Abstract
Clostridioides difficile (C. difficile) infection is still a threat to many healthcare settings worldwide. Clostridioides difficile epidemiology has changed over the last 20 years, largely due to the emergence of hypervirulent and antimicrobial-resistant C. difficile strains. The excessive use of antimicrobials, the absence of optimal antibiotic policies, and suboptimal infection control practices have fueled the development of this pressing health issue. The prudent use of antimicrobials, particularly broad-spectrum agents, and simple infection control measures, such as hand hygiene, can significantly reduce C. difficile infection rates. Moreover, the early detection of these infections and understanding their epidemiological behavior using accurate laboratory methods are the cornerstone to decreasing the incidence of C. difficile infection and preventing further spread. Although there is no consensus on the single best laboratory method for the diagnosis of C. difficile infection, the use of 2 or more techniques can improve diagnostic accuracy, and it is recommended.
Keywords: C. difficile, nosocomial infection, C. difficile infection, PCR ribotype 027, infection control
Clostridioides difficile (also known as Clostridium difficile [C. difficile]) is the primary cause of antibiotic-related diarrhea in many healthcare settings, particularly elderly centers and rehabilitation clinics.1 All classes of antimicrobials can potentially contribute to C. difficile infection (CDI) via disrupting the gut microbiota, enabling C. difficile to multiply, colonize the digestive tract, and then infect the host. As a result, resistance to many antimicrobial agents offers C. difficile a selective advantage, enhancing their survival and transmission.2,3 In addition, the ability of C. difficile spores to withstand common environmental cleaning agents and alcohol-based hand disinfection increases their survival and enhances their rapid transmission in hospital environments.1 Noteworthy, the epidemiology of C. difficile has shifted in the past 20 years, essentially due to the emergence of highly virulent and antimicrobial-resistant isolates (namely, PCR ribotype 027 and PCR ribotype 078).4,5 It is worth noting that both C. difficile ribotypes 078 and 027 were later discovered in animals such as pigs, cows, and horses. This discovery further supports the theory of transmission of C. difficile from animals to humans.6 However, there is insufficient evidence to confirm direct transmission between animals and humans.7 Nowadays, the detection of CDI presents a major challenge for clinicians and clinical laboratories, as there is no consensus on the best laboratory detection method.8,9 Preventing C. difficile infection requires implementing various measures that include using contact precautions, maintaining good hand hygiene, carrying out efficient environmental cleaning, using sporicidal cleaning agents, and practicing antimicrobial stewardship.1,10 Therefore, this review article highlights the significance of C. difficile in hospital-acquired infections and provides valuable information that may contribute to a better understanding of C. difficile epidemiology and appropriate diagnostic methods, as well as efforts to control its infection.
History
Clostridioides difficile is spore-forming, an obligate anaerobic Gram-positive bacilli bacterium. These bacteria are found in nature (soil), and they usually inhabit the gastrointestinal tract of young animals and humans without causing disease (approximately 20% in some healthcare centers for the elderly and rehabilitation centers and up to 70% of healthy human neonates). Clostridioides difficile was first identified in 1935 in the fecal microbiota of healthy newborns.8,11-13 Because the organism did not appear to be associated with a human infection during that time, it was disregarded until the advent of the era of antibiotic treatment. Pseudomembranous colitis (PMC) was reported before the antibiotics era and attributed to Staphylococcus aureus (S. aureus), which was called “Staphylococcal enterocolitis’’. Hence, oral vancomycin was prescribed as an effective option to treat this disease. Beginning in the 1970s, Tedesco et al14 found that an increase in severe diarrhea cases was associated with clindamycin treatment, which was called “clindamycin colitis’’. At that time, researchers were unable to culture S. aureus from the stool of the patients, and they became discouraged. Since the end of the 1970s, many studies have reported the presence of cytotoxin in stool specimens of pseudomembranous colitis patients.15,16 In 1978, Bartlett et al17 published the first article linking PMC disease and C. difficile cytotoxin production. The mortality rates associated with CDI were not significant until the end of the 20th century. Nevertheless, at the beginning of the 21st century, with the intersection of epidemiological factors, the health status of the hosts, the extensive use of antimicrobials, the emergence of highly virulent C. difficile isolates, and the rates of CDI increased in terms of frequency and severity. Since then, CDI has gained wide attention in the medical community in terms of treatment and laboratory diagnosis.18
Pathogenesis and virulence factors
Clostridioides difficile causes human infections that vary from mild (usually diarrhea) to severe infections, such as PMC. Clostridioides difficile is transmitted via the oral-fecal route, and its spores are highly resistant to harsh conditions such as heat, disinfectants, and antimicrobials.19 Spores can also provide additional protection for this obligate anaerobic organism from an oxygenated environment outside the host.19 Because of the ability of C. difficile to produce spores, gastric juice does not affect the bacterium and spores can reach the intestines, then turn into vegetative cells, and colonize the large intestines.20 Antimicrobials, in particular broad-spectrum antimicrobials, can disrupt the gut microbiota and thus encourage C. difficile to grow and thrive, form spores, and produce toxins. Toxigenic isolates of C. difficile usually produce 2 types of exotoxins: the first is toxin A (TcdA), which is an enterotoxin and the second is toxin B (TcdB), which is cytotoxic. Both toxins affect mucous membranes of the colon, while TcdB is a key virulence factor causing CDI.3,21-23 Additionally, C. difficile produces other virulence determinants that contribute directly and indirectly to the virulence of this pathogen. These determinants include colonization and adherence factors (namely, surface proteins, flagella, and fimbriae), biofilm formation and bacterial spread factors (namely, proteolytic enzymes).3,24 The clinical manifestations and the severity of CDI depend on the patient’s characteristics (namely, age and underlying diseases); the common symptoms include diarrhea, abdominal pain, vomiting, fatigue, and loss of appetite. In severe cases, clinical features can be life-threatening, including fatal PMC, colon perforation, septic shock, kidney failure, and death.19,25
