Abstract
Introduction and background
Rapid molecular diagnostics to predict carbapenem resistance well before the availability of routine drug sensitivity testing (DST) can serve as an antimicrobial stewardship tool in the context of high rates of Carbapenem-resistant Enterobacteriaceae (CRE).
Materials and methods
A retrospective observational study of patients more than 18 years of age on whom Xpert Carba-R (FDA approved for rectal swab specimen) was done on gram-negative bacteria (GNB) flagged blood culture samples, in an Indian intensive care unit between January 2015 and November 2018. We analyzed the performance of Xpert Carba-R in comparison with routine DST.
Results
A total of 164 GNBs were isolated from 160 patients. Klebsiella pneumoniae and Escherichia coli were the predominant isolates. Carba-R was positive in 35.36% of samples and 45.34% were carbapenem-resistant (CR) on routine DST. The distribution of the CR gene was: Oxacillinase (OXA) (50%), NDM (32.7%) followed by OXA and NDM co-expression (15.51%). The sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, positive predictive value, and negative predictive value of Carba-R were 90.74, 93.15, 13.25, 0.10, 83.58 and 96.31% for Enterobacteriaceae. The median time to obtain the Carba-R report was 30 hours 34 minutes vs 74 hours and 20 minutes for routine DST. Based on the Carba-R report, 9.72% of patients had escalation and 27.08% had de-escalation of antibiotics.
Conclusion
Xpert Carba-R serves as a rapid diagnostic tool for predicting carbapenem resistance in intensive care unit patients with bacteremia caused by Enterobacteriaceae.
How to cite this article
Rajendran S, Gopalakrishnan R, Tarigopula A, Kumar DS, Nambi PS, Sethuraman N, et al. Xpert Carba-R Assay on Flagged Blood Culture Samples: Clinical Utility in Intensive Care Unit Patients with Bacteremia Caused by Enterobacteriaceae. Indian J Crit Care Med 2023;27(9):655–662.
Keywords: Antimicrobial stewardship, Carbapenem-resistant enterobacteriaceae, Carbapenemase detection, Gram-negative resistance, Novel diagnostics for antimicrobial resistance, Rapid molecular diagnostics, Xpert Carba-R assay
Highlights
Rapid diagnostic tests to detect antibiotic resistance are the need of the hour in the context of rising antimicrobial resistance. This study finds Xpert Carba-R assay to have good performance characteristics to identify carbapenem-resistant genes in Enterobacteriacea from blood samples of critically ill patients.
Introduction and Background
Rising antimicrobial resistance (AMR) over the past 2 decades has contributed to significant morbidity and mortality across the globe and is a serious threat to health care, particularly in the context of a dry antibiotic pipeline.1 The country-specific report (2017–2018) from the Global Antimicrobial Resistance Surveillance System (GLASS) by World Health Organization (WHO) has reported up to 75% carbapenem-resistance (CR) in bacteremic isolates of Klebsiella pneumoniae (K. pneumoniae) and up to 40% CR in Escherichia coli (E.coli).2 The emergence and spread of CR in gram-negative bacteria (GNB) including Enterobacteriaceae forces the physician to choose empiric coverage for carbapenem-resistant GNB (CR-GNB) with drugs such as polymyxins, which in turn becomes an independent risk factor for polymyxin resistant infections.3–6 Rapid detection of CR in GNB sepsis, using novel molecular diagnostics well in advance of receipt of conventional culture and drug sensitivity testing (DST) would aid in antimicrobial stewardship.7 Jauréguy et al. studied the performance of Xpert Carba R [used for carbapenem-resistant Enterobacteriaceae (CRE) screening in rectal swabs] in flagged blood culture samples spiked with carbapenemase producing isolates to detect CR genes (KPC, NDM, VIM, OXA-48, and IMP-1) and found 100 percent agreement with genes already characterized.8 Knowledge of the specific carbapenemase-resistance gene could potentially enhance the rational use and stewardship of novel drugs such as ceftazidime-avibactam.9 We studied the performance of Xpert Carba-R directly on samples from flagged blood culture bottles, in comparison to standard DST, to assess its clinical utility in gram-negative bacteremia in critically ill patients.
Materials and Methods
We conducted a retrospective observational study between January 2015 and November 2018 in a Tertiary Care ICU in India, among intensive care patients aged above 18, whose blood culture flagged GNB and in whom a Carba-R test was ordered from the blood culture sample. Only one bacteremic episode was included in each patient. Case records were analyzed for demographic, clinical, and microbiological details. Performance of the test in terms of sensitivity, specificity, positive likelihood ratio (LR+), negative likelihood ratio (LR-), positive predictive value (PPV), and negative predictive value (NPV) with regards to different groups of bacteria in comparison with standard phenotypic DST were analyzed. The baseline ICU CR rates in bacteremic isolates during the study period were 27.77% in Enterobacteriaceae (57.99% in K. pneumoniae and 13.18% in E. coli) and 61.68% in pseudomonas and acinetobacter together and these values were used as prevalence in calculating PPV and NPV.
