Abstract
Link protein is encoded by the Hapln1 gene and is a prototypical protein found in the cartilage matrix. It acts as an important component of the endochondral skeleton during early development. To study its transcriptional regulation, promoter fragments derived from the link protein gene were coupled to the β-galactosidase reporter and used to study in vivo transgene expression in mice. In day 15.5 mouse embryos, a link promoter fragment spanning −1020 to +40 nucleotides demonstrated highly specific β-galactosidase staining of skeletal structures, including the appendicular and axial cartilaginous tissues. Two shorter promoter fragments, spanning −690 to +40 and −315 to +40 nucleotides, demonstrated limb- and genitalia-specific expression resembling that of homeodomain-regulated tissues. Bioinformatic analysis revealed a highly conserved, Hox-like binding site (HLBS) at approximately −220 bp of the promoter, shared by both constructs, which contained the Hox-core consensus sequence TAATTA. Electromobility shift assays demonstrated binding of Hox-B4 recombinant protein to the HLBS, which was eliminated with nucleotide substitutions within the core-binding element. Co-transfection analysis of the HLBS demonstrated a 22-fold transcriptional activation by HoxA9 expression, which was ablated with a substitution within the core HLBS element. Together these findings establish promoter regions within the Link protein gene that are important for in vivo expression and identify the potential role of homeodomain-containing proteins in controlling cartilage and limb gene expression.
Keywords: cartilage, Hox proteins, limb, Link protein, promoter, transcriptional regulation
1. Introduction
Link proteins are small proteoglycans that are encoded by four different genes (Hapln1-Hapln4) [1–3]. The major cartilage link protein encoded by the Hapln1 gene (hereafter referred to as link protein) is an important component of cartilage and forms the initial skeletal structure during embryonic development [4]. Link protein contributes to the structure of cartilaginous matrix by stabilizing interactions with other components, including aggrecan and hyaluronan [5, 6]. Highlighting its role in cartilage formation, link protein has been identified as the most highly up-regulated gene during in vivo chondrogenesis of the limb [7]. The mRNA expression of link protein is increased approximately 200-fold between embryonic day 11.5 to 13.5, a time period of mesenchymal condensation to form the precursor to the limb tibia. Besides its role in cartilage, link protein also plays an important role in the organization of extracellular matrices from other tissues, including the heart [8–10], brain [11, 12], ovaries [13] and teeth [14].
Link protein has been studied extensively, particularly for its structural role and expression in skeletal cartilage [2, 15, 16]. High levels of link protein mRNA are observed during the spatial and temporal formation of cartilaginous tissues in the chicken limb and human skeletal structures [2, 15]. Experiments using chick endochondral chondrocytes have shown that transcriptional regulation is the major mechanism controlling link gene expression and involves both a promoter and a 2.2 kb first intron enhancer [17]. Importantly, the link protein gene promoter contains multiple regulatory elements [9, 17–21]. Bioinformatic analysis of the rat, human, mouse and chicken link promoters demonstrate extensive regions of sequence homology, suggesting they likely contain a conserved, transcriptional regulatory network [17, 22]. One such regulatory element in the link promoter includes a serum and glucocorticoid responsiveness region at −918 to −906 [20]. Interestingly, another cartilage gene, collagen II, also contains a similar DNA element which is found in the enhancer region [23], possibly coordinating the expression of multiple cartilage genes through a common regulatory element. Several other regulatory sequences have also been identified and confirmed by overexpression in cell cultures of transcription factors including Sox9 [21], Runx1 and Runx2 [24], and Mef2 [25]. Experiments with preovulatory granulosa cells of the ovary found that multiple Runx1 binding sites were present in the link protein promoter, and co-expression of the Runx1 transcription factor up-regulated link protein promoter activity [24]. Studies with Mef2 also found that this transcription factor bound to two similar promoter elements. and coexpression activated the link promoter activity in heart valve interstitial cells [25]. Understanding the in vitro promoter activity of the link protein promoter has been less-well studied in chondrocytes, in part due to difficulties in maintaining the normal phenotype of these cells in cell culture [26].
Despite the in vitro studies of the link protein gene promoter, the tissue-specific activity of the promoter is not always recapitulated in cell culture. For example, Hela cells, which do not make significant amounts of link protein, show similar promoter reporter activity as chicken embryonic chondrocytes [17]. An explanation proposed for this discrepancy was that plasmids introduced by transient transfection are episomal and do not adopt the normal chromatin structure required for cell-specific expression. Moreover, little is known about the function of the link protein gene promoter in vivo and how it controls gene expression. Here, the transcriptional regulation of the link promoter was analyzed using transgenic mice in vivo to identify important DNA elements that confer spatial and restricted expression.
2. Materials and methods
2.1. Generation of transgenic mice harboring link protein promoter-LacZ
All the experiment described in this manuscript complied with National Institutes of Health guidelines for the care of laboratory animals and was approved by the ACUC of the NIDCR, NIH.
Three rat link protein gene (Hapln1) promoter sequences were engineered to drive β-galactosidase (LacZ) expression. The largest link protein promoter fragment (from accession number X55057), spanned −1020 to +40 nucleotides (hereafter referred to as −1020) and included most of the first, untranslated exon of the gene [17]. Similarly, two shorter link promoter fragments were also constructed that encompassed the −690 to +40 nucleotides (hereafter referred to as −690) and −315 to +40 nucleotides (hereafter referred to as −315) DNA regions. For construction of these promoter-LacZ expression cassettes, the link protein gene promoter fragments were amplified by PCR and then subcloned into the pCH110 vector [27].
These three promoter plasmids were linearized and purified by gel electrophoresis to contain the link protein promoter-LacZ cassette. These DNA fragments were then injected into the pronucleus of fertilized single-cell mouse oocytes. Successful integration of the transgene was determined by PCR using genomic DNA isolated from mouse tails or fetal placenta. All transgenic mice were screened for the presence of the LacZ gene sequence, and mouse lines with robust limb-specific expression were identified. The −690 promoter-LacZ and −315 promoter-LacZ were back-crossed for more than five generations; they showed Mendelian inheritance and were used for further analysis.
2.2. Analysis of transgene link gene promoter β-galactosidase activity
Timed-pregnancy mice were generated, and day 15.5 embryos were harvested. The embryos were fixed for 20 min (4% paraformaldehyde, 1X PBS), washed twice with 1X PBS, and then washed two more times for 20 min each with 0.1 M phosphate buffer (pH 7.3, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P-40). β-galactosidase activity was detected through the addition of a substrate solution containing 5-bromo-4-chloro-3-indoyl-Β-D-galactosidase (X-gal at 1.0 mg/ml), 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide and incubated overnight at 30°C. The embryos were then photographed for documentation of the X-gal staining pattern.
