Abstract
The widespread bacterial second messenger c-di-GMP is responsible for regulating many important physiological functions such as biofilm formation, motility, cell differentiation, and virulence. The synthesis and degradation of c-di-GMP in bacterial cells depend, respectively, on diguanylate cyclases and c-di-GMP-specific phosphodiesterases. Since c-di-GMP metabolic enzymes (CMEs) are often fused to sensory domains, their activities are likely controlled by environmental signals, thereby altering cellular c-di-GMP levels and regulating bacterial adaptive behaviors. Previous studies on c-di-GMP-mediated regulation mainly focused on downstream signaling pathways, including the identification of CMEs, cellular c-di-GMP receptors, and c-di-GMP-regulated processes. The mechanisms of CME regulation by upstream signaling modules received less attention, resulting in a limited understanding of the c-di-GMP regulatory networks. We review here the diversity of sensory domains related to bacterial CME regulation. We specifically discuss those domains that are capable of sensing gaseous or light signals and the mechanisms they use for regulating cellular c-di-GMP levels. It is hoped that this review would help refine the complete c-di-GMP regulatory networks and improve our understanding of bacterial behaviors in changing environments. In practical terms, this may eventually provide a way to control c-di-GMP-mediated bacterial biofilm formation and pathogenesis in general.
Keywords: c-di-GMP, diguanylate cyclase, c-di-GMP-specific phosphodiesterase, sensory domains, NO sensors, photoreceptors
The authors review the diversity of sensory domains related to the bacterial CME regulation and specifically discuss those domains that are capable of sensing gaseous or light signals and the mechanisms they use for regulating cellular c-di-GMP levels.
Introduction
Unlike intracellular bacteria that inhabit stable ecological niches, most free-living bacteria face complex and rapidly changing ecological environments. To survive, these bacteria need to be able to monitor changes in various physical or chemical parameters around them and respond quickly to adapt to various environments (Mascher et al. 2006). As a result, they have evolved complex signal transduction systems. Research in this field allowed discovering several different bacterial signal transduction systems, including, among others, the two-component signal transduction systems (Stock et al. 2000), one-component transcriptional regulators (Ulrich et al. 2005), the alternative sigma factor regulatory systems (Mascher 2013), methyl-accepting chemotaxis receptor protein (MCP)-based chemosensory systems (Miller et al. 2009, Ortega et al. 2017), and protein kinase cascades (Shi et al. 1998, Pereira et al. 2011). However, most of these signal transduction pathways act in a relatively straightforward way and typically regulate only certain classes of downstream targets. In contrast to these systems, second messenger signaling pathways are characterized by a multitude of potential receptors, comprising a new and constantly expanding field of bacterial signaling pathways (Römling et al. 2013, He et al. 2020, Lowey et al. 2020, Stülke and Krüger 2020).
The second messenger systems are important components of the signal transduction networks. In eukaryotes, they can be roughly divided into four categories: nucleotides, lipids, gases, and free radicals or ions (Newton et al. 2016). In bacteria, there are mostly nucleotide second messengers, specific nucleotide derivatives that are not used in cellular nucleic acid synthesis (Stülke and Krüger 2020). Nucleotide second messengers found in bacteria include guanosine-(penta- or tetra-)phosphate ((p)ppGpp), cyclic adenosine monophosphate (cAMP), cyclic di-adenosine monophosphate (c-di-AMP), cyclic di-guanosine monophosphate (c-di-GMP), cyclic GMP–AMP (cGAMP), and some others (Lau et al. 2020, Lowey et al. 2020). They act to bridge bacterial signal perception with cellular response(s). When the cell surface receptors (or receptor domains) receive extracellular signals (first messengers), they can affect the catalytic activities of various intracellular enzymes, including cyclic nucleotide synthases and hydrolases, resulting in changes in the concentrations of certain nucleotide molecules, which serve as the “second messengers.” Alterations of the second messenger levels affect their binding to downstream receptors, which regulate specific physiological functions of bacteria.
Signal transduction systems that rely on nucleotide second messengers have distinct advantages. First, the concentrations of nucleotide second messengers are directly controlled by their metabolic enzymes, which, upon receiving a specific signal, can rapidly change the cellular concentrations of the respective second messenger, which, in turn, would alter the properties (activity, conformation, and/or oligomeric state) of their receptor(s). Second, cellular receptors of nucleotide second messengers in bacteria are typically abundant and diverse, enabling them to regulate downstream pathways in multiple ways (expression, enzymatic activity, and binding properties). Such a signal transduction system allows the bacteria to participate in continuous sensing and/or coordinated regulation of a single biological process (Orr et al. 2016, Stülke and Krüger 2020). Therefore, bacteria can rely on the nucleotide second messenger-mediated signal transduction systems to enhance their environmental adaptability.
c-di-GMP is a cyclic dinucleotide present in a wide variety of bacteria (Römling et al. 2013). Its synthesis and degradation are regulated by diguanylate cyclases (DGCs) containing the GGDEF domain (which typically contains the Gly-Gly-Asp/Glu-Glu-Phe sequence motif) and c-di-GMP specific phosphodiesterases (PDEs) containing either EAL or HD-GYP domains, so named after their conserved sequence motifs, Glu-Ala-Leu in case of the EAL domain and His-Asp and Gly-Tyr-Pro motifs in case of the HD-GYP domain. DGCs and PDEs are collectively referred to as c-di-GMP metabolic enzymes (CMEs) (Dahlstrom and O’Toole 2017, Jenal et al. 2017).
CMEs typically act as mediators that transform environmental signals into c-di-GMP concentration changes, thereby participating in signal transduction and regulating bacterial behaviors. Signaling CMEs can be divided into two categories based on the way they sense signals.
Many signaling CMEs combine sensory domains and an enzymatic domain in a single protein, and can directly respond to various signal stimuli including gases, light, redox state, temperature, and chemical compounds; these CMEs can be referred to as one-component systems (Ulrich et al. 2005). They are thought to be the predominant mode of sensing in bacteria, and this type of CMEs is also the most easily recognized by bioinformatics.
However, some CMEs can still respond to stimuli despite having no sensory domains themselves. This usually happens in one of two ways. First, part of the CMEs receive signals through protein–protein interactions with proteins containing the sensory domains. Second, other CMEs act as downstream response regulators in two-component systems or more complex multicomponent phosphorelay systems (e.g. chemosensory cascades) and alter their enzymatic activities in response to the transfer of the phosphoryl group to upstream phosphoacceptor domains.
Although the specific mechanisms of signal sensing in signaling CMEs are different, the common feature is that after receiving signals, these CMEs adjust their catalytic activities through conformational changes, thereby altering the concentrations of c-di-GMP in bacteria. Concentration fluctuations are then sensed by specific downstream receptors, which in turn regulate multiple bacterial physiological functions (Wang et al. 2016, Valentini and Filloux 2019), including biofilm formation (Ha and O’Toole 2015, Teschler et al. 2022), motility (Sun et al. 2020), cell differentiation (Lori et al. 2015, Kaczmarczyk et al. 2020), phage resistance (Junkermeier and Hengge 2021, Sellner et al. 2021), and virulence factors expression (Fu et al. 2018, Hall and Lee 2018).
Because of the rich regulatory functions of c-di-GMP, the downstream components of c-di-GMP-dependent signaling pathways, such as the nature of c-di-GMP receptors and the organization of c-di-GMP-regulated networks, have been studied in detail for more than 35 years. However, studies of the upstream signals that the CMEs respond to are relatively limited, and relevant reviews are even rarer. The lack of comprehensive information on the kinds of signals modulating the c-di-GMP levels affects our understanding of the c-di-GMP regulatory networks and hinders the studies of the environmental adaptability of bacteria.
Considering the variety of the sensing capabilities of signaling CMEs and the paucity of comprehensive reviews of this subject, we believe that there is a need for a focused and in-depth analysis of the upstream signaling pathways that control bacterial c-di-GMP levels. Here, we have focused on the sensory domains capable of sensing gas and light to modulate the activity of CMEs. We discuss their various types, structures, and regulatory mechanisms, hoping to promote further research in this important area of signal transduction.
Diversity of sensory domains in bacterial CMEs
CMEs, which catalyze synthesis and hydrolysis of c-di-GMP, are widespread in the microbial world. While the presence of such enzymes in archaea and eukaryotes is limited to just a few cases, such as Methanocella arvoryzae MRE50 and Dictyostelium discoideum, respectively (Chen and Schaap 2012), CME genes are found in the genomes of all bacterial phyla sampled so far (Galperin 2005, Römling et al. 2013). These enzymes are encoded by most free-living bacteria and even by obligately intracellular vector-borne pathogens of the order Rickettsiales that have genome sizes under 900 kb and cause such diseases as human ehrlichiosis and Potomac horse fever. Remarkably, most of these enzymes combine the enzymatic (DGC or PDE) domains with N-terminal regions that in many cases have been recognized as ligand-binding sensory domains.
In fact, the early discovery of c-di-GMP as a component of the signal transduction machinery was partly due to the presence of the PAS domain in the Komagataeibacter xylinus CMEs (Tal et al. 1998, Chang et al. 2001). Indeed, a great majority of CMEs are coupled to such signaling domains as PAS (named after Per, ARNT, and Sim proteins) (Huang et al. 1993), GAF (the common domain in cGMP-specific and cGMP-stimulated PDEs, adenylate cyclases, and Escherichia coli FhlA) (Hurley 2003), and REC (the receiver domain of two-component response regulators). Interestingly, the PAS and GAF domains adopt similar topologies (Ho et al. 2000), and ligand-binding pockets of both can accommodate cofactors such as heme and flavin (Gilles-Gonzalez and Gonzalez 2004). These cofactors endow sensing specificity to the sensory domains, allowing proteins containing these domains to sense diatomic gases, light, redox state, and other signals (Taylor and Zhulin 1999).
