Abstract
Background
In the biological method, using nonpathogenic and extremophile bacteria systems are not only safe and highly efficient but also a trump card for synthesizing nanoparticles. Halomonas elongata QW6 IBRC-M 10,214 (He10214) and Salinicoccus iranensis IBRC-M 10,198 (Si10198), indigenous halophilic bacteria, can be used for synthesizing selenium nanoparticles (SeNPs).
Methods
SeNP biosynthesis was optimized in two halophilic bacteria and characterized by UV–Vis, Fourier transform infrared spectroscopy (FTIR), transmission electron microscopy (TEM), field emission scanning electron microscopy (FESEM), X-ray powder diffraction (XRD), zeta potential, and energy dispersive X-ray (EDX).
Results
Optimized conditions for synthesizing SeNPs was at 300 °C at 150 rpm for 72 h and 6 mM or 8 mM concentration of Na2SeO3. UV–Vis indicated a sharp absorption peak at 294 nm. Spherical-shaped nanoparticles by a diameter of 30–100 nm were observed in FESEM and TEM microscopy images. The produced SeNPs were identified by a peak in FTIR spectra. In XRD analysis, the highest peak diffraction had a relationship with SeNPs. The zeta potential analysis showed SeNP production, and elemental selenium was confirmed by EDX.
Conclusions
Halophilic bacteria, owing to easy manipulation to create optimization conditions and high resistance, could serve as appropriate organisms for the bioproduction of nanoparticles. The biological method, due to effectiveness, flexibility, biocompatibility, and low cost, could be used for the synthesis of reproducible and stable nanoparticles.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13065-023-01034-w.
Keywords: Biosynthesis, Selenium nanoparticles (SeNPs), Green synthesis, Iran
Introduction
Nanoparticle synthesis has been interested in recent years. This process is a bridge gap between bulk material and atomic or molecular structure and has much applicability in various fields such as medicine, chemistry, biology, electronics, and energy [1–7]. The importance of nanoparticles depends on their structure and functional characteristics [8].
Selenium is a trace element, sometimes a metalloid, with abundant properties and functions, including semiconductor, thermoelectric and catalytic activities to hydration and oxidation reactions [9, 10]. It acts as an antioxidant and prevents the damage of body tissues against oxidative reactions. Selenium reduces the risk of various cancers, including lung, pancreas, stomach, and intestine cancers and has an inhibitory effect on some bacteria. In spite of many advantages reported, selenium is highly toxic, and its large amounts can have adverse effect. By producing selenium nanoparticles (SeNPs), nanotechnology researchers could reduce the risk of toxicity of this element. The biological activity of SeNPs is highly dependent on their size. Compared to standard selenium compounds, these particles have high biochemical activity so that 5-200 nm of selenium could directly eliminate free radicals in the laboratory environment [11, 12].
Nanoparticle synthesis by physical and chemical methods faces difficulties and impediments that could be managed by the biological approach [13]. The synthesis of SeNPs by this method has been demonstrated to be safe, inexpensive, and eco-friendly, and there is no need for toxic materials [14, 15]. For the bioproduction of metalloid nanoparticles, various microorganisms, such as bacteria, algae, yeast, and fungi, are used [16]. These microorganisms have ability to tolerate a high concentration of toxic heavy metals [17]. The halophilic bacteria can act as bioassay indicators in a polluted environment [18]. The majority of catalytic enzymes in halophilic bacteria play a role in eliminating toxic and chemical materials and also the harmful anions and cations, without leading to the death of these bacteria [19, 20]. As a result, using halophilic bacteria influences both the environment and the survival of organisms [20]. Microorganisms can synthesize nanoparticles by the intracellular and extracellular techniques [14]. In the intracellular method, metal ions penetrate into bacterial cells, and then nanoparticles are produced by the enzymatic reactions, thus reducing agents inside the cell. However, in the extracellular method, the metal ions are put on the cell surface, and nanoparticle synthesis is performed by surface enzymes [21]. While the extracellular method has more advantages, e.g. the recovery facilitation of nanoparticles and low cost, over intracellular approach, many bacteria synthesize nanoparticles by the intracellular method [22]. In other words, the synthesis of SeNPs is carried out by the bioreduction process because reduced metal ion converts to stable biological form [23]. The biosynthesis of SeNPs is assumed to help remove heavy metals from polluted waters around industrial environments. Based on literature review, there are numerous reports on the production of SeNPs by chemical, physical, and the biological method, but scant surveys have examined the biological synthesis of these nanoparticles by halophilic bacteria in Iran. Therefore, this study for the first time biosynthesizes SeNPs using the intracellular method in two halophilic bacteria, Halomonas elongata IBRC-M 10,214 (He10214) and Salinicoccus iranensis strain QW6 IBRC-M 10,198 (Si10198), isolated from the native region of Iran. In this light, SeNPs were characterized by the aid of various analytical techniques, comprising ultraviolet-visible (UV–Vis) spectroscopy, Fourier transform infrared spectroscopy (FTIR), X-Ray diffraction (XRD), transmission electron microscopy (TEM), field emission scanning electron microscopy (FESEM), energy dispersive X-ray (EDX), and zeta potential (Additional file 1: Fig. S1).
Methods
Chemical substances and bacterial strains
For SeNP biosynthesis, sodium selenite (Na2SeO3; 172.94 g/mol) was purchased from AppliChem GmbH (Germany). The stock solution was prepared by dissolving Na2SeO3 (0.345 g) in 10 mL of distilled water, followed by sterilization using a microbiological filter (0.22 μm). He10214, a Gram-negative moderately halophilic and heavy metal resistance bacterium, was isolated from Qom Salt Lake (Qom, Iran). Likewise, the moderately halophilic, Gram-positive bacterium, Si10198, was isolated from the wastewater of textile industry in Qom [24].