Blood Culture Protocol
Blood culture was done using BacT/ALERT (Biomerieux) system. Once flagged, Gram staining of the sample was done, and the report was intimated to the physician via telephone call. After subculture, colonies were subjected to Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI TOF MS) for identification, conventional DST by the combination of methods using VITEK (Biomerieux) and Kirby Bauer disk diffusion method as recommended by Clinical Laboratory Standards Institute (CLSI) and interpreted according to CLSI guidelines for the respective years. The physician is informed of the identification (ID) of the organism on the day of the flag by MALDI-TOF MS if the flag was before 12:00 p.m. on working days. ATCC Escherichia coli 26922, ATCC Klebsiella pneumoniae 700603 were used as reference strains.10,11
DST for colistin was done using VITEK during the years 2015–2017 and by micro broth dilution from 2018. European Committee on Antimicrobial Susceptibility Testing (EUCAST) interpretive criteria (<2 mcg/L–sensitive) were used for reporting colistin sensitivity in Enterobacteriaceae, pseudomonas, and acinetobacter.12
Carba-R Protocol
The Carba-R test was done in a dedicated molecular lab with samples in the cartridge loaded on a Gene Xpert platform, which works on the principle of real-time PCR. The Carba R test has primers that could pick up the following genes – KPC, NDM, OXA-48 like (including OXA-181 and OXA-232), VIM, and IMP-1. When blood cultures flagged GNB in patients included in the study population, a Carba-R test was requested. The decision to order a Carba-R test in any ICU patient was based on the treating physician's discretion. The Carba-R report was informed telephonically or accessed with a turnaround time of two hours on the same day for all samples received before 3:00 p.m., except on Sundays. Based on the Carba-R test report, i.e., the presence or absence of CR genes in the assay, the physician decided on fine-tuning the antibiotic choice in conjunction with clinical judgment before the end of working hours the same day. Final adjustments in antibiotic regimen if necessary were made once routine DST reports are available.
Decision-making Flow
We tried to analyze the decision taken by the physician based upon the Carba-R (Table 1) and termed them as:
Table 1.
Decision taken by the physician based on Carba-R
| Empiric choice | Carba-R | Expected or possible actions | Termed | DST – CR | DST – CS |
|---|---|---|---|---|---|
| Carbapenem | +ve | Escalation to CR cover | Escalation | Appropriate action | Inappropriate action (but still can cover the organism) |
| CR cover +/– carbapenem | +ve | Continue CR cover +/– carbapenem | No action | Appropriate action | Inappropriate action (but can still cover the organism) |
| Carbapenem | –ve | Continue same | No action | Inappropriate action (will not cover the organism) | Appropriate action |
| CR cover +/– carbapenem | –ve | De-escalation | De-escalation Done |
Inappropriate (will not cover the organism) | Appropriate action |
| CR cover +/– carbapenem | –ve | De-escalation not done | De-escalation not done | Appropriate action | Inappropriate action (still may cover the organism) |
“escalation”: when CR gene is detected, the addition of an antibiotic that is usually expected to cover CR GNB (e.g., polymyxin).
“de-escalation”: when CR gene is not detected, continue/ start only carbapenem or stop coverage for CRGNB if already started.
“no action”: in patients with negative CR gene, the decision to continue the empiric carbapenem without any escalation, and in patients with positive CR gene, continuing the empiric CR cover without any de-escalation.
The above is expected, or possible actions based on the Carba-R result alone; however, the physician may choose differing options based on other factors e.g., suspicion of modes of resistance other than carbapenemase production, presence of an intrinsically carbapenem-resistant organism (known by direct MALDI ID), presence of a polymicrobial bacteremia, etc. The decision was termed appropriate when it correlated with the phenotypic DST and vice versa. The time duration between receipt of the blood sample in the lab to the release of Carba-R report, in comparison with the release of phenotypic DST was also studied.
Results
A total of 160 patients were studied, of whom 36 patients (22.5%) were solid organ transplant recipients. Of the 160 patients, 164 GNB were isolated. Baseline characteristics are shown in Table 2. Out of 160 patients, 100 (62.5%) developed index bacteremia more than 48 hours after being admitted to the hospital. This pattern aligns with the acquisition of the infection within the hospital setting rather than in the community. Xpert Carba-R was positive in 58/164 (35.36%) isolates in whom it was ordered.
Table 2.