2.3. Bioinformatic analysis of the −315 link protein gene promoter
Based on the expression pattern of the −315 bp promoter region of the rat link protein gene promoter, motif-based sequence analysis tools (http://meme-suite.org/doc/overview.html) were employed to examine homology with the transcriptional regulatory regions of other cartilage genes. The bioinformatic analyses revealed a 19 bp element was shared with two similar sequences from known regulatory elements including the enhancer region of the aggrecan gene [28] and the promoter of Col11A2 [29]. Additional searches with this element, designated the Hox-like binding sequence (HLBS), within the rat link promoter were compared to the corresponding gene from other species to understand the level of conservation of this sequence. Alignment of the conserved element from these genes was performed using the CLUSTAL program of the McVector DNA™ analysis suite.
2. 4. Electrophoretic mobility shift assays of HLBS
Transcription factor binding to the HLBS in the link protein gene promoter was analyzed by electrophoretic mobility shift assay (EMSA). HEK-293 cells were transfected with a HoxB4 mammalian expression vector (pF5a-MCS, Sigma) expressing a 3x-FLAG epitope tag to generate lysates containing HoxB4 recombinant protein. These extracts were then used to purify HoxB4 by a standard FLAG affinity purification protocol, and HxB4 recombinant protein was eluted with 200 μg/ml of 3x-FLAG peptide (Sigma). HoxB4 protein binding activity to the HLBS sequence was then examined using wild type HLBS double-stranded oligonucleotides (5’-GTTAGGCTGTAATTAGAGGA-3), and two mutant forms: HLBS-Mut1 (5’-GTTAGATCA CAATTAGAG-3’ and HLBS-Mut2 (5’-GTTAGGCTGTGGCCGGAGGA-3’). For EMSA, these different double-stranded oligonucleotides were annealed and radiolabeled with 32P-ATP using T4-polynucleotide kinase. The 32P-labeled oligonucleotide probes (approximately 20,000 CPM) were incubated with and without the recombinant HoxB4 protein at 4°C for 30 min in nuclear protein binding buffer (20 mM Tris, pH 7.8, 100 mM NaCl, 40 mM KCl, 1 mM DTT). The protein-DNA mixture was then separated on a non-denaturing 5.5% polyacrylamide gel containing 0.25x TBE and 0.5 % tween. Following electrophoresis, the gels were fixed and processed for autoradiography.
2. 5. Transcriptional activity studies with HLBS
Transcriptional activity of the HLBS was analyzed by in vitro reporter assays performed in cell culture. For these transcriptional studies, we employed a common strategy using the transactivation domain of VP16 as a fusion protein to Hox and other transcription factors [30–34]. In order to generate this reagent, a CMV-Flag epitope-tagged VP16 expression vector was first constructed. This plasmid was then used to clone the full-length, murine Hoxa9 transcription factor downstream of the VP16 activation domain, and the resulting mammalian expression vector was termed pCMV-VP16- Hoxa9. To test the activity of the HLBS in the absence of other regulatory regions of the link promoter, we used a heterologous system for analysis. Briefly, the 20 bp sequence of HLBS was concatenated to contain four direct repeats of this sequence. For analysis, the wild type HLBS (5’-GTTAGGCTGTAATT AGAGGA-3)’ and four additional mutants were constructed in a similar multimeric fashion to include HLBS-Mut1 (5’-GTTAGATCACAATTAGAGG-3’), HLBS-Mut2 (5-GTTAGGCTGT GGCCGGAGGA-3’), HLBS-Mut3 (5’-ACCGAGCTGTAATTAGAGGA-3’), and HLBS-Mut4 (5’-GTTAGGCTGTA ATTAAGAAG-3’). The multimeric target sequences were cloned in the KpnI to XhoI site upstream of the basal pGL4.23 plasmid expressing firefly luciferase as the transcriptional promoter reporter output to generate pGL4.23-4X HLBS, pGL4.23-4X HLBS-Mut1, pGL4.23-4X HLBS--Mut2, pGL4.23-4X HLBS-Mut3, and pGL4.23-4X HLBS-Mut4. DNA sequencing was used to confirm the identity of these, and other plasmids described above.
Briefly for transfection, HEK-293T cells were maintained in DMEM-5% FCS. Approximately 18 h before transfection, the culture medium was replaced with DMEM without serum and incubated overnight. The next day, the cells were trypsinized and seeded into 12-well plates at 70% confluency containing 1% FCS. Each well was then cotransfected with 1 μg of either empty vector (pGL4.23), wild type pGL4.23-4X HLBS, or the corresponding mutant targets along with 25 ng of pCMV-VP16- HoxA9, and 25 ng of CMV-Renilla luciferase. Transfections were performed in duplicate, and at 48 h post-transfection, the cells were lysed in 300 μl of passive lysis buffer (Promega). For measuring and normalizing transcriptional activity in the cell lysates, 30 μl of the cell extract was sequentially assayed for firefly and Renilla luciferase activity, respectively, with a luminometer (Berthold LB 940). Transcriptional activity was normalized to Renilla luciferase activity. Data was plotted as fold increase compared to the pGL4.23 basal promoter cotransfected with a CMV expression vector without a gene insert.
3. Results
3.1. Link protein gene promoter activity in transgenic mouse embryos
Previous studies have shown that the rat link promoter is transcriptionally active in a variety of cell types in cell culture [17, 20, 24]. Here we sought to analyze the rat link promoter in vivo to study its spatial and temporal expression during embryonic development. For these experiments, the rat link protein gene promoter region was engineered to drive β-galactosidase reporter production in mice. Initially, two different link protein gene promoter fragments were used (Fig. 1A). These two DNA constructs were then microinjected into oocytes and expression was determined in the developing mouse embryo. As shown, the −1020 link promoter region in the E15.5 day embryos demonstrated tissue-specific expression in multiple cartilaginous tissues including cervical vertebrae (C1 and C2), radius, ulna, tibia, and fibula (Fig. 1B). No significant expression was seen in other tissues. Sectioning and microscopy of the tibial tissue confirmed expression of the β-galactosidase reporter by the hypertrophic chondrocytes (data not shown). These findings highlight the marked cartilage-specific in vivo expression of this −1020 link protein gene promoter region.
Fig 1.
