Hwang et al., 2022). Thus, formation of the PEP complex is strongly controlled by light.
In mature chloroplasts, light globally activates the association of the full PEP complex with promoters to start transcription (Finster et al., 2013). PLASTID REDOX INSENSITIVE2 (PRIN2) is a key component required for PEP complex activation in response to light. PRIN2 is a redox-regulated protein that localizes to chloroplasts (Kindgren et al., 2012) and forms a dimer via its Cys68–Cys68 disulfide bond in the dark. Exposure to light activates photosynthetic electron transport and triggers the reduction of thioredoxins within chloroplasts. Thioredoxins convert the PRIN2 dimer into the active PRIN2 monomeric form. PRIN2 monomers then activate the full activity of the PEP complex, potentially via their interaction with PAP10/THIOREDOXIN Z (Diaz et al., 2018). These findings highlight a signal transduction pathway that connects light-derived electron transport in the thylakoids to activation of the PEP complex. In addition, the activity of the PEP complex and of σ factors is tightly controlled by phosphorylation events that are mediated by redox-regulated plastid transcription kinases (Baginsky et al., 1999; Schweer et al., 2010; Türkeri et al., 2012; Ibrahim et al., 2020; Puthiyaveetil et al., 2021). The redox status of the plastoquinone pool affects the phosphorylation of SIG1, which in turn regulates transcription of the photosystem reaction center genes psaA/B and psbA (Shimizu et al., 2010). Hence, light-induced modification via phosphorylation of the PEP complex and σ factors may represent another mechanism by which transcription can be initiated in the light. Because mature chloroplasts have completed the biogenesis of the PEP complex, light-induced redox changes in chloroplasts would play a major role in starting or impeding transcription upon diurnal cycles, whereas light signaling in the nucleus is necessary for initiating chloroplast biogenesis and maintaining turnover of the PEP complex.
Post-transcriptional alterations of chloroplast RNAs
In cyanobacteria, gene expression is regulated mainly at the level of transcriptional initiation, whereas post-transcriptional control is more prominent in the chloroplasts of land plants. Primary chloroplast transcripts are produced from polycistronic operons and undergo extensive processing (e.g., 5′ and 3′ trimming, as well as intercistronic cleavage). In addition, over the course of chloroplast evolution, two eukaryotic features were recruited to chloroplasts: the acquisition of introns and the modification of specific nucleotides within RNA molecules, which is termed RNA editing (Germain et al., 2013). To accommodate these new features, numerous nucleus-encoded RNA-binding proteins were acquired in the context of nucleus–chloroplast coevolution, opening up new possibilities for more precise regulation of chloroplast genes and offering greater flexibility for chloroplast biogenesis and plant adaptation.
Intercistronic cleavage and terminal RNA processing
A particularly pronounced feature of chloroplast gene expression in land plants is the great complexity of mature RNA isoforms arising from polycistronic transcript processing by ribonucleases. The chloroplast contains two types of ribonucleases: exonucleases and endonucleases (Stoppel and Meurer, 2012). Similar to those of bacteria, exonucleases in chloroplasts remove successive nucleotides from the end of an RNA molecule in either the 5′→3′ or 3′→5′ direction, whereas endonucleases carry out intercistronic RNA cleavage, yielding discrete RNA fragments. To date, three chloroplast exonucleases have been identified in Arabidopsis: RNase J with 5′→3′ specificity and RNase R and polynucleotide phosphorylase with 3′→5′ specificity (Stoppel and Meurer, 2012; Manavski et al., 2018). Among the endonucleases, RNase E and RNase J are the most likely to execute internal RNA cleavage (Stern et al., 2010; Barkan, 2011). PROTEINACEOUS RNASE P and RNase Z (TRZ2) are required for maturation of tRNAs (Stoppel and Meurer, 2012). CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 kDa a and b (CSP41a and CSP41b) are unique RNA-binding endoribonucleases in photosynthetic organisms. Arabidopsis CSP41 complexes are also thought to stabilize non-translated target mRNAs and precursor rRNAs in the absence of light (Qi et al., 2012; Leister, 2014). Two miniribonuclease III members (RNC3 and RNC4) cleave the 23S−4.5S rRNA precursor to produce the mature 5′ end of the 23S RNA and the 3′ end of the 4.5S RNA (Hotto et al., 2015). As with ribonucleases in other organisms, the substrate preferences of these chloroplast ribonucleases are determined largely by RNA structure and less by RNA sequence (Stern et al., 2010; Manavski et al., 2018).
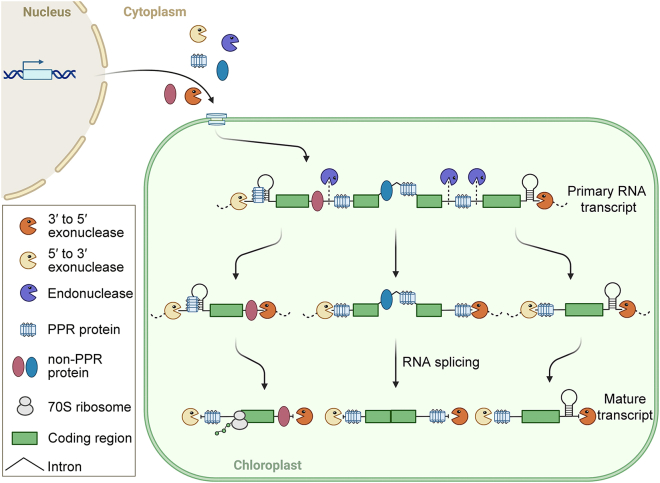
Intercistronic processing of primary transcripts is initiated by chloroplast endonucleases (such as CSP41 and RNases E and J) cleaving the RNA in unprotected and unstructured regions. Next, the resulting 5′ and 3′ termini are trimmed by exonucleases. To form proper 5′ termini, the 5′ ends of chloroplast transcripts are predominantly protected by RNA protectors that serve as barriers against 5′→3′ exonucleases. By contrast, the 3′ ends of chloroplast transcripts are often protected by secondary structures in the 3′ untranslated region (UTR) of RNAs formed by short-inverted repeat sequences (Figure 2). In particular, most RNA protectors that act on chloroplast transcripts are pentatricopeptide repeat (PPR) proteins, which passively bind to terminal RNA regions to prevent their degradation by exonucleases. Typical PPR proteins harbor repeating 35-amino-acid motifs that present a long superhelical RNA-binding surface, whereby the exact sequence of each PPR repeat dictates the nucleotide to which it will bind (the one-repeat:one-nucleotide binding mode) (Barkan et al., 2012; Yin et al., 2013; Shen et al., 2016). PPR proteins bind to the 5′ ends of transcripts to prevent 5′→3′ exonuclease invasion and sometimes also compete with stem–loop structures and release ribosomal-binding sites (i.e., the Shine–Dalgarno [SD] sequence). For instance, SUPPRESSOR OF THYLAKOID FORMATION 1 in Arabidopsis and its ortholog PPR53 in maize (Zea mays) stabilize the 5′ ends of the 23S precursor RNA and ndhA (Wu et al., 2016; Zoschke et al., 2016; Zhou et al., 2017; Li et al., 2021). In addition, some PPR proteins also protect and define the 3′ ends of transcripts that lack short inverted repeat sequences (Figure 2). For example, PPR10 binds to the intergenic regions of atpI–atpH and psaJ–rpl33 transcripts and serves as a barrier against RNA degradation from either the 5′ or 3′ direction (Pfalz et al., 2009). The PPR protein CHLOROPLAST RNA PROCESSING 1 cooperates with the S1-domain-containing RNA chaperone PETB/PETD STABILIZING FACTOR to bind to the intergenic regions of petB–petD, stabilizing the 3′ ends of petB transcripts and promoting petD translation (Jiang et al., 2019).
Figure 2.
Model for intercistronic cleavage and termini processing of chloroplast RNAs.
In chloroplasts, primary RNAs are cleaved by endonucleases (such as RNases E and J, CSP41) often in intergenic regions, giving rise to new RNA precursors. The 5′ and 3′ ends of chloroplast RNAs are protected against degradation by exonucleases via the specific binding of RNA protectors, such as PPR proteins, and/or stem–loop RNA structures. Moreover, those factors bound to 5′ termini can also remodel RNA structures, which in turn stimulates translation initiation.
To date, numerous PPR and non-PPR protectors have been found in chloroplasts, most of them specific to land plants, reflecting tight nucleus–chloroplast coevolution since the initial endosymbiosis event. A summary of known chloroplast RNA protectors, including their preferential targets and classification of their domains, is given in Table 1.
Table 1.
Summary of known chloroplast RNA protectors and their targets.
| Name | Organism | Domain | Target(s) | Reference(s) |
|---|---|---|---|---|
| AtBFA2 | Arabidopsis thaliana | PPR | atpF–atpA IGR | Zhang et al., 2019 |
| AtCRP1; ZmCRP1 | A. thaliana; Zea mays | PPR | petB–petD IGR; psaC 5′ UTR; petA 5′ UTR | Ferrari et al., 2017; Fisk et al., 1999; Schmitz-Linneweber et al., 2005 |
| AtHCF152 | A. thaliana | PPR | psbH–petB IGR; petB 5′ UTR; psbH 3′ UTR | Nakamura et al., 2003, 2004 |
| AtPGR3; ZmPGR3 | A. thaliana; Z. mays | PPR | petL 5′ UTR; rpl14–rps8 IGR; ndhA 5′ UTR | Cai et al., 2011; Rojas et al., 2018; Yamazaki et al., 2004 |
| AtPPR5; ZmPPR5 | A. thaliana; Z. mays | PPR | trnG (UCC) intron | Beick et al., 2008 |
| AtSVR7; ZmATP4 | A. thaliana; Z. mays | PPR | atpF–atpA IGR; psaJ–rpl33 IGR; atpB/E 5′ UTR | Zoschke et al., 2012, 2013b |
| ZmPPR10 | Z. mays | PPR | atpH 5′ UTR; psaJ 3′ UTR | Pfalz et al., 2009; Prikryl et al., 2011 |
| PpPPR_21 | Physcomitrium patens | PPR | psbI–ycf12 5′ UTR | Ebihara et al., 2019 |
| PpPPR_38 | P. patens | PPR | clpP–rps12 IGR | Hattori et al., 2007; Hattori and Sugita, 2009 |
| AtCRR2 | A. thaliana | PLS-class PPR-DYW | rps7-ndhB IGR | Hashimoto et al., 2003 |
| AtEMB175; ZmPPR103 | A. thaliana; Z. mays | PLS-class PPR-DYW | rpl16 5′ UTR | Hammani et al., 2016 |
| AtSOT1; ZmPPR53 | A. thaliana; Z. mays | PPR-SMR | rrn23 5′ end; ndhA 5′ UTR | Li et al., 2021; Wu et al., 2016; Zhou et al., 2017; Zoschke et al., 2016 |
| AtBSF; ZmBSF | A. thaliana; Z. mays | S1 domain | petB–petD IGR; petA 5′ UTR | Jiang et al., 2019 |
| 28RNP | Spinacea oleracea | RRM motif | psbA, rbcL, petD, rps14 3′ UTRs | Schuster and Gruissem, 1991 |
| AtCP31A | A. thaliana | RRM motif | 3′ ends of psbB, psbD, psaA/B, atpB, ndhB, and ndhF | Kupsch et al., 2012; Tillich et al., 2009 |
| AtCP29A | A. thaliana | RRM motif | 3′ ends of psbB, psbD, psaA/B, atpB, and ndhB | Kupsch et al., 2012 |
| AtCP33A | A. thaliana | RRM motif | multiple transcripts in chloroplasts | Teubner et al., 2017 |
| AtHCF107; ZmHCF107 | A. thaliana; Z. mays | HAT (half a TPR) | psbH 5′ UTR | Hammani et al., 2012; Sane et al., 2005 |
| AtHCF145; PpHCF145 | A. thaliana; P. patens | TMR domain; SRPBCC domain | psaA 5′ UTR | Lezhneva and Meurer, 2004; Manavski et al., 2015 |
| AtPrfB1/AtHCF109 | A. thaliana | ribosomal release factor; SPF and GGQ motifs | transcripts with UGA stop codons | Meurer et al., 2002 |
| AtPrfB3 | A. thaliana | homologous to PrfB1 | petB 3′ UTR | Stoppel et al., 2011 |
IGR, intergenic region
RNA splicing
RNA splicing is an essential step for chloroplast gene expression in which removal of intronic sequences from precursor RNAs and connection of the remaining sequences produces mature RNAs and tRNAs with correct genetic information. The chloroplast genomes of land plants have 17–21 introns. Based on their conserved sequences and splicing mechanisms, most chloroplast introns can be classified as group II introns, with only one group I intron in trnL (de Longevialle et al., 2010; Germain et al., 2013).