In the past several years, a wide variety of other sensory domains have been identified in CMEs. These include, among others, the globin-coupled sensor (GCS) domains (Wan et al. 2009, Patterson et al. 2021), periplasmic calcium channel and chemotaxis (CACHE) domains (Upadhyay et al. 2016, Giacalone et al. 2018), periplasmic (extracytoplasmic) cyclases/histidine kinases-associated sensory extracellular (CHASE) domain and gamma-proteobacterial periplasmic sensor (GAPES) domain series, CHASE1 through CHASE5 and GAPES1 through GAPES4 (Hengge et al. 2015), as well as a dozen of membrane-associated sensor (MASE) integral membrane domains, from MASE1 (Nikolskaya et al. 2003, Pfiffer et al. 2019) to MASE12 (Galperin and Chou 2022, Martín-Rodríguez et al. 2022). A given CME may contain a single sensory domain that would sense a specific signal (or a group of related ligands), or multiple sensory domains for signal amplification, attenuation, or integration of multiple environmental signals. The diverse combinations of various sensory domains in CMEs can be expected to help the cells to specifically and precisely regulate the c-di-GMP concentrations, thus conferring upon the bacteria high adaptability to environmental changes.
Table 1 shows the distribution of 112 sensory domains, which include 96 distinct domains and several sequence models for PAS, GAF, and tetratricopeptide repeat (TPR) domains [Table S1 (Supporting Information) contains actual domain counts but they are variable]. As sensor domains are often promiscuous, being shared by CMEs, MCPs, sensor histidine kinases, and other bacterial receptors such as adenylate cyclases and serine/threonine protein kinases (Zhulin et al. 2003), this list shows a significant overlap with the one that was compiled recently for sensor kinases, chemoreceptors and transcriptional regulators (Matilla et al. 2022). Analysis of the sensory domain composition of the signaling CMEs reveals that their distribution is quite uneven.
Table 1.
Diversity of sensory domains found in association with GGDEF, EAL, and/or HD-GYP domains.a
| Domain nameb | Pfam ID | COGc | PDB ID | PSSM, aad | Domain name origin | GGDEF only | EAL only | GGDEF + EAL | HD-GYP | GGDEF + HD-GYP | Ligands | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extracellular (periplasmic) domains | ||||||||||||
| 4HB_MCP_1 | PF12729 | – | 5XUA | 181 | Four-helix bundle of methyl-carrier proteins | + | + | + | + | + | Citrate, fumarate, succinate, pyrene | Hong et al. (2019) |
| 7TMR-DISMED2 | PF07696 | – | 3JYB | 127 | 7TM Receptors with diverse intracellular signaling modules, extracellular domain 2 | + | + | + | – | – | Ca2+, carbohydrates | Jing et al. (2010) |
| 7TMR-HDED | PF07697 | COG1480 | – | 219 | 7TM-HD extracellular domain | – | – | – | + | – | – | |
| ABC_sub_bind | PF04392 | COG2984 | 6HNI | 293 | ABC transporter substrate binding protein | + | – | + | + | – | Tyr? | Bradshaw et al. (2019) |
| Cache3/Cache2 | PF17201 | – | – | 298 | Calcium channels and chemotaxis receptors fused domains 3 and 2 | + | – | + | – | – | – | |
| CBM_2 | PF00553 | – | 2CWR | 101 | Carbohydrate binding module | – | – | – | + | – | Chitin | Nakamura et al. (2008) |
| CBM_4_9 | PF02018 | – | 1GUI | 134 | – same – | + | – | – | – | – | Cellulose, xylan | Boraston et al. (2002) |
| CHASE | PF03924 | COG3614 | 3T4J | 184 | Cyclases/histidine kinases associated sensory extracellular domain | + | + | + | + | – | Cytokinin | Hothorn et al. (2011) |
| CHASE2 | PF05226 | COG4252 | – | 264 | – same – | + | + | + | + | – | – | |
| CHASE3 | PF05227 | COG5278 | 3VA9 | 138 | – same – | + | – | + | – | – | Pyrene | Zhang et al. (unpublished data) |
| CHASE4 | PF05228 | COG3322 | – | 139 | – same – | + | – | + | + | + | – | |
| CHASE5 | PF17149 | – | – | 108 | – same – | + | – | + | – | – | Arg? | |
| CHASE7 | PF17151 | – | – | 187 | – same – | + | – | – | – | – | Taurocholate | |
| CHASE8 | PF17152 | – | – | 102 | – same – | + | + | + | – | – | – | |
| CHASE9 | PF17153 | – | – | 116 | – same – | – | + | – | – | – | – | |
| CSS-motif | PF12792 | COG4943 | – | 209 | Conserved Cys-Ser-Ser motif | – | + | – | – | – | DsbA, DsbB | |
| DAHL | PF19443 | – | – | 221 | Double all-helical ligand-binding | + | + | + | – | – | Asp, Arg, Ile, fucose, galactose, mannose | |
| dCache | Double CACHE (Calcium channel and chemotaxis receptor) domain | Dicarboxylates | ||||||||||
| dCache_1 | PF02743 | – | 3LID | 238 | – same – | + | + | + | + | + | Amino acids, pH, diamines, purines, betaine, succinate | Zhang and Hendrickson (2010) |
| dCache_2 | PF08269 | – | 5G4Z | 297 | – same – | + | – | + | – | + | C2 and C3 carboxylates | Brewster et al. (2016) |
| dCache_3 | PF14827 | – | 5IS1 | 235 | – same – | + | + | + | + | – | – | Kim et al. (2016) |
| DICTe | PF10069 | COG4250 | – | 126 | Diguanylate cyclases and two-component systems | + | + | + | + | – | Light? | |
| DUF3365 | PF11845 | – | 5B82 | 167 | Domain of unknown function | + | – | + | – | + | c-type heme, redox | Motomura et al. (2017) |
| GAPES1 | PF17155 | – | – | 274 | Gammaproteobacterial periplasmic sensor domain | + | – | – | – | – | Autoinducer-2 (AI-2) | |
| GAPES2 | PF17156 | – | – | 204 | – same – | + | – | – | – | – | – | |
| GAPES3 | PF17154 | – | – | 121 | – same – | + | + | + | – | – | – | |
| GAPES4 | PF17157 | – | – | 98 | – same – | + | – | + | – | – | – | |
| LapD_MoxY_N | PF16448 | – | 3PJV_D | 124 | N-terminal periplasmic domain of LapD and MoxY | + | + | + | – | – | LapG protein, methanol | Navarro et al. (2011) |
| LuxQ-periplasm | PF09308 | COG1879 | 3C30 | 239 | Periplasmic domain of LuxQ | – | – | + | – | – | LuxP protein | Slama and Hendrickson (unpublished data) |
| PBP1_AmiC (Peripla_BP_5) | PF13433 | COG0683 | 1PEA | 363 | Periplasmic binding protein AmiC-type | + | – | + | – | – | Acetamide, other amides | Pearl et al. (1994) |
| PBP1_ABC_LivBP (Peripla_BP_6) | PF13458 | COG0683 | 1Z15 | 342 | Periplasmic binding protein LivBP-type | + | – | + | – | – | Leu, Ile, Val | Trakhanov et al. (2005) |
| Peripla_BP_3 | PF13377 | COG1609 | 1JYE | 160 | Periplasmic binding protein-like domain | + | + | + | + | – | Ribose, galactose, glucose 6-phosphate | Bell et al. (2001) |
| Peripla_BP_4 | PF13407 | COG4203 | 3UUG | 259 | – same – | + | – | – | – | – | Fructose, galactose | Hu et al. (2013) |
| Phosphonate-bd | PF12974 | COG3221 | 5LQ5 | 243 | Phosphonate-binding | + | – | + | + | + | Phosphates, phosphonates | Bisson et al. (2017) |
| PilJ/NarX | PF13675 | COG3850 | 6GCV | 112 | Nitrate-binding domain of McpN | + | – | + | – | – | Nitrate, nitrite | Martín-Mora et al. (2019) |
| PBPb (SBP_bac_3) | PF00497 | COG0834 | 2LAO | 221 | Bacterial extracellular solute-binding proteins, family 3 | + | + | + | + | + | Amino acids | Oh et al. (1993) |
| sCache | Single CACHE (Calcium channel and chemotaxis receptor) domain | |||||||||||
| sCache_2 | PF17200 | COG4564 | 3UB6 | 153 | – same – | + | – | + | + | + | Urea, propionate, malate, pyruvate | Goers et al. (2012) |
| sCache_3_2 | PF17203 | – | – | 140 | – same – | – | – | + | – | – | Citrate, malate | |
| sCache_3_3 | PF17202 | – | – | 107 | – same – | + | – | + | + | – | – | |
| sCache_4 | PF09984 | – | 5O7J | 146 | – same – | + | – | + | – | – | – | Ali-Ahmad et al. (2017) |
| TarH | PF02203 | – | 2LIG | 152 | Ligand-binding domain of the bacterial aspartate receptor | + | – | + | – | – | Asp, Glu, Ser, citrate, 4-hydroxybenzoate | Milburn et al. (1991) |
| Integral membrane domains | ||||||||||||
| 5TM-5TMR_LYT | PF07694 | COG3275 | – | 170 | 5TM Receptors of the LytS-YhcK type, 5 TM | + | – | + | + | + | Pyruvate | |
| 7TM-7TMR_HD | PF07698 | – | – | 190 | 7TM Receptor with intracellular HD hydrolase | – | – | – | + | – | – | |
| 7TMR-DISM_7TM | PF07695 | – | – | 207 | 7TM Receptors with diverse intracellular signaling modules, 7 TM domain | + | + | + | + | – | – | |
| AA_permease (SLC12) | PF00324 | COG0531 | – | 415 | Amino acid permease, 9TM | – | – | + | – | – | Amino acids | |
| AA_permease_2 | PF13520 | COG0531 | 5J4I | 427 | Amino acid permease, 12 TM | + | – | + | + | + | Amino acids | Ilgü et al. (2016) |