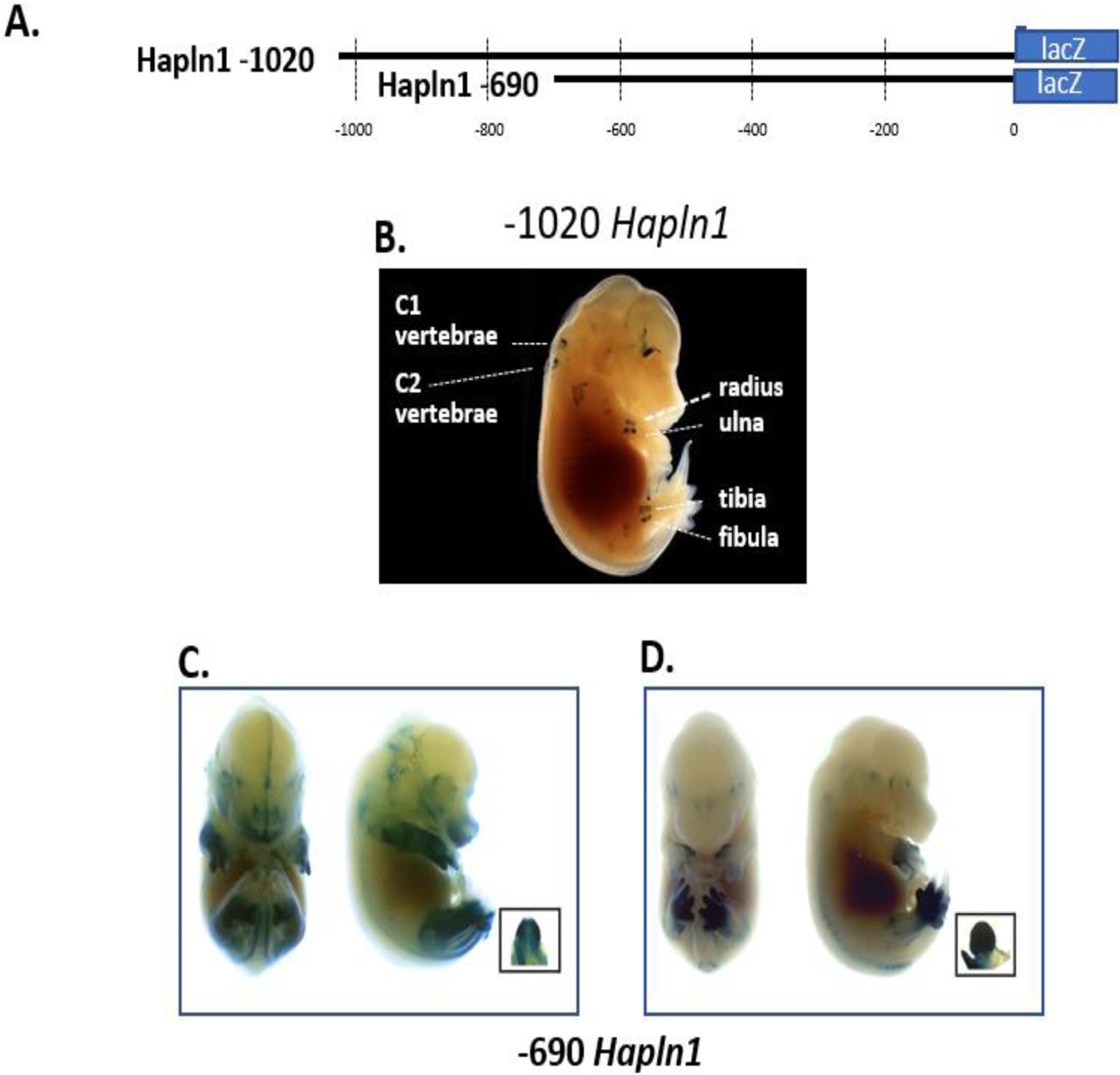
Link protein gene promoter constructs show in vivo expression patterns in either the skeleton or limb/genitalia. (A) Two different link protein promoter fragments consisting of the −1020 and −690 nt regions were tested in transgenic mice. (B) A representative E15.5 mouse transiently expressing the −1020 promoter region showed only X-gal staining in appendicular and axial cartilaginous skeletal tissues. A dark background was used to provide contrast to the photograph to allow easy visualization of the discrete expression in cartilage sites. (C) and (D) The shorter −690 promoter region demonstrated high-level expression in the limb and genitalia as shown in two representative mouse embryos. As shown, embryos showed variability of X-gal staining in the limb and hind paw regions.
To further delineate the DNA regulatory elements within the promoter responsible for this transcriptional activity, an additional shorter −690 promoter fragment was tested. As shown in Fig. 1C and Fig. 1D, the −690 promoter showed a different expression pattern (5 of the 7 mice) predominantly in the front and hind limbs and external embryonic genitalia, structures that will become fully cartilaginous tissues. However, this X-gal expression pattern showed spatial variability from mouse to mouse (Fig. 1C and Fig. 1D). Different embryos showed varying boundaries of expression in proximal-distal and anterior-posterior regions of the limbs. In particular, some embryos demonstrated more posterior than anterior expression in the paw and limb (Fig. 1C) compared to mainly in the paw (Fig. 1 D). In other embryos, the −690 promoter region demonstrated strong hind limb-specific expression compared to the forepaw (Fig. 1 D). These results highlight that the −1020 promoter shows cartilage-specific expression, while the −690 promoter region confers limb and genitalia expression.
3.2. The −315 link promoter drives high levels of limb expression in transgenic mouse lines
Mouse transgenic lines expressing the link promoter-LacZ transgenes were also generated. For the mouse line studies, the −690 region and a shorter −315 promoter were tested (Fig. 2A). Analysis of the −690 link promoter recapitulated the expression pattern seen with transient expression and showed selective limb and genital expression (Fig. 2B). However, the −315 link promoter region was even more active with high levels of X-gal staining in the limbs, genitalia and additional expression in the epithelial layer of the skin (Fig. 2C). Further deletion of the promoter region to −183 showed no X-gal expression in mice, suggesting that in vivo this region is insufficient for basal promoter activity (data not shown). These findings of both limb and genitalia-specific expression of the both the −690 and −315 promoter are reminiscent of known Hox-expression patterns [35, 36].
Fig 2.