Characteristics of the study population
| Characteristics | N = 160 |
|---|---|
| Median age (years) | 58 (18–86) |
| Male | 120 (75%) |
| Female | 40 (25%) |
| Charlson comorbidity index (mean) | 4 |
| PITT bacteremia score (mean) | 6 |
| SOFA score (mean) | 16 |
| Prior exposure to a carbapenem | 58 (36.25%) |
| Source of sepsis | |
| Line related | 38 (23.75%) |
| Unclear | 36 (22.5%) |
| Intra-abdominal | 36 (22.5%) |
| Pneumonia | 22 (13.75%) |
| Urinary | 20 (12.5%) |
| SSTI–Skin and soft tissue infection | 7 (4.37%) |
| Meningitis | 1 (0.62%) |
| Organisms (n = 164) | |
| Klebsiella pneumoniae | 77 (46.95%) |
| Escherichia coli | 39 (23.78%) |
| Enterobacter cloacae | 6 (3.65%) |
| Citrobacter koseri | 2 (1.22%) |
| Serratia marcescens | 2 (1.22%) |
| Morganella morganii | 1 (0.61%) |
| Acinetobacter baumannii | 16 (9.75%) |
| Pseudomonas aeruginosa | 7 (4.26%) |
| Pseudomonas stutzeri | 1 (0.61%) |
| Pseudomonas putida | 2 (1.22%) |
| Burkholderia pseudomallei | 2 (1.22%) |
| Burkholderia cepacia | 2 (1.22%) |
| Aeromonas hydrophila | 3 (1.65%) |
| Ralstonia mannitolilytica | 1 (0.61%) |
| Stenotrophomonas maltophilia | 2 (1.22%) |
| Elizabethkingia meningoseptica | 1 (0.61%) |
| Status on day 14 | |
| Alive | 109 (68.13%) |
| Dead | 44 (27.5%) |
| Not known | 7 (4.37%) |
| Status on day 30 | |
| Alive | 72 (45.00%) |
| Dead | 63 (39.37%) |
| Not known | 25 (15.62%) |
| No. of patients died | 67 (41.87%) |
| Median time to death from index bacteremia (days) | 9 |
| Median time to Carba-R (hours:min) | 30:34 |
| Median time to routine DST (hours:min) | 74:20 |
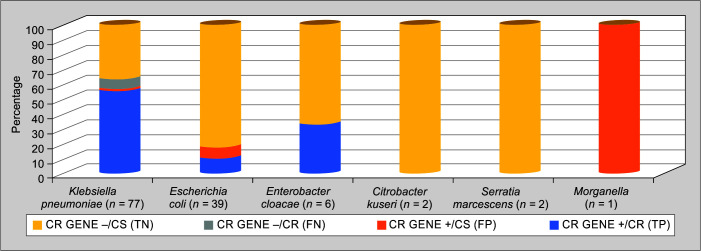
The comparison between Xpert Carba-R and routine DST among different species is depicted in Table 3 and Figure 1. Out of 161 isolates, 73 isolates (45.34%) were CR by routine DST (two patients with Stenotrophomonas and one with Elizabethkingia were excluded from the study due to intrinsic CR). Carba-R test detected CR genes in 89.58% (43/48) of CR K. pneumoniae, 100% (4/4) of CR E. coli, 50% (2/4) of CR pseudomonas species, and only 9.09% (1/11) of CR A. baumanii. Three isolates of E. coli and one isolate of K. pneumoniae had a CR gene but were phenotypically sensitive. None of the carbapenem-sensitive isolates in A. baumannii and pseudomonas species had a positive Carba-R test. The performance of the test in Enterobacteriaceae and individual isolates are also shown in Figure 1.
Table 3.
Comparison of Carba-R and conventional drug susceptibility testing (DST)
| Organism | DST | CR gene +ve | CR gene –ve | |||
|---|---|---|---|---|---|---|
| CR | CS | CR | CS | CR | CS | |
| Klebsiella pneumoniae (n = 77) | 48 (62.33%) | 29 (37.66%) | 43 (89.58%) | 1 (3.4%) | 5 (10.41%) | 28 (96.5%) |
| Escherichia coli (n = 39) | 4 (10.25%) | 35 (89.74%) | 4 (100%) | 3 (8.57%) | 0 | 32 (91.42%) |
| Enterobacter cloacae (n = 6) | 2 (33.33%) | 4 (66.66%) | 2 (100%) | 0 | 0 | 4 (100%) |
| Citrobacter koseri (n = 2) | 0 | 2 | – | – | 0 | 2 |
| Serratia marcescens (n = 2) | 0 | 2 | – | – | 0 | 2 |
| Morganella morganii (n = 1) | 0 | 1 | – | 1 | – | – |
| Acinetobacter baumannii (n = 16) | 11 (68.75%) | 5 (31.25%) | 1 (9.09%) | 0 | 10 (90.9%) | 5 (100%) |
| Pseudomonas aeruginosa (n = 7) | 4 (57.14%) | 3 (42.85%) | 2 (50%) | 0 | 2 (50%) | 3 (100%) |
| Pseudomonas stutzeri (n = 1) | 0 | 1 | – | – | 0 | 1 |
| Pseudomonas putida (n = 2) | 0 | 2 | – | – | 0 | 2 |
| Burkholderia pseudomallei (n = 2) | 0 | 2 | – | – | 0 | 2 |
| Burkholderia cepacia (n = 2) | 2 | – | – | 2 | 0 | |
| Aeromonas hydrophila (n = 3) | 2 | 1 | 1 | 0 | 1 | 1 |
| Ralstonia mannitolilytica (n = 1) | 0 | 1 | – | – | 0 | 1 |
| Stenotrophomonas maltophilia (n = 2) | – | – | – | – | – | – |
| Elizabethkingia meningoseptica (n = 1) | – | – | – | – | – | – |
*Note: Four patients who had a negative Carba-R report, had polymicrobial bacteremia from the same blood culture bottle each with two bacteria and were considered as separate isolates
Fig. 1.
