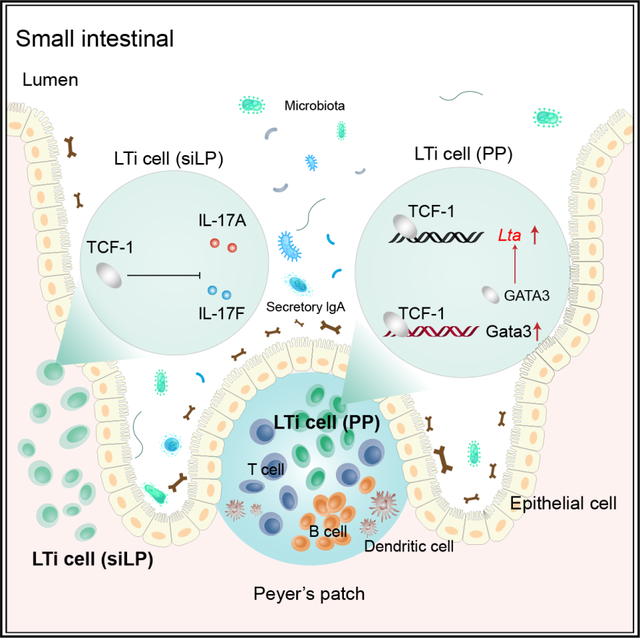
SUMMARY
Lymphoid tissue inducer (LTi) cells, a subset of innate lymphoid cells (ILCs), play an essential role in the formation of secondary lymphoid tissues. However, the regulation of the development and functions of this ILC subset is still elusive. In this study, we report that the transcription factor T cell factor 1 (TCF-1), just as GATA3, is indispensable for the development of non-LTi ILC subsets. While LTi cells are still present in TCF-1-deficient mice, the organogenesis of Peyer’s patches (PPs), but not of lymph nodes, is impaired in these mice. LTi cells from different tissues have distinct gene expression patterns, and TCF-1 regulates the expression of lymphotoxin specifically in PP LTi cells. Mechanistically, TCF-1 may directly and/or indirectly regulate Lta, including through promoting the expression of GATA3. Thus, the TCF-1-GATA3 axis, which plays an important role during T cell development, also critically regulates the development of non-LTi cells and tissue-specific functions of LTi cells.
In brief
Zheng et al. show that TCF-1 regulates the functions, but not the development, of LTi cells in different tissues. It is specifically required for optimal expression of lymphotoxin in LTi cells from Peyer’s patches and inhibits IL-17 production by LTi cells from small intestine laminal propria.
Graphical Abstract
INTRODUCTION
Lymphoid tissue inducer (LTi) cells, named after their functions in organogenesis of secondary lymphoid structures at the fetal stage, are the first reported innate lymphoid cell (ILC) population.1–3 Although CCR6-expressing LTi cells are classified into type 3 ILCs (ILC3s) characterized by the expression of the transcription factor RORγt,4 their development is distinct from the generation of the promyelocytic leukemia zinc finger protein (PLZF)/programmed cell death protein 1 (PD-1)-expressing ILC progenitors,5–7 which give rise to other non-LTi cells, including ILC1, ILC2, and conventional ILC3s, some of which express NKp46 but not CCR6 in mice.8 CCR6+ LTi/LTi-like cells reside mainly in lymphoid structures such as Peyer’s patches (PPs), crypto patches (CPs), and isolated lymphoid follicles (ILFs), whereas NKp46+ ILC3s are mostly located in the lamina propria (LP). These two RORγt-expressing ILC subsets also have different gene expression patterns and some unique functions.9,10 Recent studies have also shown that PLZF may be expressed by lymphoid tissue inducer progenitors (LTiPs)11,12; however, it is not known what is causing the discrepancy. Nevertheless, we have previously reported that the transcription factor GATA3 plays a critical role in determining the fate of LTi and non-LTi ILC progenitors.13 Consistent with the notion that LTi cells are distinct from conventional ILC3s, in the absence of GATA3, while PLZF-expressing ILC progenitors and NKp46+ ILC3s fail to develop, LTiPs seem to be normal. However, how GATA3 is regulated in this process is unknown.
T cell factor 1 (TCF-1; encoded by transcription factor 7, T cell specific [Tcf7]) was first identified as a T lymphocyte-specific transcription factor.14 Since its discovery almost 30 years ago, its critical roles in T-lineage specification and differentiation have been documented.15 Furthermore, TCF-1 upregulation defines early innate lymphoid progenitors (EILPs) in the bone marrow (BM) and is absolutely required for the generation of group 2 ILCs and NKp46+RORγt+ innate lymphocytes.16–19 However, the functions of TCF-1 in the development and functions of LTi cells, including those during the induction of multiple lymphoid organ formation, remain elusive.
PPs are important secondary lymphoid organs for the initiation of adaptive immune response in the gut, including immunoglobulin (Ig) A induction against gut microbes.20–23 PP organogenesis is tightly controlled at the fetal stage.24–26 Initially, lymphotoxin α1β2 (LT)-expressing LTi cells activate tissue-resident stroma lymphoid tissue organizer (LTo) cells through LTβR. In turn, these activated LTo cells attract more hematopoietic cells, and then the PP anlage develops with a growing mass of LTi and LTo cells, followed by the recruitment of B cells, T cells, and dendritic cells (DCs) into distinct zones of PPs. Unlike PPs, CPs and ILFs develop postnatally. CPs and ILFs develop under genetically programmed guidance24,27 as well as environmental inputs such as luminal microbial stimulation.
In this study, we describe an important role of the transcription factor TCF-1 in the development of PPs but not lymph nodes (LNs). Despite TCF-1 affecting ILC development at multiple stages, we found that LTi cells developed normally in TCF-1-deficient mice, indicating that the TCF-1-deficient LTi cells are specifically dysfunctional for the induction of PP formation. Strikingly, we found distinct gene expression patterns between LTi cells from the PPs and the small intestine laminal propria (siLP). TCF-1 specifically regulated the expression of lymphotoxin (LT) in PP LTi cells. LTβR agonist injection during the embryonic stage partially rescued the formation of PPs in TCF-1-deficient mice. Furthermore, mice with ILC3-specific TCF-1 deletion also had reduced numbers of PPs and these PP LTi cells expressed lower levels of LT compared to wild-type (WT) PP LTi cells. Further analysis of chromatin accessibility and binding targets of TCF-1 indicated that one of the mechanisms through which TCF-1 controls LT expression levels is via regulating GATA3 expression. Therefore, this conserved TCF-1-GATA3 pathway is utilized by multiple lymphocytes and their precursors in adopting cell fates and functions, and a dose effect of LT expression regulated by TCF-1 determines a differential regulation of organogenesis of distinct secondary lymphoid structures.
RESULTS
TCF-1 is indispensable only for the development of non-LTi ILCs
To study the role of TCF-1 in regulating ILC development and functions at multiple stages, we generated mice carrying Tcf7 conditional deficient allele by microinjection of targeted embryo stem cells purchased from the International Knockout Mice Consortium. We deleted Tcf7 at the hematopoietic stem cell stage by crossing Tcf7fl/fl to VavCre as we did for studying the GATA3 function in ILC development.28 The deletion efficiency was confirmed in refined common helper innate lymphoid progenitors (rChILPs)28 by flow cytometry (Figure S1A). We next examined the ILC progenitors in the BM and mature ILCs in the small intestine of WT and Tcf7fl/flVavCre mice. While rChILPs, ILC2 progenitors (ILC2Ps), and PLZF+ ILC progenitors (ILCPs) were dramatically reduced in Tcf7fl/flVavCre BM as previously reported,16,17 the percentage and cell number of CCR6+ LTiPs were increased (Figure 1A). Consistent with previous reports,17,18 NKp46+ ILC3s as well as ILC2s were dramatically reduced in the siLP of the Tcf7fl/flVavCre mice (Figure 1B). By contrast, the percentage of CCR6+ LTi-like cells was increased while the total cell number was unaltered in the small intestine (Figure 1B).
Figure 1. TCF-1 is critical for the development of non-LTi ILCs but not LTi cells.
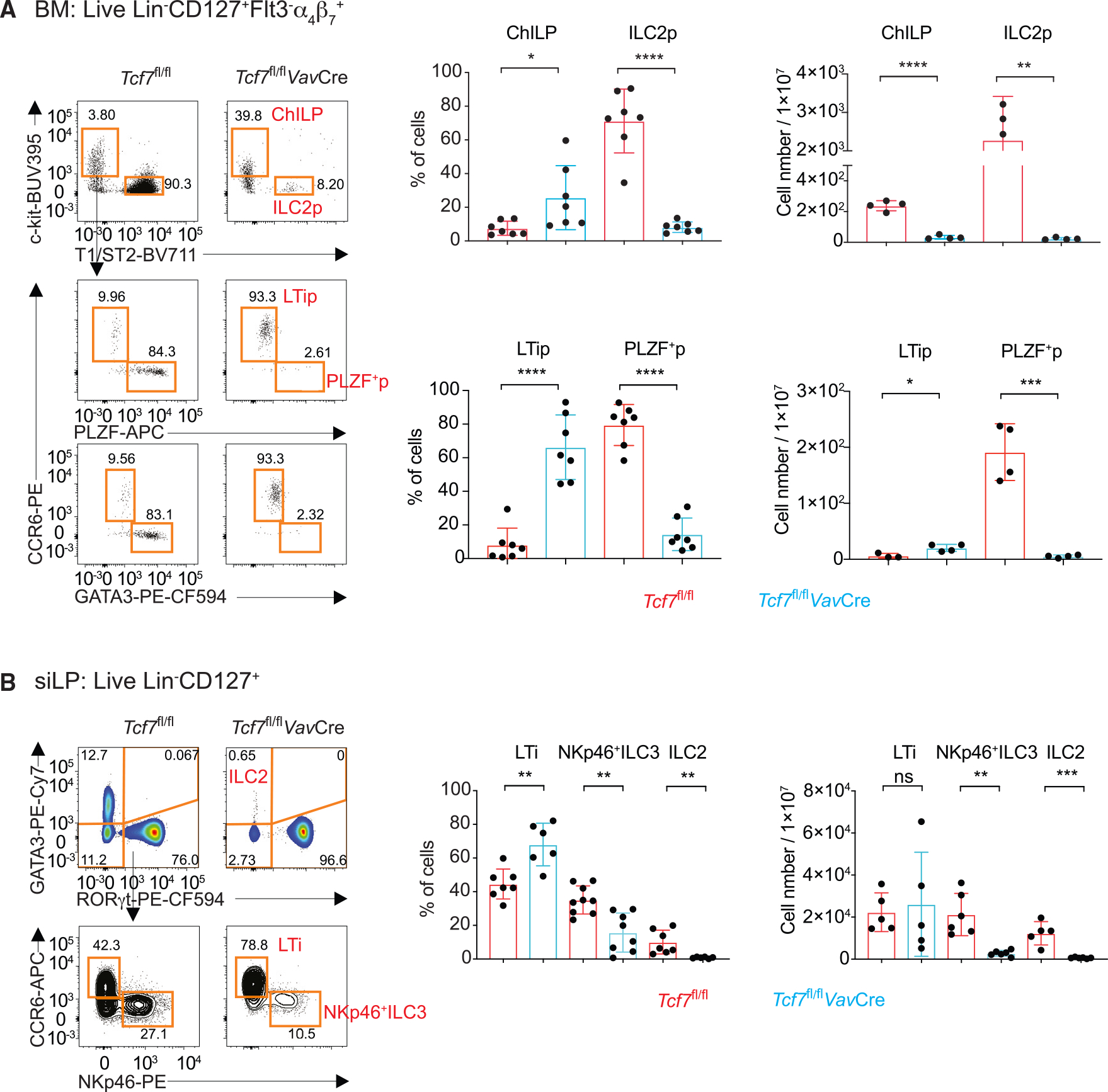
(A) BM cells were isolated from WT and Tcf7fl/flVavCre mice, and ILC2p, LTip, and PLZF+ ILC progenitor populations were analyzed by flow cytometry. Bar graphs show quantification in percentages and total cell numbers.
(B) ILC (Lin−CD127+) population in the siLP of the Tcf7fl/flVavCre mice were characterized by intracellular GATA3 and RORγt staining. The RORγt+ ILC3s were further analyzed based on their CCR6 and NKp46 expression. Bar graphs show quantification in percentages and total cell numbers. Bar graphs show quantification of total LTi cell numbers. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant, Student’s t test. Data are representative of at least three independent experiments. See also Figure S1.
We also found that TCF-1 was expressed on both CCR6+ LTiPs and ILCPs, although its expression was lower in CCR6+ LTiPs compared to ILCPs demonstrated either by antibody staining or by using the Tcf7-YFP reporter mice (Figures S1B and S1C). This expression pattern is very similar to that of GATA3 expression,13 indicating that TCF-1, just as GATA3, could have distinct functions for the development and functions of non-LTi ILCs and LTi cells.