Culture media and growth conditions
He10214 and Si10198 were cultivated aerobically at room temperature in a medium as follows (%, w/v): NaCl, 17.8; MgSO4, 0.96; CaCl2, 0.036; KC1, 0.2; NaHCO3, 0.006; NaBr, 0.0026; MgCl2, 0.7; yeast extract, 0.1; glucose, 0.1; proteose-peptone, 0.5; agar, 1.5 in 1 L of water containing 5% (w/v) NaCl, pH 7.2, adjusted with KOH [22].
Intracellular synthesis of SeNPs by halophilic strains
At first, 1 µL of the inoculum (OD600 = 0.1) was transferred to a 100-mL Erlenmeyer flask and treated with different concentrations of Na2SeO3 (Table 1). Then the production of SeNPs was optimized by incubating cultures in an orbital shaking incubator according to the conditions depicted in Table 1. For the formation of a pellet containing SeNPs and cells, the red culture, showing the formation of SeNPs, was transferred to 50-mL centrifuge tubes, followed by centrifugation at 5000 rpm for 10 min. After supernatant removal, the pellet was resuspended in NaCl (0.9%) and centrifuged (5000 rpm, 10 min); this step was repeated two times. Subsequently, liquid nitrogen was added to the precipitation. Following the formation of a uniform powder, the cell mixture containing SeNPs was ultrasonicated for 5 min. Also, the pellet was centrifuged in Tris-HCL buffer (1.5 M) containing SDS (1%) at 8000 rpm for 10 min (pH 3.8); thereafter, it was washed several times in sterile distilled water. A two-phase (organic-aqueous) system was used to separate SeNPs from cellular debris. For this purpose, following the addition of 2 mL of n-octyl alcohol to 4 mL of suspension obtained from the previous stage, the mixture was shaken to admix thoroughly the organic-aqueous phase. The tubes were centrifuged (at 3000 rpm for a period of 5 min) and incubated at 4 °C for 24 h. The organic-aqueous phase was slowly discarded, and the remaining nanoparticles were rinsed with chloroform, ethanol (70%), and distilled water, respectively. Finally, the purified nanoparticle suspension was stored at 4 °C [25]. The suspension was dried in an oven overnight for subsequent analysis.
Table 1.
Parameters for the optimization of biosynthesis of SeNPs
| No. | Parameter | Cond 1 | Cond 2 | Cond 3 | Cond4 |
|---|---|---|---|---|---|
| 1 | Na2SeO3 concentration (M) | 2 | 4 | 6 | 8 |
| 2 | Incubation temperature (°C) | 30 | 32 | 35 | 37 |
| 3 | Culturing time (h) | 24 | 48 | 72 | 96 |
| 4 | Shaker (rpm) | 150 | 180 | 210 | 240 |
Cond 1, 2, 3, 4: optimal conditions for SeNPs production
Characterization of SeNPs
UV–Vis spectrophotometry and FTIR
The first strategy to evaluate the production of nanoparticles was UV–Vis spectroscopy analysis, which was conducted by dispersing the nanoparticle suspension in distilled water. The absorption spectra of the SeNPs were recorded with wavelengths ranging between 200 and 800 nm using Varian Cary 100 UV–Vis instruments (USA). The control curve was obtained from distilled water, and the chemical structure of SeNPs was studied by FTIR analysis. The powder of the samples was mixed with KBr pellets. The FTIR spectrum was recorded in 4395–4495 cm−1 and 4388–4488 cm−1 regions on a WQF-510 A FTIR spectrometer (Rayleigh, China) at a resolution of 4 cm−1 for produced SeNPs using He10214 and Si10198.
TEM
The size and morphology of nanoparticle powder were determined using TEM. SeNPs synthesized by He10214 and Si10198 were characterized by the Zeiss EM900 transmission electron microscope (Germany) at an accelerating voltage of 200 kV.
FESEM and EDX
For the analysis of the shape of SeNPs, images were taken with SEM (FEI QUANTA 450) at 20 kV HV. EDX was used to ascertain elemental selenium in solid samples. This process was performed by BRUKER Q200 (Germany). X-ray spectrometer analysis and FESEM were simultaneously employed to indicate electron production mapping and point analysis.
XRD
The crystal structure of SeNPs was characterized by XRD on Ultima IV (Rigaku, Japan) at a voltage of 40 kV with CuKα radiation 1.22 0 A and a scanning rate of 4 degrees per minutes. The crystallite domain size of the synthesized SeNPs was obtained by Debye–Scherrer formula: .
Zeta potential
Zeta potential is one of the most important parameters for the stability of nanoparticles in suspension. The zeta potential of SeNPs was analyzed using SZ-100 (Horiba Jobin Yvon SAS, USA). For this purpose, the powder SeNPs were ultrasonicated after dissolving in double distillation water.
Results
He10214 and Si10198 as biological factories could synthesize SeNPs successfully. The first evidence in the synthesis of the SeNPs was the medium color change, which became red after the reduction of Se2O3 to Se0 (Fig. 1).
Fig. 1.

Color change of the culture from light yellow to red, indicating the biosynthesis of SeNPs
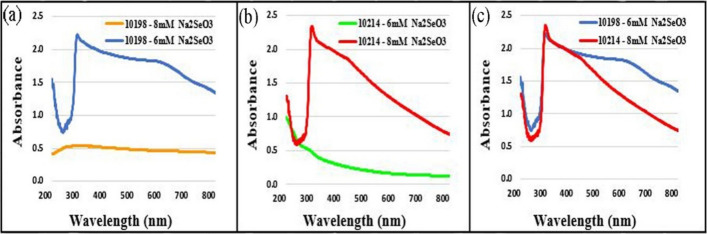
The results from UV–Vis spectroscopy demonstrated the reduction of to Se0 by the two aforementioned bacteria (Fig. 2). Identification of a strong absorption peak at 294 nm confirmed the formation of SeNPs. Moreover, the maximum production of SeNPs by Si10198 occurred at 6 mM of Na2SeO3, and the increasing concentration of Na2SeO3 did not influence the peak (Fig. 2a). To investigate the yield of SeNPs, the same above-mentioned method was used for He10214. The results indicated the maximum production of SeNPs at 8 mM of Na2SeO3 (Fig. 2b).