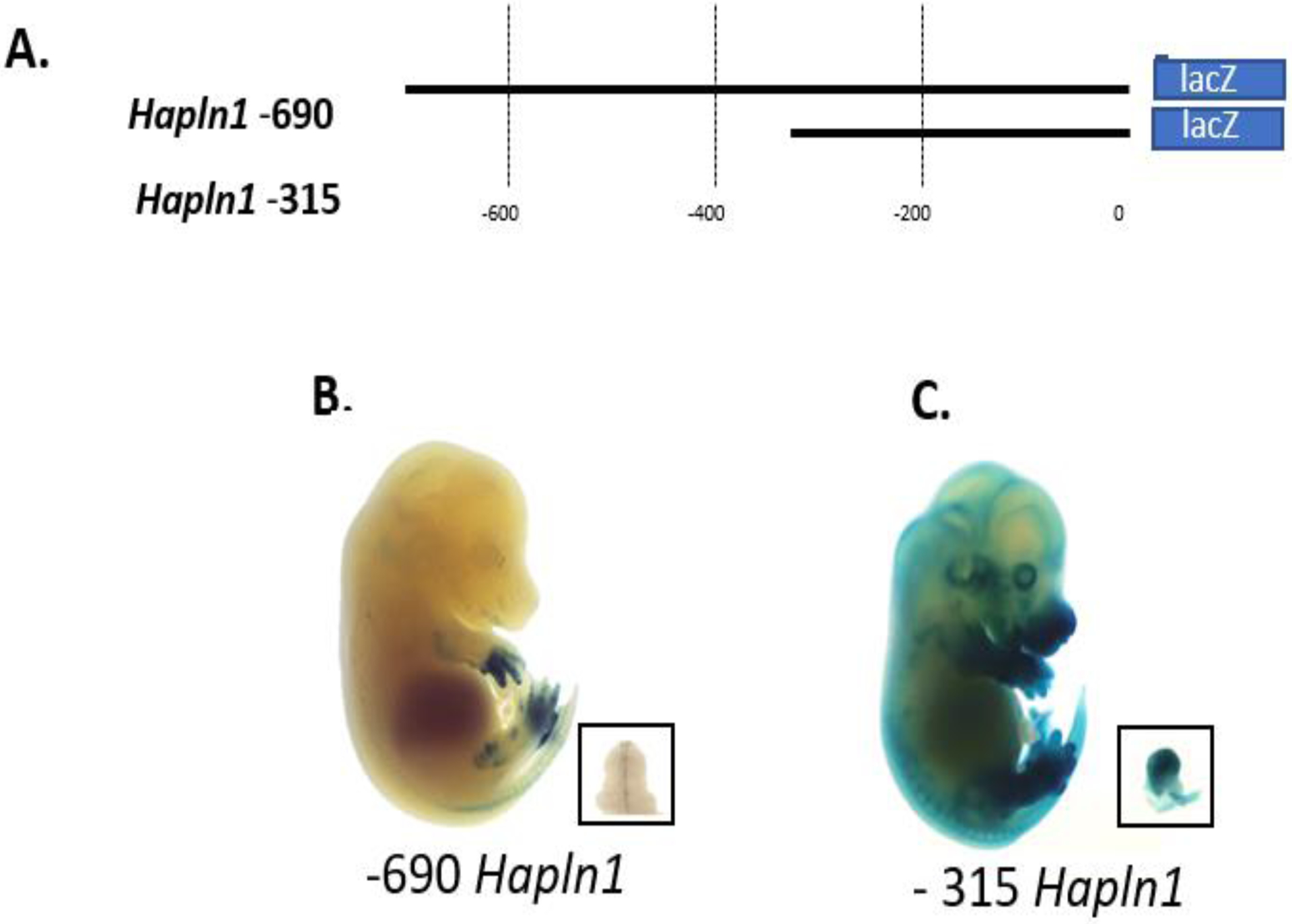
Further shortening of the link protein gene promoter reveals in vivo X-gal staining in the limb and genitalia. (A). Two link protein gene promoter regions −690 and −315 were studied in transgenic mouse lines. (B) Representative E15.5 mouse expressing the −690 promoter shows intense X-gal staining in the limb and genitalia. (C) Similarly, E15.5 mice expressing the −315 promoter also showed intense X-gal staining in the limb and genitalia with additional expression in the epidermis. Boxes indicate genitalia.
3.3. Identification of a potential homeobox binding element in the link promoter
Based on the finding that both the −690 and −315 link promoter constructs exhibited high levels of limb and genitalia expression in vivo, bioinformatic analysis was performed with this promoter fragment to determine whether shared, similar regulatory elements might be found in other cartilage genes. For this bioinformatic analysis, the −315 link promoter sequence was evaluated with the Motif-Based Sequence Analysis program and used to screen known regulatory regions from three other cartilage-specific genes including collagen II-alpha 1 (Col2A1), collagen XI-alpha 2 (Col11A2) and aggrecan (ACAN). This search revealed that a 29 bp sequence derived from the −230 to −202 region of the link promoter demonstrated significant homology with sequences found in the basal promoter region of collagen 11A2 and within the known enhancer of the aggrecan gene [28] (Fig. 3A). This highly conserved 29 bp element was a markedly AT-rich element and contained a core TAATT sequence, which resembled the key recognition motif of homeodomain transcription factors [37, 38]. Since this element located within the −315 link promoter was highly active in driving in vivo transcription in the limb and genitalia and resembled expression patterns seen by homeobox- containing proteins, we tentatively designated this element as a Hox-like binding site (HLBS). Moreover, the HLBS was highly, evolutionarily conserved within the corresponding link genes of human, chicken, frog and even, coelacanth (lungfish) further supporting its potential importance (Fig. 3A). Overall these findings suggest the possibility that the HLBS might function in coordinately regulating transcription of the link gene and multiple other cartilage genes such as aggrecan and collagen 11A2.
Fig 3.
