Abstract
An efficient method for the isolation of human immunodeficiency virus type 1 (HIV-1) nucleic acids from dry cervical swabs was developed. HIV-1 gag and env were detected in 96% (25 of 26) and 81% (21 of 26), respectively, of the samples tested by PCR from HIV-1-seropositive women in a Kenyan cohort study. Eighty-eight percent of the swabs (22 of 25) were positive for gag RNA, and 85% (17 of 20) were positive for env RNA. Fewer than 1,000 copies of HIV-1 gag RNA were detected in four swabs in which a competitive quantitative PCR assay was used. The method described here may be useful for both qualitative and quantitative analyses of HIV RNA in mucosal secretions as well as amplification and cloning of full-length viral genes for functional studies.
Human immunodeficiency virus type 1 (HIV-1) has been detected in cervical and vaginal secretions by several methods, including viral culture, antigen detection, and nucleic acid amplification by PCR. HIV-1 has been cultured from approximately one-third of cervical or cervicovaginal samples obtained from infected women (4, 6, 19, 20), and virus was cultured from four of four cervical biopsy specimens (15). HIV-1 Gag protein was detected by indirect immunofluorescence in CD4+ lymphocytes in cervicovaginal secretions in two-thirds (9 of 14) of infected women (18). More recently, HIV-1 DNA has been detected in 10 to 48% of cervical and vaginal swabs or lavage fluids following PCR amplification (2, 3, 5, 6, 8, 21). In cervical biopsy specimens examined by in situ hybridization following PCR or reverse transcriptase-PCR (RT-PCR), all of 21 specimens from lesions observed by colposcopic examination in adult women were positive for HIV-1 DNA and RNA (10). These data suggest that HIV-infected cells and virions at the cervix are shed in genital secretions. However, there are very little data available on the amount of HIV RNA present in cervical secretions, although one report detected greater than 1,000 copies of RNA per ml of cell-free cervicovaginal secretions (17). To determine the frequency with which HIV RNA is shed from the cervix, we developed an RT-PCR method for the detection of HIV-1 gag and env RNAs in cervical swab specimens.
Because dry swabs (as opposed to swabs placed in transport medium or to cervicovaginal lavages) are frequently a readily available source for analysis of genital secretions, we developed a method by which HIV-1 RNA and DNA are directly purified from dry-swab samples. Cervical secretions were collected from HIV-1-infected women in Nairobi, Kenya, during 1991 and 1992. The samples were frozen at −70°C in cryovials, shipped to Seattle on dry ice, and stored at −70°C until lysis in 1995 to 1997. Because the swabs were not stored in freezing medium, we assumed that most cells present would lyse upon thawing. Lysis solution was therefore added directly to the swabs in the cryovials to isolate all nucleic acids present. Just prior to thawing, 1 ml of guanidinium solution L6 (described by Boom et al. [1]) was added to the vials in which the swabs were shipped. After a 10-min incubation at room temperature with occasional inversion, the solution was transferred to a microfuge tube and the swab was discarded. Debris was removed by centrifugation for 2 min at 8,000 × g in an Eppendorf centrifuge, and the supernatant was transferred to a new tube. Forty microliters of either silica coarse (see Boom et al. [1]) or QIAEX (Qiagen) beads was added to each sample, followed by incubation at room temperature for 10 min with occasional inversion. The samples were then vortexed and centrifuged at 8,000 × g for 30 s, and the supernatant was discarded. The pelleted resin was washed twice with 1 ml of L2 (1) and twice with 1 ml of 70% ethanol. The resin was then air dried for 10 to 15 min to remove any traces of ethanol, and the nucleic acids were eluted twice at 56°C for 5 min with 50 μl of diethylpyrocarbonate (DEPC)-treated H2O (eluates were pooled for a total of 100 μl per sample). A 20-μl aliquot was frozen at −20°C for analysis of DNA in the sample, and the remaining 80 μl of the sample was treated with 2 U of RQ1 RNase-free DNase I (Promega) at 37°C for 30 min, followed by phenol-chloroform and chloroform extractions. The DNase I-treated RNA was then precipitated with 0.3 M sodium acetate and ethanol in the presence of 20 μg of glycogen (Boehringer Mannheim) at −20°C overnight, spun at 12,000 × g for 30 min at 4°C, and washed with 70% ethanol. The RNA pellet was air dried and resuspended in 30 μl of DEPC-treated H2O.