Cell-intrinsic effect of TCF-1 on the development of ILC subsets
To further clarify the cell intrinsic role of TCF-1 during the development of ILC subsets, we performed a mixed BM chimera experiment where CD45.2+ WT (Tcf7fl/fl) or Tcf7fl/flVavCre BMs were mixed with WT CD45.1+ BM at 1:1 ratio and co-transferred into sublethally irradiated CD45.1+ Rag2−/−Il2rg−/− mice. Eight weeks later, CD45.2+ ILCPs and ILC2Ps were absent in BM (Figures S1D and S1E), as were CD45.2+ NKp46+ ILC3s and ILC2s in the small intestine (Figures S1F and S1G) of the chimeric mice that received Tcf7fl/flVavCre BM. By contrast, although CD45.2+ LTiPs in BM (Figures S1D and S1E) and LTi-like cells in small intestine (Figures S1F and S1G) were also reduced in these mice, these cells were still present. These results demonstrated a cell-intrinsic effect of TCF-1 during the development of non-LTi ILC subsets and a competitive disadvantage of Tcf7-deficient LTi cells in cell expansion and/or maintenance compared to WT LTi cells.
Single-cell transcriptome analysis of ILC progenitors confirmed a selective requirement of TCF-1 for the generation of ILCPs but not of LTiPs
To comprehensively investigate the selective functions of TCF-1 during ILC development, we performed single-cell RNA sequencing (scRNA-seq) of sorted live Lineage−CD127+α4β7+ Flt3− T1/ST2−c-Kit+ rChILPs from WT and Tcf7fl/flVavCre mice (Figure S2A). We pooled scRNA-seq data from 9,049 WT and 2,280 Tcf7fl/flVavCre rChILPs and used Uniform Manifold Approximation and Projection (UMAP) for dimension reduction. Unsupervised clustering assigned the WT and Tcf7fl/flVavCre rChILPs into eight clusters (clusters 0–7) (Figures 2A and S2B). Clusters 0 and 2 corresponded to GATA3hiPLZFhiId2hi ILCPs and GATA3+ PLZFlowId2− earlier progenitors, respectively, whereas CCR6+ cluster 3 represented LTiPs. Cluster 1 and 5 corresponded to Flt3+ earlier progenitors, with cluster 5 largely containing proliferating cells; clusters 4 and 6 represented Eomes+ natural killer precursors and NCR1+ NK cells, respectively. Cluster 7 represented myeloid cells (Figures 2A–2D, S2B, and S2C).
Figure 2. scRNA-seq analysis reveals altered gene expression in the TCF-1-deficient LTiPs.
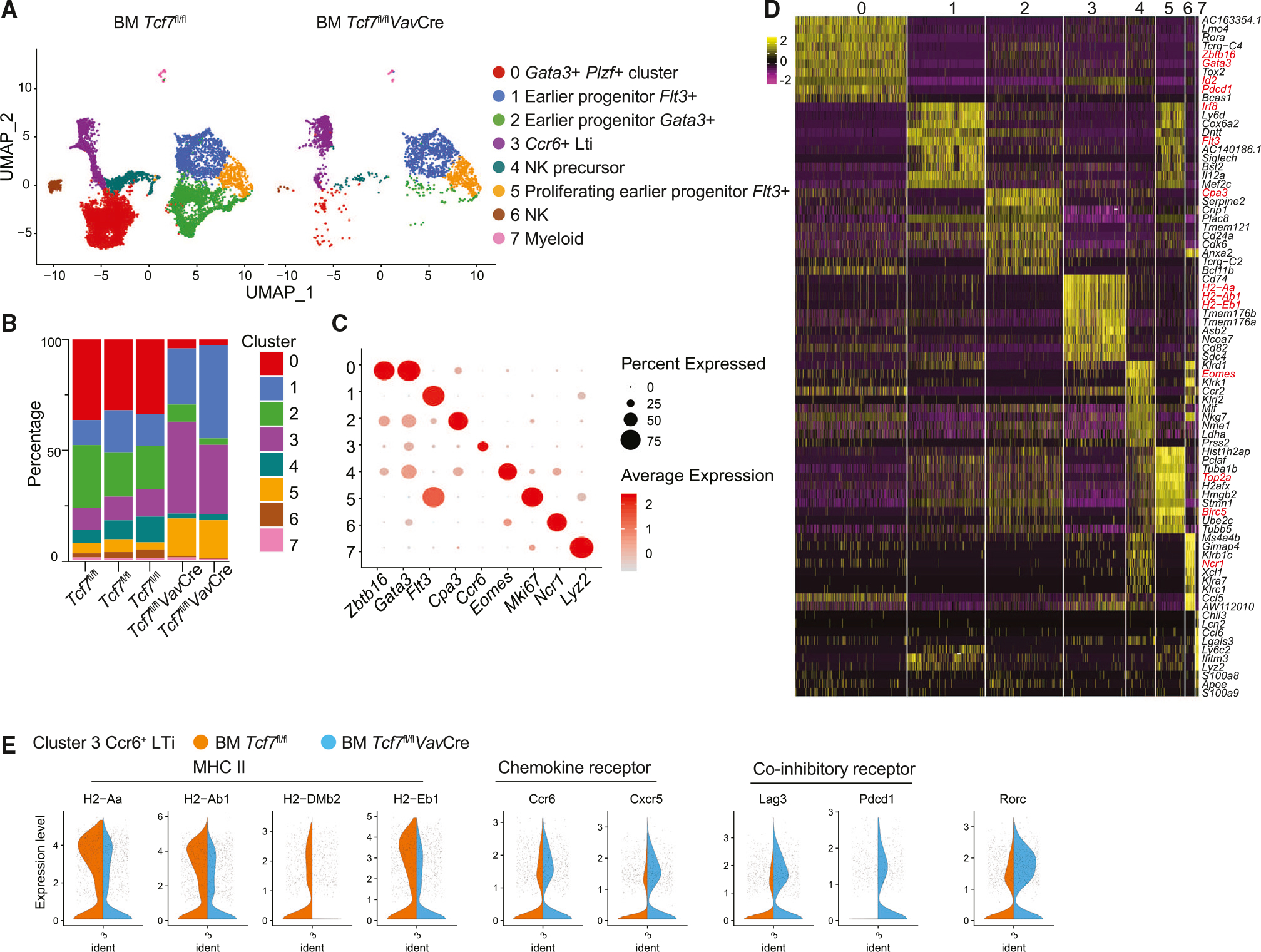
(A) The UMAP plot of ILCPs from WT and Tcf7fl/flVavCre mice. Each dot corresponds to one individual cell. A total of eight clusters (clusters 0–7) were identified and color coded.
(B) Percentages of each cluster in three WT mice and two Tcf7fl/flVavCre mice.
(C) Dot plots of gene ontology. Each row represents one cluster; each column represents a key gene. The average expression level is color-coded. The expression percentages are represented by the diameter of the circles.
(D) Heatmap of the top 10 genes expressed in each cluster as defined in (A). The columns correspond to the clusters; the rows correspond to the genes. Cells are grouped by clusters. The color scale is based on a Z score distribution from −2 (purple) to 2 (yellow).
(E) Violin plots of MHCII, chemokine receptor Ccr6 and Cxcr5, co-inhibitory receptor Lag3 and Pdcd1, and Rorc expression in WT and Tcf7fl/flVavCre LTi cells. The violin represents the probability density at each value. Data are representative of two independent experiments. See also Figure S2.
Notably, Tcf7 deficiency resulted in a dramatic reduction in the percentage of cluster 0 and 2 (ILCPs), as well as of cluster 4 and 6 (NK lineage). However, the percentages of cluster 1 and 5 (earlier progenitors) and cluster 3 (LTiPs) were increased among the Tcf7-deficient rChILPs when compared to the WT rChILPs (Figures 2A and 2B).
Interestingly, although the LTiPs are present in a relatively normal number in the Tcf7-deficient mice, these LTiPs had slightly altered gene expression pattern compared to their WT counterparts (Figures 2E and S2D). First of all, the Tcf7-deficient LTiPs had reduced expression of genes encoding MHC II, which is typically associated with antigen presentation (Figure 2E). Second, the expression of the chemokine receptors such as Ccr6 and Cxcr5 were increased in the Tcf7-deficient LTiPs, suggesting that TCF-1 might regulate the localization and migration of LTiPs. In addition, co-inhibitory molecules, including Lag3 and Pdcd1, were upregulated in Tcf7-deficient LTiPs (Figure 2E). These results suggest that, while TCF-1 is dispensable for the development of LTi cells, it may regulate their functions.
TCF-1 is required for organogenesis of PPs
Since LTi cells are important for lymphoid organogenesis, we assessed the lymphoid structures in the TCF-1-deficient mice. Strikingly, we found that PPs were almost absent in the small intestine Tcf7fl/flVavCre mice (Figures 3A and 3B), whereas other peripheral lymphoid organs were largely unaffected, including normal size of the spleen and marginally smaller mesenteric LNs and inguinal LNs (Figure S3A). All five LNs that drain different parts of intestine were found present in the mesenteric compartment of the Tcf7fl/flVavCre mice. In addition, CPs and ILFs that formed after birth detected by immunofluorescence staining were unaltered in the absence of TCF-1 (Figure S3B). We further assessed PPs by Swiss rolls histological experiment and found PPs were absent in TCF-1-deficient mice (Figure 3C). Since PPs are formed at the fetal stage, we also performed whole-mount staining of neonatal (day 1) intestine by RORγt staining, which confirmed the failure in PP formation at fetal stage in the absence of TCF-1 (Figure 3D). By analyzing digested fetal (embryonic day 16.5 [E16.5]) small intestine,29 we detected a significant but modest reduction in CCR6+ LTi cells in the Tcf7fl/flVavCre fetus (Figure 3E). By contrast, LTin cells30 were normal in number in these fetuses (Figure S3C).
Figure 3. TCF-1 is required for the formation of PPs, but not ILFs, in the small intestine.
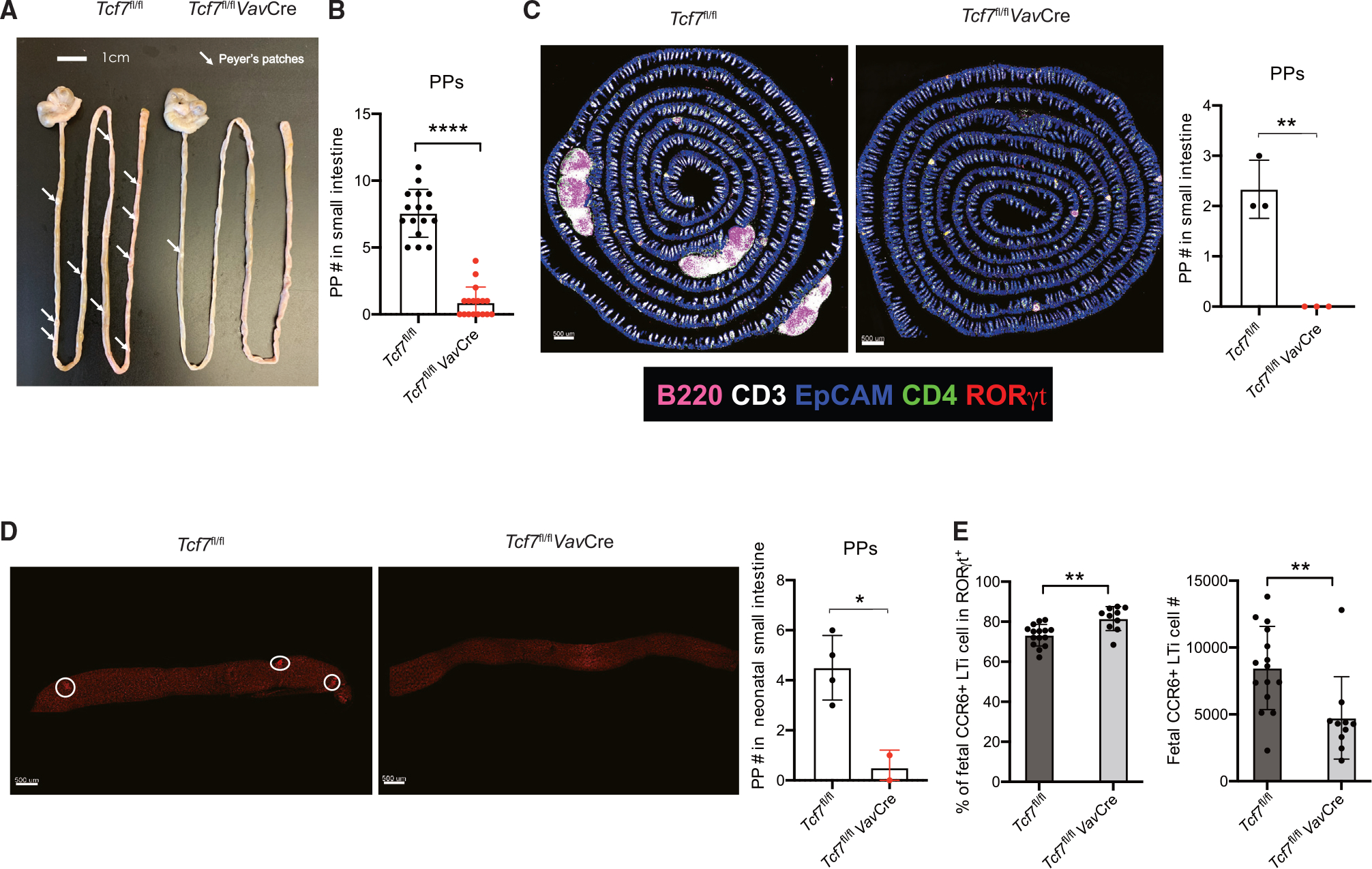
(A) Visualization of PPs from WT and Tcf7fl/flVavCre mice.