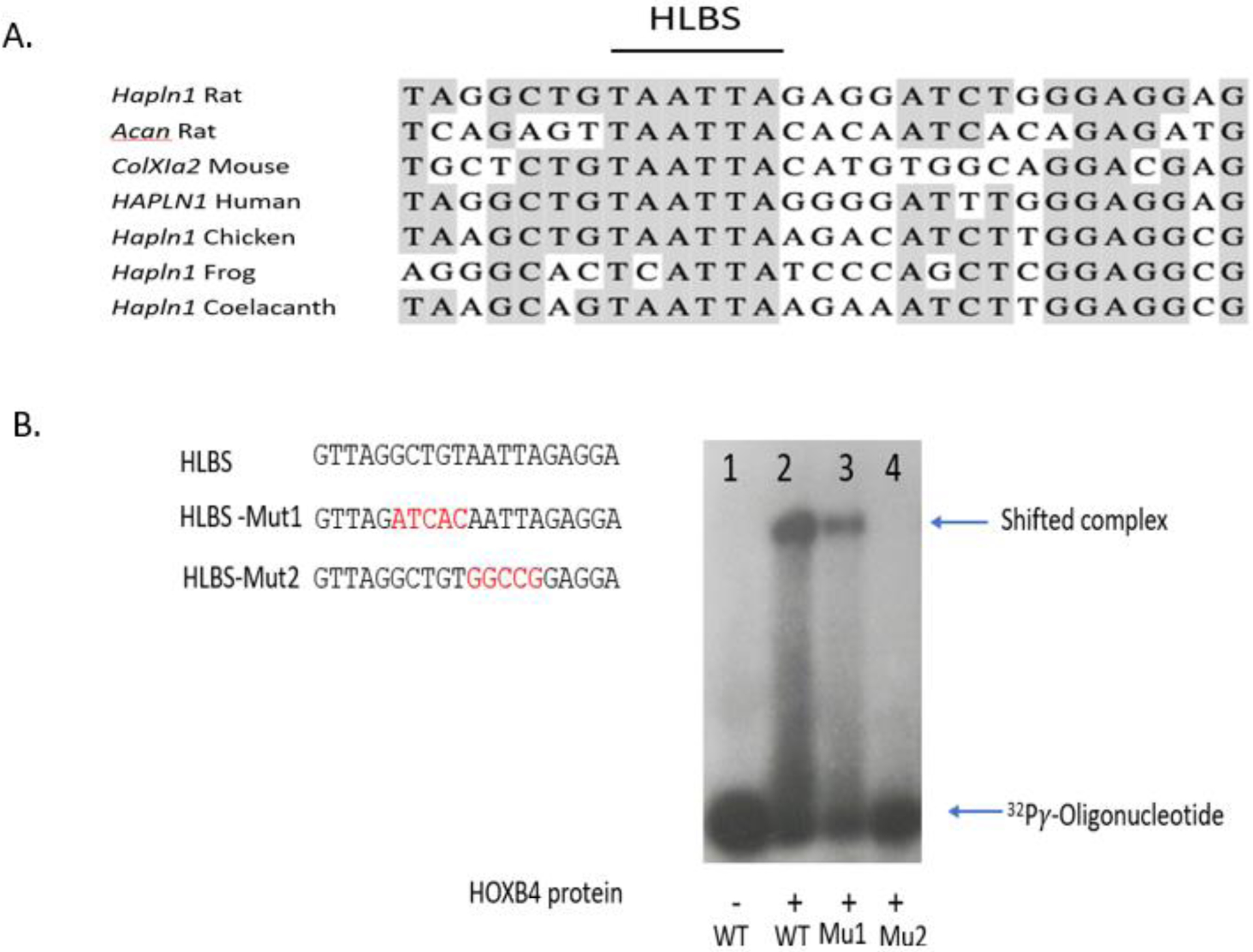
Identification of a functional Hox-like binding site (HLBS) in the link protein promoter. (A) Bioinformatic analysis of the −315 link protein promoter identified a 29 bp Hox-like binding site (HLBS) that was similar to sequences found within regulatory regions of two other cartilage genes, the aggrecan enhancer and basal Col11A2 promoter. The AT-rich core Hox-binding consensus is shown by the horizontal black line. Additional comparisons showed the HLBS was evolutionary conserved in the link protein gene promoters from frogs to humans. (B) HoxB4 binding to the HLBS in the link protein promoter. EMSA was performed with recombinant HoxB4 target protein along with wild type HLBS and mutant HLBS oligos (red font denotes nucleotide substitutions from wild type). Strong HoxB4 binding activity was observed with wild type HLBS oligonucleotide. In comparison, the HLBS-Mut1 target sequence showed diminished binding activity and the HLBS-Mut2, replacing the core AATTA sequence, was completely inactive.
3.4. HoxB4 binding to HBLS in the link promoter
To examine whether the HLBS could be bound by a homeodomain protein, electrophoretic mobility shift assays (EMSA) were performed. For this EMSA analysis, double-stranded oligonucleotides derived from HLBS were radioactively labelled. The HoxB4 protein was used as a candidate Hox protein to study potential binding to the HBLS because of its known role in regulating the murine axial skeleton, since Hoxb4 null mice show homeotic transformation of the vertebrae [39]. To generate sufficient quantities of HoxB4 protein, a mammalian expression vector for FLAG epitope-tagged-HoxB4 was overexpressed in HEK-293 cells, and then the lysate containing the recombinant protein was affinity-purified as described in Materials and Methods. As shown from the representative autoradiogram in Fig. 3B, Lane 2, addition of the recombinant HoxB4 protein to the radioactively labelled HLBS caused a marked shift up of the protein-DNA complex indicative of binding activity. In contrast, this protein-DNA complex was not seen in the absence of HoxB4 protein (Fig. 3B, Lane 1). To determine whether HoxB4 protein binding to MLBS showed specificity for the target DNA sequence, oligonucleotides containing substitutions within the HLBS were tested. One mutant, HLBS-Mut1, replacing 5 bp of flanking nucleotides adjacent to the core TAATT motif had a modest decline in HoxB4 binding (Fig. 3B, lane 3). In contrast, a second mutant oligonucleotide, HLBS-Mut2, replacing 5 bp of the core Hox-binding consensus, showed a marked loss of binding (Fig. 3B, lane 4). Overall these results confirm that HoxB4 protein bound in vitro to the HLBS of the link promoter and this binding activity was sequence-specific.
3.5. Homeobox Activation of HLBS
Based on the finding that a Hox protein bound the HLBS, we next explored whether the HLBS was involved in controlling gene expression in vitro by Hox transcription factors. Since the HoxA9 protein is expressed in the limb [40, 41], co-transfection experiments were performed to test whether this Hox transcription factor had the potential to activate transcription. Following on previous studies [30, 32–34], a chimeric Hoxa9-VP16 expression vector was utilized in order to generate a transcription factor with strong activation activity. For these studies, a concatemerized 4X HLBS target sequence was placed upstream of a basal promoter driving the reporter of firefly luciferase. In co-transfection experiments, the 4X HLBS promoter construct showed low activity of approximately 3-fold without co-expression of HoxA9 (Fig. 4). HoxA9-VP16 co-expression markedly increased MLBS promoter activity 22-fold. Co-expression of HoxA9-VP16 with 4X-HLBS-Mut1 showed a modest decrease in activity. In contrast, the HLBS-Mut2, removing the core AT-rich region which previously showed no Hox binding in EMSA, was completely transcriptionally inactive when co-expressed with HoxA9 (Fig. 4). The two additional mutant HLBS promoter constructs, HLBS-Mut3 and HLBS-Mut4 that were not tested in EMSA with altered flanking sequences had slightly similar activity to the wild type HLBS sequence (Fig. 4). Additional experiments with the −1020 link promoter luciferase reporter, rather than only HLBS by itself, showed that HoxA9 was able to increase the promoter activity by approximately 2-fold over background empty vector (data not shown). Taken together these results provide evidence that the HLBS is a target of HoxA9 and may function to control the expression of the link gene in vivo.