Comparison of Carba-R and routine drug susceptibility testing
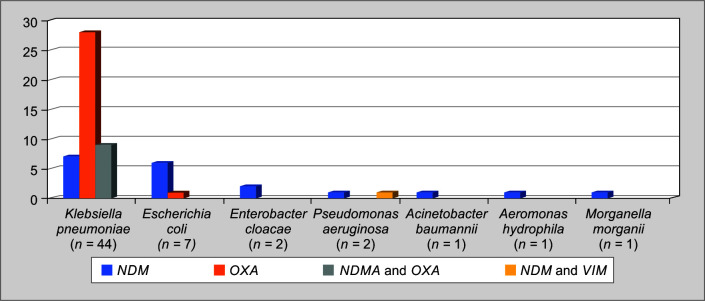
The distribution of carbapenem-resistant genes is described in Table 4 and in Figure 2. Oxacillinase was the predominant CR gene in the overall study group (38/58–65.51%), followed by NDM (29/58–50%), which includes NDM and OXA co-expression (9/58–15.51%). In K. pneumoniae, OXA-48 like was predominant (37/44–84.09%), followed by NDM (16/44–36.36%) with (9/44–20.45%) co-expression. In E. coli, NDM (6/7–85.71%) was predominant followed by OXA-48 (1/7–14.28%).
Table 4.
Distribution of carbapenem-resistant genes
| Organism | Enzymes |
|---|---|
| Klebsiella pneumoniae (n = 44) | NDM = 7 (15.90%) OXA = 28 (63.63%) NDM and OXA = 9 (20.45%) |
| Escherichia coli (n = 7) | NDM = 6 (85.71%) OXA = 1 (14.28%) |
| Enterobacter cloacae (n = 2) | NDM = 2 (100%) |
| Pseudomonas aeruginosa (n = 2) | NDM = 1 (50%) NDM and VIM = 1 (50%) |
| Acinetobacter baumannii (n = 1) | NDM = 1 (100%) |
| Aeromonas hydrophila (n = 1) | NDM = 1 (100%) |
| Morganella morgani (n = 1) | NDM = 1 (100%) |
Fig. 2.
Distribution of carbapenem-resistant genes in the study population
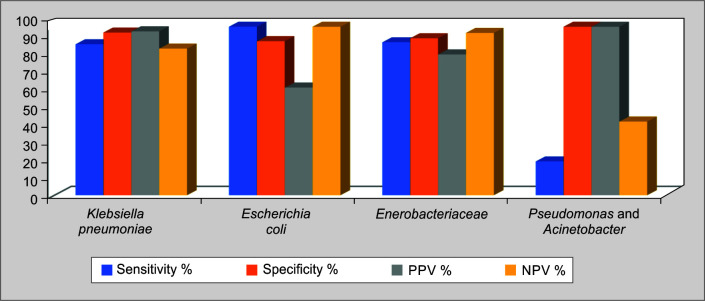
The performance of the test in terms of sensitivity, specificity, LR+/–, PPV and NPV are shown in Table 5 and Figure 3. Overall, for Enterobacteriaceae, the sensitivity, specificity, PPV, and NPV were 90.74, 93.15, 83.58, and 96.31% respectively. Pseudomonas and Acinetobacter were considered together for analysis and the sensitivity, specificity, PPV, and NVP.
Table 5.
Performance of Carba-R test in comparison with routine drug susceptibility testing
| Klebsiella pneumoniae | Escherichia coli | Enterobacteriaceae | Pseudomonas and Acinetobacter | |
|---|---|---|---|---|
| Sensitivity% (confidence interval) |
89.58 (77.34–96.53) | 100 (36.76–100) | 90.74 (79.70–96.92) | 20.00 (4.33–48.09) |
| Specificity% (confidence interval) |
96.55 (82.24–99.91) | 91.43 (76.94–98.20) | 93.15 (84.74–97.74) | 100 (71.5–100) |
| LR+ (confidence interval) |
25.98 (3.78–178.68) | 11.67 (3.95–34.42) | 13.25 (5.66–31.00) | – |
| LR– (confidence interval) |
0.10 (0.05–0.25) | 0.00 | 0.10 (0.04–0.23) | 0.8 (0.62–1.03) |
| PPV% | 97.28 | 63.91 | 83.58 | 100 |
| NPV% | 87.03 | 100 | 96.31 | 43.71 |
Fig. 3.