(B) Quantification of PPs from the mice in (A). Each symbol represents an individual mouse.
(C) Immunofluorescent staining of PPs in small intestine (left) from 6- to 8-week-old mice. Quantification of PPs (right). Each symbol represents an individual mouse.
(D) Visualization of PPs from neonatal (day 1) mice by immunofluorescent staining of RORγt. Each symbol represents an individual mouse.
(E) Cells were isolated from Tcf7fl/fl and Tcf7fl/flVavCre fetal (E16.5) intestine, and CCR6+ LTi population was analyzed; bar graphs show quantification in percentages and total cell numbers. Each symbol represents an individual mouse or fetus. mean ± SD; n = 15–16 in (B), 2–4 in (C)–(D), and 10–15 in (E); *p < 0.05, **p < 0.01, ****p < 0.0001, Student’s t test. Data are representative of at least three independent experiments. See also Figure S3.
TCF-1 is critical for T cell development and functions.15,31 Indeed, total T cell numbers were reduced in the thymus, spleen, and LN of Tcf7fl/flVavCre mice compared to the Tcf7fl/fl mice (Figures S3D–S3F). It is unlikely that the defective T cell development in the TCF-1-deficient mice resulted in the defect in PP formation in these mice since Tcrb−/−d−/− mice that lack all T cells had normal numbers of PPs compared to the WT mice (Figures S3G and S3H). These results indicate that TCF-1 is essential for the organogenesis of PPs, which is independent of its critical role during T cell development.
Impaired functions of PPs in young TCF-1-deficient mice
It has been reported that PPs are required for oral tolerance to proteins.32,33 To assess the functional consequences of missing PPs in the TCF-1-deficient mice, we transferred ovalbumin (OVA)-specific CD45.1+ Rag1−/− OT-II CD4 T cells into the WT or Tcf7fl/flVavCre mice and then fed them with drinking water with or without OVA for 1 week. Surprisingly, the proportion of OVA-specific cells that expressed Foxp3 were increased in the mesenteric LN (mLN) and spleen of both tolerized WT and Tcf7fl/flVavCre mice when compared to their untolerized counterparts (Figure S4A). The fact that Foxp3 can be efficiently induced in the TCF-1-deficient mice indicates that oral tolerance may occur outside of PPs.
PPs are also reported to be the predominant site for B cell immunoglobin class switch to generate IgA+ B220− plasma cells.34 Unexpectedly, IgA+ B220− plasma cells in the siLP of the 8-week-old WT and Tcf7fl/flVavCre mice showed no significant difference (Figure S4B). T cell-independent IgA bacteria coating35 was also similar in these mice (Figure S4C). Strikingly, even T cell-dependent fecal free IgA and serum IgA showed equivalent levels between WT and Tcf7fl/flVavCre mice (Figures S4D and S4E). Thus, IgA generation was not defective in the TCF-1-deficient adult mice.
It has been reported that ILFs can form in C57BL/6 mice 25 days after birth and ILFs may compensate for PPs in IgA production.36,37 To exclude a possible ILF compensatory effect, we analyzed IgA-producing cells in younger mice that were ~3–4 weeks old. Indeed, IgA+ B220− plasma cells in the siLP of young Tcf7fl/flVavCre mice were dramatically reduced compared to WT mice (Figure 4A). These TCF-1-deficient mice also showed a significant reduction in IgA production, as shown by reduced amounts of fecal IgA (Figure 4B) and serum IgA (Figure 4C), although bacterial coating of IgA was unaltered (Figure 4D). Therefore, the capacity of IgA production in young Tcf7fl/flVavCre mice is impaired consistent with the defect in PPs in these mice. While we cannot rule out the possibility that TCF-1 deficiency in T cells in these young mice may also contribute to the defective IgA production, the fact that the 8-week-old Tcf7fl/flVavCre mice produced normal levels of IgA makes this possibility unlikely.
Figure 4. Impaired IgA production in the small intestine of young Tcf7fl/flVavCre mice.
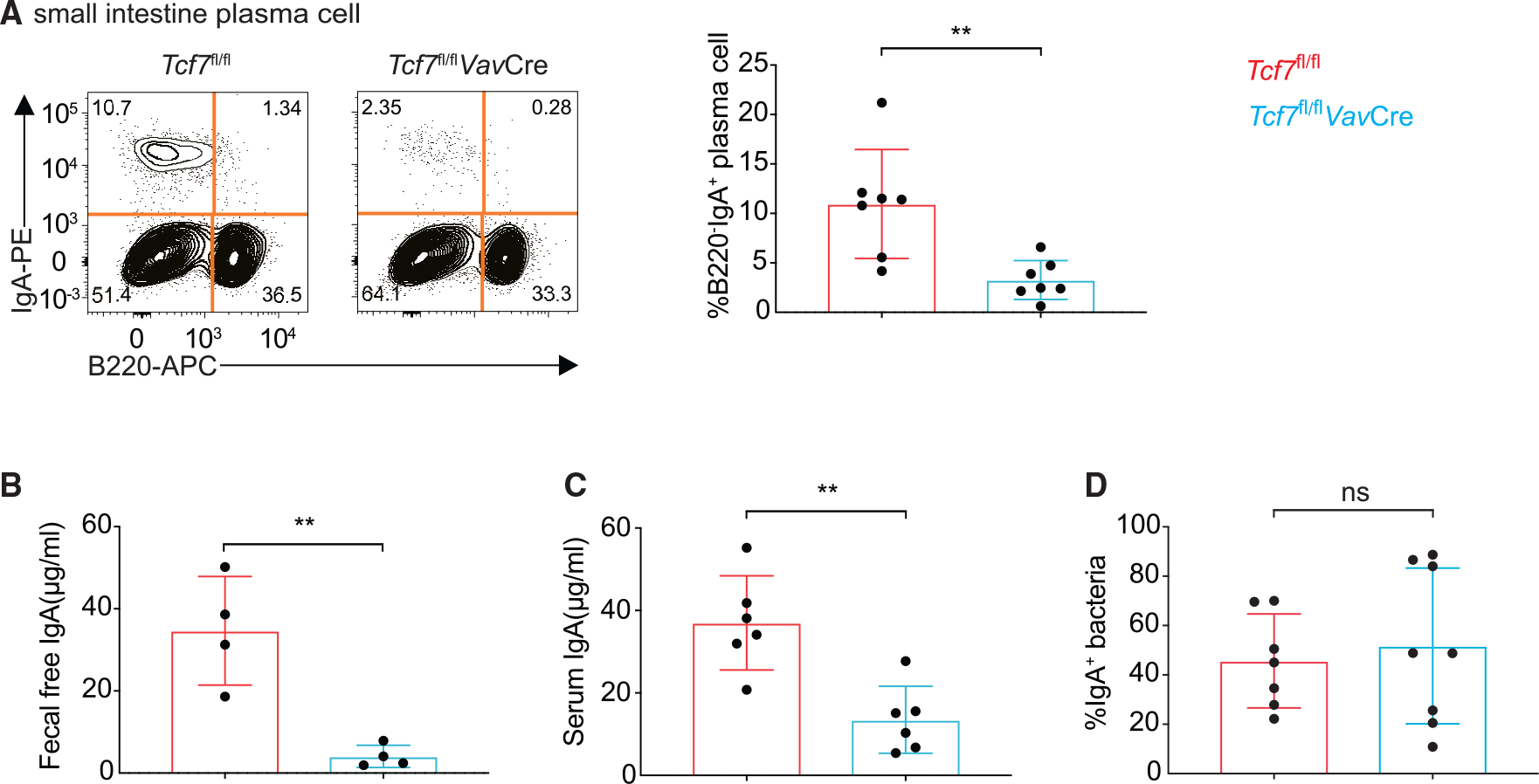
(A) Flow cytometric analysis and quantification of B220− IgA+ plasma cells in 4-week-old WT and Tcf7fl/flVavCre mice.
(B and C) Fecal free and serum IgA in above mice detected by ELISA assay.
(D) Quantification of IgA+ bacteria in above mice. Each symbol represents an individual mouse. Mean ± SD; n = 6–8; **p < 0.01; ns, not significant, Student’s t test. Data are representative of two independent experiments. See also Figure S4.
TCF-1 regulates optimal lymphotoxin expression in LTi cells
To further investigate why TCF-1 has a specific role in the PP organogenesis, we compared gene expression patterns in LTi cells from PPs, mLNs, and siLP through bulk RNA sequencing (RNA-seq) analysis38 (Figure 5A). Interestingly, gene expression analysis showed that many cytokines were differently expressed in LTi cells with different origins. Notably, while Il17a/f and Il22 were more abundant in siLP LTi cells, LTi cells from PPs and mLNs expressed higher levels of Lta and Ltb, which are critical for the formation of lymphoid tissues (Figure 5A). Principal-component analysis (PCA) also confirmed distinct gene expression patterns among LTi cells from PPs, mLNs, and siLP (Figure 5B). Flow cytometry analysis further validated that PPs LTi cells expressed higher levels of LT compared to mLN and siLP LTi cells (Figure 5C). These results indicate that gene expression patterns in LTi cells from PPs, mLNs, and siLP are highly tissue-specific. Top 100 genes that drive PC1 and PC2 specificity are listed in Table S1. Interestingly, siLP LTi cells from WT and knockout (KO) were closer to each other than to LTi cells from PPs or mLNs (Figure 5B). Nevertheless, the expression of Il17a/f and Pdcd1 was increased in Tcf7-deficient siLP LTi cells in the absence of TCF-1 (Figure 5D). In addition, a slight decrease of LT protein level was observed in TCF-1-deficient mLN LTi cells by flow cytometry analysis (Figures 5E and 5F).
Figure 5. Gene expression profiling of the LTi cells from different organs of WT and Tcf7fl/flVavCre mice.
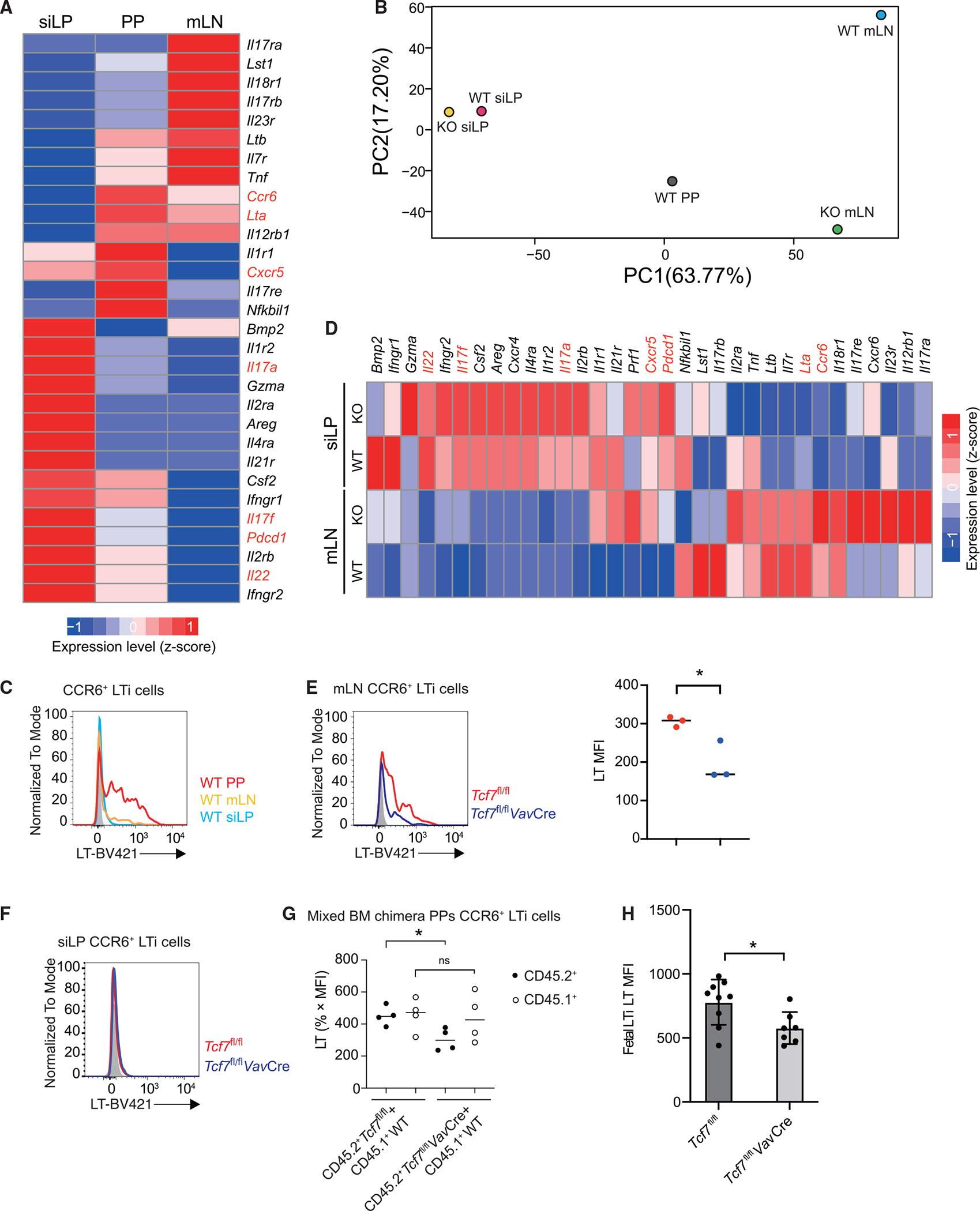
(A) RNA-seq analysis was performed to assess gene expression in LTi cells from siLP, PPs, and mLNs of WT mice.