| Ammonium_transp | PF00909 | COG0004 | 6EU6 | 399 | Ammonium channel transporter Amt, 9–11 TM | + | – | + | – | – | NH4+ | Pflüger et al. (2018) |
| DUF4084 | PF13321 | – | – | 304 | Domain of unknown function, 9–10 TM | + | – | + | – | – | – | |
| DUF4118 | PF13493 | COG2205 | 2KSF | 107 | Transmembrane domain of KdpD, 4 TM | + | + | + | – | – | – | Maslennikov et al. (2010) |
| HisKA_7TM | PF16927 | – | – | 221 | N-terminal 7TM region of histidine kinase, 7TM | + | + | + | + | + | Autoinducer-1 (AI-1) | |
| MASE1 | PF05231 | – | – | 299 | Membrane-associated sensor domain, 5 TM | + | + | + | – | – | – | |
| MASE2 | PF05230 | – | – | 89 | – same –, 6 TM | + | – | + | – | – | – | |
| MASE3 | PF17159 | – | – | 226 | – same –, 5 TM | + | + | + | + | – | – | |
| MASE4 | PF17158 | – | – | 239 | – same –, 8 TM | + | – | + | – | – | – | |
| MASE5 | PF17178 | – | – | 192 | – same –, 6 TM | + | – | – | – | – | – | |
| MHYT | PF03707 | COG3300 | – | 54 (x3) | Met-His-Tyr-Thr motif, 6 TM | + | + | + | – | – | NO, nitrate | |
| PTS_EIIC | PF02378 | COG1455 | – | 315 | Phosphotransferase system, EIIC domain, 9–10 TM | – | + | – | – | – | – | |
| Intracellular (cytoplasmic) domains | ||||||||||||
| BLUFe | PF04940 | – | 2BYC | 89 | Blue light using FAD | – | + | + | – | – | FAD | Jung et al. (2005) |
| CBSe | PF00571 | COG0517 | 2RC3 | 57 (x2) | Regulatory domain in cystathionine-beta synthase | + | + | + | + | + | Adenine derivatives | Dong et al. (unpublished data) |
| C_GCAxxG_C_C | PF09719 | – | 1H21 | 115 | Putative redox-active protein with a CGAxxG motif | – | – | + | – | – | c-type heme | Abreu et al. (2003) |
| cNMP_binding | PF00027 | COG0664 | 2ZCW | 89 | Cyclic nucleotide-binding domain | + | + | + | + | – | Cyclic NMPs | Agari et al. (2008) |
| CZBe | PF13682 | – | – | 64 | Chemoreceptor zinc-binding domain | + | + | + | – | – | Zinc | |
| Diacid_rec | PF05651 | COG3835 | – | 131 | Sugar diacid recognition domain | + | – | + | – | – | Sugar acids | |
| DUF484 | PF04340 | – | 3E98 | 219 | Domain of unknown function | + | – | – | – | – | – | JCSG (unpublished data) |
| DUF1631 | PF07793 | – | – | 742 | – same – | + | + | + | – | – | – | |
| DUF1883 | PF08980 | – | 2B1Y | 86 | – same – | + | – | – | – | – | – | Nocek et al. (unpublished data) |
| DUF2892 | PF11127 | – | – | 66 | – same – | + | – | + | + | – | – | |
| DUF3330e | PF11809 | – | – | 69 | – same – | – | + | – | – | – | – | |
| DUF3369 | PF11849 | COG3437 | – | 168 | – same – | + | + | + | + | – | – | |
| DUF3391 | PF11871 | COG2206 | – | 136 | – same – | – | – | – | + | – | – | |
| FHA | PF00498 | COG1716 | 1G6G | 66 | Forkhead-associated domain | + | + | + | + | – | pThr, pTyr | Durocher et al. (2000) |
| FIST/NosP | PF08495 | COG3287 | – | 129 | F-box and intracellular signal transduction | + | + | + | + | + | NO | |
| FIST_C/NosP | PF10442 | COG3287 | – | 135 | – same – | + | + | + | + | + | NO | |
| GAF | PF01590 | COG2203 | 5VIV | 133 | Common domain in cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA | + | + | + | + | + | Biliverdin, cGMP, phycocyanobilin (+O2, CO, NO) | Baloban et al. (2017) |
| GAF_2 | PF13185 | COG1956 | 4MN7 | 137 | – same – | + | + | + | + | + | O2, CO, NO | Kim et al. (2014) |
| GAF_3 | PF13492 | COG2203 | 3EEA | 129 | – same – | + | + | + | + | + | – | Zhang et al. (unpublished data) |
| HDODe | PF08668 | COG1639 | 3M1T | 196 | HD-related output domain | + | + | + | + | – | – | JCSG (unpublished data) |
| Hemerythrin | PF01814 | COG2703 | 4XPX | 128 | Hemerythrin HHE cation binding domain | + | + | + | + | – | O2 | Chen et al. (2015) |
| HNOBA | PF07701 | – | – | 215 | Heme NO binding associated domain | + | – | + | – | – | Oxygen, NO | |
| Laminin_G_3e | PF13385 | – | 4DQA | 151 | Laminin globular domain | + | + | + | – | – | Arabinan, O-glycans | JCSG (unpublished data) |
| MEDS | PF14417 | – | – | 160 | Methanogen/methylotroph, DcmR sensory domain | + | + | + | + | – | Dichloromethane | |
| NIT | PF08376 | – | 4AKK | 228 | Nitrate- and nitrite sensing domain | + | – | + | – | – | Nitrate, nitrite | Boudes et al. (2012) |
| NMT1 | PF09084 | COG0715 | 2X26 | 216 | NMT1/THI5 protein domain | + | – | + | – | – | Alkanesulfonate | Beale et al. (2010) |
| PAS | PF00989 | COG2202 | 1KOU | 113 | Common domain in Period circadian protein (Per), Ah receptor nuclear translocator protein (ARNT), and Single-minded protein (Sim). | + | + | + | + | + | FAD, FMN, heme, 4-hydroxycinnamic acid (+O2, CO, NO) | van Aalten et al. (2002) |
| PAS_2 | PF08446 | COG4251 | 6G20 | 107 | – same – | + | – | + | – | – | – | Schmidt et al. (2018) |
| PAS_3 | PF08447 | – | 5SY7 | 89 | – same – | + | + | + | + | + | – | Wu et al. (2016) |
| PAS_4 | PF08448 | – | – | 110 | – same – | + | + | + | + | + | Aromatic compounds | |
| PAS_7 | PF12860 | – | – | 115 | – same – | + | + | + | – | – | – | |
| PAS_8 | PF13188 | – | – | 65 | – same – | + | + | + | + | + | O2, CO, NO | |
| PAS_9/LOV | PF13426 | – | – | 102 | – same – | + | + | + | + | + | Light, O2, voltage | |
| PHYe | PF00360 | COG4251 | 2VEA | 178 | Phytochrome region | + | – | + | – | – | Red light | Essen et al. (2008) |
| PilZ | PF07238 | – | 2L74 | 102 | Type IV pili biosynthesis protein | + | + | + | + | – | c-di-GMP | Habazettl et al. (2011) |
| PocR | PF10114 | – | – | 155 | Ligand binding domain of transcriptional regulator PocR | + | + | + | + | + | 1,2-propanediol | |
| Protoglobin | PF11563 | – | 4ZVA | 149 | Globin sensor domain | + | + | + | + | – | O2, CO, NO | Tarnawski et al. (2015) |
| RsbRD_N | PF14361 | – | – | 104 | N-terminal domain of the stressosome component RsbRD | + | – | + | – | – | – | |
| SnoaL_3 | PF13474 | COG4319 | 3CNX | 121 | SnoaL-fold domain 3 | + | – | + | + | – | – | JCSG (unpublished data) |
| T2SSE_N/MshEN | PF05157 | – | 5HTL | 108 | Type II secretion system protein E, N-terminal domain | + | – | – | + | – | c-di-GMP | Wang et al. (2016) |
| TackOD1 | PF18551 | – | – | 188 | Thaumarchaeal output domain 1 | + | – | – | – | – | – | |
| TPR_1 | PF00515 | COG0457 | 2KC7 | 34 | Tetratricopeptide repeat | – | Eletsky et al. (unpublished data) | |||||
| TPR_2 | PF07719 | COG0457 | 4XI0 | – same – | + | – | – | – | – | – | Zeytuni et al. (2015) | |
| TPR_4 | PF07721 | – | – | – same – | + | – | – | – | – | – | ||
| TPR_7 | PF13176 | – | – | – same – | + | + | + | – | – | – | ||
| TPR_8 | PF13181 | – | – | 33 | – same – | + | + | + | + | – | – | |
| TPR_10 | PF13374 | – | – | – same – | + | – | + | – | – | – | ||
| TPR_12 | PF13424 | – | 3ESK | 77 | – same – | + | + | + | + | + | – | Kajander et al. (2009) |
| TPR_16 | PF13432 | – | – | 68 | – same – | + | – | + | – | – | – | |
| TPR_MalT | PF17874 | – | – | MalT-like TPR region | + | – | + | – | + | – | ||
| V4R | PF02830 | COG1719 | 2OSD | 62 | Vinyl 4 reductase | + | – | + | – | – | Hydrocarbons | JCSG (unpublished data) |
| YceI | PF04264 | COG2353 | 3HPE | 118 | YceI-like domain | – | – | + | – | – | Isoprenoids, fatty acids | Sisinni et al. (2010) |
| YkuI_Ce | PF10388 | – | 2BAS | 166 | C-terminal domain of YkuI | – | + | – | – | – | – | Minasov et al. (2009) |
| Y_Y_Y | PF07495 | – | 4A2M | 65 | Tyr-x-Tyr-x-Tyr sequence motif | + | – | + | + | – | Heparin? | Lowe et al. (2012) |
| Total number | 94 | 52 | 88 | 48 | 25 | |||||||
The table indicates presence (+) or absence (–) of the respective domain combination in the Pfam database as of 12-12-2022. Domain combinations found in a single protein in UniProt were ignored. For domain counts, additional ligands, and references, see Table S1 (Supporting Information). Additional references for some of these domains can be found in the recent review by Matilla et al. (2022). Some domain combinations in the EAL-only column are listed as absent despite being annotated as such in UniProt because all the respective entries contain a diverged GGDEF domains.
Domain names in Pfam (alternative names are in parentheses). The respective entries can be retrieved from InterPro using the https://www.ebi.ac.uk/interpro/entry/pfam/PFxxxxx/format, e.g. https://www.ebi.ac.uk/interpro/entry/pfam/PF12729/for 4HB_MCP_1
COG database (https://www.ncbi.nlm.nih.gov/research/cog/) entries that include this domain. A dash in this and other columns indicates the absence of data.