Performance of Carba-R test in comparison with routine susceptibility testing
After excluding colistin-resistant isolates (Table 6), 144 actions were observed, of which 126 (86.80%) were appropriate and 19 (13.19%) were inappropriate. Of 2 patients who had inappropriate de-escalation based on a negative CR gene, 1 was K. pneumoniae and 1 was A. baumannii. Of 6 patients in whom empiric carbapenem alone was continued inappropriately after negative Carba-R (no action), 2 were CR pseudomonas and 4 were CR A. baumannii with one poly microbial CR K. pneumoniae.
Table 6.
Decision taken by physician based on Carba-R*
| N = 144 | Positive CR gene (n = 52) | Negative CR gene (n = 92) | ||
|---|---|---|---|---|
| Action and appropriateness | Appropriate (47) | Inappropriate (5) | Appropriate (78) | Inappropriate (14) |
| Escalation done (16) | 14 (9.72%) | 2 (1.38%) ** | – | – |
| De-escalation done (41) | – | – | 39 (27.08%)*** | 2 (1.38%)$ |
| De-escalation not done (13) | – | – | 7 (4.86%) | 6 (4.16%)** |
| No action | 33 (22.91%) | 3 (2.08%)** | 32 (22.22%) | 6 (4.16%)$ |
*16 patients were excluded from this analysis including 10 patients from CR gene –ve group, i.e., 3 isolates (DST known prior to Carba R), Burkholderia sp (n = 4), Stenotrophomonas (n = 2), Elizabethkingia (n = 1) and 6 from +ve CR gene group (carbapenem and colistin-resistant Klebsiella) were excluded;
**patients received unnecessary CR over [2 + 3 + 6 = 11(7.63%)];
***27.08% de-escalated from CR cover;
$Indicates patients who did not get appropriate antibiotic cover before DST (8/144) – 5.55%
Discussion
Carbapenem resistance in bacteremic isolates of Enterobacteriaceae has risen steadily over the last decade and the prevalence varies in different geographic areas as evidenced by up to 70% CR in isolates of K. pneumoniae from Egypt and up to 40% CR in E. coli from India (GLASS WHO 2017–2018).2 A multicenter study from India by Veeraraghavan et al., shows that E. coli followed by K. pneumoniae are the most common etiologies of gram-negative infections and describes CR rates in K. pneumoniae and E. coli as 39 and 12% respectively.13 The prevalence of CR in bacteremic isolates from our hospital intensive care unit during the study period was 27.77% in Enterobacteriaceae (57.99 % in K. pneumoniae, 13.18% in E. coli) and 61.68% in Pseudomonas species and Acinetobacter combined. The antimicrobial resistance (AMR) surveillance data from India in 2017 shows that 59% of bacteremic K. pneumoniae were resistant to meropenem and 32% of bacteremic E. coli were resistant to ertapenem.14
Our study group consisted of patients who were critically ill and 62% had blood cultures drawn 48 hours after hospital admission, consistent with hospital rather than community acquisition of infection. The microbiologic profile in our study was dominated by K. pneumoniae (46.95%), followed by E. coli (23.78%), A. baumannii (9.75%), and pseudomonas spp (6.25%). The commonest source of bacteremia in our study was line related (23.75%), followed by intra-abdominal source and undetermined source (22.5% each). Pneumonia and UTI contributed only to 13.75 and 12.5 % of patients.
Carbapenemase production is the major mechanism of resistance in CRE. Tamma et al. observed a fourfold rise in the odds of dying within 14 days in carbapenemase-producing (CP-CRE) vs non-CP-CRE bacteremia which furthermore signifies the detection of CPCRE.15 Tests such as the Film Array - Blood Culture Identification Panel (BioFire) and Verigene- Gram-negative blood culture test (Nanosphere) detect carbapenemase genes, the major mechanisms of resistance in Enterobacteriaceae directly from blood culture.16 Diagnostic stewardship, whereby the laboratory facilitates appropriate antibiotic decision-making, is an increasingly emphasized component of antimicrobial stewardship.17 The epidemiology of the mechanism of carbapenem resistance has a varied geographical distribution, with KPC being the predominant mechanism in the USA, KPC followed by OXA-48 in Europe, and NDM followed by OXA-48 in India.18,19 It is essential to identify CP-CRE, including the exact enzyme produced, given the availability of newer drugs with varying activity against different carbapenemases, e.g. ceftazidime-avibactam has activity against KPC and OXA-48, but not against NDM.20,21
Mohanty S et al. screened 93 isolates of CR K. pneumoniae and CR E. coli from bloodstream infections (BSI) with PCR in a tertiary care hospital in India and observed that 76.3% were positive for at least one carbapenemase-producing gene.22 Molecular characterization of CR K. pneumoniae in BSI by Veeraraghavan et al. showed that 40/115 phenotypically CR isolates did not have an enzyme detected by multiplex PCR.18 The rate of CPCRE amongst the CRE detected by Xpert Carba-R in this study was 49/54 (90.74%) in Enterobacteriaceae, 43/48 (89.58%) in K. pneumoniae and 4/4 (100%) in E. coli.