(B) PCA analysis was performed to assess gene expression of LTi cells. Each group is in duplicates.
(C) Flow cytometric analysis of lymphotoxin expression in LTi cells from siLP, PPs, and mLNs of WT mice.
(D) RNA-seq analysis was performed to assess gene expression in LTi cells from siLP and mLNs of WT and Tcf7fl/flVavCre mice.
(E and F) Flow cytometric analysis of lymphotoxin expression in LTi cells from the mLN (E) and siLP (F) of WT and Tcf7fl/flVavCre mice. Bar graphs show quantification in mean fluorescence intensity (MFI) in (E).
(G) Flow cytometric analysis of lymphotoxin expression in LTi cells from the PPs of mix BM chimeras of CD45.1+ WT and CD45.2+ WT or Tcf7fl/flVavCre mice.
(H) Quantification of LT MFI from fetal intestinal LTi cells. Each symbol represents an individual mouse and/or embryo. Mean ± SD; n = 3 in (E), n = 4 in (G), and n = 7–9 in (H); *p < 0.05; ns, not significant, Student’s t test. Data are representative of at least three independent experiments (C–H). See also Figure S5.
Consistent with the RNA-seq results, PD-1 expression at the protein level was upregulated in the Tcf7-deficient LTi cells (Figures S5A and S5B). LTi cells can be divided into CD4+ and CD4− or major histocompatibility complex (MHC) II+ or MHCII− subsets.39–41 We verified such heterogeneity of LTi cells from siLP (Figure S5C) and found that TCF-1 deficiency affected the expression of MHCII but not CD4. However, in the absence of TCF-1, siLP LTi cells (whether MHCII+ or MHCII−) expressed higher levels of interleukin (IL)-17A (Figure S5D) and IL-17F (Figure S5E) but normal levels of IL-22 (Figure S5F). On the other hand, similar to the LT non-expressors, LT-expressing PP LTi cells consisted of both MHCII- and CD4-expressing cells (Figure S5G). These data indicate that, while TCF-1 affected MHCII-expressing LTi cells, the regulation of cytokine expression by TCF-1 (i.e., inducing LT expression in PP LTi cells but suppressing IL-17 expression in siLP LTi cells) is separated from TCF-1’s function in regulating MHCII.
To further assess LT expression by TCF-1-deficient LTi cells in the PPs, we performed a mixed BM chimera experiment. Indeed, LT expression by LTi cells in PPs from CD45.2+ Tcf7fl/flVavCre BM-reconstituted mice was reduced (Figure 5G). Furthermore, we examined the LT expression in fetal LTi cells and found it was reduced in the absence of TCF-1 (Figure 5H). Therefore, LT expression is largely limited in LTi cells within PPs and TCF-1 regulates its optimal expression.
TCF-1 expression in LTi cells is required for PP formation at an early stage
To further determine whether the lack of PP formation in the Tcf7fl/flVavCre mice is because of a defect in LTi cells, we crossed RorcCre mice with Tcf7fl/fl mice to generate ILC3-specific Tcf7 conditional KO mice. Indeed, TCF-1 was only deleted in ILC3s but not in ILC2s (Figure S6A). Interestingly, the number of PPs was significantly reduced in the Tcf7fl/flRorcCre mice (Figures 6A and 6B). LTi cell number was also decreased in the PPs of the Tcf7fl/flRorcCre mice (Figure 6C). Notably, while the LN and spleen seemed to be normal in the Tcf7fl/flRorcCre mice (Figure S6B), LT expression was significantly reduced in PP but not siLP LTi cells from the Tcf7fl/flRorcCre mice compared with those from WT controls (Figures 6C–6E).
Figure 6. The role of TCF-1 in PP formation is developmental stage specific.
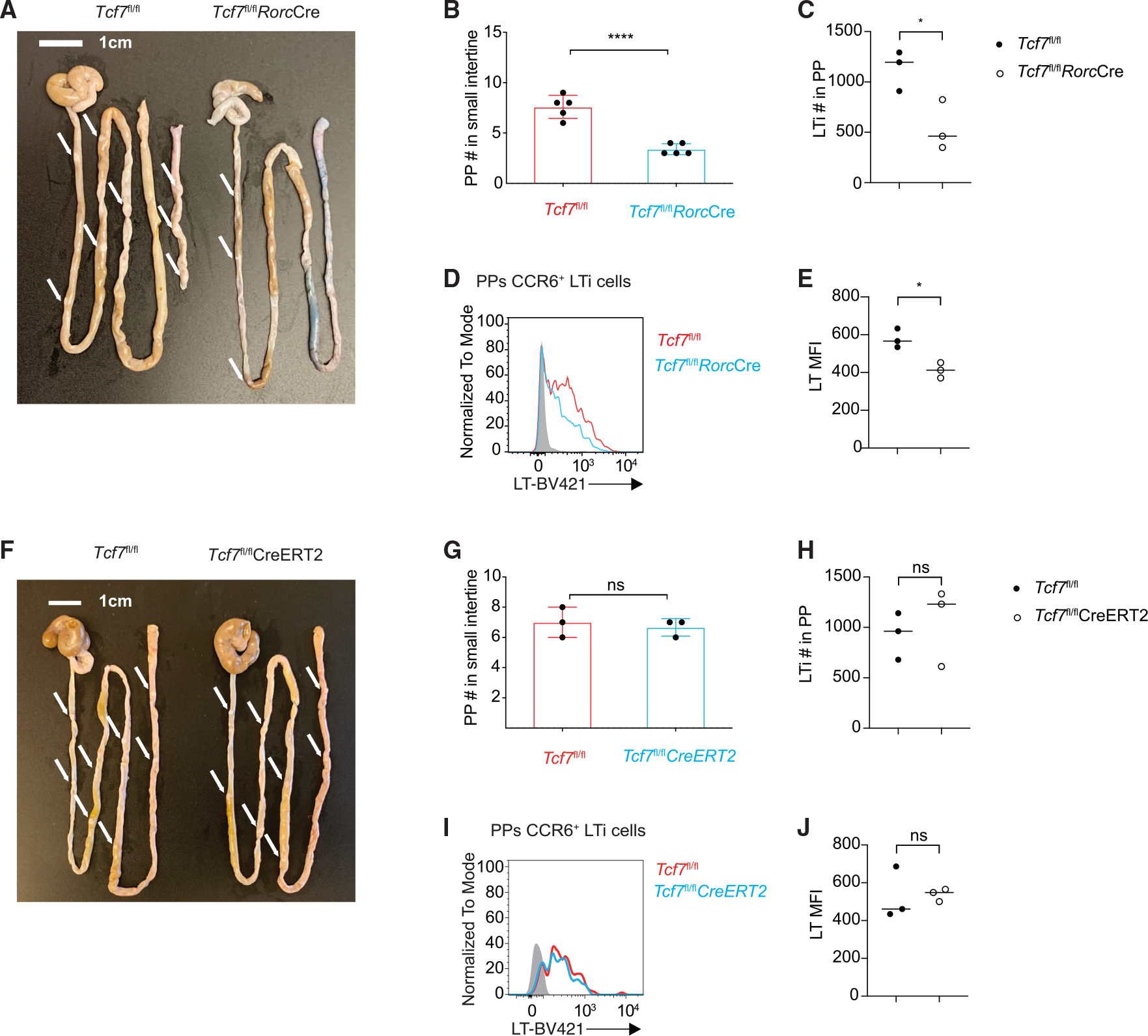
(A) Visualization of PPs from WT and Tcf7fl/flRorcCre mice.
(B) Quantification of PPs from the mice in (A). Each symbol represents an individual mouse.
(C) Bar graphs show quantification of total LTi cell numbers.
(D) Flow cytometric analysis of lymphotoxin expression in LTi cells from the PPs of WT and Tcf7fl/flRorcCre mice.
(E) Quantification of LT MFI from (D).
(F) Visualization of PPs from TMX-treated WT and Tcf7fl/flCreERT2 mice.
(G) Quantification of PPs from the mice in (F). Each symbol represents an individual mouse.
(H) Bar graphs show quantification of total LTi cell numbers.
(I) Flow cytometric analysis of lymphotoxin expression in LTi cells from the PPs of TMX-treated WT and Tcf7fl/flCreERT2.
(J) Quantification of LT MFI from (I). Each symbol represents an individual mouse. Mean ± SD; n = 4; *p < 0.05; ****p < 0.0001; ns, not significant, Student’s t test. Data are representative of two independent experiments. See also Figure S6.
To further test whether a continuous expression of TCF-1 in LTi cells is required for LTi functions, we crossed Tcf7fl/fl mice to CreERT2 mice, in which Tcf7 can be inducibly deleted at a later stage. Upon tamoxifen (TMX) injection, TCF-1 expression was abolished in both ILC3s and ILC2s (Figure S6D). Since PPs were formed at the prenatal stage, as expected, PPs, LNs, and spleen were normal when we intraperitoneally (i.p.) injected TMX to the 6- to 8-week-old mice (Figures 6F, 6G, and S6E). Also as expected, LTi cell number was unaltered in PPs of treated Tcf7fl/flCreERT2 mice (Figure 6H). Interestingly, LT expression in LTi cells from the PPs of treated WT and Tcf7fl/flCreERT2 mice was also comparable (Figures 6I, 6J, and S6F). These results strongly indicate an essential role of TCF-1 in PP organogenesis through regulating LT production at an early LTi developmental stage presumably via chromatin remodeling at the Lta/Ltb loci.
TCF-1 promotes LT expression and regulates GATA3 expression in PP LTi cells
To better understand the mechanism through which TCF-1 regulates LT expression, we performed RNA-seq analysis by comparing LTi cells from WT and Tcf7fl/flRorcCre mice, which have detectable, albeit reduced, PPs. Live lineage− CD127+ NK1.1− KLRG1− CCR6+ NKp46 LTi cells from siLP or PPs were collected by fluorescence-activated cell sorting (FACS) sorting (Figure S7). As expected, siLP LTi cells from the Tcf7fl/fl WT mice had less Lta and Ltb mRNA expression compared to PP LTi cells from the same mice (Figure 7A). In addition, a reduction in Lta but not Ltb mRNA expression by TCF-1-deficient PP LTi cells compared to WT PP LTi cells was noted. To assess whether TCF-1 directly regulates LT expression in LTi cells, we performed chromatin immunocleavage sequencing (ChIC-seq) analysis to profile TCF-1 binding sites in the genome. We also mapped open chromatin regions by using DNase sequencing (DNase-seq). ChIC-seq analysis showed a modest TCF-1 binding around the promoter of the Lta gene (Figure 7B, lower tracks). However, DNase-seq analysis did not reveal an obvious change in chromatin accessibility at the TCF-1 binding region between WT and Tcf7fl/flRorcCre PP LTi cells (Figure 7B, upper tracks). CreERT2-mediated deletion of Tcf7 did not result in a reduction in Lta expression (Figures 6I and 6J), suggesting that TCF-1 is not directly required for Lta transcription; instead, it is likely to modulate chromatin remodeling at the Lta gene, which is correlated with its significant binding to the Lta gene locus (Figure 7B).
Figure 7. TCF-1 directly and indirectly regulates Lta expression in PP LTi cells.