All of these introns have lost the ability to self-splice owing to degeneration of RNA structures and loss of intron-encoded maturases over the course of evolution. Efficient splicing of these degenerated chloroplast introns must be stimulated by a set of additional proteins. Among chloroplast introns, only the trnK intron encodes the maturase MatK that is involved in splicing of several group IIA introns (Zoschke et al., 2010). Besides this intron-coded MatK, many nucleus-encoded splicing factors have been identified that localize to the chloroplast, such as the RNase III-domain protein RNC1 of maize (Watkins et al., 2007), the plant organelle RNA recognition protein what’s this factor (WTF1), also from maize (Kroeger et al., 2009), and ACCUMULATION OF PHOTOSYSTEM ONE 1 from maize and Arabidopsis (Watkins et al., 2011). A summary of chloroplast splicing factors can be found in several detailed reviews (de Longevialle et al., 2010; Schmitz-Linneweber et al., 2015; Wang et al., 2022; Zeng et al., 2022).
Chloroplast RNA splicing and ribosome maturation (CRM) proteins are more general splicing factors and include CRS2-associated factor 1 (CAF1) and CAF2, chloroplast RNA splicing 1 (CRS1), and CRM family member 2 (CFM2) and CFM3 (de Longevialle et al., 2010). CAF1/2 bind to CRS2 to form complexes. CRS2–CAF1 complexes are necessary for the splicing of seven introns (ycf3 intron 1, trnG, rpl16, petD, rps16, clpP intron 1, and rpoC1), and CRS2–CAF2 complexes are required for the splicing of five introns (ycf3 intron 1, ndhA, ndhB, petB, and rps12 intron 1) (Ostheimer et al., 2003; Asakura and Barkan, 2006). Because CRS2–CAF1/2 complexes are required for different sets of group IIB introns with only one overlapping intron substrate, ycf3 intron 1, there must be some recognition mechanism by which CRS2–CAF1/2 complexes target distinct introns. The PPR protein PHOTOSYSTEM I BIOGENESIS FACTOR2 interacts with CAF1 and CAF2 and may contribute to the specificity of the CRS2–CAF1 and CRS2–CAF2 complexes toward ycf3 intron 1 (Wang et al., 2020). A similar mode of action was also reported for the PPR protein EMBRYO DEFECTIVE 1270 (EMB1270), which appears to be required for the splicing of clpP1 intron 2, ycf3 intron 1, ndhA, and ndhB through its specific interaction with the splicing factor CFM2 (Zhang et al., 2021). Considering that PPR proteins bind to RNA in a sequence-specific manner, interactions between specific PPR proteins and general splicing factors such as CAF1/2 and CFM2 may help target their constituent complexes to the corresponding introns to form the full splicing complexes.
Distinct from other chloroplast introns, rps12 intron 1 is the only intron that is trans-spliced in land plants, meaning that two partial rps12 transcripts transcribed from different parts of the chloroplast genome are spliced together into one functional RNA. The rps12 intron 1 is split into rps12 introns 1a and 1b; rps12 intron 1a lies between rpl20 and clpP1, whereas rps12 intron 1b is approximately 30 kb away and co-transcribed with ndhB and rps7 (Glanz and Kück, 2009). Thus, the splicing of rps12 intron 1 is more complex than that of other cis-splicing introns, requiring the rps12 precursors to be cleaved and brought together prior to regular splicing. The PPR proteins EMBRYO DEFECTIVE 2654 and PPR4 bind to the 3′ end of rps12 intron 1a and the 5′ end of rps12 intron 1b, respectively. Their binding was proposed to be an early event that is a prerequisite for the proper folding of domain 3 of rps12 intron 1, which is ready for the formation of the splicing complex via general factors such as the CRS2–CAF2 complexes (Schmitz-Linneweber et al., 2006; Aryamanesh et al., 2017; Lee et al., 2019).
Chloroplasts have a rather small genome. However, numerous splicing factors are required for chloroplast gene expression. RNA splicing in plant chloroplasts appears to be particularly complex. Compared with those in their prokaryotic ancestors, introns in chloroplasts are newly invented and no longer have the ability to self-splice (Wang et al., 2022). In order to stimulate self-splicing of introns, numerous splicing factors may have arisen to compensate for the degenerated chloroplast introns in the context of nucleus–chloroplast coevolution. It remains unknown why so many splicing factors have arisen in plant chloroplasts. Presumably, these splicing factors may be required for plant growth and development and are regulated in response to environmental stresses.
The DYW domain is the catalytic domain for C-to-U editing in plants
In plants, RNA editing mainly involves deamination reactions that target specific cytidines (Cs) in organellar RNAs and convert them to uridines (Us). This phenomenon is nearly omnipresent in land plants but entirely absent in green algae (Takenaka et al., 2013; Yan et al., 2018; Small et al., 2020). Typical flowering plants edit 20–60 sites in their chloroplast transcripts, mostly located in protein-coding transcripts with only a few sites in tRNA, introns, non-coding regions, or UTRs (Ichinose and Sugita, 2016; Edera et al., 2018). These C-to-U editing events are necessary for the proper function of organellar proteins and optimal plant growth; they change the coding sequences of RNA, introducing translation start sites (AUG) and modulating secondary structures that influence the stability and/or splicing of RNAs (Shikanai, 2015; Small et al., 2020).
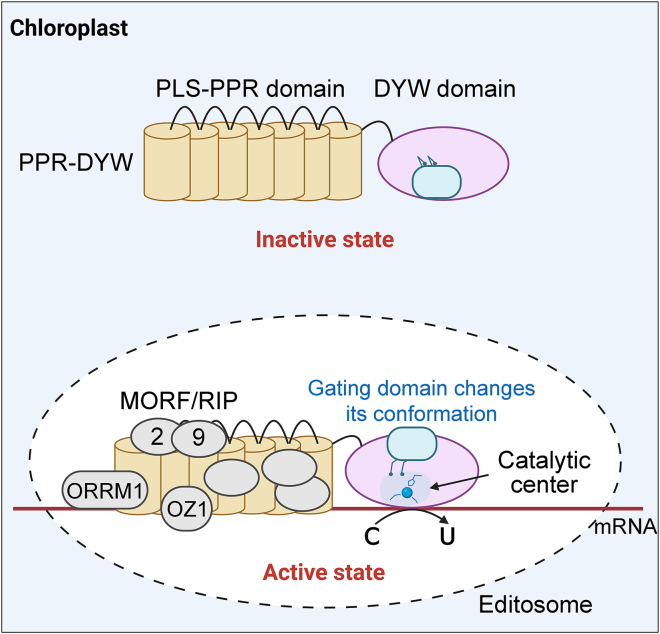
PLS-class PPR proteins are considered to be the core components for execution of RNA editing. They are distinguished from the typical PPR (P-class) proteins because they contain characteristic repeating triplets of P, L (long variants of P), and S (short variants of P) motifs (Lurin et al., 2004; Cheng et al., 2016). Approximately half of all PLS-class proteins harbor a C-terminal DYW domain, which was named after its conserved Asp-Tyr-Trp (D-Y-W) triplet residues (Lurin et al., 2004). Sequence alignments have demonstrated that the DYW domain exhibits some similarity to known cytidine deaminases from other kingdoms and contains a conserved HxE(x)nCxxX catalytic-site signature, suggesting that the DYW domain might be the deaminase catalytic entity of the organellar RNA editing complex (Salone et al., 2007; Schallenberg-Rüdinger et al., 2013). The PPR-DYW protein PPR56 from the moss Physcomitrium patens can execute effective C-to-U editing of its corresponding target ccmFC transcript on its own without additional cofactors (Oldenkott et al., 2019; Hayes and Santibanez, 2020). Considering the definitive function of PLS-class PPR domains in RNA substrate recognition, these findings show that the DYW domain serves as a cytidine deaminase during RNA editing.
Recently, the high-resolution structure of a DYW domain was resolved from the chloroplast editing factor ORGANELLE TRANSCRIPT PROCESSING 86 (OTP86DYW) of Arabidopsis (Takenaka et al., 2021). Unlike deaminases in other organisms, the deaminase fold of OTP86DYW is interrupted by an inserted gating domain, which restrains entry of the substrate cytidine into the catalytic site. Upon activation, the OTP86DYW gating domain alters its conformation and triggers the repositioning of the active site architecture, which recruits a water molecule as a fourth ligand between a zinc ion and the catalytic residue E894 to attack the cytidine for deamination (Takenaka et al., 2021). In this way, the DYW domain alone tends to remain in an autoinhibited state, and its catalytic ability is only activated after transport into the organelle (Figure 3). This autoinhibition mechanism of DYW proteins would offer protection for RNAs in the cytosol from the damage of off-target editing events. In addition, the deaminase activity exerted by the DYW domain exhibits a different preference for the nucleotides close to the RNA editing sites, indicative of a complicated interplay between RNA substrate elements and the DYW domain during RNA editing (Maeda et al., 2022). In addition to canonical C-to-U editing, “reverse” editing (U-to-C) was also detected in the organelles of three land plant clades (hornworts, lycophytes, and ferns) (Small et al., 2020). This U-to-C editing is phylogenetically correlated with a subgroup of DYW domains, referred to as DYW:KP, which display a conserved Lys-Pro (KP) dipeptide at the start of the domain (Gerke et al., 2020; Gutmann et al., 2020). Designer DYW:KP proteins can exhibit U-to-C editing activity in bacteria and human cell cultures (Ichinose et al., 2022). This result raises the interesting question of how the relatively minor sequence differences between DYW:KP and the canonical DYW domain can lead to a complete reversal of a theoretically irreversible reaction. Based on the structures of OTP86DYW, we hypothesize that minor differences in amino acid sequences between DYW:KP and canonical DYW domains affect the catalytic pocket to accept the presence of an amine acceptor or donor and execute the C-to-U deaminase or “reverse” reaction.
Figure 3.
Proposed model for C-to-U RNA editing in the chloroplasts of flowering plants.
The DYW domain alone tends to be in an inactive state, with a catalytic center closed by the gating domain. In editosomes, the deaminase activity of DYW is activated, which is accompanied by changes in the conformation of its gating domain and repositioning of the catalytic center architecture. In chloroplasts, the editosome is a large multi-protein complex that contains PLS-class PPR specificity factors and many non-PPR editing factors, such as MORF/RIP, ORRM1, and OZ1. Among these factors, MORF proteins can increase the RNA-binding activity of PLS-type PPRs, thus helping the editosome bind to the sequence upstream of its cognate target site.
In addition to PPR-DYW proteins, many essential non-PPR editing factors have been acquired by flowering plants, such as RNA-EDITING FACTOR INTERACTING PROTEIN 1/MULTIPLE ORGANELLAR RNA EDITING FACTOR (MORF), ORGANELLE RNA RECOGNITION MOTIF PROTEIN, the RNA helicase INCREASED SIZE EXCLUSION LIMIT 2, and ORGANELLE ZINC FINGER proteins (Takenaka et al., 2012; Sun et al., 2013, 2015; Bobik et al., 2017; Gipson et al., 2022). These PPR factors and non-PPR editing factors form diverse and large multi-protein complexes, also termed editosomes, that perform RNA editing (Sun et al., 2016; Sandoval et al., 2019). Among the non-PPR editing factors, MORF9 binds to the L motif close to the concave surface of the PLS triplet and therefore alters the angle between two helices of the L motif, ultimately leading to considerable compression of the PPR superhelix (Haag et al., 2017; Yan et al., 2017; Yang et al., 2018). This compressed conformational change by MORF9 markedly increases RNA-binding activity by PLS-type PPR proteins (Figure 3) (Yan et al., 2017; Royan et al., 2021). However, our understanding of the roles of other non-PPR components within editosomes remains rudimentary.