Fig 4.
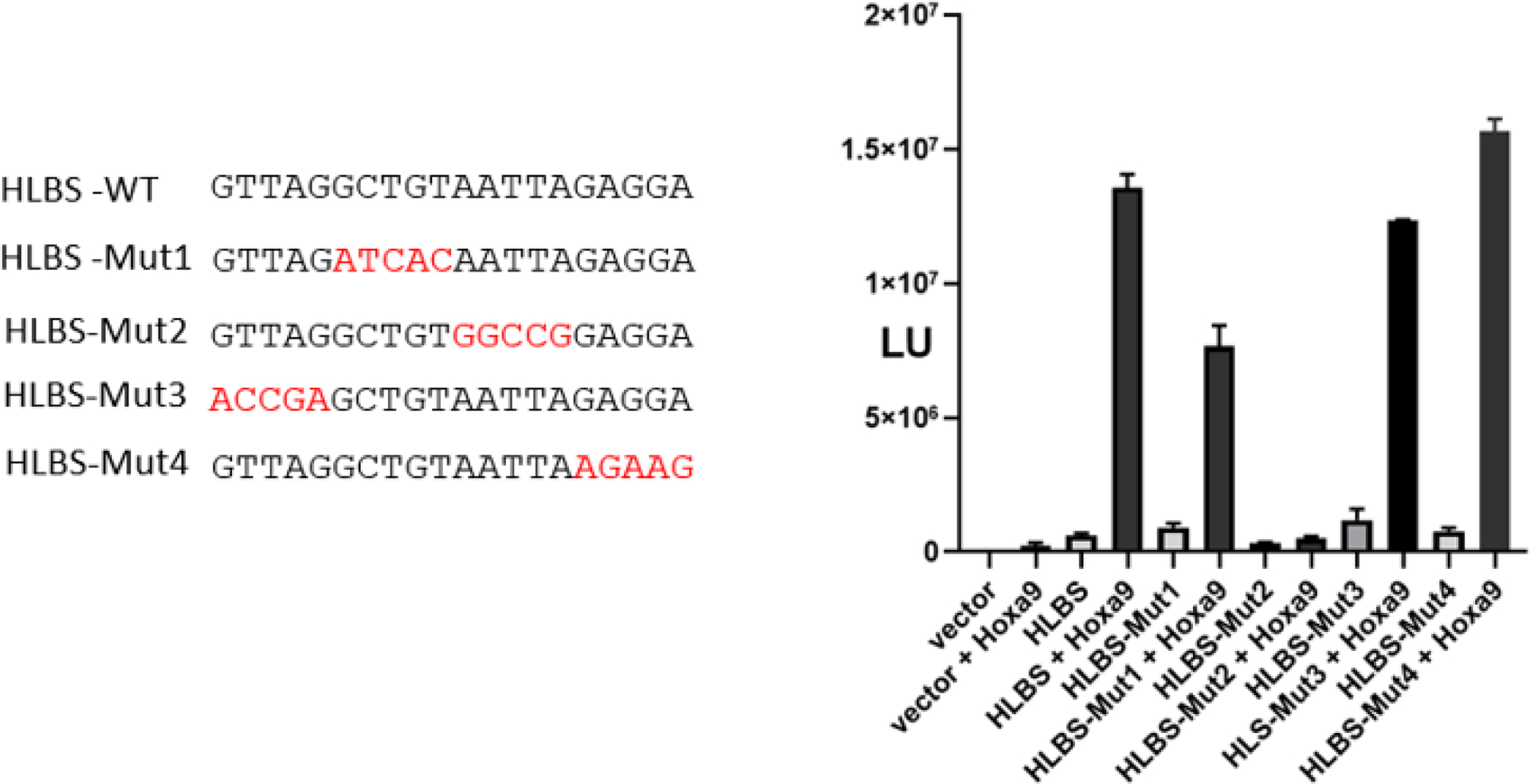
Transcriptional activation of HLBS by HoxA9-VP16 expression. Transient transfection experiments were performed in HEK-293 cells using a 4X HLBS basal promoter luciferase reporter system in conjunction with Hoxa9-VP16 expression. As shown by the left side of the figure, several mutant HLBS target sequences with nucleotide substitutions (as shown by red type) were tested. Forty hours later after transfection, luciferase activity was analyzed. Luciferase activity is plotted as fold increase over. Strong activation was observed with wild type HLBS, but the HLBS-Mut2, replacing the core AATTA sequence, was inactive.
4. Discussion
Previous analyses of the cell-specific transcriptional regulation of the link protein gene and other cartilage genes using transfection approaches have been limited, mainly due to the episomal nature of the transfected plasmid DNA and the difficulty of growing differentiated chondrocytes in cell culture. Using an in vivo approach, we show here that mouse embryos expressing the link promoter driving a β-galactosidase reporter recapitulated the known expression pattern of the endogenous link protein gene. Using this heterologous system, the largest −1020 link promoter fragment exhibited a highly tissue-specific expression pattern in axial and appendicular skeletal structures resembling the normal expression of the link protein in vivo. A striking pattern of X-gal staining was observed in skeletal tissues including maturing and hypertrophic cartilage, tissue sources enriched in link protein and aggrecan [2]. These findings are also nearly identical to the expression pattern seen with regulatory regions of aggrecan using the β-galactosidase reporter [28]. These findings highlight how the link protein gene promoter functions in vivo to control the spatial and temporal specific expression of link protein during embryonic development.