In our study, OXA-48 (50% as OXA-48 only and 65.51% including co-expression with NDM) was the most common carbapenemase gene detected followed by NDM (33% as NDM only and 50% including co-expression with OXA-48). NDM and OXA-48 were co-expressed in 15.51% of the isolates, all of which were K. pneumoniae. In K. pneumoniae, again, OXA (84.09%) was predominant followed by NDM (36.36%) including co-expressions. VIM gene was detected in only one P. aeruginosa isolate in co-expression with NDM. We did not find any IMP gene or KPC gene in the study isolates. The frequency of carbapenemase genes detected in the study by Mohanty et al. was NDM (65.6%), OXA 48 (24.7%), OXA-181 (23.6%), VIM 6.4%, and KPC (2.1%).22 In the study by Veeraraghavan et al., among klebsiella isolates, NDM and OXA co-expression was seen in 28% (32/115), NDM in 19% (22/115), OXA-48-like in 13% (15/115) and KPC was absent.17 Our study showed that OXA-48 is the predominant mechanism as seen in other studies done by Anandan S et al. and Sharma A et al.23,24
In the study by Anandan et al., Xpert Carba-R did not identify any of the OXA-48-like genes that the PCR picked up in 47/120 isolates, probably because of the absence of the primer for the missed gene variant i.e., OXA-181. There has been a subsequent update on the kit with the addition of OXA 48-like primers (OXA 181 and 232) to address this issue.23,25 Our study was able to identify OXA-48 as the predominant gene as the kit used was an updated version.
An additional utility of Carba-R is its role in predicting the activity of novel BL-BLIs such as ceftazidime-avibactam (with or without aztreonam), which may permit a “polymyxin-sparing” approach. The presence of an OXA-48 gene would predict the activity of ceftazidime-avibactam while the presence of a metalloenzyme would predict likely resistance (which may be overcome by the addition of aztreonam).9,26
The test performed well against routine DST in predicting CR with 90.74% sensitivity, 93.15% specificity, 83.58% PPV, 96.31% NPV, LR+ of 13.25, and LR- of 0.10 with regards to Enterobacteriaceae overall as well as when analyzed separately for K. pneumoniae and E. coli. Carba-R had performed well with 100% sensitivity, 77% specificity, 96% PPV and 100% NPV (excluding OXA-48 like) when compared with multiplex PCR in blood culture isolates of Enterobacteriaceae.23 When tested against DNA sequencing, in GNB isolates grown on blood culture agar and McConkey agar, carba-R had shown 100% sensitivity for blood agar isolates, 98.1% and 97.1% specificity for blood and McConkey agar respectively.27 In the study by Jauréguy et al., Xpert Carba-R detected carbapenemases including OXA-48 and OXA-181 in 100% agreement in flagged samples directly from blood culture bottles spiked with carbapenemase-producing organisms (CPO) whose carbapenemase-genes were characterized earlier.8 There was no report of Carba-R being done directly from flagged blood culture samples. The clinical utility of this test in terms of deciding on treatment depends upon the baseline prevalence of CRE and rate of CPCRE amongst CRE which were 27.77 and 90.74% (detected only by Carba-R) respectively in our study population. It did not perform well in the non-Enterobacteriaceae group (pseudomonas and Acinetobacter) due to multiple other mechanisms of resistance in these non-fermenters.28 Carba-R should not be relied upon in making treatment decisions in bacteremia caused by non-Enterobacteriaceae since a negative Carba-R test result does not predict carbapenem sensitivity in these organisms.
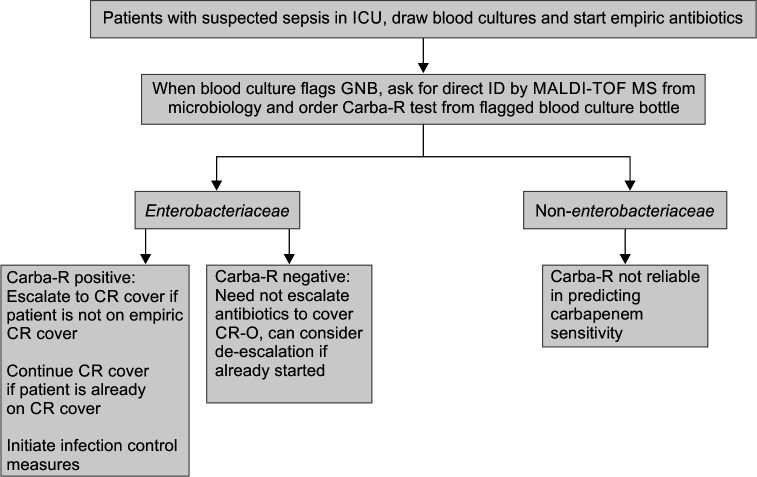
In our study, Carba-R influenced antibiotic selection on day one of the blood culture flag. The median time to receipt of the Carba-R report was 30 hours 34 minutes as against 74 hours and 20 minutes for routine DST. This assisted in both appropriate early therapy and in early de-escalation from polymyxins which may translate into less toxic and better newer therapeutic options as well as in initiating infection control measures much earlier to prevent cross-transmission.29,30 A significant minority (9.72%) of patients had escalation to CR cover after Carba-R test. Up to 27.08% of patients had de-escalation from CR cover after Carba-R test. However, 7.63% (11/144) received CR cover despite growing CS organisms. When analyzing inappropriate coverage of organisms, excluding CCRE and intrinsically colistin-resistant organisms, 5.55 % (8/144) did not have an appropriate antibiotic cover before routine DST. We suggest a workflow on the appropriate usage of Carba-R in conjunction with MALDI TOF ID as described in Figure 4.