Plastid non-coding RNAs
Plastid non-coding RNAs (pncRNAs) range in length from 15 nt to >3 kilonucleotides (knt) and are abundant in chloroplasts (Hotto et al., 2011). The majority of pncRNAs derive from intergenic, intronic, and antisense regions and seem to be generated by readthrough from the adjacent genes followed by complicated RNA processing events. A small portion of pncRNAs are thought to be primary transcripts controlled by their own promoters (Zhelyazkova et al., 2012; Castandet et al., 2019; Anand and Pandi, 2021). Ribonucleases are involved in the biogenesis of pncRNAs. The chloroplast 3′→5′ exonuclease polynucleotide phosphorylase is required for the 3′ end maturation of many pncRNAs (Germain et al., 2011; Hotto et al., 2011). Loss of the 3′→5′ exonuclease RNase R results in almost complete deletion of both sense and antisense (as) psbK RNAs as well as a considerable decrease in AS5 asRNA. Virus-induced gene silencing of RNase J causes an accumulation of nc-rbcL-accDint precursor (a pncRNA corresponding to rbcL and accD intergenic regions) and minor alterations for other pncRNAs. RNase E stabilizes the accumulation of aspsbK and nc-rbcL-accDint RNAs (Hotto et al., 2011). Some PPR proteins, such as PPR10 and SUPPRESSOR OF THYLAKOID FORMATION 1/PPR53, are involved in generating chloroplast small pncRNAs (sRNAs), also termed “PPR footprints” (Pfalz et al., 2009; Wu et al., 2016; Zoschke et al., 2016).
pncRNAs play a role in regulating various aspects of chloroplast gene expression at the post-transcriptional level. The AS5 asRNA is antisense to the chloroplast rrn5–trnR region and may have a regulatory role in 5S rRNA maturation (Hotto et al., 2011; Sharwood et al., 2011). A long transcript antisense to ndhB is suggested to regulate ndhB RNA editing or stability (Georg et al., 2010). asRNAs for psbT may interact with psbT mRNA to prevent translation of psbT and protect psbT mRNA from nucleolytic degradation (Zghidi-Abouzid et al., 2011). In addition, a systematic analysis of chloroplast sRNAs indicates that chloroplast sRNAs might serve as competitors of mRNAs for PPR proteins to regulate RNA metabolism (Ruwe et al., 2016).
Chloroplast translation
In chloroplasts, translation is performed by a bacterial-type 70S ribosome that is composed of a 50S large subunit and a 30S small subunit. The chloroplast genome encodes a full set of tRNAs, all rRNAs, one quarter of all proteins comprising the 50S large subunit, and approximately half of all proteins forming the 30S small subunit. The remaining ribosomal subunits are encoded by the nuclear genome. Although the chloroplast and bacterial 70S ribosomes share a common ancestor, their structures and operational mechanisms have diverged during evolution. In addition, chloroplast translation is highly regulated by light and coordinated with the thylakoid membrane system to achieve optimum photosynthetic performance. Here, we discuss the variations in ribosomes from bacteria to chloroplasts, translation dynamics, and the pathways that activate translation in response to light.
Differences between chloroplast and bacterial ribosomes
The structure of chloroplast ribosomes is generally similar to that of bacterial 70S ribosomes but has some distinct features (Zoschke and Bock, 2018). Chloroplast ribosomes contain a 4.5S rRNA in addition to a complete set of sequences homologous to bacterial rRNAs (23S, 16S, and 5S rRNA) with analogous functions. The 23S rRNA confers peptidyl transferase activity, and the 16S rRNA comprises the decoding center and acts as a scaffold during ribosome assembly (Shajani et al., 2011). An equivalent to chloroplast 4.5S rRNA does not exist in bacteria, but the chloroplast 4.5S rRNA is homologous to the 3′ terminus of the 23S rRNA, suggesting that the 4.5S rRNA may have arisen from fragmentation of the 23S rRNA during evolution (Edwards and Kössel, 1981; Maier et al., 2013). In addition, the chloroplast 23S rRNA is post-transcriptionally cleaved at two “hidden break” sites after assembly of the 50S subunit, generating three distinct fragments of ∼0.5, 1.2, and 1.1 kb (from the 5′ to the 3′ end). These “hidden break” cleavages are required for chloroplast translation and are produced by specific enzymes such as RNA helicase RH39 (Nishimura et al., 2010; Bieri et al., 2017).
The protein composition of the chloroplast ribosome has also changed over the course of evolution since the initial endosymbiosis. The chloroplast ribosome lacks equivalents to the bacterial large ribosomal protein subunits Rpl25 and Rpl30. All orthologs of the E. coli ribosome are present in the chloroplast 70S ribosome but exhibit N- and C-terminal chloroplast-specific extensions compared with their bacterial counterparts (Yamaguchi and Subramanian, 2000; Ahmed et al., 2016). These extensions substantially reshape the mRNA entry and exit sites on the 30S small subunit as well as the polypeptide tunnel exit and the binding site of the signal recognition particle on the 50S small subunit (Bieri et al., 2017; Perez Boerema et al., 2018). These changes reflect specific chloroplast adaptations for translation initiation and protein targeting.
Chloroplasts also have several new components with no clear E. coli homologs, designated as plastid-specific ribosomal proteins (PSRPs) 1–6. PSRP1 was first identified as the ortholog of the bacterial cold-shock protein pY (Sharma et al., 2007, 2010), but a 2.9- to 3.1-Å resolution structure of the spinach (Spinacia oleracea) 70S ribosome obtained by cryoelectron microscopy revealed that PSRP1 binds to the mRNA channel of the small subunit and is highly similar to light-repressed transcript A of cyanobacteria from the long hibernation-promoting factor family (Perez Boerema et al., 2018). PSRP2/RPS22, PSRP3/RPS23, and PSRP4/bTHXc are present in the small ribosome subunit, and PSRP5/RPL37 and PSRP6/RPL38 are present in the large ribosome subunit (Bieri et al., 2017; Perez Boerema et al., 2018). Based on the structure of the 70S ribosome, PSRP1 localizes to the mRNA channel and interacts with the 16S rRNA residues that form the decoding center, possibly preventing the binding of tRNAs, which suggests that it may have important roles in translation activity. Other PSRPs are genuine plastid-specific ribosomal proteins that structurally compensate for rRNA domains modified over evolution (Sharma et al., 2007; Bieri et al., 2017; Perez Boerema et al., 2018).
Translation mechanisms and ribosome pausing
In chloroplasts, translation is highly dynamic and entails four major steps: initiation, elongation, termination, and ribosome recycling. During initiation, the 30S small ribosome subunit is recruited to the ribosome-binding site with the help of the initiation factors IF1, IF2, and IF3 (Sijben-Müller et al., 1986; Campos et al., 2001; Miura et al., 2007; Zheng et al., 2016). About two thirds of chloroplast genes possess bacterial-type SD sequences and use them to initiate translation. The SD sequence complementarily interacts with the anti-SD sequence in the 16S rRNA to ensure proper positioning of the initiation complex (Bieri et al., 2017; Scharff et al., 2017). The remaining one third of chloroplast genes lack an SD sequence, suggestive of alternative mechanisms for chloroplast translation initiation. In these cases, a relative lack of RNA secondary structure at the 5′ UTR, which may be regulated by trans-acting factors, facilitates correct translation initiation (Scharff et al., 2011; Nakagawa et al., 2017). Binding of the 50S subunit converts the preinitiation complex into the complete 70S ribosome, which can then enter the elongation state of translation. Similar to that in bacteria, translation elongation in chloroplasts requires many prokaryotic elongation factors, such as EF-Tu, EF-G, and EF-Ts (Zoschke and Bock, 2018). During decoding of mRNA into an amino acid sequence, chloroplasts use the standard genetic code with a minimal set of tRNAs through mechanisms of wobble or superwobble base pairing (Alkatib et al., 2012). Translation termination occurs when the ribosome reaches a stop codon (UAA, UAG, or UGA) in the RNA and requires ribosome release factors RF1/PrfA (for UAA and UAG) and RF2/PrfB1 (for UAA and UGA) (Meurer et al., 2002; Motohashi et al., 2007). After termination, the chloroplast 70S ribosomes are subsequently disassembled by ribosome recycling factors to allow for their reuse in the next round of initiation (Tiller and Bock, 2014; Zoschke and Bock, 2018). However, our understanding of how these regulatory factors control translation in chloroplasts remains rudimentary.
The feedback regulation of translation represents an elegant means of robustly fine-tuning Rubisco levels. The translation of the plastid-encoded Rubisco large subunit RbcL is regulated by its nucleus-encoded partner, the Rubisco small subunit RbcS (Rodermel et al., 1988, 1996; Khrebtukova and Spreitzer, 1996). The downregulation of RbcS restrains the assembly of RbcL, and the resulting unassembled RbcL represses its own translation, possibly by direct RNA binding (Wostrikoff and Stern, 2007), thus ensuring the stoichiometry of Rubisco.
Chloroplast ribosomes are prone to pausing at conserved sites on RNAs in seed plants such as Arabidopsis and maize (Zoschke et al., 2013a; Gawronski et al., 2018). Such ribosome pausing is often induced by a combination of factors including RNA structure, internal SD sequences, and presence of positively charged amino acids. Moreover, ribosome pausing may help the insertion of transmembrane domains from photosynthetic apparatus components into the thylakoid membrane in the correct orientation, the integration of cofactors, and efficient protein folding, thereby ensuring correct assembly of photosynthetic complexes (Gawronski et al., 2018). The position features associated with ribosome pausing are conserved in photoautotrophic plants but less so in parasitic species with reduced photosynthetic capacity (Gawronski et al., 2018; Trosch et al., 2018), suggesting that ribosome pausing is a general mechanism that facilitates the biogenesis of photosynthetic complexes.
Chloroplast translation is triggered by light-induced pH changes and photosystem II (PSII) damage
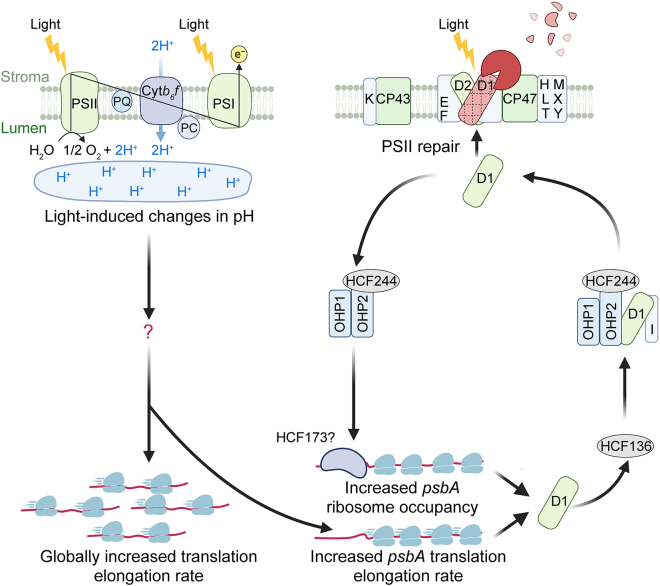
In land plants, chloroplasts can dynamically change translation rates in response to environmental stimuli, especially light, throughout their whole life cycle. Light-induced regulation of translation couples the major energy-consuming step (protein synthesis) to the major energy-producing step (photosynthesis) in chloroplasts. In addition, as light causes constant damage to the reaction center protein D1 in PSII, light-induced translation ensures the coordination of D1 protein synthesis with the need to repair PSII.
Translation elongation in chloroplasts is activated rapidly after a shift from darkness to light (Chotewutmontri and Barkan, 2018). In lysed barley (Hordeum vulgare) chloroplasts, light-activated translation elongation is regulated by light-induced changes in lumen pH (Mühlbauer and Eichacker, 1998). The pH of the thylakoid lumen is affected by the cytochrome b6f complex that pumps protons from the stroma. Light-stimulated translation elongation is considerably reduced in cytochrome b6f mutants or upon treatment with pH uncoupler agents, but this plastome-wide translational response is not obviously affected by ATP supply, reduced thioredoxin, or plastoquinone redox state (Chotewutmontri and Barkan, 2020). These findings demonstrate that light-induced pH changes globally activate chloroplast translation at the level of elongation, but how this pH change increases translation elongation is still unclear. A potential mechanism for general light-stimulated translation elongation in chloroplasts may rely on proteins that can orchestrate translation, such as the translation factor PSRP1 and/or EF-Tu (Perez Boerema et al., 2018; Zoschke and Bock, 2018).