Fig. 2.
UV–Vis spectroscopy (200–800 nm) of SeNPs biosynthesized by a Si10198 and b He10214 using different concentrations of Na2SO3. c Comparing the highest production rate of SeNPs using He10214 and Si10198
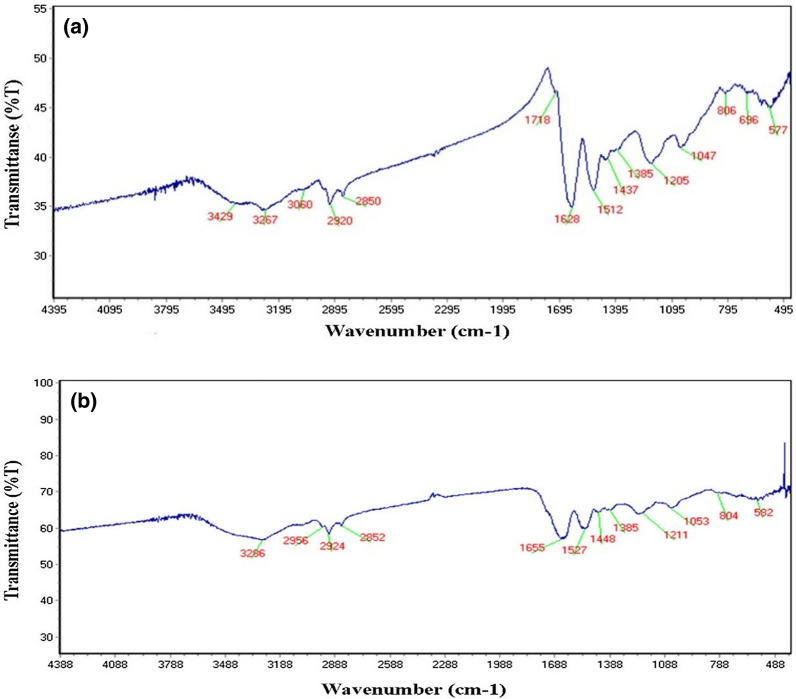
The chemical structure of SeNPs obtained from He10214 and Si10198 was investigated by FTIR. This technique was employed to study the functional groups responsible for the synthesis of SeNPs. The peaks at 2924 cm−1 and 2852 cm−1 or 2920 cm−1 and 2850 cm−1 affirmed the presence of ether-methoxy-OCH3 groups in He10214 and Si10198, respectively (Fig. 3). The other peaks associated with the functional groups observed in the FTIR spectra of both bacteria are listed in Table 2.
Fig. 3.
FTIR spectrum of SeNPs biosynthesized by a Si10198 and b He10214
Table 2.
The bonds of the functional groups of SeNPs biosynthesized by Si10198 and He10214
| Functional group |
Si10198
Wavelength (nm) |
He10214
Wavelength (nm) |
|---|---|---|
| Amide A (in protein) | 3268 | 3267 |
| C–H in –CH3 | 2956 | 3060 |
| C–H in > CH2 | 2924 | 2920 |
| C–H in > CH2 | 2852 | 2850 |
| C=O | 1655 | 1628 |
| Amide I (in protein) | 1527 | 1512 |
| Amide II (in protein) | 1448 | 1437 |
| –CH2/–CH3 | 1385 | 1385 |
| COO– | 1211 | 1205 |
| Amide III | 1053 | 1047 |
| C–O, C–C, C–O–H, C–O–C | 804 | 806 |
| Phosphoryl group | 582 | 577 |