Length (in amino acid residues) of the domain sequence model in the CDD database (https://www.ncbi.nlm.nih.gov/cdd).
This domain is usually found at the C-termini of the respective CMEs.
First, the number of signal sensory domains coupled to c-di-GMP metabolic domains is found to vary widely in different bacteria. CMEs are widely distributed in Proteobacteria, especially in γ-Proteobacteria. The number of CMEs encoded by each genome of these bacteria often reaches dozens, and most of them contain CMEs with signal sensory domains. As an example, the genome of Pseudomonas aeruginosa PAO1 encodes 43 CMEs, of which 16 CMEs contain PAS or GAF domains (Valentini and Filloux 2016). In contrast, bacteria from the phyla Firmicutes and Actinobacteria generally encode only a few CMEs. The number and type of signaling proteins contained in bacteria usually depend on the phylogenetic position, lifestyle, and environment of the bacterium (Galperin 2005). Thus, a wide diversity of signaling CMEs has been frequently seen in some opportunistic pathogens that face complex environments (Randall et al. 2022).
In addition to the differences in the number of CMEs encoded in different bacteria, many CMEs differ in the number of sensory domains per molecule. The composition of the sensory domains in a given CME appears to determine their specific role(s) in the bacterium. Table 1 clearly shows that the EAL- and HD-GYP-containing PDEs usually contain fewer sensory domains compared to their numbers in the GGDEF domain-containing DGCs and the GGDEF–EAL hybrid proteins. The relative paucity of sensory domains in EAL-only PDEs, as well as the typically lesser number of EAL-only protein-encoding genes in most genomes compared to the number of the GGDEF domain proteins (Galperin 2005), may indicate that EAL-containing PDEs serve primarily as a sink for the c-di-GMP molecules, non-specifically lowering its cellular levels. Likewise, HD-GYP-containing PDEs rarely contain multiple sensory domains (Galperin and Chou 2022). In contrast, the complex sensory domain network of GGDEF–EAL hybrid proteins may allow them to quickly respond to extracellular or intracellular signals by switching their enzymatic activities (DGC to PDE and back) and thereby control the c-di-GMP levels and c-di-GMP-mediated responses. The fountain model proposed by Sarenko et al. (2017) can well explain these results, in which some of these CMEs form cellular c-di-GMP pools while others perform specific local functions.
In conclusion, the distribution of sensory domains in CMEs is related to the complexity of the environment and specific functional characteristics of related proteins, which fully reflects the important role of signaling CMEs in bacterial signal transduction and environmental adaptation.
Regulation of c-di-GMP levels via heme-based gas-sensing domains
Gaseous molecules are ubiquitous in the environment and have high cell membrane permeability. They can serve as nutrients [e.g. carbon dioxide (CO2)], terminal electron acceptors [e.g. oxygen (O2)], or just act as signal molecules [e.g. nitric oxide (NO)] to regulate physiological processes in bacteria. Gas-sensing proteins in bacteria usually rely on cofactors such as heme, iron–sulfur cluster, or nonheme iron to capture gaseous molecules; heme-based regulators appear to predominate (Aono 2008).
Heme-based gas-sensing proteins typically use the iron-bound form of heme b (protoporphyrin IX) as a cofactor in their active site to exploit the redox-switching properties of iron for signal transduction. The iron atom present at the center of the heme porphyrin is able to coordinate six ligands: four nitrogen atoms at the center of the heme porphyrin ring, side chain of a His or Cys residue of the protein, and, finally, an exogenous ligand or another amino acid side chain (Farhana et al. 2012). When the gaseous molecule acting as an exogenous ligand associates with (or dissociates from) the sixth binding site of the heme–Fe complex, the coordination structure of the heme iron changes, causing accompanying changes in the surrounding protein. These structural changes generate a signal that can be transduced to the functional domain, ultimately enabling various crucial physiological functions to be switched on or off (Shimizu et al. 2015). The ability to switch between the coordination states of heme iron is essentially the basis for the signal transduction of heme-based gas-sensing proteins. Meanwhile, the gaseous molecules recognition specificity of heme-based gas-sensing proteins depends on the interaction of the amino acid residues around the heme group with ligands (Jain and Chan 2003).
A variety of CMEs with heme-based gas-sensing domains have been identified in bacteria that regulate cellular c-di-GMP concentrations in response to the presence of certain gases. Here, we mainly discuss the gas-sensing domains in CMEs that are sensitive to O2 and NO, both of which are of physiological significance in bacteria. We have chosen not to discuss the effects of carbon monoxide (CO) binding since (i) CO is not a physiological axial ligand in bacteria, and (ii) CO-specific sensory domains are typically found in transcription regulators, such as CooA from Rhodospirillum rubrum or RcoM from Burkholderia xenonorans, and do not include any known CMEs (Shimizu et al. 2015).
O2-sensing domains
O2 is one of the most abundant gases in the environment and has important effects on many physiological processes of bacteria, including biofilm formation (Mashruwala et al. 2017), motility (Taylor et al. 1999), respiration, chemotaxis (Muok et al. 2019), and virulence. The ability to sense changes in O2 availability is essential for many bacteria to carry out physiological switching and to counteract oxidative stress induced by reactive oxygen species (ROS). This is especially true for pathogens because many niches inside the host are hypoxic compared to the natural environment. Monitoring O2 concentrations enables these bacteria to respond quickly by readjusting gene expression programs in the face of environmental changes, thereby facilitating the switch between aerobic and anaerobic metabolism. Some CMEs containing O2-sensing domains have been identified in bacteria, and these enzymes help bacteria sense the O2 concentrations in the environment and convert it into a c-di-GMP concentration signal, thereby inducing bacterial behavioral responses and enhancing their environmental adaptability (Wan et al. 2009, Burns et al. 2017).
Currently, the reported O2-sensing domains in bacterial CMEs can be divided into two categories—one is the heme-containing PAS domain (heme-PAS domain), and the other is the heme-containing GCS domain (heme-GCS domain). There are some differences between the two O2-sensing mechanisms in bacteria as described below.
Heme-PAS domains
PAS domains are important signaling modules that are widely distributed in prokaryotes and eukaryotes (Taylor and Zhulin 1999). The core structure of the PAS domain is well-conserved across species and consists of five antiparallel β-strands (Aβ, Bβ, Gβ, Hβ, and Iβ) and four α-helices (Cα, Dα, Eα, and Fα) (Fig. 1A) (Möglich et al. 2009). Some PAS domains rely on cofactors to directly sense environmental signals such as light, gases, and redox state and subsequently transmit the signal to the functional domain; while the other PAS domains do not bind any ligands, but indirectly respond to signals through the mediation of protein–protein interaction (Huang et al. 1993).
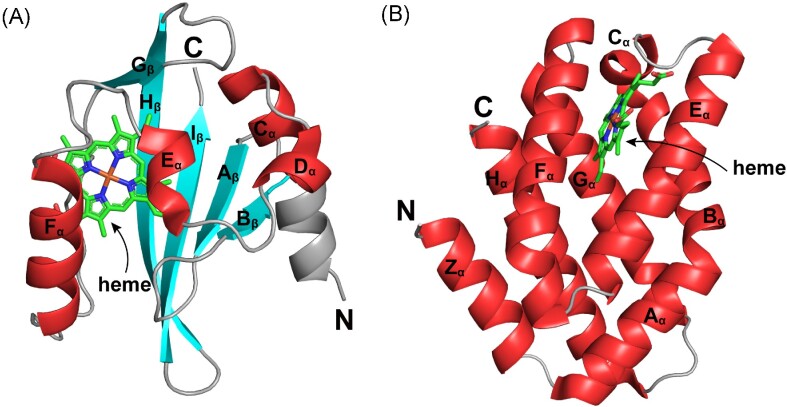
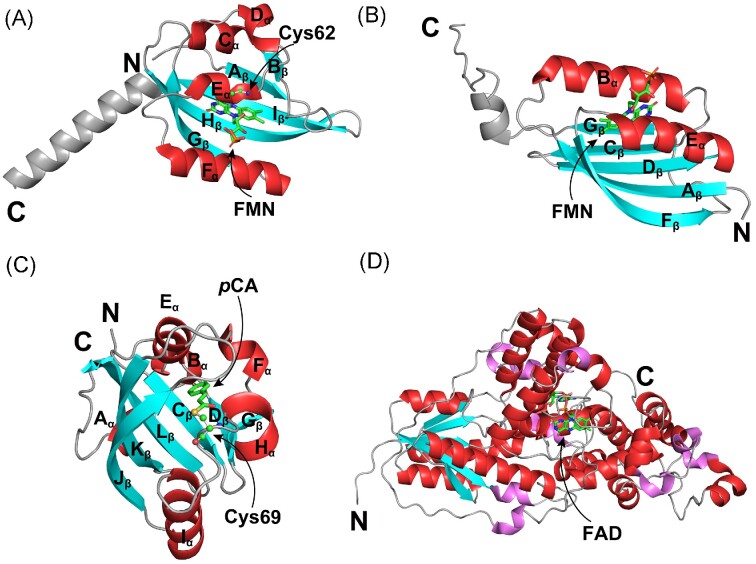
Figure 1.
Crystal structures of two types of heme-based O2 sensors. (A) Crystal structure of the heme-containing PAS domain from Escherichia coli EcDosP (PDB entry: 1V9Z) (Kurokawa et al. 2004). Its five β-strands (Aβ, Bβ, Gβ, Hβ, and Iβ) and four flanking α-helices (Cα, Dα, Eα, and Fα) are labeled as indicated. (B) Crystal structure of the heme-containing GCS domain from Escherichia coli EcDosC (PDB entry: 4ZVB) (Tarnawski et al. 2015). Each monomer contains eight α-helices, which are named Zα, Aα, Bα, Cα, Eα, Fα, Gα, and Hα according to the classical globin nomenclature. The heme ligand in each domain is indicated by arrows.
The heme-PAS domain belongs to the former group and can accomplish ligand-dependent switching of a variety of functional domains, including histidine kinases (HKs), MCPs, CMEs, and basic helix-loop-helix DNA-binding modules (Dioum et al. 2002). Among identified signaling CMEs, the heme-PAS domains were mostly reported in bacterial PDEs.