Fig. 4.
Suggested workflow with Carba-R
The limitations of our study are:
The inherent test limitation (will miss genes for which primers are not included in the assay and other modes of carbapenem resistance).
Carba-R is preferably used in conjunction with a rapid ID system such as MALDI-TOF as it is of limited usefulness in non-Enterobacteriaceae bacteremia.
Genomic sequencing was not done in the isolates which would identify the missed genes if any.
The test was done only during working hours and not around the clock.
The distribution of carbapenemase genes may not represent the real scenario in Enterobacteriaceae as the study group was very selective and dominated by K. pneumoniae.
Randomized control trials are needed further to analyze the impact of Carba-R with regard to treatment and patient outcomes.
Conclusion
Xpert Carba-R is much quicker than routine DST and reasonably accurate in identifying CP-CRE in the Indian setting. It serves as a rapid diagnostic tool in predicting carbapenem resistance in critically ill patients with bacteremia caused by Enterobacteriaceae and it should optimally be used in combination with a rapid same-day ID system such as MALDI-TOF MS. In terms of antimicrobial stewardship, it assists both in the escalation of antibiotics if patients are not already on CR cover, as well as in de-escalating therapy already initiated well before receipt of DST. It therefore reduces injudicious empiric polymyxin usage and helps in the earlier initiation of infection control measures for patients with CP-CRE. We recommend that all intensive care units which encounter CP-CRE routinely incorporate the test in their antibiotic decision-making process.
Orcid
Surendran Rajendran https://orcid.org/0000-0001-7402-5655
Ram Gopalakrishnan https://orcid.org/0000-0002-2263-1861
Anil Tarigopula https://orcid.org/0000-0001-6493-3510
D Suresh Kumar https://orcid.org/0000-0001-7643-0144
P Senthur Nambi https://orcid.org/0000-0003-0519-8768
Nandini Sethuraman https://orcid.org/0000-0002-3803-5511
Chitra Chandran https://orcid.org/0000-0002-5504-1569
Nagarajan Ramakrishnan https://orcid.org/0000-0001-5208-4013
V Ramasubramanian https://orcid.org/0000-0001-8566-2035
Footnotes
Source of support: Nil
Conflict of interest: None
References
- 1.Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. Antimicrobial resistance. JAMA. 2016;316(11):1193–1204. doi: 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- 2.Global antimicrobial resistance surveillance system (GLASS) report: Early implementation 2017–2018. Geneva: World Health Organization; 2018. Available at: https://www.who.int/publications/i/item/9789241515061. [Google Scholar]
- 3.Patel G, Bonomo RA. “Stormy waters ahead”: Global emergence of carbapenemases. Front Microbiol. 2013;4:1–17. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zak-Doron Y, Dishon Benattar Y, Pfeffer I, Daikos G, Skiada A, Antoniadou A, et al. The association between empirical antibiotic treatment and mortality in severe infections caused by carbapenem-resistant gram-negative bacteria: A prospective study. Clin Infect Dis. 2018;67(12):1815–1823. doi: 10.1093/cid/ciy371. [DOI] [PubMed] [Google Scholar]
- 5.Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: Results from a guideline-based performance improvement program. Crit Care Med. 2014;42(8):1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 6.Matthaiou DK, Michalopoulos A, Rafailidis PI, Karageorgopoulos DE, Papaioannou V, Ntani G, et al. Risk factors associated with the isolation of colistin-resistant Gram-negative bacteria: A matched case-control study. Crit Care Med. 2008;36(3):807–811. doi: 10.1097/CCM.0B013E3181652FAE. [DOI] [PubMed] [Google Scholar]
- 7.De Waele JJ, Dhaese S. Antibiotic stewardship in sepsis management: Toward a balanced use of antibiotics for the severely ill patient. Expert Rev Anti Infect Ther. 2019;17(2):89–97. doi: 10.1080/14787210.2019.1568239. [DOI] [PubMed] [Google Scholar]
- 8.Jauréguy F, Mansour H, Bigot J, Walewski V, Billard-Pomares T, Rahajamanana L, et al. Use of the XpertCarbaR assay for direct detection of carbapenemase genes from blood cultures and urine samples. J Hosp Infect. 2018;98(3):245–246. doi: 10.1016/j.jhin.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious diseases society of America guidance on the treatment of extended-spectrum β-lactamase producing enterobacterales (Esbl-E), carbapenem-resistant enterobacterales (CRE), and pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis. 2021;72(7):1109–1116. doi: 10.1093/cid/ciab295. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 12th edition. Wayne (PA): 2015. Performance standards for antimicrobial disc susceptibility testing; approved standard. CLSI document M02-A12.35(1). [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI). Wayne (PA): Clinical and Laboratory Standards Institute. CLSI document; 2015. Performance standards for antimicrobial susceptibility testing; twenty fifth informational supplement. pp. M100–S25. [Google Scholar]