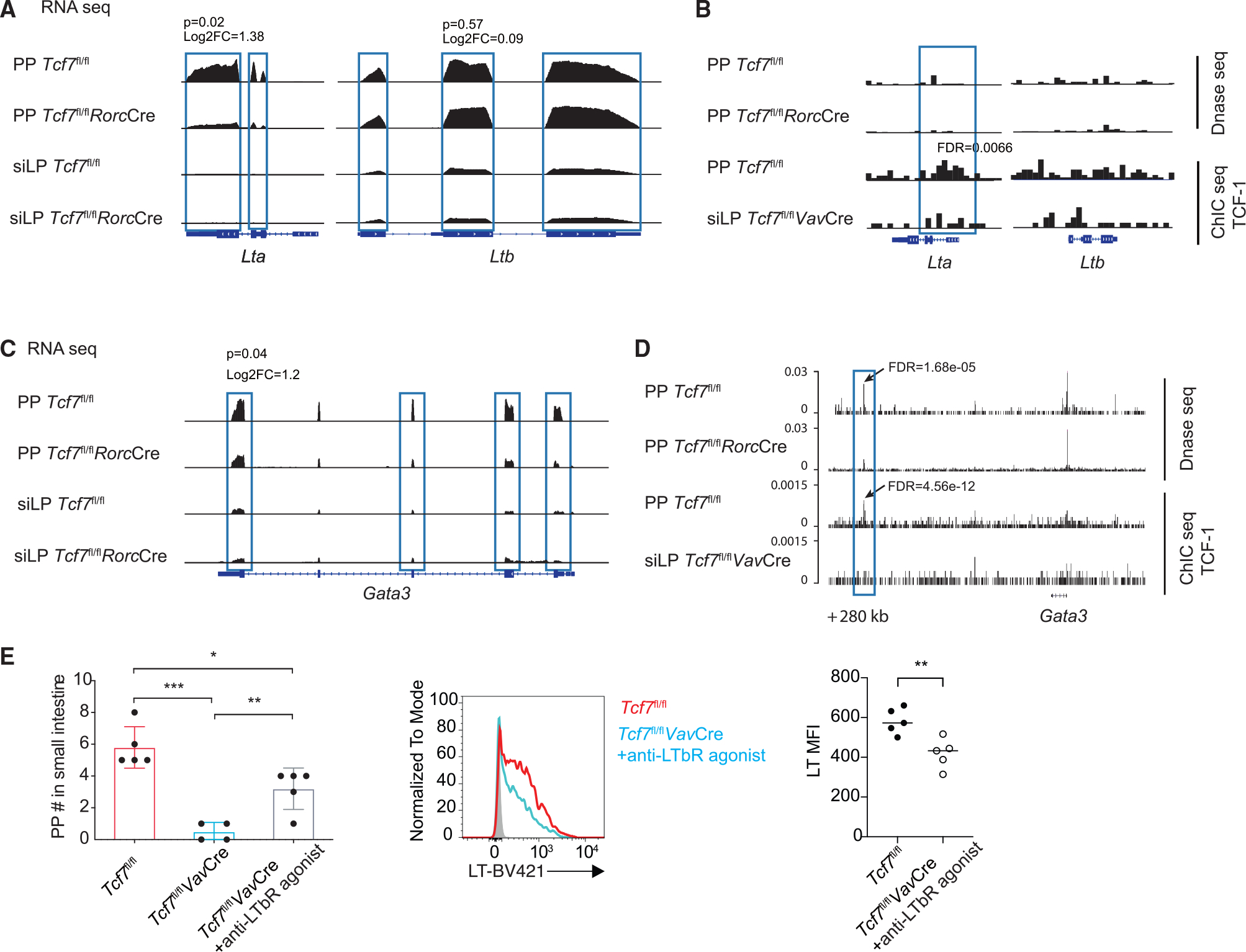
(A and C) Genome Browser snapshot showing the expression of Lta and Ltb (A) and Gata3 (C) gene in Tcf7fl/fl and Tcf7fl/flRorcCre siLP LTi cells by using RNA-seq assay.
(B and D) Genomic snapshots of the Lta/Ltb loci (B) and Gata3 (D) gene regions showing the chromatin accessibility status and density of TCF-1 ChIC-seq reads in PP LTi cells from Tcf7fl/fl and Tcf7fl/flRorcCre mice. Tcf7fl/flVavCre siLP LTi cells serve as negative control group.
(E) Quantification of PPs from LTβR agonist-treated WT and Tcf7fl/flVavCre mice. Each symbol represents an individual mouse. Flow cytometric analysis of lymphotoxin expression in LTi cells from the PPs of LTβR agonist-treated WT and Tcf7fl/flVavCre mice. Quantification of LT MFI. Mean ± SD; n = 4–5 in (E); *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test. Data are representative of two independent experiments. See also Figure S7.
Interestingly, we also observed GATA3 expression was lower in WT siLP LTi cells compared to WT PP LTi cells (Figure 7C). More importantly, GATA3 expression in PP LTi cells was reduced in the absence of TCF-1 (Figure 7C). Furthermore, TCF-1 was found to bind to the +280-kb region of the Gata3 locus in PP LTi cells (Figure 7D, lower tracks). At these same regions, a reduction in chromatin accessibility was also noted in the Tcf7fl/flRorcCre PP LTi cells compared to WT PP LTi cells (Figure 7D, upper tracks). It has been reported that GATA3 can regulate LT expression.13,42 Therefore, TCF-1 may also indirectly regulate LT expression through inducing GATA3 expression in PP LTi cells.
Since optimal LT signaling plays a critical role in PP organogenesis, we tested whether enhanced LT signaling by LTβR agonist injection39,43 can rescue the defect of PP formation in the absence of TCF-1. Indeed, PP organogenesis in the TCF-1-deficient mice was partially rescued by agonist treatment at the fetal stage, while the LT expression in PP LTi cells from these Tcf7fl/flVavCre mice was still defective (Figure 7E). Although we could not definitely prove that the defect in PP formation in TCF-1-deficient mice is solely due to an Lta defect, a partial rescue of PP formation in TCF-1-deficient mice by enhanced LT signaling is consistent with our hypothesis that quantitative regulation of optimal LT expression by TCF-1 in PP LTi cells is involved in the organogenesis of PPs.
DISCUSSION
The transcription factor TCF-1 plays an important role during the development of ILCs at multiple stages. In this study, we found that although TCF-1 is critical for the development of multiple ILC lineages, it is not required for LTi cell development. Consistent with the previous reports by Mielke and Yang that TCF-1 regulates PP organogenesis,16,18 we found that TCF-1 is specifically required for the formation of PPs but not other lymphoid structures. We further discovered that LTi cells from different tissues, including PPs, mLNs, and siLP, displayed distinct gene expression patterns. Lymphotoxins are highly expressed by PP LTi cells and TCF-1 regulates their optimal expression in these cells. Consequently, TCF-1-deficient animals specifically lack PPs but not other lymphoid structures. Mechanistically, TCF-1 may bind to the Lta gene and directly regulates its expression, possibly through chromatin remodeling. Furthermore, TCF-1 could indirectly regulate Lta expression through inducing optimal expression of GATA3 expression in PP LTi cells. A modest reduction in LTi cell number was also found in fetal intestine in the absence of TCF-1. Thus, the combination of a dose effect of LTa and GATA3 expression regulated by TCF-1 and a modest change in fetal LTi cell number may each contribute to differential regulation of organogenesis of distinct lymphoid structures.
TCF-1 is highly expressed by EILP cells and it is required for early ILC development.16 The transcription factor GATA3 has been reported as a TCF-1-regulated gene during both T cell development and ILC2 development. Although the role of GATA3 in EILP cells has not been investigated, the levels of GATA3 expression are critical for determining the LTi and non-LTi cell lineage commitment.13 In this study, we found that, like GATA3, TCF-1 is also specifically required for non-LTi ILC development but is dispensable for the development of LTi cells, which is consistent with its low levels of expression in LTiPs. Through scRNA-seq analysis, we identified the cluster 2 (TCF7hiGATA3low) as the earlier ILCPs, which may give rise to PLZF+Id2hi non-LTiPs that are also TCF7hiGATA3hi. The fact that both the TCF7hiGATA3low and TCF7hiGATA3hi populations are missing in TCF-1-deficient mice is consistent with the notion that TCF-1 acts upstream of GATA3 in regulating ILC development. Our previous work has shown that GATA3 is indispensable for all LN formation.13,28 In TCF-1 conditional KO mice, GATA3 expression was decreased but not absent. It is possible that the dose effect of GATA3 contributes to the failure of PP formation. In addition, TCF-1-deficient mice have normal ILFs which is different with the phenotype of Id2fl/flRorcCre mice,44 in which both PPs and ILFs in the gut are absent. Therefore, while Id2, GATA3, and TCF-1 have identical functions in regulating the generation of ILCPs, the interplay between these transcription factors may differ at different developmental stages of ILCs and LTi cells.
Through gene expression analysis, we also found that lymphotoxin is expressed at the highest levels in PP LTi cells compared to LTi cells from other tissues/organs. While LT is also expressed by LTi cells from mLN, LTi cells from siLP barely express lymphotoxin. By contrast, IL-17 and IL-22, which are critical for host defense, are preferably expressed by LTi cells from siLP but not PPs or mLN. It has been reported that CD4+ LTi cells are the dominant early source of IL-22 production in response to Citrobacter rodentium infection and thus promote innate immunity in the intestine.45 Thus, LTi cells from different lymphoid tissues exhibit distinct functions through the production of distinct cytokines. We also found that TCF-1 negatively regulates IL-17 expression by siLP LTi cells. The physiological importance of such regulation requires further investigation.
There is a dose effect of lymphotoxin expression for the formation of various lymphoid tissues. While the La−/− mice have neither PPs nor LNs, the Ltb−/− mice can form cervical LNs (cLNs) and mLNs but not PPs; strikingly, while all the LNs are present in the La+/−Ltb+/− mice, they lack PPs.46 The fact that TCF-1-deficient mice have a similar phenotype compared to the La+/−Ltb+/− mice is consistent with the effect of TCF-1 in regulating the optimal expression of LT in PP LTi cells.
In summary, similar to the role of GATA3 during ILC development, TCF-1 is also critical for the development of non-LTi ILCs; however, LTi cell development may occur in the absence of TCF-1. Nevertheless, TCF-1 regulates the functions of LTi cells from different lymphoid tissues. TCF-1 specifically promotes LT expression by PP LTi cells in which GATA3 is also highly expressed. On the other hand, TCF-1 represses IL-17 production by LTi cells from siLP. Therefore, the TCF-1-GATA3 axis not only determines LTi and non-LTi lineage commitment during early ILC development, it also modulates the functions of LTi cells in different tissues during lymphoid structure formation and in host defense.
Limitations of the study
Although we have provided a distinct insight into the tissue-specific regulation of LTi cell function by transcription factor TCF-1 in promoting different lymphoid tissue development, mechanistically, we cannot definitively conclude that the defect of PP formation is solely because of reduced Lta expression. It could be due to multiple reasons, including reduced Lta expression, reduced LTi cell numbers in fetal intestine, as well as reduced GATA3 expression, which has been shown to be important for regulating LTi functions. In addition, we observed a much severer defect in PP formation in the TCF-1 conditional KO mice both in adult and fetal small intestine compared to the TCF-1 germline KO mice. It is unknown what can possibly cause these differences. It is possible that the difference in animal facilities may cause such a difference; however, we have not directly compared our conditional KO with a germline KO in the same animal facility. Nevertheless, these limitations should not alter the main conclusions of our current study.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jinfang Zhu (jfzhu@niaid.nih.gov).
Materials availability
Tcf7fl/fl and Tcf7fl/fl-VavCre mouse strains are generated in this study.