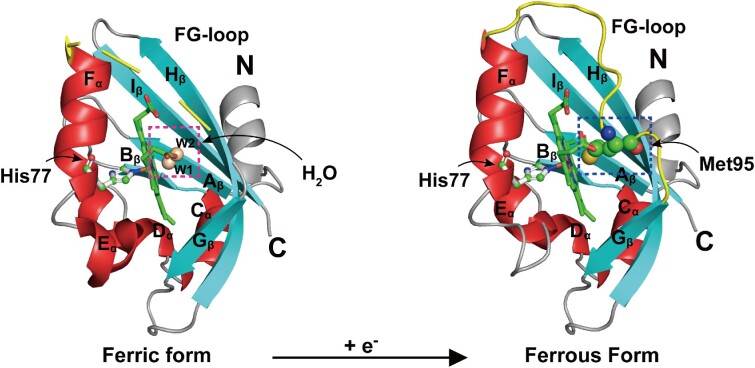
As early as 2000, the PDE EcDosP from E. coli was found to function as an O2 sensor (Delgado-Nixon et al. 2000). It contains two N-terminal PAS domains, but only the first PAS domain can bind heme. The unliganded heme Fe complex in EcDosP inhibits catalytic activity, while ligand binding to the heme–Fe(II) complex can alleviate this inhibition (Tanaka and Shimizu 2008). In the absence of any external ligands, both the heme–Fe(III) and Fe(II) complexes of EcDosP are in a six-coordinated low-spin state (Tomita et al. 2002). In the ferric form, heme iron is attached to the side chain His77 (proximal ligand) and a water molecule (distal ligand); when reduced to the ferrous form, the distal axial ligand is changed from a water molecule to Met95 on the FG-loop (encompassing residues 86–97 between the Fα-helix and Gβ-strand) (Fig. 2). Binding of exogenous axial ligand O2 molecules to the heme Fe(II) complex is dependent on the dissociation of Met95 from the heme plane (Kurokawa et al. 2004, Park et al. 2004). Thus, the ligand binding process is accompanied by a change in protein conformation, which relieves inhibition through intramolecular signal transduction to enhance the PDE activity (Shimizu 2013).
Figure 2.
Redox-induced changes at the distal heme site of EcDosP. EcDosP is O2-dependent and its activity is regulated by the transition between the ferric form (PDB entry:1V9Y) (Kurokawa et al. 2004) and ferrous form (PDB entry:1V9Z) (Kurokawa et al. 2004) of the heme-PAS domain, a process accompanied by the change of distal axial ligands. When the heme-PAS domain is in the ferric form, the distal axial ligand of its complex is a water molecule W1, stabilized by another water molecule W2 (marked by the magenta dashed box); when it is reduced to the ferrous form, the distal axial ligand of its complex is changed to Met95 (marked by the blue dashed box) of the FG-loop (shown in yellow). Also indicated is the iron-binding His77.
KxPDEA1 from K. xylinus is also a PDE containing a heme-PAS domain, although quite different from EcDosP in terms of structure and enzymatic properties. For example, the heme-free form of apo-KxPDEA1 does not retain high catalytic activity like apo-EcDosP but completely loses its activity. This suggests that the presence of the heme–Fe complex in this protein is critical for maintaining an active site structure suitable for optimal catalysis and that the PDE activity of KxPDEA1can be activated only when the ligand dissociates from the heme–Fe(II) complex, rather than binds it. In addition, the heme–Fe(II) complex of KxPDEA1 is not in the six-coordinated state; rather, it is in the five-coordinated state, i.e. more common in heme-PAS proteins. This allows O2 molecules to bind to the vacant distal site of the complex without any ligand exchange (Tomita et al. 2002). The above differences illustrate the diversity of the signal transduction mechanisms that rely on binding O2 molecules.
Heme-GCS domain
Heme-GCS domain was first discovered in Bacillus subtilis and Halobacterium salinarium as a heme sensor that controls aerotaxis and is widely distributed in bacteria, archaea, fungi, and even some protozoa (Hou et al. 2000, Vinogradov et al. 2005). The crystal structures of GCS proteins show that the GCS domain is usually in a dimer form with a canonical α-helical rich globin fold and a heme cofactor in a hydrophobic cavity formed by the fold (Fig. 1B) (Keppner et al. 2020). Compared to myoglobin and hemoglobin, GCS has a shortened globin fold that lacks a complete D-helix and partial E-helix, these changes appear to favor O2 sensing, as opposed to O2 transport (Martínková et al. 2013, Walker et al. 2017).
The heme-GCS domain is usually located at the N-terminus of the proteins and is fused to the C-terminal domains with activities such as MCPs, HKs, or CMEs. However, although the proximal histidine linked to the heme is absolutely conserved, the sequence similarity of proteins containing the heme-GCS domain is generally not very high. The currently characterized signaling CMEs with a heme-GCS domain are mainly DGCs, including EcDosC from E. coli (Fig. 2) (Tuckerman et al. 2009), DpDGC from Desulfotalea psychrophila (Sawai et al. 2010), BpGReg from Bordetella pertussis, and PcGCS from Pectobacterium carotovorum (Burns et al. 2017). These enzymes need to be in the form of multimers to exhibit catalytic activities. Some GCSs possess a middle domain that was demonstrated to adopt a four-helix bundle structure containing a short π-helix and form a dimer in the crystal structure. These GCSs transmit the ligand-binding signals sensed by the N-terminal GCS domain to the C-terminal DGC domain through this unique middle domain and orientate the three domains through the π-helix of the middle domain, resulting in a compact structure with the DGC activity (Walker et al. 2020). Like in the heme-PAS domain, the redox state and linkage of heme in the heme-GCS domain can also modulate the catalytic activities of the downstream domains. GCS proteins with a heme–Fe(II) complex (Fig. 1B) typically have some basal DGC activity, which gets enhanced by the binding of the O2 ligand. Binding of O2 to the heme causes subtle rearrangements of the heme pockets, changes in the helix flexibility, and rotation around the globin–dimer interface, and even changes in the oligomerization state of the protein, thereby regulating the DGC activity (Burns et al. 2016). Furthermore, some distal globin residues, like residues Phe42, Tyr43, Ala68 (EcGReg)/Ser68 (BpGReg), and Met69 in the distal heme pockets of EcGReg and BpGReg, exhibit certain effects on the DGC activities of GCS proteins (Wan et al. 2017).
Bacteria exploit differences in O2-sensing domains
Except for the differences in the coupling of functional domains, there are also some differences in the O2 binding properties of the heme-PAS domain and the heme-GCS domain. These are mainly manifested in (1) different binding sites. Most PAS sensors use Arg residues on the FG-loop to bind O2, while GCS sensor binds O2 mainly through Tyr residues in helix B (Martínková et al. 2013); (2) different binding affinities. The PAS sensor has a weaker binding affinity for O2 than the GCS sensor. The dissociation constants of the heme–Fe(II) complexes in the PAS sensors and GCS sensors are 12–340 μM and 0.077–14 μM, respectively (Kitanishi et al. 2010, Nakajima et al. 2012); and (3) different catalytic effects on the functional domains. Binding of O2 to the heme of the PAS domain usually reduces the PDE activity of the CME (except for EcDosP), while binding of O2 to the heme of the GCS domain can greatly enhance the DGC activity of the respective enzyme.
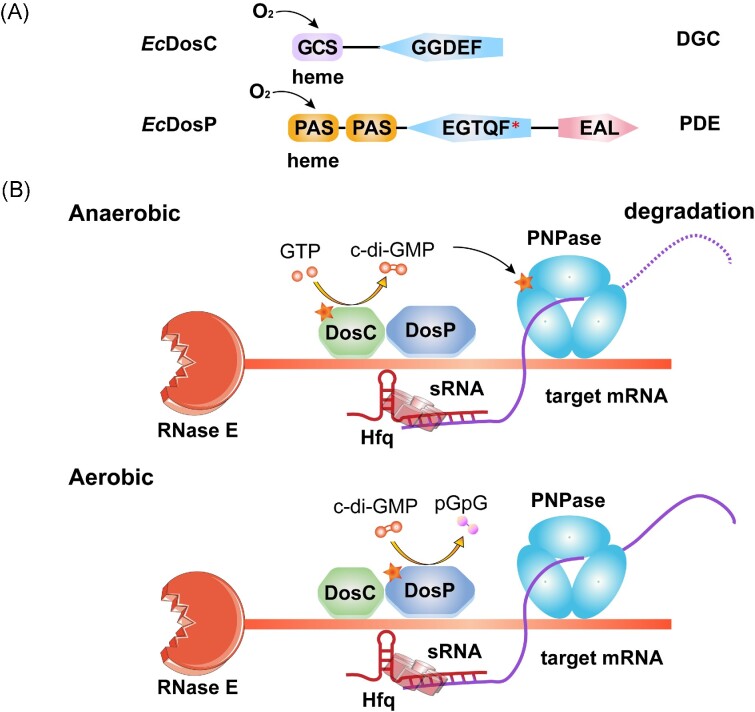
Based on the above characteristics, these heme-based sensors are able to carry out corresponding functions in different environments. PAS sensors can discern the presence of O2 and activate genes related to aerobic metabolism; while the GCS sensors can respond to hypoxia and activate associated genes (Martínková et al. 2013). The EcDosCP regulatory system serves as a very interesting model to explain how bacteria use these differences in O2 sensors to adapt to local O2 concentrations and maintain c-di-GMP homeostasis (Fig. 3). The genes encoding EcDosC and EcDosP are normally coexpressed during stationary phase by the dosCP operon (Tuckerman et al. 2009). Both EcDosC and EcDosP are heme proteins with O2-sensing domains and c-di-GMP metabolic domains. EcDosC is a DGC that fuses an N-terminal GCS domain to a C-terminal GGDEF domain, whereas EcDosP is a PDE with two PAS domains, a degenerated GGDEF domain with an EGTQF active site motif, and an EAL domain (Fig. 3A). EcDosC, EcDosP, ribonuclease E (RNase E), polynucleotide phosphorylase (PNPase), and some other components, including several degradosome-associated proteins and RNAs, were reported to form an oxy-degradosome (Gilles-Gonzalez and Sousa 2019). Bacteria selectively activate EcDosC or EcDosP according to the O2 concentrations to regulate cellular c-di-GMP levels. Changes in c-di-GMP levels affect the activity of PNPase, the receptor for c-di-GMP in this large enzyme complex, which in turn affects the processing or degradation of associated RNAs to control the effects at the post-transcriptional level (Fig.3B) (Tuckerman et al. 2011).