Reverse transcription was performed with the RNA samples in the presence of antisense primers for gag (GAG04 [13, 14]) and env (env12 [11, 16]) simultaneously. Two identical 20-μl cDNA reaction mixtures were prepared with 5 μl of each Dnase I-treated RNA sample (equivalent to 13% of the swab), but RT (superscript II; Gibco/BRL) was added to only one reaction mixture. The minus-RT reaction mixture was used to verify complete digestion of DNA in the test sample. An additional minus-RT mock cDNA reaction was performed with 5 μl of the 20-μl aliquot which had not been treated with DNase I (equivalent to 5% of the swab) in order to determine whether DNA could be detected from the swabs. Following reverse transcription, 4 μl of each reaction mixture was separately amplified in nested PCR for gag (round 1, GAG04 and GAG06 [13, 14]; round 2, Gag1 and Gag2 [9]) or env (round 1, env12 and env13; round 2, env9 and env10 [11, 16]). All PCRs were set up in our P-FREE laboratory (12), which is physically separated from any potential PCR product, plasmid, or phage contamination.
To determine the efficiency of RNA recovery by this isolation method, we spiked samples at the lysis step with a competitive gag RNA containing a deletion of 70 nucleotides (nt; similar to that used by Piatak et al. [13, 14]). The efficiency of RNA recovery was high, as judged by the fact that competitor RNA could still be detected by the RT-PCR method when as few as 100 copies were added to a swab at the time of lysis (data not shown). By the protocol described above, and assuming 100% recovery of the RNA, the cDNA synthesis reaction mixture would include 13 copies in the RT reaction mixture and approximately 3 copies of the spiked competitor would be present in the PCR (100 copies/100 μl × 80 μl/30 μl × 5 μl × 4 μl/20 μl = 3 copies). Thus, at least one-third of the spiked RNA was recovered from the swab sample.
A total of 34 swabs were tested for gag and env nucleic acids. These included 8 samples from HIV-seronegative women tested in parallel with samples from 26 seropositive women, and the laboratory investigator was blinded as to the serostatus of the subject. A reaction was scored positive when a fragment of the predicted size was detected on an ethidium bromide-stained agarose gel. The results are shown in Table 1. To distinguish between DNA and RNA present in the samples prior to DNase I treatment, PCRs were performed in the absence of RT on nucleic acid isolated directly from the swabs (Table 1 [untreated]). No HIV-1-specific PCR products were detected in 8 cervical swabs from HIV-seronegative women taken from the same Kenyan cohort and extracted at the same time as samples from HIV-seropositive women (Table 1). Among HIV-positive women, gag and env DNAs were detected in 69% (18 of 26) and 61% (14 of 23), respectively, of the samples. These percentages of samples that were DNA positive were higher in the present study than in previous studies from our group (2, 5, 6) and may reflect that as a result of purification and concentration, a larger proportion of the swab (1.25 versus 0.4%) was tested in the present analysis. Several swabs were positive for only gag (n = 4) or only env DNA (n = 4). Such sporadic detection of HIV gene products could suggest that very low copy numbers of proviruses are present in these swabs.
TABLE 1.