In chloroplasts, only psbA RNA experiences substantially increased ribosome occupancy in response to light–dark shifts and high light exposure (Chotewutmontri and Barkan, 2018; Schuster et al., 2020). In mature chloroplasts, this psbA-specific activation is triggered by signals from PSII repair and is distinct from the plastome-wide translation response triggered by light-induced pH changes (Chotewutmontri and Barkan, 2020; Chotewutmontri et al., 2020). Nascent D1 produced by psbA is transferred to the HIGH CHLOROPHYLL FLUORESCENCE 244 (HCF244)/ONE HELIX PROTEIN 1 (OHP1)/OHP2 complex with the help of HCF136. The presence of D1 in the HCF244 complex is thought to inhibit the ability of the HCF244 complex to activate the recruitment of ribosomes to psbA transcripts. It is unclear whether the interaction of the HCF244 complex with the nascent D1 occurs in the stroma or at the thylakoid membrane. Considering that OHP1 and OHP2 are tightly integrated at the thylakoid membrane (Li et al., 2019), it is most likely that the interaction of the HCF244 complex with the nascent D1 occurs at the thylakoid membrane. Light damages the PSII reaction center D1 protein, triggering delivery of nascent D1 by the HCF244/OHP1/OHP2 complex. A D1-less HCF244 complex recovers the ability to activate psbA translation (Figure 4). This autoregulatory mechanism contributes to explaining two long-standing questions in plant biology: (i) how chloroplasts perceive light to activate translation from psbA transcripts, and (ii) how the psbA translation rate is coupled to the need for nascent D1 during PSII repair.
Figure 4.
A schematic model showing chloroplast translation triggered by light-induced pH changes and PSII damage.
Chloroplast translation is rapidly activated after a shift from darkness to light. Upon illumination, the plastome-wide activation of translation elongation is triggered by light-induced changes in pH. Moreover, psbA translation is also activated by another light-induced regulatory pathway, the HCF244/OHP1/OHP2-centered autoregulatory circuit. Light causes constant damage to PSII, which requires replacement of damaged D1 by de novo synthesis of D1 proteins. HCF136 helps nascent D1 peptides insert themselves into the HCF244/OHP1/OHP2 complex, repressing its ability to activate ribosome recruitment to psbA RNA. The HCF244/OHP1/OHP2 complex delivers nascent D1 to photodamaged PSII, then releases the HCF244/OHP1/OHP2 complex to activate psbA translation. Given that HCF173 binds to the 5′ UTR of psbA RNA for translation initiation, a D1-less HCF244 complex is thought to communicate with HCF173 for the activation of psbA translation. The schematic model is drawn according to Chotewutmontri et al. (2020).
Post-translational processing and quality control
Newly synthesized precursor proteins inside chloroplasts require N-formylated methionine excision, protein cleavage, and proper folding to generate fully functional products (van Wijk, 2015; Gao et al., 2023). The N-terminal methionine excision of precursor proteins in chloroplasts is initiated by deformylation, and methionine aminopeptidase then removes N-terminal methionine from precursor proteins (Meinnel and Giglione, 2022). In particular, the D1 precursor must undergo cleavage of the C-terminal tail by an endopeptidase named carboxyl terminal peptidase (Che et al., 2013).
Proteases are key components of the protein quality control systems in chloroplasts and play important roles in promoting correct protein folding and removing damaged proteins. Regulation of protein degradation in chloroplasts has recently been reviewed (Gao et al., 2023; Sun et al., 2023). The D1 protein of the PSII reaction center is encoded by the chloroplast psbA gene; it inserts into the PSII complex with its five transmembrane helix domains that are connected by lumenal and stromal loop domains (Inagaki, 2022). D1 is always photodamaged by light. Therefore, degradation of photodamaged D1 is essential for ensuring efficient D1 turnover and PSII repair. Here, we mainly discuss the role of proteases in degradation of photodamaged D1. A series of studies have demonstrated that Deg and FtsH proteases are involved in this degradation: Deg1, Deg5, and Deg8 cleave the lumenal-side loops of D1 (Kapri-Pardes et al., 2007; Sun et al., 2007), and Deg7 cleaves its stromal-side BC loop (Sun et al., 2010). Deg7 is also involved in degradation of the chloroplast-encoded PSII subunits D2, CP43, and CP47 (Sun et al., 2010). FtsH2 has been suggested to regulate D1 degradation at the CP43 formation step during PSII repair (Kato et al., 2009). However, the detailed processes and proteolytic sites of FtsH-mediated D1 degradation remain unclear and should be investigated further in the future (Kato et al., 2009; Chi et al., 2012; Kato and Sakamoto, 2018; Yi et al., 2022).
PPR proteins could be engineered as tools for manipulating chloroplast gene expression
Characterization of the molecular mechanisms by which PPR proteins regulate chloroplast gene expression offers opportunities for designing PPR tools for RNA manipulation that could be used in chloroplast RNA research. Typical PPR proteins contain repeating 35-amino-acid motifs that stack into a long superhelical RNA-binding surface that interacts with RNA via an elegant one-repeat:one-nucleotide binding mode (Barkan et al., 2012; Yin et al., 2013; Shen et al., 2016). The combination of the 5th and 35th residues at each PPR motif is considered to be the “PPR code” that is responsible for specific RNA base recognition. PPR codes that are relatively specific for A, U, C, or G are fairly well understood, and numerous PPR codes have been deciphered (Barkan et al., 2012; Shen et al., 2016; Yan et al., 2019). Engineered PPR proteins with custom RNA-binding specificity have been created by either altering the PPR code of natural proteins (Zhou et al., 2017; Colas des Francs-Small et al., 2018) or completely redesigning the protein according to the consensus PPR motif (Coquille et al., 2014; Gully et al., 2015; Shen et al., 2015, 2016; Miranda et al., 2018; McDermott et al., 2019; Yan et al., 2019). Because natural PPRs often contain a C-terminal domain with specific functions, an engineered PPR can also be fused to a specific domain to fulfill a custom function. For example, a PPR domain with a C-terminal RNA endonuclease domain can act as an artificial sequence-specific RNA endonuclease that cleaves specific RNA targets. Therefore, engineered PPRs might be used as tools to investigate the functions of chloroplast RNAs and thereby reveal molecular mechanisms of chloroplast gene expression.
Maize PPR10 binds upstream of the atpH ribosome-binding site and activates atpH expression by preventing 5′–3′ exonucleolytic degradation and improving its translational efficiency (Pfalz et al., 2009; Prikryl et al., 2011). On the basis of PPR10 characterization, an exciting on/off switch for plastid transgenes has been developed (Rojas et al., 2019). A PPR10 variant with new sequence specificity was designed, and its corresponding binding site was inserted upstream of a chloroplast transgene encoding a green fluorescent protein (GFP). Driving expression of this PPR10 variant by an ethanol-inducible promoter resulted in ∼20-fold induction of the GFP reporter; GFP accumulated to ∼15% of the soluble protein in leaf tissues. In the absence of ethanol, there was no detectable expression of the PPR10 variant, and protection of GFP mRNA within chloroplasts was abolished, leading to only trace expression of the GFP reporter (Rojas et al., 2019). A PPR10-based system was also applied to tissue-specific expression of a plastid-encoded GFP in potato tubers, and GFP accumulated to 1.3% of the total soluble protein in the tubers (Yu et al., 2019). Modified PPR proteins can thus be exploited as tools for controlling plastid gene expression in future research.
Potential roles of new techniques for elucidating molecular mechanisms of chloroplast gene expression
The use of new techniques and approaches will be important for increasing our understanding of chloroplast gene expression. Cryoelectron microscopy can solve molecular structures with a resolution below 2 Å, and it is increasingly accessible and cost effective (Danev et al., 2019). Chloroplast gene expression machineries include PEP and NEP for transcription, “a possible editosome” for RNA editing, “a possible spliceosome” for RNA splicing, and the ribosome for translation. Elucidating the structures of these gene expression machineries by cryoelectron microscopy would reveal how they control chloroplast gene expression at the molecular level.
Various high-throughput sequencing-based techniques have rapidly emerged, providing an opportunity for quantitative genome-wide assessments of chloroplast gene expression with unprecedented depth and resolution. Global run-on sequencing and native elongating transcript sequencing are powerful tools for characterizing nascent RNA to determine the positions, relative levels, and orientation of transcriptionally engaged RNA polymerases genome wide (Zhu et al., 2018). Transcriptional pausing in chloroplasts was found to be a regulatory point for gene expression (Ding et al., 2019). Thus, global run-on sequencing and native elongating transcript sequencing can be exploited to explore the functions of transcriptional pausing in chloroplasts by monitoring the dynamic responses of paused RNA polymerase to developmental and external stimuli. Structure sequencing is a quantitative and high-throughput method that offers genome-wide details on RNA structure at single-nucleotide resolution (Deng et al., 2018). Structure sequencing could be used to map the RNA structures of chloroplast transcripts and to investigate the roles of RNA structure in regulating RNA splicing, RNA stability, and translation within chloroplasts. In addition, genome-wide association studies are used to identify genetic variants that are statistically associated with a particular trait (Abdellaoui et al., 2023). Thus, changes in chloroplast gene expression, e.g., the abundance of psbA transcripts, could be used in genome-wide association studies to determine which genetic variants are involved in regulation of chloroplast gene expression.
Single-cell omics is revolutionizing our understanding from the tissue to the single-cell level, enabling us to access individual cell modalities, such as genomes, transcriptomics, and epigenomics (Wen et al., 2022). Leaves have a complex cellular architecture with heterogeneous cell types. For example, C4 photosynthesis is performed in mesophyll and bundle sheath cells of leaves, and single-cell sequencing could be used to characterize chloroplast gene expression in these distinct cell types. Single-cell sequencing also enables us to monitor chloroplast gene expression over the course of chloroplast biogenesis in specific cell types.
Chloroplast transformation is a powerful tool to investigate many aspects of chloroplast gene expression; it can identify the regulatory elements of chloroplast genes, reveal the mechanism and evolution of plant RNA editing, and characterize the functions of pncRNAs (Ruf and Bock, 2011; Sharwood et al., 2011). A new method for chloroplast transgene introduction was recently developed. Unlike classical chloroplast transformation, this method does not require foreign transgene insertion into the chloroplast genome; instead, the foreign transgene is amplified as an independent entity named the “minichromosome” (Jakubiec et al., 2021). This method could be used to reveal the functions of regulatory elements of chloroplast genes. In addition, two protein-only base editors have been developed that can induce targeted C-to-T and A-to-G base editing in chloroplast DNA (Kang et al., 2021; Mok et al., 2022). These base editors will enable us to explore regulatory elements of chloroplast genes in situ and provide an opportunity to optimize photosynthesis and improve crop yield by altering the coding sequences of chloroplast genes, e.g., modifying RbcL to enhance Rubisco activity and CO2 assimilation.
Concluding remarks and future perspectives
Over the course of host–endosymbiont coevolution, the chloroplast has not only inherited some prokaryotic principles but also developed many new features of gene expression, such as highly complex RNA polymerases, as well as diverse RNA cleavage, splicing, and editing mechanisms. These combined factors ensure correct chloroplast gene expression. Over the past few years, extensive studies focused on chloroplast gene expression have advanced from describing phases to exploring their underlying gene expression mechanisms. Here, we discuss the next wave of questions that will improve our understanding in this field.
Chloroplast genes are mainly transcribed by the PEP complex that consists of bacterial-like core subunits and at least 12 additional components, the PAPs. All PAPs are required for the transcription activity of the PEP complex. However, our understanding of PAPs remains rudimentary. Future studies should therefore address two major issues: where PAPs are located within the PEP complex, and how they coordinately control the transcriptional activity of the PEP complex. In addition, transcriptional pausing is widespread in chloroplasts and has an important role in regulating chloroplast transcription. As this mechanism is newly emerging, many gaps in our understanding remain. Most importantly, future research should aim to explore the causes and biological functions of transcriptional pausing by PEP.
The maturation of RNAs in chloroplasts requires a set of post-transcriptional processing events, such as intercistronic cleavage, RNA termini trimming, RNA splicing, and RNA editing (Zhang and Lu, 2019). Intercistronic cleavage and termini trimming of RNAs in chloroplasts are accomplished by ribonucleases and RNA-binding proteins, but how chloroplasts initiate the endonucleolytic cleavage and the interplay between ribonucleases and RNA-binding proteins remain largely unclear. In addition, the ion imbalance of K+ and Na+ significantly impairs chloroplast rRNA maturation by altering their secondary structure and reducing helper protein interactions with chloroplast rRNA transcripts (DeTar et al., 2021). Hence, how chloroplast ion homeostasis directly modulates RNA folding and RNA–protein interaction should be considered in the future.