The electron microscopic images of SeNPs is represented in Fig. 4 and shows a spherical shape with a mean diameter of around 50–100 nm and 30–100 nm in 6 and 8 mM of Na2SeO3, synthesized by He10214 and Si10198, respectively. Images obtained from FESEM confirmed the spherical shape of SeNPs biosynthesized by the two bacteria (Fig. 5).
Fig. 4.

TEM image of SeNPs biosynthesized by Si10198 (a and b; 6 mM) and He10214 (c and d; 8 mM)
Fig. 5.
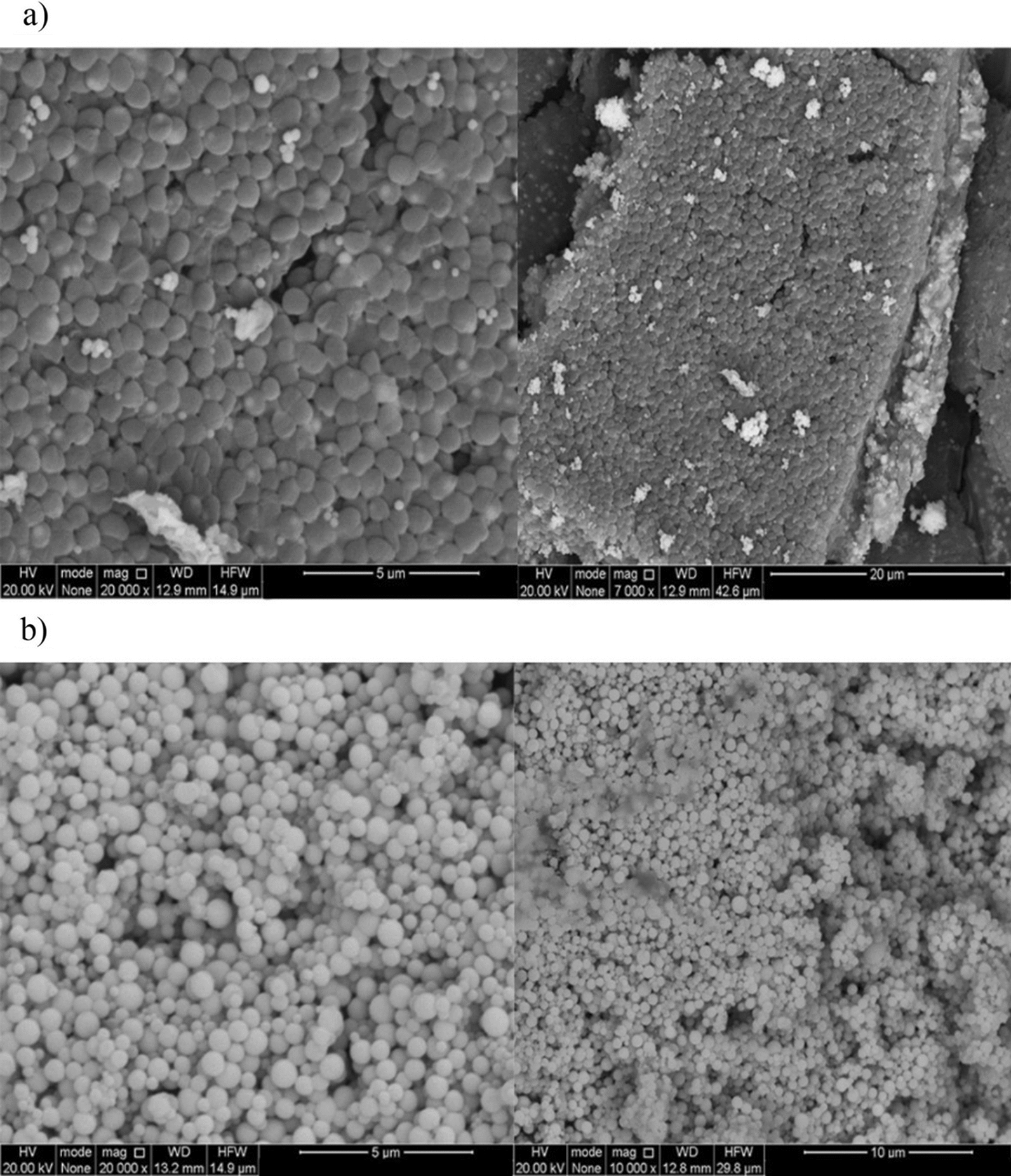
FESEM image of SeNPs biosynthesized by a Si10198 and b He10214
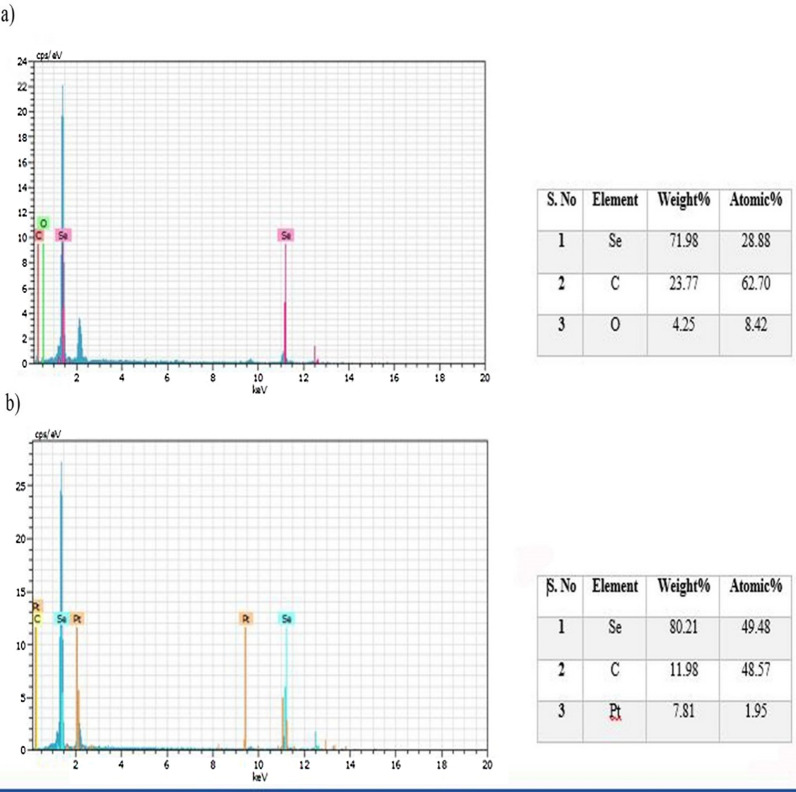
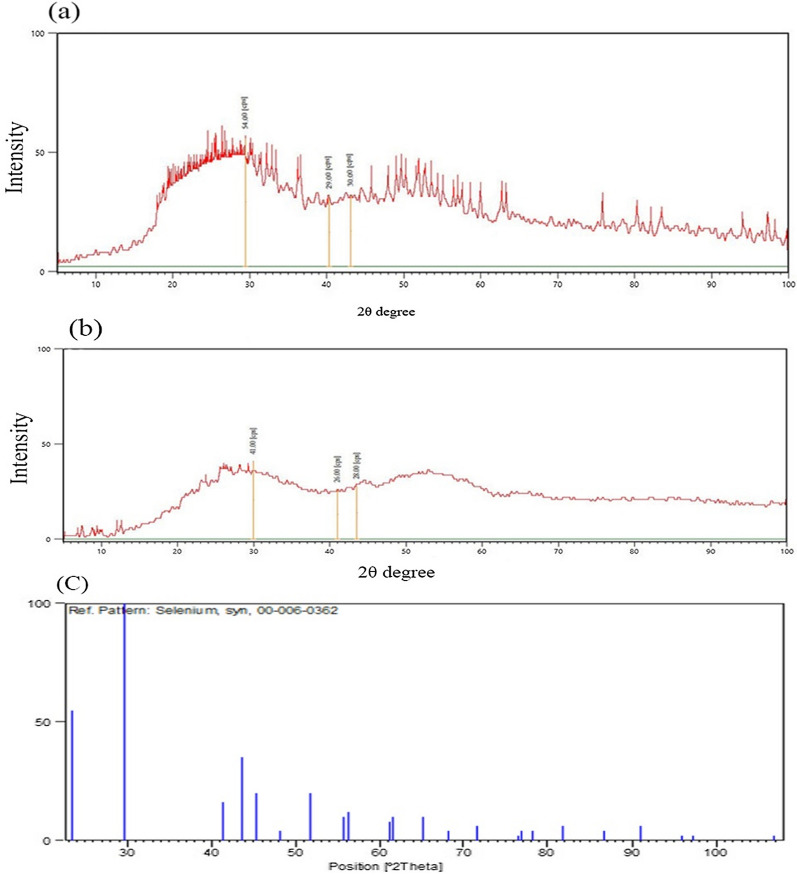
The EDX analysis of SeNPs synthesized by He10214 and Si10198, as shown in Fig. 6, exhibited that absorption peaks in both bacteria were 1.4 (SeLα), 11.7 (SeKα), and 12.5 KeV (SeKβ). XRD analysis also confirmed the synthesis of SeNPs by the two bacteria (Fig. 7). The highest intense peak was detected at 54 cps, and the diffraction peak was about 29.5 (2θ). Owing to the impurity of nanoparticles and adherence of bacteria, the peak in our study was lower than other studies [12, 26], and the standard diffraction peak at 2θ was observed at ~ 30, with the intensity of 101. The zeta potential of the synthesized SeNPs by Si10198 was found to be − 60.6 mV (Fig. 8a), while that of He10214 was − 51.2 mV (Fig. 8b).