Detection of gag and env sequences in cervical swabs
| Sample | Gag detection
|
Env detection
|
CD4 count | ||||
|---|---|---|---|---|---|---|---|
| Untreated (−RT) | DNasea
|
Untreated (−RT) | DNasea
|
||||
| +RT | −RT | +RT | −RT | ||||
| HIV-seropositive women | |||||||
| YO117 | − | + | − | + | + | − | NT |
| YO121 | − | + | − | − | − | − | 944 |
| YO137 | − | + | − | + | + | − | 140 |
| YO174 | − | + | − | + | + | − | 168 |
| YO175 | − | + | − | − | + | − | NT |
| YO182 | − | + | − | − | + | − | NT |
| YO183 | − | − | − | − | − | − | NT |
| YO184 | + | + | − | − | + | − | 109 |
| YO189 | + | + | − | + | + | − | 155 |
| YO215 | + | + | − | − | NT | NT | 379 |
| YO235 | + | − | − | + | NT | NT | NT |
| YO236 | + | + | − | + | + | − | 49 |
| YO240 | + | + | − | + | NT | NT | 262 |
| YO247 | + | + | − | − | + | − | NT |
| YO252 | + | + | − | + | + | − | 169 |
| YO253 | + | + | − | − | NT | NT | 104 |
| YO264 | + | + | − | + | NT | NT | 348 |
| YO274 | + | + | − | NT | + | − | 482 |
| YO275 | + | − | − | NT | − | − | 213 |
| YO278 | + | + | + | NT | + | − | NT |
| YO287 | + | + | − | + | + | + | NT |
| YO291 | + | + | − | + | + | − | 294 |
| YO298 | + | + | − | + | + | − | 462 |
| YO299 | + | + | − | + | + | − | 266 |
| YO317 | + | + | − | − | + | − | 692 |
| YO321 | − | + | − | + | + | − | 495 |
| Total(+)b | 18/26 | 23/26 | 1/26 | 14/23 | 18/21 | 1/21 | |
| HIV-seronegative women | |||||||
| YO161 | − | − | − | − | − | − | NT |
| YO163 | − | − | − | − | − | − | NT |
| YO164 | − | − | − | − | − | − | NT |
| YO169 | − | − | − | − | − | − | NT |
| YO171 | − | − | − | − | − | − | NT |
| YO173 | − | − | − | − | − | − | NT |
| YO178 | − | − | − | − | − | − | NT |
| YO179 | − | − | − | − | − | − | NT |
| Total(+)b | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | |
DNase, DNaseI treatment of sample prior to reverse transcription; NT, not tested.
Total(+), number of positive samples/number of samples tested.
To detect RNA, samples were first treated with DNase I, and complete digestion of DNA was verified by performing PCR in the absence of RT. Two samples remained positive for DNA (YO278 for gag and YO287 for env) and, hence, could not be specifically evaluated for RNA. Therefore, excluding those samples, gag and env RNAs were detected in 88% (22 of 25) and in 85% (17 of 20) of samples from HIV-seropositive women. None of the seronegative women was positive for HIV RNA. Several samples were positive for gag or env RNA but negative for DNA in the untreated fraction. Because we did not separate cells from free virus prior to nucleic acid extraction, we cannot determine whether the HIV RNA detected was from virions or from a few infected cells actively expressing multiple viral transcripts. Combining the DNA and RNA data, 96% (25 of 26) of the swabs tested positive for gag nucleic acid and 81% (21 of 26, including samples not tested [Table 1, untreated or DNase treated]) were positive for env. Thus, our results indicate that HIV-1 is present and is expressed in cervical secretions in a majority of infected women.
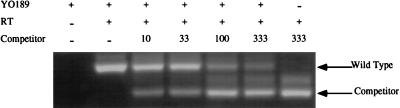
To determine the quantity of RNA in dry swab samples, four RNAs from positive samples were subjected to a quantitative competitive RT-PCR method as previously reported (7), but with nested-PCRs as described above. Briefly, in vitro-transcribed competitor RNA (a 550-nt transcript containing a 70-nt internal deletion of nt 966 to 1,027 of the wild-type sequence as described in reference 7) was quantitated by spectrophotometry, and 5-μl aliquots of dilutions containing the indicated copy numbers were frozen at −70°C until use in PCR tubes, to which the test samples were directly added. Addition of the gag competitor RNA during the RT reaction, followed by nested PCR, revealed fewer than 1,000 copies of RNA per swab. Figure 1 illustrates the results for one subject, YO189. The wild-type and competitor bands are indicated, and the intermediate-size band is of unknown origin but appears with competitor alone, suggesting that it is an artifact of the amplification of competitor. Each swab was extracted into a final volume of 100 μl of H2O. Eighty microliters was DNase I treated and resuspended in a final volume of 30 μl. Therefore, the 1.5 μl of test sample RNA added to each competitor PCR tube translates to 1/25 of the swab sample (1.5/30 × 80% = 4%). The intensity of the wild-type gag product was equivalent to that of the competitor RNA between 10 and 33 copies, indicating the presence of 10 to 33 copies of gag RNA in the 1.5 μl tested. Thus, the initial swab contained 250 (10 copies/0.04 swab) to 825 (33 copies/0.04 swab) copies of gag RNA. Similar results were obtained for YO252, YO298, and YO299 (data not shown). Because we did not distinguish between RNA present in virions or RNA present in cells, these copy numbers could correspond to free virus and/or cell-associated RNA.