Based on the results with the large link promoter fragment, this in vivo expression system was further exploited to map and understand the regulatory networks involved in this highly regulated gene pattern using smaller link promoter fragments. The −690 link promoter fragment-β−galactosidase reporter, deleting about 500 bp from the promoter, showed a different expression pattern and demonstrated a visible loss of the skeletal-specific staining and a dramatic increase in expression in the limb, particularly at the distal limb margins. These findings are likely due to the removal of a repressor element and/or the loss of highly-selective regulatory sequence driving cartilage expression. One possibility is that one or more such suppressive elements are located within the −1020 to −690 region, possibly involving a serum-like response element at −923 [20]. Moreover, MEF2c binding to a −923 AT-rich element and a similar sequence at −720 are required for maximum activation by the MEF2c in heart valve interstitial cells, where alteration in these target sequences resulted in loss of activity (Lockhart, 2013).
A second deletion mutant containing −315 bp of the link promoter was visibly the most active exhibiting high levels of β-galactosidase in the limb, genitalia and with diffuse epithelial expression. Bioinformatic analysis of this small −313 promoter region revealed a potential Hox-like binding sequence that was evolutionarily conserved in link protein promoters and was shared with two other regulatory regions of cartilage genes. We explored this possibility further, demonstrating that the HLBS bound Hox protein in vitro and could activate gene transcription in a highly sequence-specific fashion.
Interestingly, contemporaneous whole genome chromatin immunoprecipitation (ChIP) analysis identified the in vivo DNA binding regions for HoxA13 and HoxD13 transcription factors using the distal portion of the limb from the E13.5-day mouse embryo [42]. Inspection of this dataset revealed that the link promoter was the target of these transcription factors, and the maximum peak HoxA13 and HoxD13 binding activity was found close to the −220 nt location of HLBS that we characterized in our study. Moreover, the HLBS region in the aggrecan gene was also found to bind HoxA13 transcription factor by ChIP analysis [42]. These results further validate that HLBS functions to control coordinated cartilage gene expression by at least HoxA13. Since Hox transcription factors normally function in regulatory networks by either heterotypic DNA associations and/or in hierarchical competition for cis-acting regulatory elements, this interplay of interactions may allow for fine tuning of the spatial and temporal expression of link protein. Due to the fact that the ChIP analysis only examined HoxA13 and HoxD13 transcription factors, we do know the normal physiologic contribution of other Hox proteins that might be involved in cartilage skeletal formation. Lastly, it is likely there are other Hox-binding sequences located in distant enhancers that also function to regulate link protein and other cartilage genes.
Based on the highly conserved link promoter regions between coelacanth, frogs and humans, there is likely to be a large amount of transcriptional information imbedded within this region. Our current findings along with our previous studies support a hierarchical model for gene regulation, in which Hox transcription factors play a substantial role in the developmental expression of this gene. The observation that collagen X1a2 promoter, and aggrecan enhancer also bind Hox transcription factors, in a similar fashion, presents a mechanistic pathway for the coregulation of these genes during the formation of the endochondral skeleton.
Supplementary Material
Highlights.
A 1.2 kb DNA fragment of the link protein gene promoter directed in vivo heterologous reporter expression in mouse embryonic endochondral skeleton including vertebrae and appendicular structures.
Two promoter deletion fragment, −690 and −315 nucleotides, demonstrated strong and selective expression in the limb and genitalia suggesting Hox transcription factor regulation.
A Hox-like binding sequences (HLBS) was identified at −220 in the link promoter and was also found in the regulatory regions of other cartilage genes, such as aggrecan and type XI collagen.
In vitro binding experiments demonstrated that HLBS bound Hox protein and transfection analysis revealed activation of the HLBS element by Hoxa9.
These findings highlight an important role of Hox proteins in the transcriptional regulation of link protein and other cartilage genes.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Dental and Craniofacial Research. We thank Dr. Peter D. Burbelo for critical reading of the manuscript.
Abbreviations:
- HLBS
Hox-like binding site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no competing or financial interests.
References
- [1].Dudhia J, Aggrecan, aging and assembly in articular cartilage, Cell Mol Life Sci, 62 (2005) 2241–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mundlos S, Meyer R, Yamada Y, Zabel B, Distribution of cartilage proteoglycan (aggrecan) core protein and link protein gene expression during human skeletal development, Matrix, 11 (1991) 339–346. [DOI] [PubMed] [Google Scholar]
- [3].Watanabe H, Yamada Y, Chondrodysplasia of gene knockout mice for aggrecan and link protein, Glycoconj J, 19 (2002) 269–273. [DOI] [PubMed] [Google Scholar]
- [4].Spicer AP, Joo A, Bowling RA Jr., A hyaluronan binding link protein gene family whose members are physically linked adjacent to chondroitin sulfate proteoglycan core protein genes: the missing links, J Biol Chem, 278 (2003) 21083–21091. [DOI] [PubMed] [Google Scholar]
- [5].Kimura JH, Hardingham TE, Hascall VC, Assembly of newly synthesized proteoglycan and link protein into aggregates in cultures of chondrosarcoma chondrocytes, J Biol Chem, 255 (1980) 7134–7143. [PubMed] [Google Scholar]
- [6].Neame PJ, Barry FP, The link proteins, Experientia, 49 (1993) 393–402. [DOI] [PubMed] [Google Scholar]
- [7].Cameron TL, Belluoccio D, Farlie PG, Brachvogel B, Bateman JF, Global comparative transcriptome analysis of cartilage formation in vivo, BMC Dev Biol, 9 (2009) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Binette F, Cravens J, Kahoussi B, Haudenschild DR, Goetinck PF, Link protein is ubiquitously expressed in non-cartilaginous tissues where it enhances and stabilizes the interaction of proteoglycans with hyaluronic acid, J Biol Chem, 269 (1994) 19116–19122. [PubMed] [Google Scholar]