Fig. 6.
EDX analysis confirming selenium element in a Si10198 and b He10214
Fig. 7.
X-ray analysis confirming the presence of SeNPs in a Si10198 and b He10214, and c SeNP standard pattern on XRD
Fig. 8.
Zeta potential measurement of the SeNPs using a Si10198 and b He10214
Discussion
The present study explored that He10214 and Si10198 as a biological factory could successfully synthesize SeNPs. The high compatibility of these bacteria in specific environmental conditions and their structural characteristics shows that halophilic bacteria have a prominent role in the production of SeNPs. While the high amount of selenium is toxic for humans, plants, and microorganisms, owing to the antioxidant and antimicrobial features, this element has a leading function in the protection of the ecosystem. As a result, SeNP biosynthesis in optimized conditions using bacteria resistant to heavy metals and harsh circumstances would be cost-effective. In this survey, SeNP production was optimized by 6 or 8 mM of Na2SeO3 for He10214 and Si10198, respectively at 150 rpm, 30 °C for 72 h [15]. The red color of the culture affirmed the presence of SeNPs, as indicated in several studies [4, 10, 15, 27].
UV–Vis analysis
In the present study, the UV–Vis spectroscopy results exhibited that decreased to Se0 by both He10214 and Si10198, which is consistent with a finding achieved in the Srivastava et al.’s [26] study, in which a peak at 300 nm was obtained from the synthesized SeNPs using some species of Lactobacillus. Overall, the observation of the peak between 200 and 300 nm indicates the presence of SeNPs [9, 28–32]. Based on the UV–Vis spectra of biosynthesized SeNPs by both bacteria in different concentrations of Na2SeO3 (6 mM and 8 mM), He10214 could tolerate the higher concentration of selenium compared to Si10198. Thus, it seems that He10214 outperform Si10198 in refining the selenium-rich wastewater of factories.
FTIR analysis
The same as fingerprints in humans, FTIR can be used to identify nanoparticles. This work compared the FTIR patterns and matched all the adsorbents in the two molecules spectrum. The peaks indicated the existence of a biopolymer associated with the SeNPs, which is found in cell walls as a reducing factor for the synthesis of SeNPs [27, 31]. Fritea et al. [32] found an association between the absorption peak of 2918 cm−1 and SeNPs, which probably implies the presence of a biopolymer in the cell wall. On the other hand, He10214 is Gram-negative and Si10198 Gram-positive bacteria; the differences in the type and thickness of their cell wall have caused slight variations between the absorption pattern of these two bacteria.
TEM analysis
The morphology of SeNPs by TEM revealed that these nanoparticles had a spherical shape with a mean diameter of about 50–100 nm and 30–100 nm in 6 mM and 8 mM Na2SeO3, which were synthesized by He10214 and Si10198, respectively. According to previous studies, the spherical shape of the produced nanoparticles confirms the presence of SeNPs [30, 33]. While these results are comparable with a study in which the size of these nanoparticles was observed at 50–150 nm [34], the size of the biosynthesized nanoparticles varies depending on the host producing these particles and also the conditions of production. The sizes of biosynthesized SeNPs obtained by TEM was in the range of 20–80 nm [26]. In this regard, Avendaño et al. [33] and Forootanfar et al. [35] have observed SeNPs in the sizes 100–500 nm and 80–220 nm, respectively.
FESEM analysis
An electron microscope FESEM is more suitable for imaging nanoparticles due to providing high resolution at a low voltage. As reported in numerous studies, the spherical and uniform shape of synthesized SeNPs is observable through the FESEM technique [36].
EDX analysis
The EDX analysis determines the concentrations of the constituent elements and their composition by scanning via an electron microscope [37–43]. Quantitative analysis of SeNPs synthesized by He10214 and Si10198, as demonstrated in Fig. 6, reflected that absorption peaks in He10214 and Si10198 bacteria were 1.4 (SeLα), 11.7 (SeKα), and 12.5 KeV (SeKβ). The highest peaks were related to the selenium element, and their weights in He10214 and Si10198 bacteria were 71.98% and 80.21%, respectively. The presence of carbon (C), oxygen (O), and platin (Pt) elements in EDX analysis might be due to capping with proteins and cell membranes, reactions of reduction, and also the stability of produced SeNPs. Potassium element was identified in the He10214 strain and the oxygen element in the Si10198 strain, which is likely due to different types of cell walls. In multiple investigations, such as Fernández-Llamosas et al. [10], Srivastava et al. [26], Cremonini et al. [44], Sharma et al. [12], and Dhanjal et al. [45], the selenium peak was explored at 1.4 (SeLα), which is the highest peak related to selenium synthesis.