FIG. 1.
An ethidium bromide-stained 3% agarose gel of the competitive RT-PCR for YO189. The numbers of competitor RNA molecules added to the initial RT reaction are indicated.
In conclusion, we have described an efficient and sensitive method for the isolation of HIV-1 nucleic acids, including RNA, from dry cervical swabs. By this method, the selective use of DNase I and RT treatment can be used to distinguish between RNA and DNA in a single sample. We have found that the majority of infected women have HIV-1 nucleic acids in their cervical secretions. Moreover, we could amplify a 1.2-kb fragment representing the majority of the coding sequences for the extracellular glycoprotein, suggesting that this method could be used to clone and analyze full-length viral genes in swab samples. The HIV-1-seropositive women in this study had CD4 cell counts ranging from 50 to 950, with 7 of the 18 women tested having counts of fewer than 200. Thus, these women represented a spectrum of HIV infection, suggesting that this method may be applicable for analysis of women at all stages of disease. Because so few samples were negative by this method, we cannot determine any correlation between a positive reaction and the clinical status of the subject. These issues will likely require a large cohort study by the quantitative method. Because dry cervical swabs can be obtained relatively easily during routine physical examination, this method will be particularly useful for archived samples as well as samples collected in field settings with limited access to laboratory equipment.
Acknowledgments
We thank members of the Overbaugh lab for helpful discussions, Stephanie Jackson and Dana DeVange for technical assistance, Barbra Richardson for archival sample retrieval, and the Nairobi STD/AIDS project for their support. We also thank Beth Moorefield and Michelle Long for helpful comments on the manuscript.
This work was supported by grant AI 38518. A.M.H. was supported by NIH research scientist development award RR00079.
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-Van Dillen P M E, Van Der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemetson D B, Moss G B, Willerford D M, Hensel M, Emonyi W, Holmes K K, Plummer F, Ndinya-Achola J, Roberts P L, Hillier S, Kreiss J K. Detection of HIV DNA in cervical and vaginal secretions. Prevalence and correlates among women in Nairobi, Kenya. JAMA. 1993;269:2860–2864. [PubMed] [Google Scholar]
- 3.Ghys P D, Fransen K, Diallo M O, Ettiegne-Traore V, Coulibaly I-M, Yeboue K M, Kalish M L, Maurice C, Whitaker J P, Greenberg A E, Laga M. The associations between cervicovaginal HIV shedding, sexually transmitted diseases and immunosuppression in female sex workers in Abidjan, Cote d’Ivoire. AIDS. 1997;11:F85–F93. doi: 10.1097/00002030-199712000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Henin Y, Mandelbrot L, Henrion R, Pradinaud R, Coulaud J P, Montagnier L. Virus excretion in the cervicovaginal secretions of pregnant and nonpregnant HIV-infected women. J AIDS. 1993;6:72–75. [PubMed] [Google Scholar]
- 5.John G C, Nduati R W, Mbori-Ngacha D, Overbaugh J, Welch M, Richardson B A, Ndinya-Achola J, Bwayo J, Krieger J, Onyango F, Kriess J K. Genital shedding of human immunodeficiency virus type 1 DNA during pregnancy: association with immunosuppression, abnormal cervical or vaginal discharge, and severe vitamin A deficiency. J Infect Dis. 1997;175:57–62. doi: 10.1093/infdis/175.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreiss J K, Willerford D, Hensel M, Emony W, Plummer F, Ndinya-Achola J, Roberts P, Hoskyn J, Hillier S, Kiviat N, Holmes K. Association between cervical inflammation and cervical shedding of human immunodeficiency virus DNA. J Infect Dis. 1994;170:1597–1601. doi: 10.1093/infdis/170.6.1597. [DOI] [PubMed] [Google Scholar]