Antimicrobial resistance
Nowadays, antimicrobial-resistant C. difficile is a primary concern for the healthcare community. Furthermore, the emergence of C. difficile infection is usually associated with antibiotic use.26 Although C. difficile is a spore-forming bacterium capable of surviving during antimicrobial therapy, it is also resistant to several antibiotics, particularly broad-spectrum antimicrobials such as aminoglycosides, tetracyclines, erythromycin, clindamycin, penicillin, cephalosporins, and fluoroquinolones.26 Fortunately, fidaxomicin and vancomycin are still effective against C. difficile, the former is recommended as the first-line choice for treating initial and first recurrence CDI, while the latter is used for severe CDI.27-29 Moreover, multidrug resistance is a common feature of many clinical C. difficile isolates, and this is attributed to the accumulation of resistance due to extensive exposure to frequently used antimicrobials.2,30 Antimicrobial resistance of C. difficile is multifactorial and ascribable to the acquisition of resistance genes, alteration in the antimicrobial target, gene mutation, and biofilm formation.2,26 Many studies have shown that the rate of antimicrobial resistance in C. difficile varies from study to another one (Table 1). Although C. difficile isolates resistant to metronidazole and vancomycin are still rare, some isolates have decreased susceptibility to both antimicrobials.31,32 Metronidazole and vancomycin resistance mechanisms in C. difficile are not fully understood. However, recent evidence suggests that metronidazole resistance may arise as a result of alteration in metabolic pathways (namely, the activity of nitroreductases), DNA repair, and biofilm formation, while the vancomycin resistance is attributed to changes in amino acid of peptidoglycan biosynthesis-associated proteins (Table 1).2,26,33 Biofilm may also contribute to resistance to high concentrations of vancomycin.34 A multicenter study from the United States of America revealed that approximately 36% of C. difficile isolates are resistant against clindamycin.35 Nevertheless, some countries in Europe, Asia, and Africa have a higher rate of resistance to clindamycin, 80.3% in Kenya, 74% in Spain, 73.3% in China, and 65% in Poland.36-39 Clostridioides difficile isolates also showed a relatively high rate of resistance against erythromycin (range: 13-100%).26,32,36 Clindamycin and erythromycin belong to the macrolide-lincosamide-streptogramin B family, which act on the ribosomal 50S subunit to inhibit protein synthesis.32 Many bacterial pathogens can resist clindamycin and erythromycin by modifying their ribosomal target or active efflux. Erythromycin resistance in C. difficile is commonly due to ribosomal methylation encoded by the ermB gene.32,40 In addition, C. difficile is resistant to most cephalosporins, with a resistance rate between 11-100%. This resistance might be due to the production of β-lactamases and modified penicillin-binding proteins (PBPs).2,26,30 Fluoroquinolones (namely, ciprofloxacin) are broad-spectrum antimicrobials associated with CDI. Global studies on the antimicrobial resistance of C. difficile showed that 77-99% of isolates were resistant to ciprofloxacin.41 Fluoroquinolones resistance in C. difficile is due to alteration in DNA gyrase subunits GyrA or GyrB.26,36,42 Clostridioides difficile’s resistance to tetracyclines varies between different countries ranging from 2.4-62.7%.26,30,32 Resistance to tetracycline is achieved by generating a ribosomal protective protein (TetM) and an active efflux pump system.2,26,42 Rifamycin and fidaxomicin have been recommended as alternatives to treat recurrent CDI associated with treatment failure of metronidazole and vancomycin.30 Rifamycins inhibit bacterial DNA transcription by targeting the β-subunit of DNA-dependent RNA polymerase (RpoB).26 Clostridioides difficile has also developed resistance to rifamycins, with high levels of resistance being reported in several studies. Krutova et al43 reported that 65% of C. difficile isolates were rifampicin-resistant. Very recently, Mutai et al36 determined the prevalence of CDI and evaluated antimicrobial resistance among C. difficile cases in Kenya and found that 91.5% of C. difficile isolates were resistant against rifampicin. The primary resistance mechanism to rifamycins is mutations in the β-subunit of the rpoB gene.26,34 Although there is evidence that biofilm formation plays a vital role in the antimicrobial resistance of C. difficile, more research is needed to clarify its specific contribution mechanism.
Table 1.
- Mechanism of action and resistance of different antimicrobials and resistance rates against Clostridioides difficile.
| Antimicrobials | Targets | Putative resistance mechanism (S) | Resistances (%) | Refrences |
|---|---|---|---|---|
| Metronidazole | Bacterial DNA | Alterations in some metabolic pathways, biofilm formation | 0-20.25 | 32,33,36,41 |
| Vancomycin | Binding to D-alanyl-D-alanine residues precursor of peptidoglycan | Mutations in peptidoglycan biosynthesis-required proteins, biofilm formation | 0-41 | 32,33,36,41 |
| Rifampicin | Bacterial DNA-dependent RNA polymerase | Mutations in rpoB | 0-91 | 32,36,41 |
| Clindamycin | 50s ribosomal subunit of bacteria | Alterations in ribosomal target or active efflux pumps | 0-100 | 26,32,33,36,41 |
| Erythromycin | 50s subunit of the bacteria | Alterations in ribosomal target or active efflux pumps | 0-100 | 26,32,33,36,41 |
| Tetracycline | 30S ribosomal subunit of bacteria | Tetracycline resistance protein (tetM) and the active efflux pumps | 0-62.7 | 26,32,33,36,41 |
| Moxifloxacin | Bacterial DNA gyrase | Alteration of the drug target | 0-100 | 32,33,41 |
| Fusidic acid | Blocking the ribosome by binding to “factor G” | Mutations in fusA | 0-40 | 41 |
| Ciprofloxacin | Bacterial DNA gyrase | Alteration of the drug target and the active efflux pumps | 23-42 | 33,41 |
Clostridioides difficile’s genome
Its genome is ~ 4.3 Mbp in length with a G + C content of 29%. Furthermore, a circular plasmid of 7,881 bp with a G + C content of 27% was identified. Clostridioides difficile contains a large pan-genome with 9000 coding sequences (CDS) and a large proportion of mobile genetic elements (MGEs, 11% in C. difficile strain 630).44,45 However, C. difficile has an ultra-low level of core genome (between 16-24%), while other bacterial species have more than 60%.45-48 Many CDS identified in C. difficile’s genome are responsible for its adaptation to the gastrointestinal tract and survival in the harsh environment through endospore formation.49 Mobile genetic elements of C. difficile are mainly carried on conjugative transposons and insertion sequences that encode antimicrobial resistance, 2 key virulence factors, toxins A (TcdA) and B(TcdB), and the production of surface structures.49,50 These mobile elements are exchanged with high frequency and contribute to the diversity and plasticity of the C. difficile genome. The genomic plasticity and variability of C. difficile contribute to its extraordinary capacity to adapt to different growth conditions for a long time in the gut and recurrent infection.49