XRD analysis
XRD analysis confirmed the synthesis of SeNPs by He10214 and Si10198 bacteria. The highest intensity peak was 54 cps, and the diffraction peak was about 29.5 (2θ), which supports the study conducted by Alagesan and Venugopal [46]. Because of nanoparticles impurity and bacteria adherence in the current study, the peak was lower than that in previous studies [26, 27, 47], and standard diffraction peak at 2θ was observed at ~ 30, with the intensity of 101. In a surface absorption study related to particle morphology and suspension stability, zeta potential analysis is practical [48].
Zeta potential analysis
The zeta potential of the synthesized SeNPs suspended was found to be − 60.6 mV by Si10198 and − 51.2 mV by He10214. These results of our study are in conformity with the findings of Mollania et al.’s [49] investigation in which the zeta potential reflected a high negative charge on the SeNPs (− 46.86 mV). However, in previous studies, the zeta potential of this nanoparticle has been reported in different rates; for instance, Srivastava and Mukhopadhyay [30] observed the zeta potential of SeNPs in − 7.7 mV, and Vekariya et al. [50] reported a zeta potential of − 28.8 mV. Also, the zeta potential value for fabricated SeNPs using fungi was reported as − 22.9 mV by Zare et al. [51]. Capping and stabilization of nanoparticles by bacterial proteins cause a high negative charge on the SeNPs. Stabilized particles are not transformed to black amorphous form when were kept for a long time [34, 45]. Since the zeta potential of SeNPs synthesized by Si10198 was more negative; therefore, this strain had a higher effective role in stabilizing SeNPs. Research is being completed to produce, evaluate and apply nanoparticles, and human knowledge in this regard is increasing day by day. One of the limitations of nanoparticle production is storage and particle size maintenance. After nanoparticle synthesis, an important matter is the synthesis of nanoparticles that are more stable and remain in the nanoscale for a longer period, or in other words, the accumulation of nanostructures is prevented. The protection of body balance has been indicated to be dependent on the normal function of antioxidant compounds and enzymes [52]. The generation of oxidative stress has a relationship with the altered function of the electron transport chain in the inner membrane of mitochondria [53]. The growth of biologic pollution, particularly certain drugs, heavy metals, and radiation, can eliminate the body’s hemostasis and increase the free radical production. Oxidative stress development results in destructing not only body cells but also biological molecules [52].
Conclusions
In the present study, SeNPs were synthesized in a bacterial system using the biological method, which is safe, effective, eco-friendly, inexpensive, and flexible. The findings of this survey suggest that halophilic bacteria He10214 and Si10198, found in the native region of Iran, have capability of producing nanoparticles. Under optimal conditions, the two bacteria could produce SeNPs with 100 − 30 nm. These nanoparticles, in virtue of biocompatibility and high potential, could be used as antioxidant, anticancer, and antimicrobial agents in future studies.
Supplementary Information
Additional file 1: Fig. 1. Synthesis of SeNPs using bacteria.
Acknowledgements
The authors would like to thank Dr. R. Zand Monfared (at the Department of Analytical Chemistry) and Dr. A. Javadi (at Microbiological Research Center), Qom Islamic Azad University, for their assistance in chemical and microbial experiments.
Abbreviations
- EDX
Energy dispersive X-ray
- FESEM
Field emission scanning electron microscopy
- FTIR
Fourier transform infrared spectroscopy
- Na2SeO3
Sodium selenite
- SENPS
Selenium nanoparticles
- TEM
Transmission electron microscopy
- UV–Vis
Ultraviolet–visible
- XRD
X-ray diffraction
Author contributions
MT conceived and designed the evaluation, drafted the manuscript, collected the data, reanalyzed the statistical data, revised the manuscript; SA participated in designing the evaluation, performed parts of the statistical analysis, helped to draft the manuscript, reevaluated the data, revised the manuscript, and performed the statistical analysis; MAA re-evaluated the data, revised the manuscript, and performed the statistical analysis; RN revised the manuscript and supervised the study; MRZ revised the manuscript and supervised the study. All authors have read, reviewed and approved the final version of manuscript.
Funding
This research was conducted at Qom Azad University, but all costs were provided privately.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Soheil Aghaei, Email: soheilaghaee@yahoo.com.
Mohammad Ali Amoozegar, Email: amoozegar@ut.ac.ir.
References
- 1.Mushtaq MM, Peerzada S, Ishtiaq S. Pharmacognostic features, preliminary phytochemical screening and in vitro antioxidant studies of indigenous plant of Lahore. Int J Adv Biol Biomed Res. 2021;9(2):190–201. [Google Scholar]
- 2.Ukwubile CA, Odugu JA, Njidda S, Umeokoli BO, Bababe AB, Bingari MS, et al. Survey of plants used in folk medicine in Bali, Gashaka and Sarduana local government areas Taraba state Nigeria for the treatment of cancers. Int J Adv Biol Biomed Res. 2020;8(4):322–39. [Google Scholar]
- 3.Chavoshi E, Labbafi M, Zadeh AT. Selenuim-enriched probiotic fermented milk drink (DOOGH): characteristics and survival microorganisms. Int J Adv Biol Biomed Res. 2014;2(4):397–403. [Google Scholar]
- 4.Khaledian S, Noroozi-Aghideh A, Kahrizi D, Moradi S, Abdoli M, Ghasemalian AH, et al. Rapid detection of diazinon as an organophosphorus poison in real samples using fluorescence carbon dots. Inorg Chem Commun. 2021;130:108676. [Google Scholar]