To date, numerous splicing factors (e.g., PPR, CRM, plant organellar RNA recognition proteins) in chloroplasts have been identified that help remove introns from precursor RNAs (Wang et al., 2022). These splicing factors regulate the splicing of distinct but overlapping intron sets. Although chloroplast splicing events have been described over several decades, the specific mechanisms by which these splicing factors activate the self-splicing ability of introns have not yet been uncovered.
In flowering plants, C-to-U RNA editing is executed by multi-protein editosome complexes in which the PPR protein confers the RNA-binding specificity, MORF proteins regulate the binding activity of PPR domains, and the DYW domain catalyzes the deaminase activity (Yan et al., 2017; Small et al., 2020; Royan et al., 2021). However, the functions of other editosome components, such as the RNA helicase INCREASED SIZE EXCLUSION LIMIT 2 or ORGANELLE RNA RECOGNITION MOTIF PROTEIN and ORGANELLE ZINC FINGER proteins, remain largely unknown, and determination of their precise roles is an important future direction. In addition, a recent study revealed that there are complex interplays between RNA substrates and editosomes, whereby the DYW domains show distinct preferences for the neighboring nucleotides of RNA editing sites (Maeda et al., 2022). In light of these questions, the field anxiously awaits structural studies of RNA bound to a DYW domain and/or even the complete editosome.
Although chloroplast ribosomes show substantial deviations from the bacterial ribosome, a number of studies on their function and structure have demonstrated that the basic function and catalytic mechanism of the chloroplast translation apparatus are conserved. However, our understanding of the molecular mechanisms by which chloroplast translation is regulated remains rudimentary. In the future, we should focus on identifying novel RNA cis-elements and proteinaceous trans-factors and exploring their roles in regulating translation in chloroplasts.
In contrast to that of bacteria, the coupling of translation and transcription in the chloroplasts of land plants is believed to be relaxed, perhaps because chloroplast RNAs are stable when not covered by ribosomes and exhibit a longer half-time than bacterial RNAs, and translation initiation of numerous mRNAs is activated by post-transcriptional processing. However, there is also evidence that transcription–translation coupling has not been fully lost in chloroplasts. Electron micrographs from 1982 showed ribosomes associated with nascent transcripts (Rose and Lindbeck, 1982). In addition, several ribosomal proteins co-purify with the PEP complex (Pfalz et al., 2006), and many translation-related factors are enriched in plastid nucleoids in a ribonuclease-sensitive manner, which is a hallmark for proteins tethered to nascent transcripts (Majeran et al., 2011). Moreover, pTAC13/NusG associates with the PEP complex via PAP9 as well as two 30S ribosome subunits, RPS5 and RPS10. Hence, pTAC13/NusG might link the translational apparatus to the main chloroplast RNA polymerase, PEP (Xiong et al., 2022). Taken together, these observations suggest that further analysis of the relationships between the transcriptional and translational apparatus will be necessary to improve our understanding of chloroplast gene expression.
Light is one of the most important environmental cues required to start transcription. Despite several fresh insights into the signaling steps of the activation of transcription and translation by light, our knowledge of how light switches on gene expression within chloroplasts is still limited. Therefore, identifying new signaling components that link light and chloroplast gene expression would be a very important step forward. Another complementary approach could be to explore the influence on chloroplast gene expression mediated by other internal and external triggers, such as temperature, ion balance, osmotic status, phytohormones, and circadian rhythms.
Recent studies suggest that manipulation of RbcL and D1 with enhanced abundance can improve crop yield and stress tolerance. Supplementing RbcL of nuclear origin together with the RbcS and Rubisco assembly chaperone RAF1 results in a >30% increase in Rubisco content, a 15% increase in CO2 assimilation, and a 27% increase in fresh weight in maize (Salesse-Smith et al., 2018). The artificial nuclear D1 imported into the chloroplast through the chimeric RbcS signal peptide leads to enhanced thermotolerance and a considerable increase in biomass accumulation in Arabidopsis, tobacco, tomato, and rice (Chen et al., 2020). Presumably, this artificial nuclear D1 synthesis complements nascent D1, which helps the rapid repair of damaged PSII under conditions of heat and high-light stress. In addition, as discussed above, modified PPR proteins might be used as a tool to improve crop yield and environmental stress resistance by regulating the expression of chloroplast genes. Clearly, it will be a challenge to elucidate the role of chloroplast gene expression in improving crop yield and stress, and these topics should be investigated extensively in future work.
Funding
This work was supported by the National Key Research and Development Program of China (grant no. 2020YFA0907600), the National Natural Science Foundation of China (grant nos. 31730102 and 32000184), the Natural Science Foundation of Shandong Province (grant no. ZR2020QC023), and the China Postdoctoral Science Foundation (grant no. 2020M672093).
Author contributions
Y.Z. and C.L. designed the review. Y.Z. and L.T. wrote the first draft, and Y.Z. and C.L. contributed to the revision and improvement of the text. All authors read and approved the final version of the manuscript.
Acknowledgments
The authors are grateful to the authors of all excellent papers discussed in this review and apologize to those not cited due to space limitations. No conflict of interest is declared.
Published: May 4, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Abdellaoui A., Yengo L., Verweij K.J.H., Visscher P.M. 15 years of GWAS discovery: realizing the promise. Am. J. Hum. Genet. 2023;110:179–194. doi: 10.1016/j.ajhg.2022.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T., Yin Z., Bhushan S. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci. Rep. 2016;6 doi: 10.1038/srep35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar J., Kreim N., Marini F., Mohana G., Brüne D., Binder H., Roignant J.Y. Promoter-proximal pausing mediated by the exon junction complex regulates splicing. Nat. Commun. 2019;10:521. doi: 10.1038/s41467-019-08381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkatib S., Scharff L.B., Rogalski M., Fleischmann T.T., Matthes A., Seeger S., Schöttler M.A., Ruf S., Bock R. The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A., Pandi G. Noncoding RNA: an insight into chloroplast and mitochondrial gene expressions. Life. 2021;11:49. doi: 10.3390/life11010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J.M. Genomic perspectives on the birth and spread of plastids. Proc. Natl. Acad. Sci. USA. 2015;112:10147–10153. doi: 10.1073/pnas.1421374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsova B., Hoja U., Wimmelbacher M., Greiner E., Ustün S., Melzer M., Petersen K., Lein W., Börnke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell. 2010;22:1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryamanesh N., Ruwe H., Sanglard L.V.P., Eshraghi L., Bussell J.D., Howell K.A., Small I., des Francs-Small C.C. The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 2017;173:1164–1176. doi: 10.1104/pp.16.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S., Tiller K., Pfannschmidt T., Link G. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol. Biol. 1999;39:1013–1023. doi: 10.1023/a:1006177807844. [DOI] [PubMed] [Google Scholar]
- Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Gil J., Bouza-Morcillo L., Espinosa-Cores L., Piñeiro M., Jarillo J.A. H4 acetylation by the NuA4 complex is required for plastid transcription and chloroplast biogenesis. Nat. Plants. 2022;8:1052–1063. doi: 10.1038/s41477-022-01229-4. [DOI] [PubMed] [Google Scholar]
- Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell Biol. 2008;28:5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017;36:475–486. doi: 10.15252/embj.201695959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik K., McCray T.N., Ernest B., Fernandez J.C., Howell K.A., Lane T., Staton M., Burch-Smith T.M. The chloroplast RNA helicase ISE2 is required for multiple chloroplast RNA processing steps in Arabidopsis thaliana. Plant J. 2017;91:114–131. doi: 10.1111/tpj.13550. [DOI] [PubMed] [Google Scholar]
- Börner T., Aleynikova A.Y., Zubo Y.O., Kusnetsov V.V. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta. 2015;1847:761–769. doi: 10.1016/j.bbabio.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Cahoon A.B., Harris F.M., Stern D.B. Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep. 2004;5:801–806. doi: 10.1038/sj.embor.7400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Okuda K., Peng L., Shikanai T. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011;67:318–327. doi: 10.1111/j.1365-313X.2011.04593.x. [DOI] [PubMed] [Google Scholar]
- Campos F., García-Gómez B.I., Solórzano R.M., Salazar E., Estevez J., León P., Alvarez-Buylla E.R., Covarrubias A.A. A cDNA for nuclear-encoded chloroplast translational initiation factor 2 from a higher plant is able to complement an infB Escherichia coli null mutant. J. Biol. Chem. 2001;276:28388–28394. doi: 10.1074/jbc.M100605200. [DOI] [PubMed] [Google Scholar]
- Castandet B., Germain A., Hotto A.M., Stern D.B. Systematic sequencing of chloroplast transcript termini from Arabidopsis thaliana reveals >200 transcription initiation sites and the extensive imprints of RNA-binding proteins and secondary structures. Nucleic Acids Res. 2019;47:11889–11905. doi: 10.1093/nar/gkz1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon L., Gillet F.X., Chieb M., Cobessi D., Pfannschmidt T., Blanvillain R. PAP8/pTAC6 is part of a nuclear protein complex and displays RNA recognition motifs of viral origin. Int. J. Mol. Sci. 2022;23:3059. doi: 10.3390/ijms23063059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Lee S., Um T.Y., Kim J.K., Do Choi Y., Jang G. pTAC10, a key subunit of plastid-encoded RNA polymerase, promotes chloroplast development. Plant Physiol. 2017;174:435–449. doi: 10.1104/pp.17.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che Y., Fu A., Hou X., McDonald K., Buchanan B.B., Huang W., Luan S. C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2013;110:16247–16252. doi: 10.1073/pnas.1313894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Chen S.T., He N.Y., Wang Q.L., Zhao Y., Gao W., Guo F.Q. Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants. 2020;6:570–580. doi: 10.1038/s41477-020-0629-z. [DOI] [PubMed] [Google Scholar]
- Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M.F., Bai F., Castleden I., Song Y., Song B., Huang J., et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016;85:532–547. doi: 10.1111/tpj.13121. [DOI] [PubMed] [Google Scholar]
- Chi W., He B., Mao J., Jiang J., Zhang L. Plastid sigma factors: their individual functions and regulation in transcription. Biochim. Biophys. Acta. 2015;1847:770–778. doi: 10.1016/j.bbabio.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Chi W., Sun X., Zhang L. The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim. Biophys. Acta. 2012;1817:239–246. doi: 10.1016/j.bbabio.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. Light-induced psbA translation in plants is triggered by photosystem II damage via an assembly-linked autoregulatory circuit. Proc. Natl. Acad. Sci. USA. 2020;117:21775–21784. doi: 10.1073/pnas.2007833117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Williams-Carrier R., Barkan A. Exploring the link between photosystem II assembly and translation of the chloroplast psbA mRNA. Plants. 2020;9:152. doi: 10.3390/plants9020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C., Vincis Pereira Sanglard L., Small I. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol. 2018;1:166. doi: 10.1038/s42003-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquille S., Filipovska A., Chia T., Rajappa L., Lingford J.P., Razif M.F.M., Thore S., Rackham O. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014;5:5729. doi: 10.1038/ncomms6729. [DOI] [PubMed] [Google Scholar]
- Core L., Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R., Yanagisawa H., Kikkawa M. Cryo-electron microscopy methodology: current aspects and future directions. Trends Biochem. Sci. 2019;44:837–848. doi: 10.1016/j.tibs.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Daniell H., Lin C.S., Yu M., Chang W.J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle A.F., Small I.D., Lurin C. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant. 2010;3:691–705. doi: 10.1093/mp/ssq025. [DOI] [PubMed] [Google Scholar]