Clostridioides difficile colonization
It is defined as the presence of an organism or its toxins without any symptoms of CDI, which can vary from mild symptoms (usually diarrhea) to severe infections such as PMC.51,52 The ingestion of C. difficile spores is the first stage of colonization; the spores can survive in gastric acid and reach the colon to germinate into vegetative cells. Colonization of C. difficile is prevented by the fecal microbiota barrier, which can be disrupted by antimicrobial use.20 The incidence of C. difficile colonization varies depending on various factors, such as the host, pathogen, and environment.53 Previous hospitalization, antimicrobial exposure (cephalosporins), use of proton-pump inhibitors, immunosuppressive drugs, and the presence of antibodies against toxin B are the most commonly risk factors linked with C. difficile colonization.54,55 Although the colonization rate by C. difficile is still low among healthy adults, it increases significantly in individuals with previous hospitalization, particularly among rehabilitation center patients. Moreover, underlying diseases also contribute to the high percentage of asymptomatic C. difficile colonization.52 Unlike adults, a high rate of C. difficile colonization has been reported in infants and neonates.56 The colonization process involves many factors, including immune evasion of the host defenses and adhesion to the host mucous membrane, usually facilitated by surface proteins. These surface layer proteins (SLPs) of C. difficile play a crucial role in their adherence to the mucus layer in the intestine.24 Moreover, flagella of C. difficile are important for colonization and host invasion.57,58 Although the relationship between CDI and C. difficile colonization is not fully understood, the former is not directly associated with the development of the latter.52 Noteworthy, colonization by toxigenic strains can increase the risk of developing CDI compared to being colonized by non-toxigenic strains.51
Molecular epidemiology of C. difficile
Until the end of the last century, C. difficile infection was a leading cause of antimicrobial-associated diarrhea among the elderly in healthcare settings. However, over the last 20 years, there has been a dramatic increase in reports of C. difficile infection, describing a high number of cases with considerable changes in clinical manifestations, including more severe cases, many outbreaks, and novel risk factors.59,60 These changes have led to the emergence of new hypervirulent isolates of C. difficile, such as the fluoroquinolone-resistant PCR ribotype 027/North American pulsed-field gel electrophoresis type I (NAPI) strains in North America and PCR ribotype 078 strain in Europe.4,5 Although PCR ribotype 027 was linked to severe cases and higher mortality, other C. difficile ribotypes have also been responsible for many outbreaks worldwide.59 The hypervirulence of PCR ribotype 027/NAPI strains was attributed to the polymorphisms in the receptor-binding domain of tcdB, resulting in a hypertoxic form of TcdB. Additionally, these strains produce a binary toxin that is linked to higher clinical severity.61 Noteworthy, the earliest report of the PCR ribotype 027 was from a Parisian hospital in 1985, and the next record was from a Minneapolis hospital, on a non-epidemic strain specified BI-1 in 1988.62,63 A few years later, many CDI outbreaks with severe cases were reported in hospitals across the United States and in Canada.63,64 The initial reports of PCR ribotype 027 outbreaks in Europe came from the Netherlands in 2005 and England in 2006, and afterward, there were several reports from various other European countries.65 Nevertheless, the emergence of this clone and its rapid global dissemination remained unknown until a global set of C. difficile ribotype 027 clinical isolates collected between 1985-2010 were sequenced. Genomic analysis revealed that 2 different epidemic lineages of ribotype 027 (FQR1 and FQ2) with a similar fluoroquinolone resistance mutation, had emerged, and disseminated in North America within a short period.66,67 The 2 lineages showed different spread patterns. One spread throughout North America, Chile, Switzerland, and South Korea, while the other was reported broadly across Europe and Australia (Figure 1).66,68 These findings shed new light on the importance of fluoroquinolone use as a selective pressure in the evolution and spread of the FQR1 and FQ2 lineages within healthcare settings.67 Clostridioides difficile ribotype 027 strains have been sporadically detected in China and Japan, but the Japanese strains are susceptible to fluoroquinolones, in contrast to the fluoroquinolone-resistant ribotype 027 strains spreading in North America and Europe.60,69 Also, in the United States and Europe, CDI clusters are often caused by ribotypes 001, 002, and 014/020.19 Moreover, ribotypes 017, 018, 014, 002, and 001 are the most prevalent ribotypes in Asian countries (Japan, Korea, Hong Kong, Singapore, Taiwan, and China).70,71 There are few molecular epidemiological studies on C. difficile in Africa and the Middle East. Although many epidemiological studies on the prevalence of CDI in Saudi Arabia have already been carried out72-75, only one study characterized the ribotypes of isolated strains; they detected the hypervirulent ribotype 027 in 4 elderly patients in Riyadh.76 Furthermore, Al-Thani et al77 found 258, 001, 014, 046, 011, 053, 056, and 107 were the most prevalent ribotypes in Qatar, and the hypervirulent RT027 was detected in only one isolate. Another study in Kuwait showed that ribotypes 139, 014, 056, 070, 097, and 179 were the most common ribotypes.78 A recent study from Iran has revealed that ribotypes 001 and 126 were the most frequent ribotypes, and none of the hypervirulent ribotypes (027, 078) were reported.79 Moreover, the genotype of hypervirulent isolates is not as clearly apparent in the Middle East as it is in Europe and the United States. This can be attributed to the lack of information on the ribotypes in circulation in the Middle East. Therefore, further studies are required to pinpoint the prevalent hypervirulent strains in this geographical region.79
Figure 1.
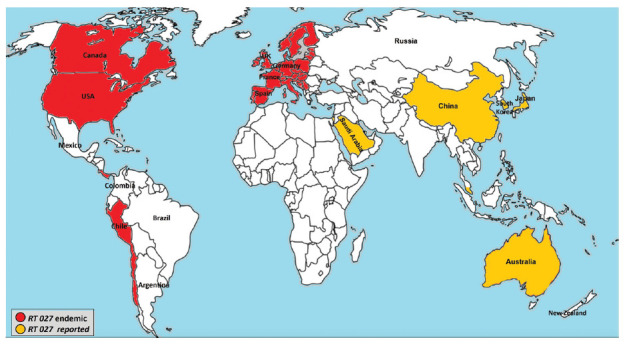
- Worldwide distribution of hypervirulent Clostridioides difficile PCR ribotype (RT) 027.