- [9].Lockhart M, Wirrig E, Phelps A, Wessels A, Extracellular matrix and heart development, Birth Defects Res A Clin Mol Teratol, 91 (2011) 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wirrig EE, Snarr BS, Chintalapudi MR, O’Neal J L, Phelps AL, Barth JL, Fresco VM, Kern CB, Mjaatvedt CH, Toole BP, Hoffman S, Trusk TC, Argraves WS, Wessels A, Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development, Dev Biol, 310 (2007) 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ripellino JA, Margolis RU, Margolis RK, Immunoelectron microscopic localization of hyaluronic acid-binding region and link protein epitopes in brain, J Cell Biol, 108 (1989) 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kwok JC, Carulli D, Fawcett JW, In vitro modeling of perineuronal nets: hyaluronan synthase and link protein are necessary for their formation and integrity, J Neurochem, 114 (2010) 1447–1459. [DOI] [PubMed] [Google Scholar]
- [13].Kobayashi H, Sun GW, Hirashima Y, Terao T, Identification of link protein during follicle development and cumulus cell cultures in rats, Endocrinology, 140 (1999) 3835–3842. [DOI] [PubMed] [Google Scholar]
- [14].Pemberton TJ, Li FY, Oka S, Mendoza-Fandino GA, Hsu YH, Bringas P Jr., Chai Y, Snead ML, Mehrian-Shai R, Patel PI, Identification of novel genes expressed during mouse tooth development by microarray gene expression analysis, Dev Dyn, 236 (2007) 2245–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stirpe NS, Goetinck PF, Gene regulation during cartilage differentiation: temporal and spatial expression of link protein and cartilage matrix protein in the developing limb, Development, 107 (1989) 23–33. [DOI] [PubMed] [Google Scholar]
- [16].Shibata S, Fukada K, Imai H, Abe T, Yamashita Y, In situ hybridization and immunohistochemistry of versican, aggrecan and link protein, and histochemistry of hyaluronan in the developing mouse limb bud cartilage, J Anat, 203 (2003) 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rhodes C, Savagner P, Line S, Sasaki M, Chirigos M, Doege K, Yamada Y, Characterization of the promoter for the rat and human link protein gene, Nucleic Acids Res, 19 (1991) 1933–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Deak F, Barta E, Mestric S, Biesold M, Kiss I, Complex pattern of alternative splicing generates unusual diversity in the leader sequence of the chicken link protein mRNA, Nucleic Acids Res, 19 (1991) 6083–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dudhia J, Bayliss MT, Hardingham TE, Human link protein gene: structure and transcription pattern in chondrocytes, Biochem J, 303 ( Pt 1) (1994) 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rhodes C, Yamada Y, Characterization of a glucocorticoid responsive element and identification of an AT-rich element that regulate the link protein gene, Nucleic Acids Res, 23 (1995) 2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kou I, Ikegawa S, SOX9-dependent and -independent transcriptional regulation of human cartilage link protein, J Biol Chem, 279 (2004) 50942–50948. [DOI] [PubMed] [Google Scholar]
- [22].Deak F, Mates L, Krysan K, Liu Z, Szabo PE, Mann JR, Beier DR, Kiss I, Characterization and chromosome location of the mouse link protein gene (Crtl1), Cytogenet Cell Genet, 87 (1999) 75–79. [DOI] [PubMed] [Google Scholar]
- [23].Krebsbach PH, Nakata K, Bernier SM, Hatano O, Miyashita T, Rhodes CS, Yamada Y, Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein gene, J Biol Chem, 271 (1996) 4298–4303. [DOI] [PubMed] [Google Scholar]
- [24].Liu J, Park ES, Curry TE Jr., Jo M, Periovulatory expression of hyaluronan and proteoglycan link protein 1 (Hapln1) in the rat ovary: hormonal regulation and potential function, Mol Endocrinol, 24 (2010) 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lockhart MM, Wirrig EE, Phelps AL, Ghatnekar AV, Barth JL, Norris RA, Wessels A, Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart, PLoS One, 8 (2013) e57073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheng T, Maddox NC, Wong AW, Rahnama R, Kuo AC, Comparison of gene expression patterns in articular cartilage and dedifferentiated articular chondrocytes, J Orthop Res, 30 (2012) 234–245. [DOI] [PubMed] [Google Scholar]
- [27].Hall CV, Jacob PE, Ringold GM, Lee F, Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells, J Mol Appl Genet, 2 (1983) 101–109. [PubMed] [Google Scholar]
- [28].Doege K, Hall LB, McKinnon W, Chen L, Stephens DT, Garrison K, A remote upstream element regulates tissue-specific expression of the rat aggrecan gene, J Biol Chem, 277 (2002) 13989–13997. [DOI] [PubMed] [Google Scholar]
- [29].Tsumaki N, Kimura T, Matsui Y, Nakata K, Ochi T, Separable cis-regulatory elements that contribute to tissue- and site-specific alpha 2(XI) collagen gene expression in the embryonic mouse cartilage, J Cell Biol, 134 (1996) 1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Alexandre C, Vincent JP, Requirements for transcriptional repression and activation by Engrailed in Drosophila embryos, Development, 130 (2003) 729–739. [DOI] [PubMed] [Google Scholar]
- [31].Grueneberg DA, Natesan S, Alexandre C, Gilman MZ, Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor, Science, 257 (1992) 1089–1095. [DOI] [PubMed] [Google Scholar]
- [32].Rundle CH, Macias MP, Yueh YG, Gardner DP, Kappen C, Transactivation of Hox gene expression in a VP16-dependent binary transgenic mouse system, Biochim Biophys Acta, 1398 (1998) 164–178. [DOI] [PubMed] [Google Scholar]
- [33].Shimizu T, Bae YK, Hibi M, Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue, Development, 133 (2006) 4709–4719. [DOI] [PubMed] [Google Scholar]
- [34].Wang GG, Pasillas MP, Kamps MP, Persistent transactivation by meis1 replaces hox function in myeloid leukemogenesis models: evidence for co-occupancy of meis1-pbx and hox-pbx complexes on promoters of leukemia-associated genes, Mol Cell Biol, 26 (2006) 3902–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zakany J, Duboule D, The role of Hox genes during vertebrate limb development, Curr Opin Genet Dev, 17 (2007) 359–366. [DOI] [PubMed] [Google Scholar]
- [36].Kmita M, Duboule D, Organizing axes in time and space; 25 years of colinear tinkering, Science, 301 (2003) 331–333. [DOI] [PubMed] [Google Scholar]
- [37].Burglin TR, Affolter M, Homeodomain proteins: an update, Chromosoma, 125 (2016) 497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pearson JC, Lemons D, McGinnis W, Modulating Hox gene functions during animal body patterning, Nat Rev Genet, 6 (2005) 893–904. [DOI] [PubMed] [Google Scholar]
- [39].Horan GS, Kovacs EN, Behringer RR, Featherstone MS, Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function, Dev Biol, 169 (1995) 359–372. [DOI] [PubMed] [Google Scholar]
- [40].Raines AM, Magella B, Adam M, Potter SS, Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development, BMC Dev Biol, 15 (2015) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Woltering JM, Noordermeer D, Leleu M, Duboule D, Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits, PLoS Biol, 12 (2014) e1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sheth R, Barozzi I, Langlais D, Osterwalder M, Nemec S, Carlson HL, Stadler HS, Visel A, Drouin J, Kmita M, Distal Limb Patterning Requires Modulation of cis-Regulatory Activities by HOX13, Cell Rep, 17 (2016) 2913–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