- 5.Sheikhshoaie I, Tohidiyan Z. A novel facile synthesis route for nano-sized zn (ii) schiff base complex and nano-sized znse/zno. Chem Methodol. 2019;3(1):30–42. [Google Scholar]
- 6.Dehno Khalaji A, Ghorbani M, Dusek M, Eigner V. The bis (4-methoxy-2-hydroxybenzophenone) copper (ii) complex used as a new precursor for preparation of CuO nanoparticles. Chem Methodol. 2020;4(2):143–51. [Google Scholar]
- 7.Alavi M, Hamblin MR, Kennedy JF. Antimicrobial applications of lichens: secondary metabolites and green synthesis of silver nanoparticles: a review. Nano Micro Biosystems. 2022;1(1):15–21. [Google Scholar]
- 8.Alkherraz AM, Ali AK, Elsherif KM. Equilibrium and thermodynamic studies of pb (II), zn (II), Cu (II) and cd (II) adsorption onto mesembryanthemum activated carbon. J Med Chem Sci. 2020;3(1):1–10. [Google Scholar]
- 9.Zhanga W, Chena Z, Liua H, Zhangb L, Gaoa P, Li D. Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonasalcaliphila. Colloids Surf B: Biointerfaces. 2011;88(1):196–202. doi: 10.1016/j.colsurfb.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Llamosas H, Castro L, Blázquez ML, Díaz E, Carmona M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci Rep. 2017;7:16046. doi: 10.1038/s41598-017-16252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkel LHE, Annette Johnson C, Lenz M, Grundl T, Leupin OX, Amini M, Charlet L. Environmental selenium research: from microscopic processes to global understanding. Environ Sci Technol. 2012;46(2):571–9. doi: 10.1021/es203434d. [DOI] [PubMed] [Google Scholar]
- 12.Sharma G, Sharma A, Bhavesh R, Park J, Ganbold B, Nam JS, et al. Biomolecule-mediated synthesis of selenium nanoparticles using dried Vitis vinifera (raisin) extract. Molecules. 2014;19(3):2761–70. doi: 10.3390/molecules19032761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava P, Bragança J, Ramanan SR, Kowshik M. Synthesis of silver nanoparticles using haloarchaeal isolate Halococcus salifodinae BK 3. Extremophiles. 2013;17(5):821–31. doi: 10.1007/s00792-013-0563-3. [DOI] [PubMed] [Google Scholar]
- 14.Wadhwani SA, Shedbalkar UU, Singh R, Chopade BA. Biogenic selenium nanoparticles: current status and future prospects. Appl Microbiol Biotechnol. 2016;100(6):2555–66. doi: 10.1007/s00253-016-7300-7. [DOI] [PubMed] [Google Scholar]
- 15.Shoeibi S, Mashreghi M. Biosynthesis of selenium nanoparticles using Enterococcus faecalis and evaluation of their antibacterial activities. J Trace Elem Med Biol. 2017;39:135–9. doi: 10.1016/j.jtemb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Alavi M, Rai M, Alencar Menezes I. Therapeutic applications of lactic acid bacteria based on the nano and micro biosystems. Nano Micro Biosyst. 2022;1:8–14. [Google Scholar]
- 17.Olfati A, Kahrizi D, Balaky ST, Sharifi R, Tahir MB, Darvishi E. Green synthesis of nanoparticles using Calendula officinalis extract from silver sulfate and their antibacterial effects on Pectobacterium caratovorum. Inorg Chem Commun. 2021;125:108439. [Google Scholar]
- 18.Nieto J, Fernandez-Castillo R, Marquez M, Ventosa A, Quesada E, Ruiz-Berraquero F. Survey of metal tolerance in moderately halophilic eubacteria. Appl Environ Microbiol. 1989;55(9):2385–90. doi: 10.1128/aem.55.9.2385-2390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava P, Kowshik M. Biosynthesis of nanoparticles from halophiles. Halophiles: Springer; 2015.
- 20.Mirmoeini T, Pishkar L, Kahrizi D, Barzin G, Karimi N. Phytotoxicity of green synthesized silver nanoparticles on Camelina sativa L. Physiol Mol Biol Plants. 2021;27:417–27. doi: 10.1007/s12298-021-00946-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma D, Kanchi S, Bisetty K. Biogenic synthesis of nanoparticles: a review. Arab J Chem. 2019;12(8):3578–600. [Google Scholar]
- 22.Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvan PT, Venketesan R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram-negative bacteria. Nanomed. 2010;6(1):103–9. doi: 10.1016/j.nano.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Pantidos N, Horsfall LE. Biological synthesis of metallic nanoparticles by bacteria, fungi and plants. J Nanomed Nanotechnol. 2014;5(5):1. [Google Scholar]
- 24.Amoozegar MA, Hajighasemi M, Ashengroph M, Razav MR. Salinicoccus iranensis sp. nov., a novel moderate halophile. Int J Syst Evol Microbiol. 2008;58(1):178–83. doi: 10.1099/ijs.0.65221-0. [DOI] [PubMed] [Google Scholar]
- 25.Shakibaie M, Khorramizadeh MR, Faramarzi MA, Sabzevari O, Shahverdi AR. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase-2 expression. Appl Biochem Biotechnol. 2010;56(1):7–15. doi: 10.1042/BA20100042. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava N, Mukhopadhyay M. Biosynthesis and structural characterization of selenium nanoparticles using Gliocladium roseum. J Clust Sci. 2015;26:1473–82. [Google Scholar]
- 27.Hernández-Díaz JA, Garza-García JJ, León-Morales JM, Zamudio-Ojeda A, Arratia-Quijada J, Velázquez-Juárez G, et al. Antibacterial activity of biosynthesized selenium nanoparticles using extracts of Calendula officinalis against potentially clinical bacterial strains. Molecules. 2021;26(19):5929. doi: 10.3390/molecules26195929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava P, Braganca JM, Kowshik M. In vivo synthesis of selenium nanoparticles by Halococcus salifodinae BK18 and their anti-proliferative properties against HeLa cell line. Biotechnol Prog. 2014;30(6):1480–7. doi: 10.1002/btpr.1992. [DOI] [PubMed] [Google Scholar]