Laboratory detection of C. difficile
Its symptoms vary from asymptomatic colonization, and mild symptoms to severe infections such as PMC and toxic megacolon. Clostridioides difficile infection is defined by the emergence of more than 3 unformed stools within 24 hours and is approved through a laboratory diagnosis for the presence of toxin-producing C. difficile isolate.80 For many clinicians and clinical laboratories, the identification of CDI poses a significant problem, and there is still no agreement on the one approach that is the optimal laboratory detection method.8,9 Currently, there are many laboratory tests for CDI detection, including toxigenic culture (TC), cell cytotoxicity assay (CTA), enzyme immunoassay (EIA) for detecting glutamate dehydrogenase (GDH), EIA for detecting toxins A or B, and nucleic acid amplification test (NAAT; Table 2).
Table 2.
- Simplified comparison of various laboratory detection methods for the detection of Clostridioides difficile infection.
| Techniques | Criteria | Advantages | Limitations | References | |||||
|---|---|---|---|---|---|---|---|---|---|
| Simple to perform | Simple to interpret | Specimen type | Turnaround time | Sensitivity (%) | Specificity (%) | ||||
| TC | Good | Good | Stool | 2-7 days | High | Low | Due to its high sensitivity, it is recommended as a reference method; it allows performing strain typing and antimicrobial suitability test | Long turnaround time; need to be combined with another method | 8,18,85 |
| CTA | Fair | Good | Stool filtrate | 48-72 hours | High | High | Has acceptable sensitivity and specificity, and it is inexpensive | Labor-intensive, long turnaround time, and lack of harmonization | 18,85,86 |
| EIA for detecting GDH | Good | Good | Stool | Rapid 2-6 hours | Low | Low | Quick to produce results, and simple to use | Lacks specificity, high false-positive rate, and should be used in combination with another method | 8,18,85,87 |
| EIA for detecting toxins A or B | Good | Good | Stool | Rapid 2-6 hours | Low | Moderate | Quick to produce results, and simple to use | Insensitive enough in the detection of toxin-producing isolates of C. difficile in comparison to other techniques such as TC or CTA. High false-negative rate | 8,18,85,87 |
| NAAT | Good | Good | Stool | Rapid 2 hours | High | Low/moderate | Rapid and highly sensitive | Identify carriers of toxigenic C. difficile who have no symptoms. Genetic variation in tcdB or tcdA genes might lead to false-negative results. Expensive | 8,85,87,90 |
TC: toxigenic culture, CTA: cell cytotoxicity assay, GDH: glutamate dehydrogenase, NAAT: nucleic acid amplification test, EIA: enzyme immunoassay
Toxigenic culture
It is a 2-step technique based on isolating C. difficile from stool and determining whether the isolated strain is a toxin producer or not. There are many approaches for accomplishing this purpose, including stool culture on selective and differential media under prolonged anaerobic incubation (~ 48 hours), identifying suspected colonies, and finally, the identified C. difficile isolates are subjected to toxin testing.8,18 Although there is no consensus on the optimal method for C. difficile culture from stool, a heat shock or alcohol treatment before direct plating on a selective media (namely, cycloserine-cefoxitin-fructose agar or its variant) is recommended by many investigators.18,81-83 This is mainly to inhibit or minimize the growth of stool organisms and enhance C. difficile spores.18 However, other studies revealed that using an enrichment broth containing antimicrobials, carbohydrates, lysozyme, and taurocholate can increase the recovery of C. difficile without requiring heat shock or ethanol pretreatment.8 Additionally, several chromogenic media have been developed to isolate C. difficile from stool samples. Those chromogenic media showed variable sensitivity, and they include chromID C. difficile TCCA, chromID C. difficile TCCFA, chromID C. difficile CDSA, chromID C. difficile CCFA, and chromID C. difficile CDSA.84 Thereafter, identification of isolate can be achieved by conventional methods such as the morphology of colonies, Gram staining, and biochemical tests such as latex agglutination for GDH or using Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS).84,85 After identification of an isolate, its ability to produce toxins must be confirmed by testing the supernatant for bacterial growth using a cell cytotoxicity neutralization assay or by a more rapid method such as PCR to detect toxin genes.8,18,84,85 Although the TC method requires a relatively long time (>2 days), it is the recommended reference method for detecting toxigenic strains of C. difficile and is regarded as the gold standard technique in evaluation studies.18,85
Cell cytotoxicity assay
It is regarded a standard technique for the detection of C. difficile toxins in a stool sample. This technique involves incubating stool filtrate with a proper cell line for 1-2 days and observing any cytopathic effects of toxins using a neutralization assay with an antiserum against C. difficile toxin B or against the Clostridium sordellii toxins, which have similar antigens.18,85 Although CTA has acceptable sensitivity and specificity and is inexpensive, it is currently used only by a small number of laboratories due to its labor-intensive, long turnaround time and lack of harmonization (cell type, dilution of stool specimen, and incubation period).18,85,86
Enzyme immunoassays
It uses specific monoclonal or polyclonal antibodies to detect toxins A and B, and GDH antigens. Many commercial EIAs are readily available, quick to produce results, and simple to use. For many years, EIAs that detect toxins A or toxin B were among the most detective tests for C. difficile isolates. They include solid-phase microwell, lateral flow membranes, immunoassays in chromatographic cassettes, and chemiluminescent immunoassays.8,18,85 However, EIAs are not sensitive enough to identify toxigenic isolates of C. difficile compared with techniques such as toxigenic culture or cytotoxicity assays.85,87 Recently, an ultrasensitive immunoassay for toxin A and toxin B was developed by Song et al88 that can serve as a standalone diagnostic test for CDI. Thereafter, many ultrasensitive immunoassays were introduced for the detection of C. difficile toxins. Those assays can detect disease-specific markers at extremely low concentrations with good accuracy.89 However, these ultrasensitive immunoassays require further validation.8,89 Glutamate dehydrogenase immunoassays identify a metabolic enzyme that is highly conserved and abundant in all C. difficile isolates. Since GDH is found in both toxigenic and nontoxigenic isolates, this test lacks specificity and should be combined with another method (namely, toxin detection test).87
Nucleic acid detection techniques