- de Vries J., Archibald J.M. Plastid genomes. Curr. Biol. 2018;28 doi: 10.1016/j.cub.2018.01.027. R336–r337. [DOI] [PubMed] [Google Scholar]
- Deng H., Cheema J., Zhang H., Woolfenden H., Norris M., Liu Z., Liu Q., Yang X., Yang M., Deng X., et al. Rice in vivo RNA structurome reveals RNA secondary structure conservation and divergence in plants. Mol. Plant. 2018;11:607–622. doi: 10.1016/j.molp.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTar R.A., Barahimipour R., Manavski N., Schwenkert S., Höhner R., Bölter B., Inaba T., Meurer J., Zoschke R., Kunz H.H. Loss of inner-envelope K+/H+ exchangers impairs plastid rRNA maturation and gene expression. Plant Cell. 2021;33:2479–2505. doi: 10.1093/plcell/koab123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz M.G., Hernández-Verdeja T., Kremnev D., Crawford T., Dubreuil C., Strand Å. Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat. Commun. 2018;9:50. doi: 10.1038/s41467-017-02468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Zhang Y., Hu Z., Huang X., Zhang B., Lu Q., Wen X., Wang Y., Lu C. mTERF5 acts as a transcriptional pausing factor to positively regulate transcription of chloroplast psbEFLJ. Mol. Plant. 2019;12:1259–1277. doi: 10.1016/j.molp.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T., Matsuda T., Sugita C., Ichinose M., Yamamoto H., Shikanai T., Sugita M. The P-class pentatricopeptide repeat protein PpPPR_21 is needed for accumulation of the psbI-ycf12 dicistronic mRNA in Physcomitrella chloroplasts. Plant J. 2019;97:1120–1131. doi: 10.1111/tpj.14187. [DOI] [PubMed] [Google Scholar]
- Edera A.A., Gandini C.L., Sanchez-Puerta M.V. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018;97:215–231. doi: 10.1007/s11103-018-0734-9. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981;9:2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier A., Gans P., Boeri Erba E., Signor L., Muthukumar S.S., Pfannschmidt T., Blanvillain R., Cobessi D. The plastid-encoded RNA polymerase-associated protein PAP9 is a superoxide dismutase with unusual structural features. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.668897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Tadini L., Moratti F., Lehniger M.K., Costa A., Rossi F., Colombo M., Masiero S., Schmitz-Linneweber C., Pesaresi P. CRP1 Protein: (dis)similarities between Arabidopsis thaliana and Zea mays. Front. Plant Sci. 2017;8:163. doi: 10.3389/fpls.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster S., Eggert E., Zoschke R., Weihe A., Schmitz-Linneweber C. Light-dependent, plastome-wide association of the plastid-encoded RNA polymerase with chloroplast DNA. Plant J. 2013;76:849–860. doi: 10.1111/tpj.12339. [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.X., Zhang J., Zhang G.Q., Liu Z.J., Chen Z.D. Insights into the origin and evolution of plant sigma factors. J. Systemat. Evol. 2021;59:326–340. [Google Scholar]
- Gao L.L., Hong Z.H., Wang Y., Wu G.Z. Chloroplast proteostasis: a story of birth, life, and death. Plant Commun. 2023;4 doi: 10.1016/j.xplc.2022.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z.P., Yu Q.B., Zhao T.T., Ma Q., Chen G.X., Yang Z.N. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 2011;157:1733–1745. doi: 10.1104/pp.111.184762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., Myouga F., Takechi K., Sato H., Nabeshima K., Nagata N., Takio S., Shinozaki K., Takano H. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 2008;53:924–934. doi: 10.1111/j.1365-313X.2007.03379.x. [DOI] [PubMed] [Google Scholar]
- Gawroński P., Jensen P.E., Karpiński S., Leister D., Scharff L.B. Pausing of chloroplast ribosomes is induced by multiple features and is linked to the assembly of photosynthetic complexes. Plant Physiol. 2018;176:2557–2569. doi: 10.1104/pp.17.01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J., Honsel A., Voss B., Rennenberg H., Hess W.R. A long antisense RNA in plant chloroplasts. New Phytol. 2010;186:615–622. doi: 10.1111/j.1469-8137.2010.03203.x. [DOI] [PubMed] [Google Scholar]
- Gerke P., Szövényi P., Neubauer A., Lenz H., Gutmann B., McDowell R., Small I., Schallenberg-Rüdinger M., Knoop V. Towards a plant model for enigmatic U-to-C RNA editing: the organelle genomes, transcriptomes, editomes and candidate RNA editing factors in the hornwort Anthoceros agrestis. New Phytol. 2020;225:1974–1992. doi: 10.1111/nph.16297. [DOI] [PubMed] [Google Scholar]
- Germain A., Herlich S., Larom S., Kim S.H., Schuster G., Stern D.B. Mutational analysis of Arabidopsis chloroplast polynucleotide phosphorylase reveals roles for both RNase PH core domains in polyadenylation, RNA 3′-end maturation and intron degradation. Plant J. 2011;67:381–394. doi: 10.1111/j.1365-313X.2011.04601.x. [DOI] [PubMed] [Google Scholar]
- Germain A., Hotto A.M., Barkan A., Stern D.B. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA. 2013;4:295–316. doi: 10.1002/wrna.1161. [DOI] [PubMed] [Google Scholar]
- Gipson A.B., Hanson M.R., Bentolila S. The RanBP2 zinc finger domains of chloroplast RNA editing factor OZ1 are required for protein-protein interactions and conversion of C to U. Plant J. 2022;109:215–226. doi: 10.1111/tpj.15569. [DOI] [PubMed] [Google Scholar]
- Glanz S., Kück U. Trans-splicing of organelle introns--a detour to continuous RNAs. Bioessays. 2009;31:921–934. doi: 10.1002/bies.200900036. [DOI] [PubMed] [Google Scholar]
- Gully B.S., Shah K.R., Lee M., Shearston K., Smith N.M., Sadowska A., Blythe A.J., Bernath-Levin K., Stanley W.A., Small I.D., Bond C.S. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. D Biol. Crystallogr. 2015;71:196–208. doi: 10.1107/S1399004714024869. [DOI] [PubMed] [Google Scholar]
- Gutmann B., Royan S., Schallenberg-Rüdinger M., Lenz H., Castleden I.R., McDowell R., Vacher M.A., Tonti-Filippini J., Bond C.S., Knoop V., Small I.D. The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant. 2020;13:215–230. doi: 10.1016/j.molp.2019.11.002. [DOI] [PubMed] [Google Scholar]
- Haag S., Schindler M., Berndt L., Brennicke A., Takenaka M., Weber G. Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9. Nucleic Acids Res. 2017;45:4915–4928. doi: 10.1093/nar/gkx099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Cook W.B., Barkan A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. USA. 2012;109:5651–5656. doi: 10.1073/pnas.1200318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Takenaka M., Miranda R., Barkan A. A PPR protein in the PLS subfamily stabilizes the 5′-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 2016;44:4278–4288. doi: 10.1093/nar/gkw270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sugita M. A moss pentatricopeptide repeat protein binds to the 3' end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 2009;276:5860–5869. doi: 10.1111/j.1742-4658.2009.07267.x. [DOI] [PubMed] [Google Scholar]
- Hattori M., Miyake H., Sugita M. A Pentatricopeptide repeat protein is required for RNA processing of clpP Pre-mRNA in moss chloroplasts. J. Biol. Chem. 2007;282:10773–10782. doi: 10.1074/jbc.M608034200. [DOI] [PubMed] [Google Scholar]
- Hayes M.L., Santibanez P.I. A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 2020;295:3497–3505. doi: 10.1074/jbc.RA119.011790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel J., Duttke S.H., Benner C., Chory J. Nascent RNA sequencing reveals distinct features in plant transcription. Proc. Natl. Acad. Sci. USA. 2016;113:12316–12321. doi: 10.1073/pnas.1603217113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto A.M., Castandet B., Gilet L., Higdon A., Condon C., Stern D.B. Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell. 2015;27:724–740. doi: 10.1105/tpc.114.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto A.M., Schmitz R.J., Fei Z., Ecker J.R., Stern D.B. Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 (Bethesda) 2011;1:559–570. doi: 10.1534/g3.111.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y., Han S., Yoo C.Y., Hong L., You C., Le B.H., Shi H., Zhong S., Hoecker U., Chen X., Chen M. Anterograde signaling controls plastid transcription via sigma factors separately from nuclear photosynthesis genes. Nat. Commun. 2022;13:7440. doi: 10.1038/s41467-022-35080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim I.M., Wu H., Ezhov R., Kayanja G.E., Zakharov S.D., Du Y., Tao W.A., Pushkar Y., Cramer W.A., Puthiyaveetil S. An evolutionarily conserved iron-sulfur cluster underlies redox sensory function of the Chloroplast Sensor Kinase. Commun. Biol. 2020;3:13. doi: 10.1038/s42003-019-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M., Sugita M. RNA editing and its molecular mechanism in plant organelles. Genes. 2016;8:5. doi: 10.3390/genes8010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M., Kawabata M., Akaiwa Y., Shimajiri Y., Nakamura I., Tamai T., Nakamura T., Yagi Y., Gutmann B. U-to-C RNA editing by synthetic PPR-DYW proteins in bacteria and human culture cells. Commun. Biol. 2022;5:968. doi: 10.1038/s42003-022-03927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N. Processing of D1 protein: a mysterious process carried out in thylakoid lumen. Int. J. Mol. Sci. 2022;23:2520. doi: 10.3390/ijms23052520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A., Sarokina A., Choinard S., Vlad F., Malcuit I., Sorokin A.P. Replicating minichromosomes as a new tool for plastid genome engineering. Nat. Plants. 2021;7:932–941. doi: 10.1038/s41477-021-00940-y. [DOI] [PubMed] [Google Scholar]
- Jiang J., Chai X., Manavski N., Williams-Carrier R., He B., Brachmann A., Ji D., Ouyang M., Liu Y., Barkan A., et al. An RNA chaperone-like protein plays critical roles in chloroplast mRNA stability and translation in Arabidopsis and Maize. Plant Cell. 2019;31:1308–1327. doi: 10.1105/tpc.18.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.C., Bae S.J., Lee S., Lee J.S., Kim A., Lee H., Baek G., Seo H., Kim J., Kim J.S. Chloroplast and mitochondrial DNA editing in plants. Nat. Plants. 2021;7:899–905. doi: 10.1038/s41477-021-00943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapri-Pardes E., Naveh L., Adam Z. The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell. 2007;19:1039–1047. doi: 10.1105/tpc.106.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. FtsH protease in the thylakoid membrane: physiological functions and the regulation of protease activity. Front. Plant Sci. 2018;9:855. doi: 10.3389/fpls.2018.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 2009;151:1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrebtukova I., Spreitzer R.J. Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. USA. 1996;93:13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Strand Å. Chloroplast transcription, untangling the gordian knot. New Phytol. 2015;206:889–891. doi: 10.1111/nph.13388. [DOI] [PubMed] [Google Scholar]
- Kindgren P., Ivanov M., Marquardt S. Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis. Nucleic Acids Res. 2020;48:2332–2347. doi: 10.1093/nar/gkz1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Kremnev D., Blanco N.E., de Dios Barajas López J., Fernández A.P., Tellgren-Roth C., Kleine T., Small I., Strand A. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 2012;70:279–291. doi: 10.1111/j.1365-313X.2011.04865.x. [DOI] [PubMed] [Google Scholar]
- Kleine T. Arabidopsis thaliana mTERF proteins: evolution and functional classification. Front. Plant Sci. 2012;3:233. doi: 10.3389/fpls.2012.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger T.S., Watkins K.P., Friso G., van Wijk K.J., Barkan A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA. 2009;106:4537–4542. doi: 10.1073/pnas.0812503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell. 2012;24:4266–4280. doi: 10.1105/tpc.112.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. Transcriptional pausing as a mediator of bacterial gene regulation. Annu. Rev. Microbiol. 2021;75:291–314. doi: 10.1146/annurev-micro-051721-043826. [DOI] [PubMed] [Google Scholar]
- Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne M.D., Konstantopoulos D., Ntakou-Zamplara K.Z., Liakos A., Fousteri M. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat. Commun. 2017;8:2076. doi: 10.1038/s41467-017-02145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Park S.J., Colas des Francs-Small C., Whitby M., Small I., Kang H. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 2019;100:1193–1207. doi: 10.1111/tpj.14509. [DOI] [PubMed] [Google Scholar]
- Leister D. Complex(iti)es of the ubiquitous RNA-binding CSP41 proteins. Front. Plant Sci. 2014;5:255. doi: 10.3389/fpls.2014.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L., Meurer J. The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 2004;38:740–753. doi: 10.1111/j.1365-313X.2004.02081.x. [DOI] [PubMed] [Google Scholar]