- 12.The European Committee on Antimicrobial Susceptibility Testing. Routine internal quality control as recommended by EUCAST. 2017 Version 7.0. Available from: http://www.eucast.org/ [Google Scholar]
- 13.Veeraraghavan B, Jesudason MR, Prakasah JA, Anandan S, Sahni RD, Pragasam AK, et al. Antimicrobial susceptibility profiles of gram-negative bacteria causing infections collected across India during 2014–2016: Study for monitoring antimicrobial resistance trend report. Indian J Med Microbiol. 2018;36(1):32–36. doi: 10.4103/ijmm.IJMM_17_415. [DOI] [PubMed] [Google Scholar]
- 14.AMR surveillance network Indian Council of Medical Research 2017. Annual report Antimicrobial Resistance Surveillance Network January 2017–December 2017. Retrieved from: https://icmr.nic.in/sites/default/files/reports/annual_report_amr_jan2017-18.pdf. [Google Scholar]
- 15.Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, et al. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis. 2016;64(3):257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee R, Humphries R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence. 2016;8(4):427–439. doi: 10.1080/21505594.2016.1185577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barlam TF, Cosgrove SE, Abbo LM, MacDougall C, Schuetz AN, Septimus EJ, et al. Implementing an antibiotic stewardship program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clini Infect Dis. 2016;62(10):e51–e77. doi: 10.1093/cid/ciw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veeraraghavan B, Shankar C, Karunasree S, Kumari S, Ravi R, Ralph R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Patho Glob Health. 2017;111(5):240–246. doi: 10.1080/20477724.2017.1340128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duin DV, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8(4):460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutgring JD, Limbago BM. The problem of carbapenemase producing- carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol. 2016;54(3):529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castanheira M, Farrell SE, Krause KM, Jones RN, Sader HS. Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine U.S. census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob Agents Chemother. 2014;58(2):833–838. doi: 10.1128/AAC.01896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohanty S, Gajanand M, Gaind R. Identification of carbapenemase-mediated resistance among Enterobacteriaceae bloodstream isolates: A molecular study from India. Indian J Med Microbiol. 2017;35(3):421–425. doi: 10.4103/ijmm.IJMM_16_386. [DOI] [PubMed] [Google Scholar]
- 23.Anandan S, Damodaran S, Gopi R, Bakthavatchalam YD, Veeraraghavan B. Rapid screening for carbapenem resistant organisms: Current results and future approaches. J Clin Diagn Res. 2015;9(9):DM01–3. doi: 10.7860/JCDR/2015/14246.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Bakthavatchalam YD, Gopi R, Anandan S, Verghese VP, Veeraraghavan B. Mechanisms of carbapenem resistance in K. pneumoniae and E. Coli from bloodstream infections in India. J Infect Dis Ther. 2016;4:293. doi: 10.4172/2332-0877.1000293. [DOI] [Google Scholar]
- 25.Dortet L, Fusaro M, Naas T. Improvement of the Xpert Carba-R kit for the detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2016;60(6):3832–3837. doi: 10.1128/AAC.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagvekar V, Shah A, Unadkat VP, Chavan A, Kohli R, Hodgar S, et al. Clinical outcome of patients on Ceftazidime – Avibactam and combination therapy in carbapenem-resistant enterobacteriaceae. Indian J Crit Care Med. 2021;25(7):780–784. doi: 10.5005/jp-journals-10071-23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traczewski MM, Carretto E, Canton R, Moore NM. Multicenter evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in gram-negative isolates. J Clin Microbiol. 2018;56(8):e00272–e002718. doi: 10.1128/JCM.00272-18. Carba-R Study Team. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonomo R, Szabo D. Mechanisms of multidrug resistance in acinetobacter species and pseudomonas aeruginosa. Clinical Infectious Diseases. 2006;43(Suppl 2):S49–S56. doi: 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- 29.Wangchinda W, Pati N, Maknakhon N, Seenama C, Tiengrim S, Thamlikitkul V. Collateral damage of using colistin in hospitalized patients on emergence of colistin-resistant Escherichia coli and Klebsiella pneumoniae colonization and infection. Antimicrob Resist Infect Control. 2018;7(1):84. doi: 10.1186/s13756-018-0375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prayag PS, Patwardhan SA, Panchakshari S, Sambasivam R, Dhupad S, Soman RN, et al. Ceftazidime-avibactam with or without Aztreonam vs Polymyxin-based combination therapy for carbapenem-resistant Enterobacteriaceae: A retrospective analysis. Indian J Crit Care Med. 2023;27(6):444–450. doi: 10.5005/jp-journals-10071-24481. [DOI] [PMC free article] [PubMed] [Google Scholar]