Nucleic acid amplification assays (NAATs) are mainly based on real-time PCR, and there are several commercial platforms targeting a diversity of genes, involving tcdA, tcdB and 16S ribosomal RNA (rRNA).85,87 Of note, the first FDA-approved NAAT platform was not accessible in the United States until 2009, despite NAATs for detecting C. difficile in stool first appearing in the literature in the early 1990s.8,18,85 Nucleic acid amplification assays are less sensitive than TC but more sensitive than toxin-based EIAs for C. difficile detection. However, depending on the prevalence of the disease and the assay’s limit of detection, the positive predictive value of NAATs for CDI ranges from low to moderate.87 Nucleic acid amplification assays, on the other hand, only identify the existence of toxin genes and, consequently, the ability of C. difficile to produce toxins. Consequently, the major drawback of NAATs is that they can also identify carriers of toxigenic C. difficile who have no symptoms in addition to CDI cases.90 Moreover, genetic variation in the tcdB or tcdA genes is another potential issue (Table 2), which might lead to false-negative results. Therefore, the European Society of Clinical Microbiology and Infectious Diseases guidelines advise against using NAAT as a single test to diagnose C. difficile and instead suggest using it as a screening test.85 Ultimately, NAATs are still more expensive than other detection methods such as toxigenic culture or EIAs.8
Infection control of C. difficile
Clostridioides difficile is considered the primary cause of nosocomial infections and gastroenteritis-associated deaths.1 In the United States alone, C. difficile accounts for approximately 500,000 healthcare infections and approximately 29,000 deaths every year.91,92 The European Centre for Disease Prevention and Control has reported that approximately 124,000 patients in the European Union develop CDI each year.19 In addition, C. difficile has been linked to many outbreaks, some of which are caused by new, highly virulent strains (namely, RT027 [NAP1/BI]) that leads to more severe disease and worse patient outcomes. Furthermore, C. difficile spores increase their ability to survive in hospital environments for a long time and to resist common environmental cleaning agents and standard alcohol-based hand disinfection; together, these factors contribute to the rapid spread of C. difficile in hospitals.1 Although C. difficile initially appeared as a hospital-associated infection, its epidemiology is constantly changing, and currently, one-third of C. difficile cases are community-acquired infections.92 This pathogen spreads mainly via the fecal-oral route, and old age, antimicrobial therapy, and hospitalization are the most significant risk factors associated with CDI.25 Non-antimicrobial medications such as acid-suppressing drugs (namely, proton pump inhibitors and histamine-2 receptor antagonists) have also been linked to an increased risk of CDI.93 In addition to morbidity and mortality rates, CDI adds considerable financial burdens to healthcare systems. Therefore, C. difficile has gained global attention from the healthcare community.94 Healthcare workers also play a role in transferring these bacteria among patients and between different units.
Clostridioides difficile is endemic to many hospital departments, elderly centers, and rehabilitation clinics, especially under poor hygiene conditions, as well as with the use of broad-spectrum antimicrobials. Therefore, the best way to prevent CDI is to apply barrier precautions such as effective hand washing before and after contact with all patients and wearing gloves, and the appropriate personal protective equipment.1 However, the use of alcohol-based hand gels alone does not destroy C. difficile spores.87 In addition, effective cleaning and disinfection of the hospital environment and medical instruments such as rectal thermometers, colonoscopy devices, toilet chairs, handles, and any potential source of C. difficile are very important.1,10 Moreover, asymptomatic individuals of C. difficile are a crucial source of pathogen spread to other patients who are more susceptible to the infection.95 Therefore, the early detection of carriers is vital to ensure rapid patient treatment and to avoid the further spread of C. difficile.8 Additionally, restricting the use of antimicrobials (particularly broad-spectrum forms that disrupt the balance of gut microbiota, promote C. difficile growth, and cause infection) is considered highly significant way to reduce C. difficile infections.96
Fecal microbiota transplantation as a treatment option for CDI
Fecal microbiota transplantation (FMT) is the introducing of a liquid solution containing fecal matter from a healthy individual into the recipient’s intestinal tract to alter his microbiota and confer benefit. The process typically involves selecting a donor who has no family history of autoimmune diseases or cancer and then screening for any possible blood-borne and fecal pathogens.97 It is not a modern method; it was utilized in the 4th century in China to treat several maladies, including diarrhea.97 In the 1950s, FMT was first described as an option for the treatment of pseudomembranous colitis, and in the last 10 years, there has been a lot of interest in FMT to treat CDI. This is mainly due to the limitations of other treatment options (usually metronidazole or vancomycin) and their inability to effectively treat recurrent CDI.80,98-100
Recurrent and refractory CDI cases have been successfully treated with FMT, with an efficacy of 85-90% for recurrent CDI and 55% for refractory CDI.80 However, neither the successes achieved by FMT in treating CDI nor its potentially protective mechanisms are not fully described. However, the most likely scenario is microbial antagonism through the competitive exclusion of the pathogen by other microbes that outcompete C. difficile, depriving it of nutrients and creating an unfavorable environment for its growth.85 Based on many studies, FMT seems to be both safe and effective, especially in treating recurrent CDI, compared to other therapeutic methods using antimicrobials.99 Fecal microbiota transplantation and its mechanism of action against C. difficile warrants further investigation.
In conclusion, C. difficile infections are still a threat to many healthcare settings worldwide. Clostridioides difficile epidemiology has changed over the last 20 years, largely due to the emergence of highly virulent, antimicrobial-resistant C. difficile isolates. The excessive use of antimicrobials, the absence of optimal antibiotic policies, and suboptimal infection control practices have fueled the development of this pressing health issue. Therefore, the prudent use of antimicrobials, particularly broad-spectrum agents, and the application of simple infection control measures, such as washing hands with soap, can significantly reduce the rates of C. difficile infection. Moreover, the early detection of these infections and an understanding of their epidemiological behavior based on accurate laboratory diagnostic methods are the cornerstones to decreasing the prevalence of C. difficile infection and preventing its further spread. Although there is no agreement on the single best laboratory method for the diagnosis of C. difficile infection, the use of 2 or more techniques can improve diagnostic accuracy. Molecular genotyping of C. difficile isolates is also an important step in controlling C. difficile outbreaks to recognize the differences between sporadic and epidemic strains, and to monitor their epidemic spread.