- Li X., Luo W., Zhou W., Yin X., Wang X., Li X., Jiang C., Zhang Q., Kang X., Zhang A., et al. CAF proteins help SOT1 regulate the stability of chloroplast ndhA transcripts. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222312639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu B., Zhang J., Kong F., Zhang L., Meng H., Li W., Rochaix J.D., Li D., Peng L. OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiol. 2019;179:195–208. doi: 10.1104/pp.18.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macadlo L.A., Ibrahim I.M., Puthiyaveetil S. Sigma factor 1 in chloroplast gene transcription and photosynthetic light acclimation. J. Exp. Bot. 2020;71:1029–1038. doi: 10.1093/jxb/erz464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Takenaka S., Wang T., Frink B., Shikanai T., Takenaka M. DYW deaminase domain has a distinct preference for neighboring nucleotides of the target RNA editing sites. Plant J. 2022;111:756–767. doi: 10.1111/tpj.15850. [DOI] [PubMed] [Google Scholar]
- Maier U.G., Zauner S., Woehle C., Bolte K., Hempel F., Allen J.F., Martin W.F. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol. Evol. 2013;5:2318–2329. doi: 10.1093/gbe/evt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–189. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Schmid L.M., Meurer J. RNA-stabilization factors in chloroplasts of vascular plants. Essays Biochem. 2018;62:51–64. doi: 10.1042/EBC20170061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Torabi S., Lezhneva L., Arif M.A., Frank W., Meurer J. HIGH CHLOROPHYLL FLUORESCENCE145 binds to and stabilizes the psaA 5′ UTR via a newly defined repeat motif in Embryophyta. Plant Cell. 2015;27:2600–2615. doi: 10.1105/tpc.15.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J.J., Watkins K.P., Williams-Carrier R., Barkan A. Ribonucleoprotein capture by in vivo expression of a designer pentatricopeptide repeat protein in Arabidopsis. Plant Cell. 2019;31:1723–1733. doi: 10.1105/tpc.19.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Giglione C. N-terminal modifications, the associated processing machinery, and their evolution in plastid-containing organisms. J. Exp. Bot. 2022;73:6013–6033. doi: 10.1093/jxb/erac290. [DOI] [PubMed] [Google Scholar]
- Méteignier L.V., Ghandour R., Meierhoff K., Zimmerman A., Chicher J., Baumberger N., Alioua A., Meurer J., Zoschke R., Hammani K. The Arabidopsis mTERF-repeat MDA1 protein plays a dual function in transcription and stabilization of specific chloroplast transcripts within the psbE and ndhH operons. New Phytol. 2020;227:1376–1391. doi: 10.1111/nph.16625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Lezhneva L., Amann K., Gödel M., Bezhani S., Sherameti I., Oelmüller R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda R.G., McDermott J.J., Barkan A. RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res. 2018;46:2613–2623. doi: 10.1093/nar/gkx1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E., Kato Y., Matsushima R., Albrecht V., Laalami S., Sakamoto W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell. 2007;19:1313–1328. doi: 10.1105/tpc.106.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok Y.G., Hong S., Bae S.J., Cho S.I., Kim J.S. Targeted A-to-G base editing of chloroplast DNA in plants. Nat. Plants. 2022;8:1378–1384. doi: 10.1038/s41477-022-01279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi R., Yamazaki T., Myouga F., Ito T., Ito K., Satou M., Kobayashi M., Nagata N., Yoshida S., Nagashima A., et al. Chloroplast ribosome release factor 1 (AtcpRF1) is essential for chloroplast development. Plant Mol. Biol. 2007;64:481–497. doi: 10.1007/s11103-007-9166-7. [DOI] [PubMed] [Google Scholar]
- Mühlbauer S.K., Eichacker L.A. Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J. Biol. Chem. 1998;273:20935–20940. doi: 10.1074/jbc.273.33.20935. [DOI] [PubMed] [Google Scholar]
- Muniz L., Nicolas E., Trouche D. RNA polymerase II speed: a key player in controlling and adapting transcriptome composition. EMBO J. 2021;40 doi: 10.15252/embj.2020105740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F., Hosoda C., Umezawa T., Iizumi H., Kuromori T., Motohashi R., Shono Y., Nagata N., Ikeuchi M., Shinozaki K. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell. 2008;20:3148–3162. doi: 10.1105/tpc.108.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Niimura Y., Gojobori T. Comparative genomic analysis of translation initiation mechanisms for genes lacking the Shine-Dalgarno sequence in prokaryotes. Nucleic Acids Res. 2017;45:3922–3931. doi: 10.1093/nar/gkx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Meierhoff K., Westhoff P., Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2003;270:4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Schuster G., Sugiura M., Sugita M. Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem. Soc. Trans. 2004;32:571–574. doi: 10.1042/BST0320571. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Ashida H., Ogawa T., Yokota A. A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J. 2010;63:766–777. doi: 10.1111/j.1365-313X.2010.04276.x. [DOI] [PubMed] [Google Scholar]
- Noe Gonzalez M., Blears D., Svejstrup J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2021;22:3–21. doi: 10.1038/s41580-020-00308-8. [DOI] [PubMed] [Google Scholar]
- Oldenkott B., Yang Y., Lesch E., Knoop V., Schallenberg-Rüdinger M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019;2:85. doi: 10.1038/s42003-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortelt J., Link G. Plastid gene transcription: promoters and RNA polymerases. Methods Mol. Biol. 2014;1132:47–72. doi: 10.1007/978-1-62703-995-6_3. [DOI] [PubMed] [Google Scholar]
- Ortelt J., Link G. Plastid gene transcription: an update on promoters and RNA polymerases. Methods Mol. Biol. 2021;2317:49–76. doi: 10.1007/978-1-0716-1472-3_2. [DOI] [PubMed] [Google Scholar]
- Ostheimer G.J., Williams-Carrier R., Belcher S., Osborne E., Gierke J., Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomar V.M., Jaksich S., Fujii S., Kuciński J., Wierzbicki A.T. High-resolution map of plastid-encoded RNA polymerase binding patterns demonstrates a major role of transcription in chloroplast gene expression. Plant J. 2022;111:1139–1151. doi: 10.1111/tpj.15882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Boerema A., Aibara S., Paul B., Tobiasson V., Kimanius D., Forsberg B.O., Wallden K., Lindahl E., Amunts A. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants. 2018;4:212–217. doi: 10.1038/s41477-018-0129-6. [DOI] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Holtzegel U., Barkan A., Weisheit W., Mittag M., Pfannschmidt T. ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol. 2015;206:1024–1037. doi: 10.1111/nph.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Liere K., Kandlbinder A., Dietz K.J., Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T., Blanvillain R., Merendino L., Courtois F., Chevalier F., Liebers M., Grübler B., Hommel E., Lerbs-Mache S. Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 2015;66:6957–6973. doi: 10.1093/jxb/erv415. [DOI] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S., McKenzie S.D., Kayanja G.E., Ibrahim I.M. Transcription initiation as a control point in plastid gene expression. Biochim. Biophys. Acta. Gene Regul. Mech. 2021;1864 doi: 10.1016/j.bbagrm.2021.194689. [DOI] [PubMed] [Google Scholar]
- Qi Y., Armbruster U., Schmitz-Linneweber C., Delannoy E., de Longevialle A.F., Rühle T., Small I., Jahns P., Leister D. Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J. Exp. Bot. 2012;63:1251–1270. doi: 10.1093/jxb/err347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S., Haley J., Jiang C.Z., Tsai C.H., Bogorad L. A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc. Natl. Acad. Sci. USA. 1996;93:3881–3885. doi: 10.1073/pnas.93.9.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S.R., Abbott M.S., Bogorad L. Nuclear-organelle interactions: nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell. 1988;55:673–681. doi: 10.1016/0092-8674(88)90226-7. [DOI] [PubMed] [Google Scholar]
- Rojas M., Ruwe H., Miranda R.G., Zoschke R., Hase N., Schmitz-Linneweber C., Barkan A. Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10. Nucleic Acids Res. 2018;46:10448–10459. doi: 10.1093/nar/gky737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M., Yu Q., Williams-Carrier R., Maliga P., Barkan A. Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants. 2019;5:505–511. doi: 10.1038/s41477-019-0412-1. [DOI] [PubMed] [Google Scholar]
- Rose R.J., Lindbeck A.G.C. Morphological studies on the transcription of spinach chloroplast DNA. Z. Pflanzenphysiol. 1982;106:129–137. [Google Scholar]
- Royan S., Gutmann B., Colas des Francs-Small C., Honkanen S., Schmidberger J., Soet A., Sun Y.K., Vincis Pereira Sanglard L., Bond C.S., Small I. A synthetic RNA editing factor edits its target site in chloroplasts and bacteria. Commun. Biol. 2021;4:545. doi: 10.1038/s42003-021-02062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruedas R., Muthukumar S.S., Kieffer-Jaquinod S., Gillet F.X., Fenel D., Effantin G., Pfannschmidt T., Couté Y., Blanvillain R., Cobessi D. Three-dimensional envelope and subunit interactions of the plastid-encoded RNA polymerase from Sinapis alba. Int. J. Mol. Sci. 2022;23:9922. doi: 10.3390/ijms23179922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Bock R. In vivo analysis of RNA editing in plastids. Methods Mol. Biol. 2011;718:137–150. doi: 10.1007/978-1-61779-018-8_8. [DOI] [PubMed] [Google Scholar]
- Ruwe H., Wang G., Gusewski S., Schmitz-Linneweber C. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016;44:7406–7417. doi: 10.1093/nar/gkw466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salesse-Smith C.E., Sharwood R.E., Busch F.A., Kromdijk J., Bardal V., Stern D.B. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants. 2018;4:802–810. doi: 10.1038/s41477-018-0252-4. [DOI] [PubMed] [Google Scholar]
- Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- Sandoval R., Boyd R.D., Kiszter A.N., Mirzakhanyan Y., Santibańez P., Gershon P.D., Hayes M.L. Stable native RIP9 complexes associate with C-to-U RNA editing activity, PPRs, RIPs, OZ1, ORRM1 and ISE2. Plant J. 2019;99:1116–1126. doi: 10.1111/tpj.14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sane A.P., Stein B., Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- Saponaro M., Kantidakis T., Mitter R., Kelly G.P., Heron M., Williams H., Söding J., Stewart A., Svejstrup J.Q. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallenberg-Rüdinger M., Lenz H., Polsakiewicz M., Gott J.M., Knoop V. A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol. 2013;10:1549–1556. doi: 10.4161/rna.25755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff L.B., Childs L., Walther D., Bock R. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff L.B., Ehrnthaler M., Janowski M., Childs L.H., Hasse C., Gremmels J., Ruf S., Zoschke R., Bock R. Shine-Dalgarno sequences play an essential role in the translation of plastid mRNAs in tobacco. Plant Cell. 2017;29:3085–3101. doi: 10.1105/tpc.17.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Lampe M.K., Sultan L.D., Ostersetzer-Biran O. Organellar maturases: a window into the evolution of the spliceosome. Biochim. Biophys. Acta. 2015;1847:798–808. doi: 10.1016/j.bbabio.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 59 region of mRNAs whose translation it activates. Plant Cell. 2005;7:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter Y., Steiner S., Matthäi K., Pfannschmidt T. Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics. 2010;10:2191–2204. doi: 10.1002/pmic.200900678. [DOI] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991;10:1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Gao Y., Schöttler M.A., Bock R., Zoschke R. Limited responsiveness of chloroplast gene expression during acclimation to high light in tobacco. Plant Physiol. 2020;182:424–435. doi: 10.1104/pp.19.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J., Türkeri H., Link B., Link G. AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J. 2010;62:192–202. doi: 10.1111/j.1365-313X.2010.04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z., Sykes M.T., Williamson J.R. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- Sharma M.R., Dönhöfer A., Barat C., Marquez V., Datta P.P., Fucini P., Wilson D.N., Agrawal R.K. PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G) J. Biol. Chem. 2010;285:4006–4014. doi: 10.1074/jbc.M109.062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M.R., Wilson D.N., Datta P.P., Barat C., Schluenzen F., Fucini P., Agrawal R.K. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. USA. 2007;104:19315–19320. doi: 10.1073/pnas.0709856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood R.E., Hotto A.M., Bollenbach T.J., Stern D.B. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA. 2011;17:230–243. doi: 10.1261/rna.2336611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang X., Liu Y., Li Q., Yang Z., Yan N., Zou T., Yin P. Specific RNA recognition by designer pentatricopeptide repeat protein. Mol. Plant. 2015;8:667–670. doi: 10.1016/j.molp.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Shen C., Zhang D., Guan Z., Liu Y., Yang Z., Yang Y., Wang X., Wang Q., Zhang Q., Fan S., et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016;7 doi: 10.1038/ncomms11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. RNA editing in plants: machinery and flexibility of site recognition. Biochim. Biophys. Acta. 2015;1847:779–785. doi: 10.1016/j.bbabio.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Kato H., Ogawa T., Kurachi A., Nakagawa Y., Kobayashi H. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl. Acad. Sci. USA. 2010;107:10760–10764. doi: 10.1073/pnas.0911692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijben-Müller G., Hallick R.B., Alt J., Westhoff P., Herrmann R.G. Spinach plastid genes coding for initiation factor IF-1, ribosomal protein S11 and RNA polymerase alpha-subunit. Nucleic Acids Res. 1986;14:1029–1044. doi: 10.1093/nar/14.2.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I.D., Schallenberg-Rüdinger M., Takenaka M., Mireau H., Ostersetzer-Biran O. Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 2020;101:1040–1056. doi: 10.1111/tpj.14578. [DOI] [PubMed] [Google Scholar]