Figure 3.
O2 sensors EcDosC and EcDosP mediate c-di-GMP-dependent RNA processing in Escherichia coli. (A) Domain organization of EcDosC and EcDosP. EcDosP contains a degenerate GGDEF domain with the EGTQF motif at its active site. (B) Possible scheme for O2-dependent RNA degradation in the EcDosCP complex (based on Tuckerman et al. 2011). Under anaerobic conditions (upper panel), EcDosP and EcDosC are unliganded (deoxygenated), and the DGC activity of EcDosC is activated (marked with an asterisk), producing c-di-GMP to activate the receptor PNPase in the RNA degradation complex. Under aerobic conditions (lower panel), EcDosP and EcDosC are liganded (oxygenated) and the PDE activity of EcDosP is induced (marked with an asterisk). EcDosP hydrolyzes c-di-GMP to pGpG, which drastically decreases PNPase activity. mRNAs that depend on O2 for preservation and degradation may be selected by a mechanism involving sRNAs and Hfq, where sRNAs serve as mediators to recognize target mRNAs, and the RNA chaperone Hfq catalyzes this hybridization.
A similar regulatory mechanism may take place in bifunctional CMEs. The enzymatic activities of these proteins are determined by the ligands binding to their sensory domains. For example, PdDcpG from Paenibacillus dendritiformis possesses GCS, GGDEF, and EAL domains and exhibits dual functions of DGC and PDE (Patterson et al. 2021). PdDcpG relies on the GCS domain to bind different gaseous molecules to achieve differential regulation of downstream PDE/DGC activity: when the GCS domain is in the Fe(II)–NO state, its DGC activity is activated; whereas when the GCS domain is in the Fe(II)–O2 state, the PDE activity is activated, allowing bacteria to control biofilm formation in response to different gaseous environments (Patterson et al. 2021). Bifunctional CMEs like PdDcpG containing multiple sensory domains are abundant in bacteria, but the signal transduction mechanisms behind them remain obscure. It is tempting to speculate that similar regulatory mechanisms might function in other bifunctional CMEs.
Sensing O2 through changes in the redox state
While heme-containing proteins discussed above sense O2 through direct binding of O2 molecules, some proteins can monitor the change in the environmental O2 concentrations indirectly, by sensing the change of the redox state of the electron transport chains.
KxDGC2 from K. xylinus (Qi et al. 2009), AvNifL from Azotobacter vinelandii (Hill et al. 1996), and EcAer from E. coli (Taylor 2007) are all capable of sensing O2 concentrations indirectly by utilizing a PAS domain that binds a redox-sensitive flavin adenine dinucleotide (FAD) cofactor. The redox state of FAD determines the signaling output of these PAS sensors. In addition to the FAD-binding PAS domains, there are other domains that can help bacteria sense changes in O2 concentrations, such as the bacterial hemerythrin domain that may be present in either DGCs (Schaller et al. 2012) or PDEs (Kitanishi et al. 2020).
Hemerythrin domains typically have characteristic sequence motifs that provide ligand residues for the nonheme diiron site that binds O2 molecules and undergoes autoxidation. The diiron site is capable of cycling between diferric and diferrous forms depending on the O2 concentrations, thereby affecting the catalytic activities of the downstream domains. Existing research data show that such CMEs generally have higher catalytic activities in the reduced ferrous form compared to the oxidized ferric form (Kitanishi 2022).
Several years ago, a new class of PDEs that can respond to redox state has been described. These proteins combine the EAL domain with the periplasmic Cys-Ser-Ser (CSS)-motif domain that contains two highly conserved Cys residues flanked by two transmembrane segments. Such PDEs appear to use the disulfide–dithiol transition in the CSS domain as a redox switch that regulates the PDE activity of the EAL domain (Herbst et al. 2018). The CSS–EAL domain combination is encoded in five copies in both E. coli and Salmonella enterica and in three copies in P. aeruginosa. A recently described variant of the CSS-motif domain, referred to as the CSS_CxxC domain, contains two extra Cys residues. PDEs combining this domain with the EAL domain are encoded in a single copy in Shewanella, Vibrio, and some other species (Martín-Rodríguez et al. 2022). The discovery of such signaling CMEs illustrates the complexity of bacterial signal transduction pathways and suggests that there might be additional sensors of O2 and/or redox state.
NO-sensing domains
NO has been previously referred to as a double-edged sword in many physiological and pathological processes in a variety of organisms (Mocellin et al. 2007). That characterization is even more true for bacteria, so their ability to sense NO has a clear physiological significance. On the one hand, as a toxic gas, NO can diffuse freely and has a wide range of sources. It may come from bacteria, e.g. as an intermediate in the denitrification process of bacterial reduction of nitrate and nitrite, or from the oxidation of l-arginine by bacterial NO synthases; at the same time, NO may be produced by host macrophages as a line of defense against bacterial infection (Spiro 2007, Crane et al. 2010). This requires some bacteria, especially pathogens facing chronic exposure to high concentrations of NO, to have a mechanism to monitor and eliminate NO (Williams et al. 2018). On the other hand, low concentrations of NO have been demonstrated to act as a signaling molecule to regulate bacterial community behaviors, such as biofilm formation (Hossain et al. 2017), quorum sensing (Urbano et al. 2018), and symbiotic relationships (Wang et al. 2010).
C-di-GMP has been shown to be involved in regulating NO-responsive bacterial behaviors through some heme-based NO sensors (Rinaldo et al. 2018). These NO sensors can sense NO concentrations to regulate the catalytic activity of signaling CMEs. They are mainly divided into two categories: the heme-nitric oxide/oxygen (H-NOX) protein family, which has been intensively studied, and the NO-sensing protein (NosP) family that has been discovered in recent years (Bacon et al. 2017, Williams and Boon 2019). They are described in detail below.
H-NOX domain
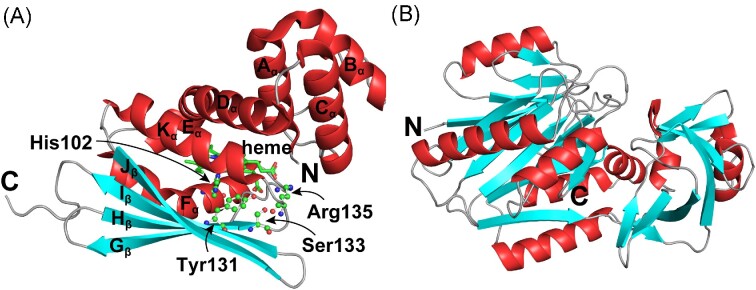
The H-NOX protein, a heme-protein identified in bacteria, is homologous to the eukaryotic NO sensor soluble guanylyl cyclase and can bind diatomic gaseous molecules (Cary et al. 2006). Its crystal structure was first solved in Caldanaerobacter subterraneus, which showed that the H-NOX family possesses a novel fold comprising an N-terminal helical subdomain and a C-terminal subdomain. Among them, the N-terminal subdomain is composed of five helices (Aα–Dα and Kα), the C-terminal subdomain is composed of a four-stranded antiparallel β-sheet and two helices (Eα and Fα) (Fig. 4), with the heme cofactor buried deep between the two subdomains and stabilized by a conserved His residue (His102) and three highly conserved residues (Tyr131, Ser133, and Arg135) in the YxSxR motif of Fα (Nioche et al. 2004, Pellicena et al. 2004).
Figure 4.
Crystal structures of two classes of heme-based NO sensors. (A) Crystal structure of the H-NOX domain from Caldanaerobacter subterraneus (PDB entry: 5JRU) (Hespen et al. 2016). The H-NOX fold consists of seven α-helices (Aα–Fα, and Kα) and a four-stranded antiparallel β-sheet (Gβ, Hβ, Iβ, and Jβ). Located on the α-helix Fα, His102 is the proximal axial ligand for heme iron and is highly conserved across all H-NOX domains. Tyr131, Ser133, and Arg135 are strictly conserved residues in the YXSXR motif. The heme ligand is also indicated by arrows. (B) Predicted structure of Pseudomonas aeruginosa NosP (PA1975), obtained from the AlphaFold website (https://alphafold.ebi.ac.uk/entry/Q9I2D0). This model contains 10 α-helices and 21 β-sheets.
H-NOX can appear as a domain in signaling proteins or as a free protein adjacent to other signaling partners. In the obligate anaerobic bacteria, H-NOX with a distal pocket hydrogen-bonding network often appears as a domain at the C-terminus of MCPs, which can not only bind to NO and CO molecules but also has the ability to bind to O2 tightly. In aerobic or facultative anaerobic bacteria, H-NOX is often used as an independent protein associated with HKs or CMEs, which can only stably bind NO and CO, but not O2 (Bacon et al. 2017, Guo and Marletta 2019). This strict discrimination against O2 ligands is necessary for a selective NO sensor because under aerobic conditions, O2 concentrations in cells typically far exceed the NO concentrations (Boon and Marletta 2005, Plate and Marletta 2013a). Differences in ligand-binding capacity also provide the basis for bacteria to selectively transmit signals in cells. It is worth mentioning that all currently identified bacterial H-NOXs have high affinity for NO at nanomolar to femtomolar levels, and most of the known physiological functions are related to NO sensing, highlighting their important role as NO sensors in signal transduction (Table 2).
Table 2.