- 29.Asghari-Paskiabi F, Imani M, Razzaghi-Abyaneh M, Rafii-Tabar H. Fusarium oxysporum, a bio-factory for nano selenium compounds: synthesis and characterization. Sci Iran. 2015;25(3):1857–63. [Google Scholar]
- 30.Srivastava N, Mukhopadhyay M. Green synthesis and structural characterization of selenium nanoparticles and assessment of their antimicrobial property. Bioprocess Biosyst Eng. 2015;38:1723–30. doi: 10.1007/s00449-015-1413-8. [DOI] [PubMed] [Google Scholar]
- 31.Shubharani R, Mahesh M. Biosynthesis and characterization, antioxidant and antimicrobial activities of selenium nanoparticles from ethanol extract of bee propolis. J Nanomed Nanotechnol. 2019;10(1):2. [Google Scholar]
- 32.Fritea L, Laslo V, Cavalu S, Costea T, Vicas SI. Green biosynthesis of selenium nanoparticles using parsley (Petroselinum crispum) leaves extract. Stud Univ Vasile Goldis Arad Ser Stiint Vietii. 2017;27(3):203–8. [Google Scholar]
- 33.Avendaño R, Chaves N, Fuentes P, Sánchez E, Jiménez JI, Chavarría M. Production of selenium nanoparticles in Pseudomonas putida KT2440. Sci Rep. 2016;6:37155. doi: 10.1038/srep37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kora AJ. Bacillus cereus, selenite-reducing bacterium from contaminated lake of an industrial area: a renewable nanofactory for the synthesis of selenium nanoparticles. Bioresour Bioprocess. 2018;5:30. [Google Scholar]
- 35.Forootanfar H, Adeli-Sardou M, Nikkhoo M, Mehrabani M, Amir-Heidari B, Shahverdi AR, et al. Antioxidant and cytotoxic effect of biologically synthesized selenium nanoparticles in comparison to selenium dioxide. J Trace Elem Med Biol. 2014;28(1):75–9. doi: 10.1016/j.jtemb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Kazemi M, Akbari A, Soleimanpour S, Feizi N, Darroudi M. The role of green reducing agents in gelatin-based synthesis of colloidal selenium nanoparticles and investigation of their antimycobacterial and photocatalytic properties. J Clust Sci. 2019;30:767–775. [Google Scholar]
- 37.Sengar MS, Saxena S, Satsangee SP, Jain R. Silver nanoparticles decorated functionalized multiwalled carbon nanotubes modified screen printed sensor for the voltammetric determination of butorphanol. J Appl Organometal Chem. 2021;1(2):95–108. [Google Scholar]
- 38.Sharma G, Sharma SB. Synthetic impatienol analogues as potential cyclooxygenase-2 inhibitors: a preliminary study. J Appl Organometal Chem. 2021;1(2):66–75. [Google Scholar]
- 39.Hatami A, Azizi Haghighat Z. Evaluation of application of drug modeling in treatment of liver and intestinal cancer. Prog Chem Biochem Res. 2021;4(2):220–33. [Google Scholar]
- 40.Saadatmand M, Abbasi F. Evaluating factors affecting reproductive success by intrauterine fertilization: the cases in Qom University Jihad Center, 2018–2019. Prog Chem Biochem Res. 2021;4(2):148–59. [Google Scholar]
- 41.Oladipupo AR, Alaribe CS, Akintemi TA, Coker HAB. Effect of Phaulopsis falcisepala (Acanthaceae) leaves and stems on mitotic arrest and induction of chromosomal changes in meristematic cells of Allium Cepa. Prog Chem Biochem Res. 2021;4(2):134–47. [Google Scholar]
- 42.Korir BK, Kibet J, Mosonik BC. A review of the current trends in the production and consumption of bioenergy. Prog Chem Biochem Res. 2021;4(2):109–33. [Google Scholar]
- 43.Khosravanian A, Moslehipour A, Ashrafian H. A review on bioimaging, biosensing, and drug delivery systems based on graphene quantum dots. Prog Chem Biochem Res. 2021;4(1):44–56. [Google Scholar]
- 44.Cremonini E, Zonaro E, Donini M, Lampis S, Boaretti M, Dusi S, et al. Biogenic selenium nanoparticles: characterization, antimicrobial activity and effects on human dendritic cells and fibroblasts. Microb Biotechnol. 2016;9(6):758–71. doi: 10.1111/1751-7915.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhanjal S, Cameotra SS. Aerobic biogenesis of selenium nanospheres by Bacillus cereus isolated from coalmine soil. Microb Cell Fact. 2010;9:52. doi: 10.1186/1475-2859-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alagesan V, Venugopal S. Green synthesis of selenium nanoparticle using leaves extract of Withania somnifera and its biological applications and photocatalytic activities. Bionanoscience. 2019;9:105–16. [Google Scholar]
- 47.Kannan S, Mohanraj K, Prabhu K, Barathan S, Sivakumar G. Synthesis of selenium nanorods with assistance of biomolecule. Bull Mater Sci. 2014;37:1631–5. [Google Scholar]
- 48.Clogston JD, Patri AK. Zeta potential measurement. Characterization of nanoparticles intended for drug delivery. Methods Mol Biol. 2011;697:63–70. doi: 10.1007/978-1-60327-198-1_6. [DOI] [PubMed] [Google Scholar]
- 49.Mollania N, Tayebee R, Narenji-Sani F. An environmentally benign method for the biosynthesis of stable selenium nanoparticles. Res Chem Intermed. 2016;42:4253–71. [Google Scholar]
- 50.Vekariya KK, Kaur J, Tikoo K. ERα signaling imparts chemotherapeutic selectivity to selenium nanoparticles in breast cancer. Nanomed. 2012;8(7):1125–32. doi: 10.1016/j.nano.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Zare B, Babaie S, Setayesh N, Shahverdi AR. Isolation and characterization of a fungus for extracellular synthesis of small selenium nanoparticles. Nanomed J. 2013;1(1):13–9. [Google Scholar]
- 52.Zoidis E, Seremelis I, Kontopoulos N, Danezis GP. Selenium-dependent antioxidant enzymes: actions and properties of selenoproteins. Antioxidants. 2018;7(5):66. doi: 10.3390/antiox7050066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao RZ, Jiang S, Zhang L, Yu ZB. Mitochondrial electron transport chain, ROS generation and uncoupling. Int J Mol Med. 2019;44(1):3–15. doi: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. 1. Synthesis of SeNPs using bacteria.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.