- Steiner S., Schröter Y., Pfalz J., Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 2011;157:1043–1055. doi: 10.1104/pp.111.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- Stoppel R., Meurer J. The cutting crew - ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 2012;63:1663–1673. doi: 10.1093/jxb/err401. [DOI] [PubMed] [Google Scholar]
- Stoppel R., Lezhneva L., Schwenkert S., Torabi S., Felder S., Meierhoff K., Westhoff P., Meurer J. Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell. 2011;23:2680–2695. doi: 10.1105/tpc.111.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Bentolila S., Hanson M.R. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 2016;21:962–973. doi: 10.1016/j.tplants.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Sun T., Germain A., Giloteaux L., Hammani K., Barkan A., Hanson M.R., Bentolila S. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. USA. 2013;110:E1169–E1178. doi: 10.1073/pnas.1220162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Shi X., Friso G., Van Wijk K., Bentolila S., Hanson M.R. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 2010;152:1263–1273. doi: 10.1104/pp.109.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell. 2007;19:1347–1361. doi: 10.1105/tpc.106.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li J., Zhang L., Lin R. Regulation of chloroplast protein degradation. J. Genet. Genomics. 2023 doi: 10.1016/j.jgg.2023.02.010. [DOI] [PubMed] [Google Scholar]
- Takenaka M., Takenaka S., Barthel T., Frink B., Haag S., Verbitskiy D., Oldenkott B., Schallenberg-Rüdinger M., Feiler C.G., Weiss M.S., et al. DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis. Nat. Catal. 2021;4:510–522. doi: 10.1038/s41929-021-00633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA. 2012;109:5104–5109. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teubner M., Fuß J., Kühn K., Krause K., Schmitz-Linneweber C. The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 2017;89:472–485. doi: 10.1111/tpj.13396. [DOI] [PubMed] [Google Scholar]
- Tiller N., Bock R. The translational apparatus of plastids and its role in plant development. Mol. Plant. 2014;7:1105–1120. doi: 10.1093/mp/ssu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Hardel S.L., Kupsch C., Armbruster U., Delannoy E., Gualberto J.M., Lehwark P., Leister D., Small I.D., Schmitz-Linneweber C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA. 2009;106:6002–6007. doi: 10.1073/pnas.0808529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Barahimipour R., Gao Y., Badillo-Corona J.A., Gotsmann V.L., Zimmer D., Mühlhaus T., Zoschke R., Willmund F. Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. Plants. 2018;4:564–575. doi: 10.1038/s41477-018-0211-0. [DOI] [PubMed] [Google Scholar]
- Türkeri H., Schweer J., Link G. Phylogenetic and functional features of the plastid transcription kinase cpCK2 from Arabidopsis signify a role of cysteinyl SH-groups in regulatory phosphorylation of plastid sigma factors. FEBS J. 2012;279:395–409. doi: 10.1111/j.1742-4658.2011.08433.x. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J. Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu. Rev. Plant Biol. 2015;66:75–111. doi: 10.1146/annurev-arplant-043014-115547. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang J., Li S., Lu C., Sui N. An overview of RNA splicing and functioning of splicing factors in land plant chloroplasts. RNA Biol. 2022;19:897–907. doi: 10.1080/15476286.2022.2096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang Z., Zhang Y., Zhou W., Zhang A., Lu C. Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of ycf3. J. Integr. Plant Biol. 2020;62:1741–1761. doi: 10.1111/jipb.12936. [DOI] [PubMed] [Google Scholar]
- Watkins K.P., Kroeger T.S., Cooke A.M., Williams-Carrier R.E., Friso G., Belcher S.E., van Wijk K.J., Barkan A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19:2606–2623. doi: 10.1105/tpc.107.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K.P., Rojas M., Friso G., van Wijk K.J., Meurer J., Barkan A. APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell. 2011;23:1082–1092. doi: 10.1105/tpc.111.084335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A., Börner T. Transcription and the architecture of promoters in chloroplasts. Trends Plant Sci. 1999;4:169–170. doi: 10.1016/s1360-1385(99)01407-7. [DOI] [PubMed] [Google Scholar]
- Wen L., Li G., Huang T., Geng W., Pei H., Yang J., Zhu M., Zhang P., Hou R., Tian G., et al. Single-cell technologies: from research to application. Innovation. 2022;3 doi: 10.1016/j.xinn.2022.100342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink E.M., Vihervaara A., Tippens N.D., Lis J.T. Nascent RNA analyses: tracking transcription and its regulation. Nat. Rev. Genet. 2019;20:705–723. doi: 10.1038/s41576-019-0159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Stern D. Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA. 2007;104:6466–6471. doi: 10.1073/pnas.0610586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Liu S., Ruwe H., Zhang D., Melonek J., Zhu Y., Hu X., Gusewski S., Yin P., Small I.D., et al. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 2016;85:607–621. doi: 10.1111/tpj.13126. [DOI] [PubMed] [Google Scholar]
- Xiong H.B., Pan H.M., Long Q.Y., Wang Z.Y., Qu W.T., Mei T., Zhang N., Xu X.F., Yang Z.N., Yu Q.B. AtNusG, a chloroplast nucleoid protein of bacterial origin linking chloroplast transcriptional and translational machineries, is required for proper chloroplast gene expression in Arabidopsis thaliana. Nucleic Acids Res. 2022;50:6715–6734. doi: 10.1093/nar/gkac501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Shiina T. Recent advances in the study of chloroplast gene expression and its evolution. Front. Plant Sci. 2014;5:61. doi: 10.3389/fpls.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Ishizaki Y., Nakahira Y., Tozawa Y., Shiina T. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. USA. 2012;109:7541–7546. doi: 10.1073/pnas.1119403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Subramanian A.R. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast) J. Biol. Chem. 2000;275:28466–28482. doi: 10.1074/jbc.M005012200. [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Tasaka M., Shikanai T. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004;38:152–163. doi: 10.1111/j.1365-313X.2004.02035.x. [DOI] [PubMed] [Google Scholar]
- Yan J., Yao Y., Hong S., Yang Y., Shen C., Zhang Q., Zhang D., Zou T., Yin P. Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 2019;47:3728–3738. doi: 10.1093/nar/gkz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang Q., Yin P. RNA editing machinery in plant organelles. Sci. China Life Sci. 2018;61:162–169. doi: 10.1007/s11427-017-9170-3. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang Q., Guan Z., Wang Q., Li L., Ruan F., Lin R., Zou T., Yin P. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants. 2017;3 doi: 10.1038/nplants.2017.37. [DOI] [PubMed] [Google Scholar]
- Yang E.J., Yoo C.Y., Liu J., Wang H., Cao J., Li F.W., Pryer K.M., Sun T.P., Weigel D., Zhou P., Chen M. NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat. Commun. 2019;10:2630. doi: 10.1038/s41467-019-10517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang M., Wang X. Crystal structure of the chloroplast RNA editing factor MORF2. Biochem. Biophys. Res. Commun. 2018;495:2038–2043. doi: 10.1016/j.bbrc.2017.12.044. [DOI] [PubMed] [Google Scholar]
- Yang Z., Liu M., Ding S., Zhang Y., Yang H., Wen X., Chi W., Lu C., Lu Q. Plastid deficient 1 is essential for the accumulation of plastid-encoded RNA polymerase core subunit β and chloroplast development in Arabidopsis. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222413648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Liu B., Nixon P.J., Yu J., Chen F. Recent advances in understanding the structural and functional evolution of FtsH proteases. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.837528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W., et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- Yoo C.Y., Pasoreck E.K., Wang H., Cao J., Blaha G.M., Weigel D., Chen M. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun. 2019;10:2629. doi: 10.1038/s41467-019-10518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Barkan A., Maliga P. Engineered RNA-binding protein for transgene activation in non-green plastids. Nat. Plants. 2019;5:486–490. doi: 10.1038/s41477-019-0413-0. [DOI] [PubMed] [Google Scholar]
- Zeng C., Jiao Q., Jia T., Hu X. Updated progress on group II intron splicing factors in plant chloroplasts. Curr. Issues Mol. Biol. 2022;44:4229–4239. doi: 10.3390/cimb44090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghidi-Abouzid O., Merendino L., Buhr F., Malik Ghulam M., Lerbs-Mache S. Characterization of plastid psbT sense and antisense RNAs. Nucleic Acids Res. 2011;39:5379–5387. doi: 10.1093/nar/gkr143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen J., Zhang L., Wei Y., Li Y., Xu X., Wu H., Yang Z.N., Huang J., Hu F., et al. The pentatricopeptide repeat protein EMB1270 interacts with CFM2 to splice specific group II introns in Arabidopsis chloroplasts. J. Integr. Plant Biol. 2021;63:1952–1966. doi: 10.1111/jipb.13165. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhou W., Che L., Rochaix J.D., Lu C., Li W., Peng L. PPR protein BFA2 is essential for the accumulation of the atpH/F transcript in chloroplasts. Front. Plant Sci. 2019;10:446. doi: 10.3389/fpls.2019.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu C. The enigmatic roles of PPR-SMR proteins in plants. Adv. Sci. 2019;6 doi: 10.1002/advs.201900361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang A., Li X., Lu C. The role of chloroplast gene expression in plant responses to environmental stress. Int. J. Mol. Sci. 2020;21:6082. doi: 10.3390/ijms21176082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Sharma C.M., Förstner K.U., Liere K., Vogel J., Börner T. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24:123–136. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Liu X., Liang S., Fu S., Qi Y., Zhao J., Shao J., An L., Yu F. Chloroplast translation initiation factors regulate leaf variegation and development. Plant Physiol. 2016;172:1117–1130. doi: 10.1104/pp.15.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lu Q., Li Q., Wang L., Ding S., Zhang A., Wen X., Zhang L., Lu C. PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. USA. 2017;114:E1554–E1563. doi: 10.1073/pnas.1612460114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Liu M., Liu X., Dong Z. RNA polymerase II activity revealed by GRO-seq and pNET-seq in Arabidopsis. Nat. Plants. 2018;4:1112–1123. doi: 10.1038/s41477-018-0280-0. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Bock R. Chloroplast translation: structural and functional organization, operational control, and regulation. Plant Cell. 2018;30:745–770. doi: 10.1105/tpc.18.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Kroeger T., Belcher S., Schöttler M.A., Barkan A., Schmitz-Linneweber C. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012;72:547–558. doi: 10.1111/j.1365-313X.2012.05081.x. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Liere K., Börner T. From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 2007;50:710–722. doi: 10.1111/j.1365-313X.2007.03084.x. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA. 2010;107:3245–3250. doi: 10.1073/pnas.0909400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Qu Y., Zubo Y.O., Börner T., Schmitz-Linneweber C. Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J. Plant Res. 2013;126:403–414. doi: 10.1007/s10265-012-0527-1. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Watkins K.P., Barkan A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013;25:2265–2275. doi: 10.1105/tpc.113.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Watkins K.P., Miranda R.G., Barkan A. The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 2016;85:594–606. doi: 10.1111/tpj.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]