Data and code availability
The RNA-Seq, DNase-Seq, ChIC-Seq and scRNA-Seq datasets have been deposited and are available at the Gene Expression Omnibus (GEO) database under the accession number GSE190302, which is also listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-CD117 (c-kit) BUV395 (2B8) | BD Biosciences | Cat# 564011; RRID: AB_2738541 |
| anti-LPAM-1 (α4β7) BV421 (DATK32) | BD Biosciences | Cat# 566294 |
| anti-CD335 (NKp46) PE (29A1.4) | BD Biosciences | Cat# 560757; RRID: AB_1727466 |
| anti-PLZF Alexa Fluor 647 (R17–809) | BD Biosciences | Cat# 563490; RRID: AB_2738238 |
| anti-ST2 BV711 (U29–93) | BD Biosciences | Cat# RRID: 745549; AB_2743075 |
| anti-GATA3 PE-CF594 (L50–823) | BD Biosciences | Cat# 563510; RRID: AB_2738248 |
| anti-GATA3 PE-Cy7 (L50–823) | BD Biosciences | Cat# 560405; RRID: AB_1645544 |
| anti-RORγt BV650 (Q31–378) | BD Biosciences | Cat# 564722; RRID: AB_2738915 |
| anti-RORγt PE-CF594 (Q31–378) | BD Biosciences | Cat# 562684; RRID: AB_2651150 |
| anti-TCF-1 PE (S33–966) | BD Biosciences | Cat# 564217; RRID: AB_2687845 |
| anti-CD196 (CCR6) APC (29–2L17) | Biolegend | Cat# 129814; RRID: AB_1877147 |
| anti-CD196 (CCR6) PE (29–2L17) | Biolegend | Cat# 129804; RRID: AB_1279137 |
| anti-CD127(IL-7Rα) PE-Cy7 (A7R34) | Biolegend | Cat# 135014; RRID: AB_1937265 |
| anti-CD127(IL-7Rα) BV421 (A7R34) | Biolegend | Cat# 135024; RRID: AB_11218800 |
| anti-CD45.1 BV711 (A20) | Biolegend | Cat# 110739; RRID: AB_2562605 |
| anti-CD45.2 Alexa Fluor 700 (104) | Biolegend | Cat# 109822; RRID: AB_493731 |
| anti-CD279 (PD-1) APC-Cy7 (29F.1A12) | Biolegend | Cat# 135224; RRID: AB_2563523 |
| anti-NK1.1 BV650(PK136) | Biolegend | Cat# 108735; RRID: AB_2563159 |
| anti-KLRG1 (MAFA) PE-Cy7 (2F1/KLRG1) | Biolegend | Cat# 138416; RRID: AB_2561736 |
| anti-CD3e PerCP-Cy5.5 (145–2C11) | Biolegend | Cat# 100328; RRID: AB_893318 |
| anti-CD4 BV421 (GK1.5) | Biolegend | Cat# 100438; RRID: AB_11203718 |
| anti-CD8a Alexa Fluor 700 (53–6.7) | Biolegend | Cat# 100730; RRID: AB_493703 |
| anti-B220 APC (RA3–6B2) | Biolegend | Cat# 103212; RRID: AB_312997 |
| anti-CD19 BV786 (6D5) | Biolegend | Cat# 115543; RRID: AB_11218994 |
| anti-CD3 Biotin (145–2C11) | Biolegend | Cat# 100304; RRID: AB_312669 |
| anti-CD19 Biotin (6D5) | Biolegend | Cat# 115504; RRID: AB_313639 |
| anti-Ter-119 Biotin (TER-119) | Biolegend | Cat# 116204; RRID: AB_313705 |
| anti-CD11b Biotin (M1/70) | Biolegend | Cat# 101204; RRID: AB_312787 |
| anti-CD11c Biotin (N418) | Biolegend | Cat# 117304; RRID: AB_313773 |
| anti-Ly-6G/Ly-6C (Gr-1) Biotin (RB6–8C5) | Biolegend | Cat# 108404; RRID: AB_313369 |
| anti-FcεRIα Biotin (MAR-1) | Biolegend | Cat# 134304; RRID: AB_1626106 |
| Streptavidin Brilliant Violet 785 (N/A) | Biolegend | Cat# 405249 |
| Streptavidin FITC (N/A) | Biolegend | Cat# 405202 |
| anti-γδ TCR Biotin (eBioGL3 (GL-3, GL3) | Thermofisher | Cat# 13–5711-82; RRID: AB_466668 |
| anti-CD135 (Flt3) PerCP-eFluor 710 (A2F10) | Thermofisher | Cat# 46–1351-82; RRID: AB_10733392 |
| anti-IgA PE (mA-6E1) | Thermofisher | Cat# 12–4204-82; RRID: AB_465917 |
| Fixable Viability Dye eFluor™ 506 (N/A) | Thermofisher | Cat# 65–0866-18 |
| LTβRIg antibody (AF.H6) | Biogen MA, Inc. | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 100 bp DNA Ladder | Thermofisher | Cat#15628019 |
| SYBRTM Safe DNA Gel Stain | ThermoFisher | Cat#S33102 |
| PlatinumTM Hot Start PCR Master Mix (2X) | ThermoFisher | Cat#13001012 |
| ACK Lysing Buffer | ThermoFisher | Cat#A1049201 |
| HBSS, calcium, magnesium | ThermoFisher | Cat#24020117 |
| PBS, pH 7.4 | ThermoFisher | Cat#10010031 |
| RPMI 1640 Medium | ThermoFisher | Cat#21870–076 |
| Fetal Bovine Serum | R&D Systems | Cat#S12450 |
| Penicillin-Streptomycin | ThermoFisher | Cat#15140122 |
| L-Glutamine (200 mM) | ThermoFisher | Cat#25030081 |
| MEM Non-Essential Amino Acids Solution (100X) | ThermoFisher | Cat#11140050 |
| Sodium Pyruvate | ThermoFisher | Cat#11360 |
| 2-Mercaptoethanol | ThermoFisher | Cat#21985023 |
| Phorbol 12-myristate 13-acetate | Sigma-Aldrich | Cat#P8139–5MG |
| Ionomycin | Sigma-Aldrich | Cat#407952–5MG |
| Brefeldin A Solution | BioLegend | Cat#420601 |
| eBioscienceTM Foxp3/Transcription Factor Staining Buffer Set | eBioscience | Cat#00–5523-00 |
| DNase I recombinant grade I, from bovine pancreas, expressed in Pichia pastoris | Roche | Cat#4536282001 |
| Liberase DH | Roche | Cat#5401020001 |
|
| ||
| Deposited data | ||
|
| ||
| Raw Data Files for RNA Sequencing | This paper | GEO: GSE190302 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Tcf7tm1a(EUCOMM)Wtsi | the University of California, Davis, KOMP Repository | www.mmrrc.org |
| Mouse: VavCre | JAX mice | Stock No: 008610 |
| Mouse: RorcCre | JAX mice | Stock No: 022791 |
| Mouse: CreERT2 | Taconic | line 10471 |
| Mouse: CD45.1+ | NIAID-Taconic repository | line 7 |
| Mouse: CD45.1+CD45.2+ | NIAID-Taconic repository | line 8422 |
| Mouse: CD45.1+ Rag2−/−il2rg−/− | NIAID-Taconic repository | line 8494 |
| Mouse: CD45.1+ Rag1−/− OT-II | NIAID-Taconic repository | line 361 |
| Mouse: Tcrb−/−d−/− | Dr. Amy Palin of NCI | N/A |
| Mouse: Tcf7-YFP mice | Dr. Bhandoola Avinash | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo v10 | Tree Star | https://www.flowjo.com |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
The embryonic stem (ES) cells carrying the Tcf7tm1a(EUCOMM)Wtsi allele on C57BL/6 background were obtained from the University of California, Davis, KOMP Repository (now also available at www.mmrrc.org). The ES cells were injected into C57BL/6 blastocysts and the resulting chimeric mice were genotyped per instructions of the IKMC project 37596. After successful germline transmission, the Tcf7+/flox-frt-neo mice were crossed with the Flpe mice (Taconic line 7089) to delete the FRT-lacZ-neo-FRT cassette in vivo. The resulting Tcf7fl/fl mice were then crossed to the VavCre mice (JAX mice Stock No: 008610), RorcCre mice (JAX mice Stock No: 022791) or CreERT2 (Taconic line 10471) to generate Tcf7fl/fl-VavCre, Tcf7fl/fl-RorcCre or Tcf7fl/fl-CreERT2, respectively. CD45.1+ congenic mice (line 7), CD45.1+CD45.2+ congenic mice (line 8422), CD45.1+Rag2−/−Il2rg−/− (line 8494) and CD45.1+Rag1−/−OT-II (line 361) were obtained from the NIAID-Taconic repository. Tcrb−/−d−/− mice were provided by Dr. Amy Palin of NCI, and Tcf7-YFP mice were previously described.16 All mice were bred and/or maintained in the National Institute of Allergy and Infectious Diseases specific pathogen-free animal facility. All experiments without specification used 6–16 weeks old female and male mice under an animal study protocol which was approved by the National Institute of Allergy and Infectious Diseases Animal Care and Use Committee.
METHOD DETAILS
Cell preparation
For isolation of lamina propria cells, small intestines from euthanized mice were emptied of the contents, excised of Peyer’s patches, opened longitudinally and cut into 1 cm pieces. The intraepithelial lymphocytes (IEL) were dissociated from the intestine fragments by first shaking the fragments for 20 min at 37°C in RPMI-1640 medium containing 3% FBS, 5 mM EDTA and 1 mM dithiothreitol, and then vortexing the fragments three times with RPMI-1640 medium containing 2 mM EDTA. To isolate lamina propria cells, the remaining fragments were minced and digested at 37°C for 30 min in RPMI-1640 medium containing 0.1 mg/mL Liberase and 10 U/ml DNase I. The digestion suspension was then filtered through a 40 μm strainer, centrifuged at 1,600 r.p.m. for 6 min, the cell pellet was washed twice and resuspended in HBSS containing 3% FBS for further analysis.
Single cell suspensions were generated by mashing the organs, including Peyer’s patches, mesenteric lymph nodes, spleen and thymus, through 40-μm strainers. Erythrocytes in the spleen single cell suspensions were lysed by resuspending the cell pellets in 1 mL ACK buffer for 5 min at room temperature (RT). The reaction was stopped by adding 10 mL of PBS with 2% FBS, cells were spun down immediatedly after washed in 10 mL ice-cold PBS. Dead cells were removed by filtering the lysed cells suspensions through 40-μm strainers.
Flow cytometry
Single-cell suspensions were preincubated with anti-CD16/32 (clone 2.4G2) for 10 min to block the surface Fc receptors. Then, cell-surface molecules were stained with different antibody combinations for 30 min in HBSS with 3% FBS. The stained cells were centrifuged at 1,600 r.p.m. for 6 min and resuspended in PBS with 3% FBS for flow cytometry analyses on an LSR Fortessa (BD Biosciences). For detection of LT by flow cytometry, cells were incubated with MOPC21 or LTβRIg (kindly provided by Biogen MA, Inc.) at 2 mg/mL for 60 min at 37°C in RPMI-1640 (ThermoFisher), 2% FBS (R&D), 1mM sodium pyruvate (ThermoFisher), 2mM L-glutamine (ThermoFisher), 100 IU/mL penicillin (ThermoFisher), 100 mg/mL, streptomycin (ThermoFisher), 1×non-essential amino acids (ThermoFisher) at no more than 5×106 cells per mL. Cells were washed twice in the RPMI solution, then stained for 30 min with surface antibodies including Rat anti-Mouse IgG1k at 200 ng/mL for 30 min at RT. Cells were then washed twice in RPMI buffer and were fixed with 1×fix/perm buffer (Invitrogen) at RT in the dark for 60 min, washed, stained by intracellular antibodies, washed, run on an LSR Fortessa flow cytometer (BD Biosciences). The flow cytometry data were analyzed with FlowJo software (Tree Star).
Antibodies
Antibodies specific to mouse γδ TCR Biotin (eBioGL3 (GL-3, GL3)), CD135 (Flt3) PerCP-eFluor 710 (A2F10), IgA PE (mA-6E1) amd Fixable Viability Dye eFluor506 were purchased from ThermoFisher; antibodies specific to mouse CD196 (CCR6) APC (29–2L17),CD196 (CCR6) PE (29–2L17), CD127(IL-7Rα) PE-Cy7 (A7R34), CD127(IL-7Rα) BV421 (A7R34), CD45.1 BV711 (A20), CD45.2 Alexa Fluor 700 (104), CD279 (PD-1) APC-Cy7 (29F.1A12), NK1.1 BV650 (PK136), KLRG1 (MAFA) PE-Cy7 (2F1/KLRG1), CD3e PerCP-Cy5.5 (145–2C11), CD4 BV421 (GK1.5), CD8a Alexa Fluor 700 (53–6.7), B220 APC (RA3–6B20), CD19 BV786 (6D5), CD3 Biotin (145–2C11), CD19 Biotin (6D5), Ter119 Biotin (TER119), CD11b Biotin (M1/70), CD11c Biotin (N418), Ly-6G/Ly-6C (Gr-1) Biotin (RB6–8C5), FcεRIα Biotin (MAR-1), Streptavidin Brilliant Violet 785 and Streptavidin FITC were purchased from BioLegend; and antibodies specific to mouse anti-CD117 (c-kit) BUV395 (2B8), LPAM-1αα4β7βBV421 (DATK32), CD335 (NKp46) PE (29A1.4), PLZF Alexa Fluor 647 (R17–809), ST2 BV711 (U29–93), GATA3 PE-CF594 (L50–823), GATA3 PE-Cy7 (L50–823), RORγt BV650 (Q31–378), RORγt PE-CF594 (Q31–378) and TCF-1 PE (S33–966) were purchased from BD Biosciences.
Bone marrow transplant
The congenically marked WT bone marrow cells (CD45.1+) were mixed with WT or Tcf7fl/flVavCre (CD45.2+) bone marrow cells at 1:1 ratio and intravenously injected into 6–8 week-old sublethally irradiated (550 rad) CD45.1+Rag2−/−Il2rg−/− or CD45.1+CD45.2+ WT recipients. Repopulated T cells, B cells and ILCs in the recipients were analyzed six to eight weeks after bone marrow transplant.
Immunofluorescent staining
The ileum of the small intestine was prepared as a ‘Swiss roll’ and was treated for 12 h with fixation and permeabilization solution (554722; BD Bioscience), followed by dehydration in 30% sucrose, before being embedded in OCT freezing medium (Sakura Fine-tek). Sections 18 μm in thickness were cut on a CM3050S cryostat (Leica) and were made to adhere to Superfrost Plus slides (VWR). Frozen sections were permeabilized and then blocked in PBS containing 0.1% Triton X-100 (Sigma) and 10% normal mouse serum (Jackson Immunoresearch), followed by staining in PBS containing 0.01% Triton X-100 and 5% normal mouse serum. RORγt was stained with monoclonal antibody to human and/or mouse RORγt (AFKJS-9; eBioscience), followed by Alexa Fluor 555-conjugated goat antibody to rat (anti-rat; A11006; Molecular Probes). Mouse epithelial cell adhesion molecule (EpCAM) and CD3ε, B220 were stained with Alexa Fluor 647-conjugated rat anti-mouse EpCAM (G8.8; BD Pharmingen) and BV421 rat anti-mouse CD3ε (17A2; BioLegend), Alexa Fluor 488-conjugated anti-B220 (RA3–6B2; BioLegend) respectively. After staining, slides were mounted with Fluormount G (Southern Biotech) and examined on a Leica TCS SP8 confocal microscope. Images were analyzed by Imaris software (Bitplane).