- 7.Lewis P, Nduati R, Kreiss J K, John G C, Richardson B A, Mbori-Ngacha D, Ndinya-Achola J, Overbaugh J. Cell-free human immunodeficiency virus type 1 in breast milk. J Infect Dis. 1998;177:34–39. doi: 10.1086/513816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loussert-Ajaka I, Mandelbrot L, Delmas M-C, Bastian H, Benifla J-L, Farfara I, de Vincenzi I, Matheron S, Simon F, Brun-Vezinet F. HIV-1 detection in cervicovaginal secretions during pregnancy. AIDS. 1997;11:1575–1581. doi: 10.1097/00002030-199713000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Moss G B, Overbaugh J, Welch M, Reilly M, Bwayo J, Plummer F A, Ndinya-Achola J O, Malisa M A, Kreiss J K. Human immunodeficiency virus DNA in urethral secretions in men: association with gonococcal urethritis and CD4 cell depletion. J Infect Dis. 1995;172:1469–1474. doi: 10.1093/infdis/172.6.1469. [DOI] [PubMed] [Google Scholar]
- 10.Nuovo G J, Forde A, MacConnell P, Fahrenwald R. In situ detection of PCR-amplified HIV-1 nucleic acids and tumor necrosis factor cDNA in cervical tissues. Am J Pathol. 1993;143:40–48. [PMC free article] [PubMed] [Google Scholar]
- 11.Overbaugh J, Anderson R J, Ndinya-Achola J O, Kreiss J K. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–115. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- 12.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 14.Piatak M J, Luk K-C, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–80. [PubMed] [Google Scholar]
- 15.Pomerantz R J, de la Monte S M, Donegan S P, Rota T R, Vogt M W, Craven D E, Hirsch M S. Human immunodeficiency virus (HIV) infection of the uterine cervix. Ann Intern Med. 1988;108:321–327. doi: 10.7326/0003-4819-108-3-321. [DOI] [PubMed] [Google Scholar]
- 16.Poss M, Martin H L, Kreiss J K, Granville L, Chohan B, Nyange P, Mandaliya K, Overbaugh J. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasheed S, Li Z, Xy D, Kovacs A. Presence of cell-free human immunodeficiency virus in cervicovaginal secretions is independent of viral load in the blood of human immunodeficiency virus-infected women. Am J Obstet Gynecol. 1996;175:122–130. doi: 10.1016/s0002-9378(96)70261-2. [DOI] [PubMed] [Google Scholar]
- 18.Van de Perre P, De Clercq A, Cogniaux-Leclerc J, Nzaramba D, Butzler J P, Sprecher-Goldberger S. Detection of HIV p17 antigen in lymphocytes but not epithelial cells from cervicovaginal secretions of women seropositive for HIV: implications for heterosexual transmission of the virus. Genitourin Med. 1988;64:30–33. doi: 10.1136/sti.64.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogt M W, Witt D J, Craven D E, Byington R, Crawford D F, Schooley R T, Hirsch M S. Isolation of HTLV-III/LAV from cervical secretions of women at risk for AIDS. Lancet. 1986;1:525–527. doi: 10.1016/s0140-6736(86)90884-6. [DOI] [PubMed] [Google Scholar]
- 20.Wofsy C B, Cohen J B, Hauer L B, Padian N S, Michaelis B A, Evans L A, Levy J A. Isolation of AIDS-associated retrovirus from genital secretions of women with antibodies to the virus. Lancet. 1986;1:527–529. doi: 10.1016/s0140-6736(86)90885-8. [DOI] [PubMed] [Google Scholar]
- 21.Zorr B, Schafer A P, Dilger I, Habermehl K-O, Kosh M. HIV-1 detection in endocervical swabs and mode of HIV-1 infection. Lancet. 1994;343:852. [PubMed] [Google Scholar]