Acknowledgment
The author gratefully acknowledge AME for English language editing.
Footnotes
References
- 1.Vonberg RP, Kuijper EJ, Wilcox MH, Barbut F, Tüll P, Gastmeier P, et al. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect 2008; 14: 2–20. [DOI] [PubMed] [Google Scholar]
- 2.Spigaglia P, Mastrantonio P, Barbanti F.. Antibiotic resistances of Clostridium difficile. Adv Exp Med Biol 2018; 1050: 137–159. [DOI] [PubMed] [Google Scholar]
- 3.Borriello SP. Pathogenesis of Clostridium difficile infection. J Antimicrob Chemother 1998; 41: 13–19. [DOI] [PubMed] [Google Scholar]
- 4.Kuijper EJ, Coignard B, Tüll P.. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 2006; 12: 2–18. [DOI] [PubMed] [Google Scholar]
- 5.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 2008; 47: 1162–1170. [DOI] [PubMed] [Google Scholar]
- 6.Bolton D, Marcos P.. The environment, farm animals and foods as sources of Clostridioides difficile infection in humans. Foods 2023; 12: 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weese JS. Clostridium (Clostridioides) difficile in animals. J Vet Diagn Invest 2020; 32: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll KC, Mizusawa M.. Laboratory tests for the diagnosis of Clostridium difficile. Clin Colon Rectal Surg 2020; 33: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraft CS, Parrott JS, Cornish NE, Rubinstein ML, Weissfeld AS, McNult P, et al. A laboratory medicine best practices systematic review and meta-analysis of nucleic acid amplification tests (NAATs) and algorithms including NAATs for the diagnosis of Clostridioides (Clostridium) difficile in adults. Clin Microbiol Rev 2019; 32: e00032–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayfield JL, Leet T, Miller J, Mundy LM.. Environmental control to reduce transmission of Clostridium difficile. Clin Infect Dis 2000; 31: 995–1000. [DOI] [PubMed] [Google Scholar]
- 11.Hall IC, O’Toole E.. Intestinal flora in new-born infants with a description of a new pathogenic anaerobe, bacillus difficilis. Am J Dis Child 1935; 49: 390–402. [Google Scholar]
- 12.Montoya A, Mody L.. Common infections in nursing homes: a review of current issues and challenges. Aging health 2011; 7: 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinlen L, Ballard JD.. Clostridium difficile infection. Am J Med Sci 2010; 340: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tedesco FJ, Barton RW, Alpers DH.. Clindamycin-associated colitis. A prospective study. Ann Intern Med 1974; 81: 429–433. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett JG. Clostridium difficile infection: historic review. Anaerobe 2009; 15: 227–229. [DOI] [PubMed] [Google Scholar]
- 16.Larson HE, Price AB.. Pseudomembranous colitis: presence of clostridial toxin. Lancet 1977; 2: 1312–1314. [DOI] [PubMed] [Google Scholar]
- 17.Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB.. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med 1978; 298: 531–534. [DOI] [PubMed] [Google Scholar]
- 18.Burnham CA, Carroll KC.. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 2013; 26: 604–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ.. Clostridium difficile infection. Nat Rev Dis Primers 2016; 2: 16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leffler DA, Lamont JT.. Clostridium difficile Infection. N Engl J Med 2015; 373: 287–288. [DOI] [PubMed] [Google Scholar]
- 21.Denève C, Janoir C, Poilane I, Fantinato C, Collignon A.. New trends in Clostridium difficile virulence and pathogenesis. Int J Antimicrob Agents 2009; 33: S24–S28. [DOI] [PubMed] [Google Scholar]
- 22.Ling Z, Liu X, Jia X, Cheng Y, Luo Y, Yuan L, et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep 2014; 4: 7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, et al. Toxin B is essential for virulence of Clostridium difficile. Nature 2009; 458: 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janoir C. Virulence factors of Clostridium difficile and their role during infection. Anaerobe 2016; 37: 13–24. [DOI] [PubMed] [Google Scholar]
- 25.Czepiel J, Dróżdż M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, et al. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis 2019; 38: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Z, Jin D, Kim HB, Stratton CW, Wu B, Tang YW, et al. Update on antimicrobial resistance in Clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. J Clin Microbiol 2017; 55: 1998–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debast SB, Bauer MP, Kuijper EJ.. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 2014; 20: 1–26. [DOI] [PubMed] [Google Scholar]
- 28.Petrosillo N, Granata G, Cataldo MA.. Novel antimicrobials for the treatment of Clostridium difficile infection. Front Med (Lausanne) 2018; 5: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson S, Lavergne V, Skinner AM, Gonzales-Luna AJ, Garey KW, Kelly CP, et al. Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin Infect Dis 2021; 73: 755–757. [DOI] [PubMed] [Google Scholar]
- 30.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis 2016; 3: 23–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tkhawkho L, Nitzan O, Pastukh N, Brodsky D, Jackson K, Peretz A.. Antimicrobial susceptibility of Clostridium difficile isolates in Israel. J Glob Antimicrob Resist 2017; 10: 161–164. [DOI] [PubMed] [Google Scholar]
- 32.Banawas SS. Clostridium difficile infections: a global overview of drug sensitivity and resistance mechanisms. Biomed Res Int 2018; 2018: 8414257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dilnessa T, Getaneh A, Hailu W, Moges F, Gelaw B.. Prevalence and antimicrobial resistance pattern of Clostridium difficile among hospitalized diarrheal patients: a systematic review and meta-analysis. PLoS One 2022; 17: e0262597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Grady K, Knight DR, Riley TV.. Antimicrobial resistance in Clostridioides difficile. Eur J Clin Microbiol Infect Dis 2021; 40: 2459–2478. [DOI] [PubMed] [Google Scholar]