NO dissociation rate constants for H-NOX and NosP proteins
| Protein | Species | k off (NO) (× 10−4 s−1) | Reference |
|---|---|---|---|
| TtH-NOX | Thermoanaerobacter tengcongensis | 5.6 ± 0.5 | Boon et al. (2005) |
| LpH-NOX1 | Legionella pneumophila | 10.3 ± 1.4 | Boon et al. (2006) |
| LpH-NOX2 | Legionella pneumophila | 21.8 ± 0.5 | Boon et al. (2006) |
| VfH-NOX | Vibrio fischeri | 21 ± 0.6 | Wang et al. (2010) |
| SwH-NOX | Shewanella woodyi | 15.2 ± 3.5 | Liu et al. (2012) |
| PaH-NOX | Pseudoalteromonas atlantica | 8.9 ± 3.6 | Arora and Boon (2012) |
| VhH-NOX | Vibrio harveyi | 4.6 ± 0.9 | Henares et al. (2012) |
| VpH-NOX | Vibrio parahaemolyticus | 4.3 ± 0.5 | Ueno et al. (2019) |
| SdH-NOX1 (Sde_3804) | Saccharophagus degradans | 97.0 ± 1.8 | Guo et al. (2018) |
| SdH-NOX2 (Sde_3557) | Saccharophagus degradans | 3.3 ± 0.6 | Guo et al. (2018) |
| PaNosP | Pseudomonas aeruginosa | 1.8 ± 0.5 | Hossain and Boon (2017) |
| LpNosP | Legionella pneumophila | < 2 | Bacon et al. (2018) |
| VcNosP | Vibrio cholerae | 4.6 ± 0.1 | Hossain et al. (2018) |
| Soluble guanylyl cyclase | Bovine lung | 3.6 ± 0.8 | Stone and Marletta (1994) |
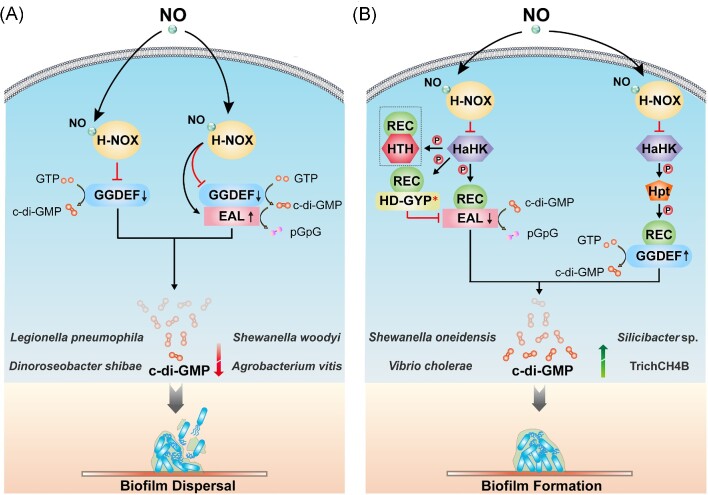
A possible mechanism by which NO activates H-NOX proteins can be described as follows: due to the special interaction of some amino acid residues of H-NOX with heme, the heme cofactor of H-NOX is in a severely distorted conformation before binding the NO molecules. Upon binding NO, the Fe–His bond between the heme–Fe(II) complex and the histidine ligand is broken, followed by relaxation of the heme cofactor and conformational rearrangement, ultimately causing activity changes of the functional domain or a partner protein (Olea et al. 2010, Herzik et al. 2014, Hespen et al. 2016). As common signaling partners of H-NOX, some CMEs are, therefore, also regulated by NO, and we refer to these CMEs as H-NOX-associated c-di-GMP metabolic enzymes (HaCMEs). The genes encoding HaCMEs are mostly found in the genomes of γ-Proteobacteria. Some HaCME-encoding genes are adjacent to hnoX, and the two genes are cotranscribed in the same operon; some HaCME-encoding genes are not in the hnoX operons, but their encoded products serve as response regulators that form two-component systems with H-NOX-associated histidine kinase (HaHK), whose gene is adjacent to hnoX (Plate and Marletta 2013a). Due to the different composition of the hnoX operons, there are certain differences in the specific mechanisms of how NO/H-NOX regulates c-di-GMP, so they can be divided into the following two categories (Fig. 5):
Figure 5.
Mechanisms of NO-induced biofilm dispersal or formation via H-NOX domain. CMEs in bacteria function normally in the absence of NO. However, when bacteria are exposed to a certain concentration of NO, it would affect the activities of some CMEs, changing the cellular c-di-GMP concentrations and affecting the formation of bacterial biofilm. (A) NO may directly affect the protein–protein interaction between H-NOX proteins and HaCMEs, thereby altering their catalytic activities. When such HaCME has only a separate GGDEF domain (in some bacteria, it may have an additional degenerated EAL domain), the binding of NO to H-NOX inhibits the DGC activity of such HaCME. When the HaCME contains both GGDEF and EAL domains, binding of NO to H-NOX will maintain or even down-regulate the DGC activity of this HaCME, or activate its PDE activity. These signal events would reduce the cellular c-di-GMP concentrations in bacteria, ultimately leading to biofilm dispersal. (B) NO may also affect the interaction of H-NOX protein with HaHK, thereby affecting the transfer of the phosphoryl group to indirectly regulate the activity of response regulator HaCME. Such HaCME proteins usually fuse the phospho-signaling receptor REC domain and the GGDEF/EAL domain. Binding of NO to H-NOX protein inhibits the autophosphorylation of HaHK, hindering the downstream transmission of the phosphoryl group, and changes the phosphorylation state of HaCME, thereby inhibiting the PDE activity or activating the DGC, resulting in an elevated c-di-GMP level, which ultimately promotes the formation of bacterial biofilms. * indicates domain degeneration and a lack of catalytic activity. The arrows on the c-di-GMP metabolic domains represent an increase or decrease in activities of the corresponding enzymes. The protein shown in the dashed box in Fig. 5(B) is HnoC, which may not be present in the signaling networks of some bacteria.
When the hnoX gene is adjacent to the gene encoding HaCME, binding of NO to the H-NOX protein can directly affect the protein–protein interaction between H-NOX and HaCME to either inhibit the DGC activity of HaCME or stimulate its PDE activity, causing the down-regulation of the cellular c-di-GMP concentrations, eventually leading to biofilm dispersal (Fig. 5A).
When the hnoX gene is adjacent to the gene encoding HahK, binding of NO to the H-NOX protein first inhibits the autophosphorylation activity of HaHK, and then blocks the downstream transfer of the phosphoryl group (the process in a hybrid two-component system is also dependent on histidine phosphotransfer protein (Hpt) as a mediator), changing the phosphorylation state of the downstream HaCME, thereby inhibiting the PDE activity of HaCME or activating its DGC activity, which results in increased cellular c-di-GMP concentrations in bacteria and promotes biofilm formation (Fig. 5B). Since many hnoX operons contain a hahK gene, this multicomponent signaling system has been reported in a variety of bacteria (Table 3). In addition, HaHK may also have other phosphate transfer acceptors than HaCME in some bacteria. Taking Shewanella oneidensis as an example, its H-NOX/HaHK system has two other phosphotransfer acceptors besides SoHnoB (HaCME), namely SoHnoD and SoHnoC. SoHnoD contains a degenerated HD-GYP domain and does not itself have the ability to hydrolyze c-di-GMP. However, it can fine-tune the catalytic activity of SoHnoB through allosteric effects in different phosphorylation states to control the cellular concentrations of c-di-GMP; SoHnoC is a transcriptional regulator that controls the expression of some genes in the NO-signaling network, thus creating a transcriptional feedback loop, which could further modulate NO-response dynamics (some bacteria, such as Vibrio cholerae, have this multicomponent signaling network, except for the HnoC homolog) (Plate and Marletta 2013b). Compared with the classical H-NOX/HaHK system containing a unique response regulator HaCME, the proteins mentioned above form a more complex multicomponent phosphotransfer regulatory network for NO signaling (Fig. 5B) (Plate and Marletta 2012).
Table 3.
H-NOX- and NosP-mediated regulation of c-di-GMP signaling.
| Domains composition of CME | NO effect | ||||||
|---|---|---|---|---|---|---|---|
| Species | Pathway | HK activity | CME activity | c-di-GMP level | Biofilm | Reference | |
| Legionella pneumophila | H-NOX/HaCME | GGDEF and an inactive EAL | None | DGC↓ | ↓ | ↓ | Carlson et al. (2010) |
| Dinoroseobacter shibae | H-NOX/HaCME | GGDEF | None | DGC↓ | ↓ | ND | Bedrunka et al. (2018) |
| Shewanella woodyi | H-NOX/HaCME | GGDEF and EAL | None | DGC↓ PDE↑ |
↓ | ↓ | Liu et al. (2012) |
| Agrobacterium vitis | H-NOX/HaCME | GGDEF and EAL | None | DGC− PDE↑ |
↓ | ND | Williams et al. (2018) |
| Shewanella oneidensis | H-NOX/HaHK/HaCME | REC, PAS and EAL | ↓ | PDE↓ | ↑ | ↑ | Plate and Marletta (2012) |
| Vibrio cholerae | H-NOX/HaHK/HaCME | REC and EAL | ↓ | ND | ND | ND | Mukhopadyay et al. (2016) |
| Pseudoalteromonas atlantica | H-NOX/HaHK/HaCME | REC and an inactive HD-GYP | ↓ | May alter its interaction with other CMEs | ND | ND | Arora and Boon (2012) |
| Silicibacter sp. TrichCH4B | H-NOX/HaHK/Hpt/HaCME | REC and GGEDF | ↓ | DGC↑ | ↑ | ↑ | Rao et al. (2015) |
| Legionella pneumophila | NosP/NaHK/NaCME | REC, PAS, GGDEF and EAL | ↑ | DGC↓ PDE↑ |
↓ | ↓ | Fischer et al. (2019) |
| Shewanella oneidensis | NosP/NaHK/H-NOX/HaHK/HaCME | REC, PAS and EAL |
SoNaHK↓ SoHaHK↑ |
PDE↑ | ↓ | ↓ | Nisbett et al. (2019) |
↑ indicates an increase in enzyme activity, c-di-GMP concentration, or biofilm formation, while ↓ indicates the opposite and - indicates no change.
ND, not determined.
From the available data, NO seems to mediate the dispersion of bacterial biofilm through the simpler H-NOX system, while the multilevel regulation of the more complicated H-NOX system helps bacteria increase adhesion and biofilm formation (Table 3). However, considering that the H-NOX system model is still in its early stage, and some experiments were also performed extracellularly or under conditions of excessive NO, whether this is true in bacterial physiological settings remains to be further explored. Furthermore, with the discovery of orphan H-NOX, bifunctional H-NOX, and other NO sensors, there is a growing realization that the NO regulatory networks in bacteria might be quite complicated (Mukhopadyay et al. 2016, Guo et al. 2018).