Bacterial flow cytometry and antibodies
The small intestine contents were removed by running forceps along a given intestinal segment and placed in a 1.5 mL Eppendorf tube on ice. Contents were then weighed and resuspended at 0.1 mg/μL in phosphate-buffered saline (PBS; Corning) with protease inhibitors (SIGMAFAST Protease Inhibitor Tablets). Contents were homogenized by taping the Eppendorf tube horizontally on a vortex and vortexing vigorously for 5 min. Tubes were centrifuged at 400g for 5 min to pellet large debris and the supernatant was filtered through a sterile 70 μm cell strainer (Fisher) and transferred to a new tube. This tube was spun at 8000g for 5 min to pellet bacteria. Supernatant was saved and frozen at −20°C for free IgA analysis by ELISA. Bacterial pellets were resuspended in PBS containing 20% Normal Rat Serum and SYTO BC (Life Technologies) for 15 min on ice. Samples were washed and resuspended in PBS with PE-conjugated anti-mouse IgA (mA-6E1; ThermoFisher) for 20 min on ice. Samples were washed and resuspended in PBS for flow cytometry analysis. Flow cytometry was performed on a Fortessa Flow Cytometer (Becton Dickinson) with a low FSC and SSC threshold to allow bacterial detection. FSC and SSC were set to a Log scale and samples were gated FSC+SSC+SYTO BC+ and then assessed for IgA staining.
Free IgA measurement and ELISA
Intestinal supernatants containing free IgA were isolated as described above in Bacterial Flow Cytometry and Antibodies. ELISA’s were performed according to the protocol included in the Bethyl Mouse IgA Quantitation Set. Briefly, ELISA plates (Bethyl) were coated with goat anti-mouse IgA (Bethyl) and washed with an ELISA plate washer. Plates were then blocked and washed. Experimental samples and standards were added to wells and incubated for 1 h at RT. All samples were run in duplicates and were serially diluted along with the plate. Plates were then washed and incubated with goat anti-mouse IgA HRP (Bethyl). Plates were washed again and developed in the dark using TMB substrate (Bethyl) and 0.18M H2SO4 to stop the reaction. Plates were read using an ELISA plate reader (Bio-Tek) and sample values quantified by comparison to standards fit to a four-parameter logistic curve.
Visualization and induction of PPs
Small intestines were submerged for 5 min in a 7% (v/v) solution of acetic acid in PBS. To induce PPs in the progenies of the TCF-1 deficient mice, pregnant females were injected with 250 μg of agonist anti-LTβ-R MAb AF.H6 (kindly provided by Biogen MA, Inc.) via the tail vein beginning on gd 12 or 13, followed by additional injections i.v. every 48 h.39,43,47 In all experiments, we avoided daily dosing to alleviate stress on the pregnant mice. Representative progenies were examined for the presence of Peyer’s patches at 4 weeks of age.
Adoptive transfer and antigen specific treg assessment
Naive CD4+ T cells were isolated from CD45.1+Rag1−/−OT-II mice and 1×106 of these cells were injected i.v. into WT or Tcf7fl/flVavCre recipients. One day later, mice were provided with OVA at 4 mg/mL ad libitum in drinking water; controls received normal drinking water. After 7 days, tissues were harvested and analyzed by flow cytometry.
RNA-seq analysis
RNA-Seq was performed as described48 with some modifications. Total RNA from 3K sorted cells was extracted with the miRNAeasy Micro kit (1071023; Qiagen). Total RNAs from 1K cells were reverse transcribed and pre-amplified by PCR using KAPA HiFi Hotstart Ready Mix (Roche) with IS PCR for 12 cycles. PCR products were purified by Ampure XP beads (Beckman Coulter) and eluted with 17.5 μl nuclease-free water as described in Smart-seq2 method. cDNA was sonicated to 200–400 bp by Bioruptor Pico (Diagenode) for 20 cycles (30 s on and 30 s off). Sonicated cDNA was blunt-ended, ligated to the Solexa adaptors, amplified, and sequenced with an Illumina HiSeq-3000 system, and pair end 50-bp reads were generated by the National Heart, Lung, and Blood Institute DNA Sequencing and Computational Biology Core. RNA-Seq reads were mapped to the mm10 genome (UCSC Genome Browser). Gene expression levels were measured by reads per kilobase of exon per million reads (RPKM). Differentially expressed genes were identified by edgeR49 with the following criteria: false discovery rate (FDR) < 0.05, fold change ≥2.
DNase-seq analysis
DNase-Seq assays were performed as described.19,50 Briefly, 300 cells from each cell type were collected by fluorescence-activated cell sorting. Then, 40 μl lysis buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3nM MgCl2 0.1% (v/v) Triton X-100) contain 0.3 U DNase I (Cat#: 04716728001; Roche) was added to each samples and incubated at 37°C for 5 min. Reactions were stopped by adding 80 μL of stop buffer (10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 0.15% SDS and 10 mM EDTA) containing 1 μL of 20 mg/mL proteinase K. Samples were incubated at 55°C overnight, and DNA was purified by phenol–chloroform extraction followed by precipitation with ethanol in the presence of 20 μg glycogen. DNA was further processed for sequence library preparation.
Small cell number ChIC-Seq analysis
ChIC-Seq was performed as described.19,51 500 cells for each sample were fixed by adding a 1:15 volume of 16% w/v formaldehyde solution (ThermoFisher Scientific) and incubating at RT with rotation for 10 min. Reactions were terminated by adding a 1:10 volume of 1.25M glycine and incubating on ice for 5 min. The fixed cells were washed with 1 mL ice-cold phosphate buffered saline (PBS), and collected by centrifugation. Then, 1 mL RIPA buffer (10 mM Tris-Cl,1 mM EDTA, 0.1% Sodium Deoxycholate, 0.2% Sodium Dodecyl sulfate and 1% Triton X-100) was added to each sample, and incubated at RT for 5 min with rotation. Then, cell pellets were rinsed with 500 μL binding buffer (10 mM Tris-Cl, 1 mM EDTA, 150 mM Sodium Dhloride and 0.1% Triton X-100) twice and resuspended in 50 μL binding buffer. To prepare anti-TCF-1-bound protein A–micrococcal nuclease (pA-MNase; antibody + PA-MNase), anti-TCF-1 (C46C7; Cell Signaling) and the PA-MNase at a molecular ratio of 1:2 were pre-incubated at 4°C for 30 min in 50 μL binding buffer. TCF-1 + PA-MNase were added to 50 μL binding buffer-resuspended cells and incubated for 1 h at 4°C with rotation. Cells were washed using 200 μL wash buffer (10 mM Tris-Cl, 1 mMEDTA, 150 mM Sodium Chloride, 0.1% w/v SDS, 0.1% w/v Sodium Deoxycholateand 1% v/v Triton X-100) three times and pelleted by centrifugation at 600g for 2 min. Next, cells were rinsed using 200μL rinsing buffer (10 mM Tris-Cl, 10 mM Sodium Chloride and 0.1% v/v Triton X-100). The MNase digestion was initiated by resuspending the rinsed cells in 40 μL reaction solution buffer (10 mM Tris-Cl,10 mM Sodium Chloride, 0.1% v/vTritonX-100 and 2 mMCaCl2) and incubating at 37°C for 3 min. The reaction was stopped by adding 80 μL stop buffer (20 mM Tris-Cl (pH 8.0), 10 mM Ethylenedioxy-bis-(ethylenenitrilo)-tetraacetic Acid, 20 mM Sodium Chloride and 0.2% w/v SDS) and 1 μL proteinase K (Sigma–Aldrich), then incubating at 65°C overnight. DNA was purified using phenol–chloroformextraction and ethanol precipitation. The purified DNA was further processed for HiSeq-3000 library construction.
scRNA-seq analysis
The scRNA-Seq libraries were generated using Chromium Single Cell 3′ Library & Gel BeadKit v.2 (10x Genomics) according to the manufacturer’s protocol as previous described.52 Briefly, cells were FACS-sorted and used to generate single-cell gel-beads in emulsion. After reverse transcription, gel-beads in emulsion were disrupted. Barcoded complementary DNA was isolated and amplified by PCR (12 cycles). Following fragmentation, end repair and A-tailing, sample indexes were added during index PCR (8 cycles). The purified libraries were sequenced on a NovaSeq 6000 (Illumina) with 26 cycles of read 1, 8 cycles of i7 index and 98 cycles of read 2. Alignment, filtering, barcode counting and unique molecular identifier counting were performed using Cell Ranger v.3.0.2. Data were further analyzed using Seuratv.3.3.2.Briefly, cells with a percentage of mitochondrial genes below 0.05% were included. Cells with the highest (top0.2%) or lowest (bottom 0.2%) numbers of detected genes were considered as outliers and excluded from the downstream analyses. Raw unique molecular identifier counts were normalized to unique molecular identifier count per million total counts and log-transformed. Principal component analysis was performed using top 2,000 variable genes. Clusters and UMAP plots were generated based on selected principal component analysis dimensions. Marker genes were identified by the Seurat function FindAllMarkers. Scaled expression data of these marker genes were used for creating the heatmaps. Normalized data are shown in the form of feature plots or violin plots.
Accession numbers. RNA-Seq, DNase-Seq, ChIC-Seq and scRNA-Seq data are available in the Gene Expression Omnibus (GEO) database under accession number GSE190302.
QUANTIFICATION AND STATISTICAL ANALYSIS
Samples were compared with Prism 7 software (GraphPad) by a two-tailed unpaired Student’s t-test. Data were presented as mean ± s.d. A p value <0.05 was considered statistically significant and indicated as *; p < 0.01 was indicated as **; p < 0.001 was indicated as ***; and p < 0.0001 was indicated as ****. Not statistically significant was indicated as ns.
Supplementary Material
Highlights.
TCF-1 is required for LTi’s function selectively in Peyer’s patch (PP) formation
TCF-1 promotes optimal expression of lymphotoxin and GATA3 in PP LTi cells
LTi cells from PPs and small intestine laminal propria (siLP) are transcriptionally distinct
TCF-1 negatively regulates IL-17 expression in siLP LTi cells
ACKNOWLEDGMENTS
This work is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. M.Z. is also supported by the National Natural Science Foundation of China (82171717), the Fundamental Research Funds for the Central Universities (2242022K4007), and Jiangsu Distinguished Professor Foundation. G.H. is supported by a WV-INBRE grant (P20 GM103434) and NIGMS grant (U54 GM-104942). We thank Yu Zhang from Southeast University for assistance in preparing the graphical abstract.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112924.