- 35.Tickler IA, Goering RV, Whitmore JD, Lynn AN, Persing DH, Tenover FC.. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011-2013. Antimicrob Agents Chemother 2014; 58: 4214–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mutai WC, Mureithi MW, Anzala O, Revathi G, Kullin B, Burugu M, et al. High prevalence of multidrug-resistant Clostridioides difficile following extensive use of antimicrobials in hospitalized patients in Kenya. Front Cell Infect Microbiol 2021; 10: 604986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Pardo D, Almirante B, Bartolomé RM, Pomar V, Mirelis B, Navarro F, et al. Epidemiology of Clostridium difficile infection and risk factors for unfavorable clinical outcomes: results of a hospital-based study in Barcelona, Spain. J Clin Microbiol 2013; 51: 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong D, Zhang L, Chen X, Jiang C, Yu B, Wang X, et al. Antimicrobial susceptibility and resistance mechanisms of clinical Clostridium difficile from a Chinese tertiary hospital. Int J Antimicrob Agents 2013; 41: 80–84. [DOI] [PubMed] [Google Scholar]
- 39.Huang H, Fang H, Weintraub A, Nord CE.. Distinct ribotypes and rates of antimicrobial drug resistance in Clostridium difficile from Shanghai and Stockholm. Clin Microbiol Infect 2009; 15: 1170–1173. [DOI] [PubMed] [Google Scholar]
- 40.Isidro J, Menezes J, Serrano M, Borges V, Paixão P, Mimoso M, et al. Genomic study of a Clostridium difficile multidrug resistant outbreak-related clone reveals novel determinants of resistance. Front Microbiol 2018; 9: 2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chow VCY, Kwong TNY, So EWM, Ho YII, Wong SH, Lai RWM, et al. Surveillance of antibiotic resistance among common Clostridium difficile ribotypes in Hong Kong. Sci Rep 2017; 7: 17218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sholeh M, Krutova M, Forouzesh M, Mironov S, Sadeghifard N, Molaeipour L, et al. Antimicrobial resistance in Clostridioides (Clostridium) difficile derived from humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control 2020; 9: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krutova M, Matejkova J, Tkadlec J, Nyc O.. Antibiotic profiling of Clostridium difficile ribotype 176--A multidrug resistant relative to C. difficile ribotype 027. Anaerobe 2015; 36: 88–90. [DOI] [PubMed] [Google Scholar]
- 44.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 2006; 38: 779–786. [DOI] [PubMed] [Google Scholar]
- 45.Knight DR, Squire MM, Collins DA, Riley TV.. Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front Microbiol 2017; 7: 2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scaria J, Ponnala L, Janvilisri T, Yan W, Mueller LA, Chang YF.. Analysis of ultra low genome conservation in Clostridium difficile. PLoS One 2010; 5: e15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knight DR, Kullin B, Androga GO, Barbut F, Eckert C, Johnson S, et al. Evolutionary and genomic insights into Clostridioides difficile sequence type 11: a diverse zoonotic and antimicrobial-resistant lineage of global one health importance. mBio 2019; 10: e00446–e00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulecka M, Waker E, Ambrozkiewicz F, Paziewska A, Skubisz K, Cybula P, et al. Higher genome variability within metabolism genes associates with recurrent Clostridium difficile infection. BMC Microbiol 2021; 21: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV.. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev 2015; 28: 721–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sommermeyer H, Piątek J.. Clostridioides difficile: infections, risk factors, prevention and treatment. [Updated 2021; 2022. Dec 20]. Available from: https://link.springer.com/book/10.1007/978-3-030-81100-6
- 51.Crobach MJT, Vernon JJ, Loo VG, Kong LY, Péchiné S, Wilcox MH, et al. Understanding Clostridium difficile colonization. Clin Microbiol Rev 2018; 31: e00021–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schäffler H, Breitrück A.. Clostridium difficile - from colonization to infection. Front Microbiol 2018; 9: 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furuya-Kanamori L, Marquess J, Yakob L, Riley TV, Paterson DL, Foster NF, et al. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis 2015; 15: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin JH, Chaves-Olarte E, Warren CA.. Clostridium difficile infection. Microbiol Spectr 2016; 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez E, Taminiau B, Rodriguez C, Daube G.. Gut microbiota composition associated with Clostridioides difficile colonization and infection. Pathogens 2022; 11: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, Collignon A.. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 2012; 55: 1209–1215. [DOI] [PubMed] [Google Scholar]
- 57.Baban ST, Kuehne SA, Barketi-Klai A, Cartman ST, Kelly ML, Hardie KR, et al. The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One 2013; 8: e73026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D.. Clostridium difficile virulence factors: insights into an anaerobic spore-forming pathogen. Gut Microbes 2014; 5: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 2011; 377: 63–73. [DOI] [PubMed] [Google Scholar]
- 60.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 2010; 23: 529–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lanis JM, Heinlen LD, James JA, Ballard JD.. Clostridium difficile 027/BI/NAP1 encodes a hypertoxic and antigenically variable form of TcdB. PLoS Pathog 2013; 9: e1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popoff MR, Rubin EJ, Gill DM, Boquet P.. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun 1988; 56: 2299–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valiente E, Cairns MD, Wren BW.. The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move. Clin Microbiol Infect 2014; 20: 396–404. [DOI] [PubMed] [Google Scholar]
- 64.Labbé AC, Poirier L, Maccannell D, Louie T, Savoie M, Béliveau C, et al. Clostridium difficile infections in a Canadian tertiary care hospital before and during a regional epidemic associated with the BI/NAP1/027 strain. Antimicrob Agents Chemother 2008; 52: 3180–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, Lambert ML, et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill 2008; 13: 18942. [PubMed] [Google Scholar]
- 66.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 2013; 45: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiuff C, Banks AL, Fitzpatrick F, Cottom L.. The need for European surveillance of CDI. Adv Exp Med Biol 2018; 1050: 13–25. [DOI] [PubMed] [Google Scholar]
- 68.Hernández-Rocha C, Barra-Carrasco J, Pizarro-Guajardo M, Ibáñez P, Bueno SM, Sarker MR, et al. Epidemic Clostridium difficile ribotype 027 in Chile. Emerg Infect Dis 2012; 18: 1370–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang RF, Man YX, Bai YY, Shao CH, Liu CM, Wang CH, et al. Molecular characterization of Clostridioides difficile ribotype 027 in a major Chinese hospital. J Microbiol Immunol Infect 2021; 54: 1179–1183. [DOI] [PubMed] [Google Scholar]