REFERENCES
- 1.Diefenbach A, Colonna M, and Koyasu S (2014). Development, differentiation, and diversity of innate lymphoid cells. Immunity 41, 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artis D, and Spits H (2015). The biology of innate lymphoid cells. Nature 517, 293–301. [DOI] [PubMed] [Google Scholar]
- 3.Spits H, and Di Santo JP (2011). The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27. [DOI] [PubMed] [Google Scholar]
- 4.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, and Littman DR (2004). An essential function for the nuclear receptor ROR gamma t in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73. [DOI] [PubMed] [Google Scholar]
- 5.Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A (2014). A committed precursor to innate lymphoid cells. Nature 508, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Y, Tsang JCH, Wang C, Clare S, Wang J, Chen X, Brandt C, Kane L, Campos LS, Lu L, et al. (2016). Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature 539, 102–106. [DOI] [PubMed] [Google Scholar]
- 7.Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, and Golub R (2016). Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 17, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klose CSN, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, and Diefenbach A (2013). A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature 494, 261–265. [DOI] [PubMed] [Google Scholar]
- 9.Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS, Mack M, Cella M, and Colonna M (2015). Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212, 1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong C, Cui K, Wilhelm C, Hu G, Mao K, Belkaid Y, Zhao K, and Zhu J (2016). Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 17, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, Liu P, Golub R, and Di Santo JP (2019). An Id2(RFP)-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity 50, 1054–1068.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker JA, Clark PA, Crisp A, Barlow JL, Szeto A, Ferreira ACF, Rana BMJ, Jolin HE, Rodriguez-Rodriguez N, Sivasubramaniam M, et al. (2019). Polychromic Reporter Mice Reveal Unappreciated Innate Lymphoid Cell Progenitor Heterogeneity and Elusive ILC3 Progenitors in Bone Marrow. Immunity 51, 104–118.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong C, Zheng M, Cui K, Martins AJ, Hu G, Li D, Tessarollo L, Kozlov S, Keller JR, Tsang JS, et al. (2020). Differential Expression of the Transcription Factor GATA3 Specifies Lineage and Functions of Innate Lymphoid Cells. Immunity 52, 83–95.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Wetering M, Oosterwegel M, Dooijes D, and Clevers H (1991). Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber BN, Chi AWS, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, and Bhandoola A (2011). A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, Wang H, Wang X, Yu S, Zhou X, et al. (2015). TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16, 1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Monticelli LA, Saenz SA, Chi AWS, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, et al. (2013). T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38, 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, and Belz GT (2013). TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 191, 4383–4391. [DOI] [PubMed] [Google Scholar]
- 19.Harly C, Kenney D, Ren G, Lai B, Raabe T, Yang Q, Cam MC, Xue HH, Zhao K, and Bhandoola A (2019). The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat. Immunol. 20, 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reboldi A, and Cyster JG (2016). Peyer’s patches: organizing B-cell responses at the intestinal frontier. Immunol. Rev. 271, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fagarasan S, and Honjo T (2003). Intestinal IgA synthesis: regulation of front-line body defences. Nat. Rev. Immunol. 3, 63–72. [DOI] [PubMed] [Google Scholar]
- 22.Pabst O (2012). New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 12, 821–832. [DOI] [PubMed] [Google Scholar]
- 23.Lycke NY, and Bemark M (2012). The role of Peyer’s patches in synchronizing gut IgA responses. Front. Immunol. 3, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buettner M, and Lochner M (2016). Development and Function of Secondary and Tertiary Lymphoid Organs in the Small Intestine and the Colon. Front. Immunol. 7, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van de Pavert SA, and Mebius RE (2010). New insights into the development of lymphoid tissues. Nat. Rev. Immunol. 10, 664–674. [DOI] [PubMed] [Google Scholar]
- 26.Eberl G, and Lochner M (2009). The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2, 478–485. [DOI] [PubMed] [Google Scholar]
- 27.Mowat AM, and Agace WW (2014). Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14, 667–685. [DOI] [PubMed] [Google Scholar]
- 28.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, et al. (2014). The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity 40, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stehle C, Rückert T, Fiancette R, Gajdasik DW, Willis C, Ulbricht C, Durek P, Mashreghi MF, Finke D, Hauser AE, et al. (2021). T-bet and RORalpha control lymph node formation by regulating embryonic innate lymphoid cell differentiation. Nat. Immunol. 22, 1231–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veiga-Fernandes H, Coles MC, Foster KE, Patel A, Williams A, Natarajan D, Barlow A, Pachnis V, and Kioussis D (2007). Tyrosine kinase receptor RET is a key regulator of Peyer’s patch organogenesis. Nature 446, 547–551. [DOI] [PubMed] [Google Scholar]
- 31.Escobar G, Mangani D, and Anderson AC (2020). T cell factor 1: A master regulator of the T cell response in disease. Sci. Immunol. 5, eabb9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujihashi K, Dohi T, Rennert PD, Yamamoto M, Koga T, Kiyono H, and McGhee JR (2001). Peyer’s patches are required for oral tolerance to proteins. Proc. Natl. Acad. Sci. USA 98, 3310–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunis MC, Dawod B, Carson KR, Veinotte LL, and Marshall JS (2015). Toll-like receptor 2 activators modulate oral tolerance in mice. Clin. Exp. Allergy 45, 1690–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, et al. (2015). Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 43, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiyama Y, Hamada H, Nonaka S, Yamamoto H, Nanno M, Katayama Y, Takahashi H, and Ishikawa H (2002). Homeostatic regulation of intestinal villous epithelia by B lymphocytes. J. Immunol. 168, 2626–2633. [DOI] [PubMed] [Google Scholar]
- 37.McDonald KG, McDonough JS, Dieckgraefe BK, and Newberry RD (2010). Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues. Am. J. Pathol. 176, 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, and Colonna M; Immunological Genome Consortium (2015). Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, and Littman DR (2004). An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73. [DOI] [PubMed] [Google Scholar]
- 40.Mebius RE, Miyamoto T, Christensen J, Domen J, Cupedo T, Weissman IL, and Akashi K (2001). The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3-cells, as well as macrophages. J. Immunol. 166, 6593–6601. [DOI] [PubMed] [Google Scholar]
- 41.Simic M, Manosalva I, Spinelli L, Gentek R, Shayan RR, Siret C, Girard-Madoux M, Wang S, de Fabritus L, Verschoor J, et al. (2020). Distinct Waves from the Hemogenic Endothelium Give Rise to Layered Lymphoid Tissue Inducer Cell Ontogeny. Cell Rep. 32, 108004. [DOI] [PubMed] [Google Scholar]
- 42.Zheng M, Mao K, Fang D, Li D, Lyu J, Peng D, Chen X, Cannon N, Hu G, Han J, et al. (2021). B cell residency but not T cell-independent IgA switching in the gut requires innate lymphoid cells. Proc. Natl. Acad. Sci. USA 118, e2106754118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rennert PD, James D, Mackay F, Browning JL, and Hochman PS (1998). Lymph node genesis is induced by signaling through the lymphotoxin beta receptor. Immunity 9, 71–79. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Li Y, Hao J, He Y, Dong X, Fu YX, and Guo X (2020). The Interaction between Lymphoid Tissue Inducer-Like Cells and T Cells in the Mesenteric Lymph Node Restrains Intestinal Humoral Immunity. Cell Rep. 32, 107936. [DOI] [PubMed] [Google Scholar]
- 45.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, and Artis D (2011). CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mebius RE (2003). Organogenesis of lymphoid tissues. Nature reviews 3, 292–303. [DOI] [PubMed] [Google Scholar]
- 47.Brinkman CC, Iwami D, Hritzo MK, Xiong Y, Ahmad S, Simon T, Hippen KL, Blazar BR, and Bromberg JS (2016). Treg engage lymphotoxin beta receptor for afferent lymphatic transendothelial migration. Nat. Commun. 7, 12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. [DOI] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, and Smyth GK (2010). a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper J, Ding Y, Song J, and Zhao K (2017). Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat. Protoc. 12, 2342–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ku WL, Nakamura K, Gao W, Cui K, Hu G, Tang Q, Ni B, and Zhao K (2019). Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nat. Methods 16, 323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, et al. (2019). Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat. Immunol. 20, 890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq, DNase-Seq, ChIC-Seq and scRNA-Seq datasets have been deposited and are available at the Gene Expression Omnibus (GEO) database under the accession number GSE190302, which is also listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-CD117 (c-kit) BUV395 (2B8) | BD Biosciences | Cat# 564011; RRID: AB_2738541 |
| anti-LPAM-1 (α4β7) BV421 (DATK32) | BD Biosciences | Cat# 566294 |
| anti-CD335 (NKp46) PE (29A1.4) | BD Biosciences | Cat# 560757; RRID: AB_1727466 |
| anti-PLZF Alexa Fluor 647 (R17–809) | BD Biosciences | Cat# 563490; RRID: AB_2738238 |
| anti-ST2 BV711 (U29–93) | BD Biosciences | Cat# RRID: 745549; AB_2743075 |
| anti-GATA3 PE-CF594 (L50–823) | BD Biosciences | Cat# 563510; RRID: AB_2738248 |
| anti-GATA3 PE-Cy7 (L50–823) | BD Biosciences | Cat# 560405; RRID: AB_1645544 |
| anti-RORγt BV650 (Q31–378) | BD Biosciences | Cat# 564722; RRID: AB_2738915 |
| anti-RORγt PE-CF594 (Q31–378) | BD Biosciences | Cat# 562684; RRID: AB_2651150 |
| anti-TCF-1 PE (S33–966) | BD Biosciences | Cat# 564217; RRID: AB_2687845 |
| anti-CD196 (CCR6) APC (29–2L17) | Biolegend | Cat# 129814; RRID: AB_1877147 |
| anti-CD196 (CCR6) PE (29–2L17) | Biolegend | Cat# 129804; RRID: AB_1279137 |
| anti-CD127(IL-7Rα) PE-Cy7 (A7R34) | Biolegend | Cat# 135014; RRID: AB_1937265 |
| anti-CD127(IL-7Rα) BV421 (A7R34) | Biolegend | Cat# 135024; RRID: AB_11218800 |
| anti-CD45.1 BV711 (A20) | Biolegend | Cat# 110739; RRID: AB_2562605 |
| anti-CD45.2 Alexa Fluor 700 (104) | Biolegend | Cat# 109822; RRID: AB_493731 |
| anti-CD279 (PD-1) APC-Cy7 (29F.1A12) | Biolegend | Cat# 135224; RRID: AB_2563523 |
| anti-NK1.1 BV650(PK136) | Biolegend | Cat# 108735; RRID: AB_2563159 |
| anti-KLRG1 (MAFA) PE-Cy7 (2F1/KLRG1) | Biolegend | Cat# 138416; RRID: AB_2561736 |
| anti-CD3e PerCP-Cy5.5 (145–2C11) | Biolegend | Cat# 100328; RRID: AB_893318 |
| anti-CD4 BV421 (GK1.5) | Biolegend | Cat# 100438; RRID: AB_11203718 |
| anti-CD8a Alexa Fluor 700 (53–6.7) | Biolegend | Cat# 100730; RRID: AB_493703 |
| anti-B220 APC (RA3–6B2) | Biolegend | Cat# 103212; RRID: AB_312997 |
| anti-CD19 BV786 (6D5) | Biolegend | Cat# 115543; RRID: AB_11218994 |
| anti-CD3 Biotin (145–2C11) | Biolegend | Cat# 100304; RRID: AB_312669 |
| anti-CD19 Biotin (6D5) | Biolegend | Cat# 115504; RRID: AB_313639 |
| anti-Ter-119 Biotin (TER-119) | Biolegend | Cat# 116204; RRID: AB_313705 |
| anti-CD11b Biotin (M1/70) | Biolegend | Cat# 101204; RRID: AB_312787 |
| anti-CD11c Biotin (N418) | Biolegend | Cat# 117304; RRID: AB_313773 |
| anti-Ly-6G/Ly-6C (Gr-1) Biotin (RB6–8C5) | Biolegend | Cat# 108404; RRID: AB_313369 |
| anti-FcεRIα Biotin (MAR-1) | Biolegend | Cat# 134304; RRID: AB_1626106 |
| Streptavidin Brilliant Violet 785 (N/A) | Biolegend | Cat# 405249 |
| Streptavidin FITC (N/A) | Biolegend | Cat# 405202 |
| anti-γδ TCR Biotin (eBioGL3 (GL-3, GL3) | Thermofisher | Cat# 13–5711-82; RRID: AB_466668 |
| anti-CD135 (Flt3) PerCP-eFluor 710 (A2F10) | Thermofisher | Cat# 46–1351-82; RRID: AB_10733392 |
| anti-IgA PE (mA-6E1) | Thermofisher | Cat# 12–4204-82; RRID: AB_465917 |
| Fixable Viability Dye eFluor™ 506 (N/A) | Thermofisher | Cat# 65–0866-18 |
| LTβRIg antibody (AF.H6) | Biogen MA, Inc. | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 100 bp DNA Ladder | Thermofisher | Cat#15628019 |
| SYBRTM Safe DNA Gel Stain | ThermoFisher | Cat#S33102 |
| PlatinumTM Hot Start PCR Master Mix (2X) | ThermoFisher | Cat#13001012 |
| ACK Lysing Buffer | ThermoFisher | Cat#A1049201 |
| HBSS, calcium, magnesium | ThermoFisher | Cat#24020117 |
| PBS, pH 7.4 | ThermoFisher | Cat#10010031 |
| RPMI 1640 Medium | ThermoFisher | Cat#21870–076 |
| Fetal Bovine Serum | R&D Systems | Cat#S12450 |
| Penicillin-Streptomycin | ThermoFisher | Cat#15140122 |
| L-Glutamine (200 mM) | ThermoFisher | Cat#25030081 |
| MEM Non-Essential Amino Acids Solution (100X) | ThermoFisher | Cat#11140050 |
| Sodium Pyruvate | ThermoFisher | Cat#11360 |
| 2-Mercaptoethanol | ThermoFisher | Cat#21985023 |
| Phorbol 12-myristate 13-acetate | Sigma-Aldrich | Cat#P8139–5MG |
| Ionomycin | Sigma-Aldrich | Cat#407952–5MG |
| Brefeldin A Solution | BioLegend | Cat#420601 |
| eBioscienceTM Foxp3/Transcription Factor Staining Buffer Set | eBioscience | Cat#00–5523-00 |
| DNase I recombinant grade I, from bovine pancreas, expressed in Pichia pastoris | Roche | Cat#4536282001 |
| Liberase DH | Roche | Cat#5401020001 |
|
| ||
| Deposited data | ||
|
| ||
| Raw Data Files for RNA Sequencing | This paper | GEO: GSE190302 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Tcf7tm1a(EUCOMM)Wtsi | the University of California, Davis, KOMP Repository | www.mmrrc.org |
| Mouse: VavCre | JAX mice | Stock No: 008610 |
| Mouse: RorcCre | JAX mice | Stock No: 022791 |
| Mouse: CreERT2 | Taconic | line 10471 |
| Mouse: CD45.1+ | NIAID-Taconic repository | line 7 |
| Mouse: CD45.1+CD45.2+ | NIAID-Taconic repository | line 8422 |
| Mouse: CD45.1+ Rag2−/−il2rg−/− | NIAID-Taconic repository | line 8494 |
| Mouse: CD45.1+ Rag1−/− OT-II | NIAID-Taconic repository | line 361 |
| Mouse: Tcrb−/−d−/− | Dr. Amy Palin of NCI | N/A |
| Mouse: Tcf7-YFP mice | Dr. Bhandoola Avinash | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo v10 | Tree Star | https://www.flowjo.com |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com |