- 70.Collins DA, Hawkey PM, Riley TV.. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2013; 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collins DA, Sohn KM, Wu Y, Ouchi K, Ishii Y, Elliott B, et al. Clostridioides difficile infection in the Asia-Pacific region. Emerg Microbes Infect 2019; 9: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Al-Tawfiq JA, Abed MS.. Clostridium difficile-associated disease among patients in Dhahran, Saudi Arabia. Travel Med Infect Dis 2010; 8: 373–376. [DOI] [PubMed] [Google Scholar]
- 73.Alzouby S, Baig K, Alrabiah F, Shibl A, Al-Nakhli D, Senok AC.. Clostridioides difficile infection: incidence and risk factors in a tertiary care facility in Riyadh, Saudi Arabia. J Infect Public Health 2020; 13: 1012–1017. [DOI] [PubMed] [Google Scholar]
- 74.Al-Tawfiq JA, Rabaan AA, Bazzi AM, Raza S, Noureen M.. Clostridioides (Clostridium) difficile-associated disease: epidemiology among patients in a general hospital in Saudi Arabia. Am J Infect Control 2020; 48: 1152–1157. [DOI] [PubMed] [Google Scholar]
- 75.Aljafel NA, Al-Shaikhy HH, Alnahdi MA, Thabit AK.. Incidence of Clostridioides difficile infection at a Saudi tertiary academic medical center and compliance with IDSA/SHEA, ACG, and ESCMID guidelines for treatment over a 10-year period. J Infect Public Health 2020; 13: 1156–1160. [DOI] [PubMed] [Google Scholar]
- 76.Alzahrani N, Johani SA.. Emergence of a highly resistant Clostridium difficile strain (NAP/BI/027) in a tertiary care center in Saudi Arabia. Ann Saudi Med 2013; 33: 198–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Al-Thani AA, Hamdi WS, Al-Ansari NA, Doiphode SH, Wilson GJ.. Polymerase chain reaction ribotyping of Clostridium difficile isolates in Qatar: a hospital-based study. BMC Infect Dis 2014; 14: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jamal W, Pauline E, Rotimi V.. A prospective study of community-associated Clostridium difficile infection in Kuwait: epidemiology and ribotypes. Anaerobe 2015; 35: 28–32. [DOI] [PubMed] [Google Scholar]
- 79.Baghani A, Mesdaghinia A, Kuijper EJ, Aliramezani A, Talebi M, Douraghi M.. High prevalence of Clostridiodes diffiicle PCR ribotypes 001 and 126 in Iran. Sci Rep 2020; 10: 4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee HS, Plechot K, Gohil S, Le J.. Clostridium difficile: diagnosis and the consequence of over diagnosis. Infect Dis Ther 2021; 10: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marler LM, Siders JA, Wolters LC, Pettigrew Y, Skitt BL, Allen SD.. Comparison of 5 cultural procedures for isolation of Clostridium difficile from stools. J Clin Microbiol 1992; 30: 514–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hink T, Burnham CA, Dubberke ER.. A systematic evaluation of methods to optimize culture-based recovery of Clostridium difficile from stool specimens. Anaerobe 2013; 19: 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biasizzo M, Vadnjal S, Henigman U, Krizman M, Kirbis A, Jamnikar-Ciglenecki U.. Development and validation of a new protocol for detecting and recovering Clostridium difficile from meat samples. J Food Prot 2018; 81: 561–568. [DOI] [PubMed] [Google Scholar]
- 84.Perry JD. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin Microbiol Rev 2017; 30: 449–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gateau C, Couturier J, Coia J, Barbut F.. How to: diagnose infection caused by Clostridium difficile. Clin Microbiol Infect 2018; 24: 463–468. [DOI] [PubMed] [Google Scholar]
- 86.Musher DM, Manhas A, Jain P, Nuila F, Waqar A, Logan N, et al. Detection of Clostridium difficile toxin: comparison of enzyme immunoassay results with results obtained by cytotoxicity assay. J Clin Microbiol 2007; 45: 2737–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66: 987–994. [DOI] [PubMed] [Google Scholar]
- 88.Song L, Zhao M, Duffy DC, Hansen J, Shields K, Wungjiranirun M, et al. Development and validation of digital enzyme-linked immunosorbent assays for ultrasensitive detection and quantification of Clostridium difficile toxins in stool. J Clin Microbiol 2015; 53: 3204–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sandlund J, Davies K, Wilcox MH.. Ultrasensitive Clostridioides difficile toxin testing for higher diagnostic accuracy. J Clin Microbiol 2020; 58: e01913–e01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crobach JTM, Baktash A, Duszenko N, Kuijper EJ.. Diagnostic guidance for C. difficile infections. [Updated 2018; 2023. Jan 15]. Available from: https://link.springer.com/chapter/10.1007/978-3-319-72799-8_3
- 91.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372: 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner NA, Anderson DJ.. Hospital infection control: Clostridioides difficile. Clin Colon Rectal Surg 2020; 33: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tawam D, Baladi M, Jungsuwadee P, Earl G, Han J.. The positive association between proton pump inhibitors and Clostridium difficile infection. Innov Pharm 2021; 12: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang D, Prabhu VS, Marcella SW.. Attributable healthcare resource utilization and costs for patients with primary and recurrent Clostridium difficile infection in the United States. Clin Infect Dis 2018; 66: 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perumalsamy S, Riley TV.. Molecular epidemiology of Clostridioides difficile infections in children. J Pediatric Infect Dis Soc 2021; 10: S34–S40. [DOI] [PubMed] [Google Scholar]
- 96.Hassoun A. Clostridium difficile associated disease. BMJ 2018; 363: k4369. [DOI] [PubMed] [Google Scholar]
- 97.Gupta S, Allen-Vercoe E, Petrof EO.. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol 2016; 9: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, De Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368: 407–415. [DOI] [PubMed] [Google Scholar]
- 99.Rao K, Safdar N.. Fecal microbiota transplantation for the treatment of Clostridium difficile infection. J Hosp Med 2016; 11: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baunwall SMD, Andreasen SE, Hansen MM, Kelsen J, Høyer KL, Rågård N, et al. Faecal microbiota transplantation for first or second Clostridioides difficile infection (EarlyFMT): a randomised, double-blind, placebo-controlled trial. Lancet Gastroenterol Hepatol 2022; 7: 1083–1091. [DOI] [PubMed] [Google Scholar]