Orphan H-NOXs whose genes are not adjacent to any partner genes, found in a few bacteria, also have the potential to regulate the activities of the components in another typical H-NOX system in the same bacteria. For example, in addition to the typical H-NOX/HaHK pair, the genome of the marine bacterium Saccharophagus degradans encodes an orphan H-NOX protein SdH-NOX2, which also has the function of binding gaseous molecules and inhibiting HaHK activities, but compared with the conventional H-NOX (SdH-NOX1), this protein has a smaller NO dissociation rate and a weaker binding to the kinase (Guo et al. 2018). This property may help increase the duration of intracellular NO-induced signaling and prolong kinase inhibition. Therefore, S. degradans may use the dual H-NOX system to help bacteria more flexibly regulate downstream responses in the face of complex environments. Since this type of orphan H-NOX can affect the autophosphorylation activity of HaHK in the H-NOX/HaHK pair, it may also affect the activities of the downstream response regulators (presumably HaCMEs) in this signaling system. In conclusion, some bacteria encoding both an orphan H-NOX and an H-NOX/HaHK pair may have more complex mechanisms of NO-responsive regulation of the c-di-GMP concentrations.
In recent years, some H-NOX proteins have also been found to act as both heme-dependent NO sensors and heme-independent redox sensors, realizing the regulation of downstream signaling protein activity under the dual conditions of NO binding and cysteine oxidation (Mukhopadyay et al. 2016, Mukhopadhyay et al. 2020). For example, in the genomes of γ-Proteobacteria, which include many well-known pathogens, some conserved Cys residues are present in about half of the H-NOX domains, although such H-NOXs still remain to be experimentally characterized (Bacon et al. 2017). Considering that the partners of these bifunctional H-NOXs may be HaCMEs, further research would be required to figure out whether their bifunctionality affects the concentrations of bacterial c-di-GMP and exhibits physiological significance in regulating bacterial biofilm formation.
NosP (FIST) domain
Although H-NOX is the primary NO sensor for many bacteria, there are still many bacteria that lack the H-NOX domain but are still able to respond to low concentrations of NO. Therefore, there appear to be other, additional NO sensors in these bacteria. P. aeruginosa belongs to this group of bacteria, which does not encode any H-NOX protein, but still responds to NO for regulating biofilm formation (Barraud et al. 2006, Cutruzzolà and Frankenberg-Dinkel 2016).
In the past, a variety of proteins, including MCPs such as PaBdlA (Morgan et al. 2006, Barraud et al. 2009) and some CMEs such as PaDipA (Roy et al. 2012), PaNbdA (Li et al. 2013), and PaGcbA (Petrova et al. 2015) among others, have been identified in P. aeruginosa that were implicated in NO-mediated biofilm dispersal; however, none of them was confirmed as the primary NO sensor. Later, a new NO-sensing heme protein, named NosP, was identified. Upon mutating the relevant components of the NosP signaling pathway in P. aeruginosa, the biofilms formed by the mutant were found to no longer disperse in response to NO, confirming that NosP is a bacterial NO sensor (Hossain and Boon 2017).
The NosP protein is currently annotated as the F-box intracellular signal transduction (FIST) protein (Borziak and Zhulin 2007), and its predicted structure consists of more than 20 β-strands and several α-helices) (Fig. 4B). Compared to H-NOX, which often occurs in eukaryotes, NosP has been seen almost exclusively in bacteria (Williams and Boon 2019). Most NosPs are encoded as stand-alone proteins, although some NosPs appear as domains that are coupled to well-known MCP, HK, and CME signaling domains (Bacon et al. 2017, Hughes et al. 2022). The binding characteristics of NosP domains are currently not well-known, although previous studies have shown that the ligand-binding properties of NosP are consistent with it being a dedicated NO sensor, which can bind NO and CO molecules but cannot form stable ferrous–oxy complexes (Hossain and Boon 2017, Hossain et al. 2018).
The NO-sensing mechanism of NosP is similar to that of H-NOX to a certain extent: when bound to NO, it replaces the original histidine ligand with a NO molecule as an axial ligand to form a five-coordinated heme complex (Olea et al. 2010, Bacon et al. 2018). The dissociation rate of NO from NosP is usually much slower than that from H-NOX (Table 2). This difference indicates that H-NOX and NosP may be sensitive to different concentrations of NO or may play different roles in bacterial physiology.
The NO/NosP system can also regulate cellular c-di-GMP concentrations in a manner similar to the second mode of regulation of NO/H-NOX/c-di-GMP: as a component of a multicomponent signaling system, NosP binding of NO molecules affects the kinase activity of NosP-associated histidine kinase (NaHK), thereby controlling the phosphate flux of related signaling pathways, ultimately regulating the activities of NaCMEs (NosP-associated c-di-GMP metabolic enzymes) and affect cellular c-di-GMP concentrations (Williams et al. 2018, Fischer et al. 2019). But unlike H-NOX, iron-free NosP has a strong inhibitory effect on NaHK (Rao et al. 2017, Fischer et al. 2019). When NO binds to NosP, it weakens the original inhibition of NaHK’s autophosphorylation activity by NosP, allowing the phosphoryl group to be delivered to the downstream components (Fig. 6) (Price et al. 2007, Nisbett et al. 2019). It is worth noting that the current research on the NO/NosP system is limited, so this conclusion may be revised or expanded when more cases appear in the future.
Figure 6.
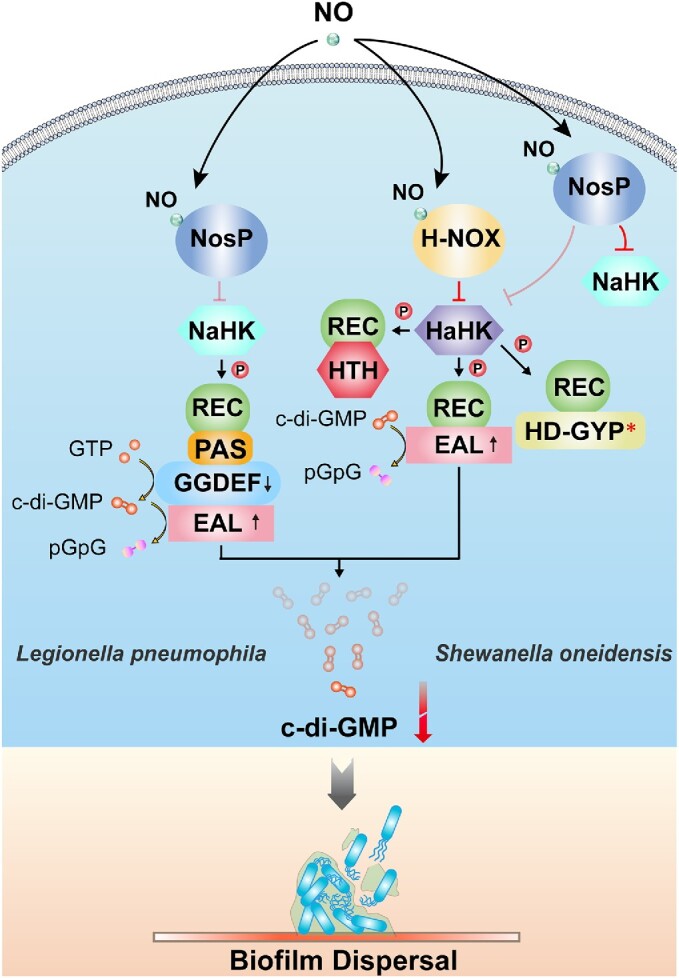
Mechanisms of NO-induced biofilm dispersal via the NosP domain. Possible NosP signaling pathway in Legionella pneumophila (left, based on Fischer et al. 2019). Binding of NO to LpNosP weakens the interaction between LpNosP and LpNahK and diminishes the inhibitory effect of LpNosP on the autophosphorylation of LpNaHK. LpNaHK can thus transfer the phosphoryl group to the downstream bifunctional LpNaCME, which exhibits reduced DGC activity and increased PDE activity, causing a decrease in the cellular c-di-GMP concentrations, and ultimately leading to biofilm dispersal. SoNosP is a master regulator of the multicomponent No/c-di-GMP signaling network in Shewanella oneidensis (right, based on Nisbett et al. 2019). When the bacteria are not exposed to NO, iron-free SoNosP strongly inhibits the autophosphorylation activity of SoNaHK and SoHaHK, thereby preventing downstream components of the phosphate transport chain from being phosphorylated. However, when NO is present, SoNosP attenuates the inhibitory effect on SoHaHK, enabling the transfer of the phosphoryl group to SoHaCME and enhancing the PDE activity of SoHaCME to induce biofilm dispersion. * indicates that the domain is degraded and lacks catalytic activity. The arrows on the c-di-GMP metabolic domains represent an increase or decrease in activities of the corresponding enzymes.
Cross-talk between H-NOX and NosP systems
Although studies of H-NOX and NosP systems are still limited to only a few bacteria, they already detected a cross-talk between the H-NOX and NosP systems. For example, the NosP system was found to add regulation upstream of the H-NOX system in S. oneidensis (Nisbett et al. 2019). When bacteria were not exposed to NO, SoNosP without the linking ferrous ion was able to bind to SoNaHK or even SoHaHK, thereby strongly inhibiting their autophosphorylation activities and resulting in the inability of downstream components of the phosphate transport system to be phosphorylated. When SoHnoB cannot be phosphorylated, its PDE activity cannot be activated, and unphosphorylated SoHnoD will simultaneously inhibit the activity of SoHnoB, resulting in an increase in the cellular concentrations of c-di-GMP. In contrast, when bacteria are exposed to NO, although SoNosP still maintains its inhibitory effect on SoNaHK and SoHaHK after binding NO, the addition of NO would weaken the control effect of SoNosP on SoHaHK. Moreover, SoH-NOX does not significantly inhibit the autophosphorylation activity of SoHaHK in the absence of a significant stoichiometric excess of NO-bound SoH-NOX (Price et al. 2007). Therefore, this achieves a certain degree of relief of SoHaHK inhibition compared to the absence of NO, resulting in increased phosphate flux to downstream targets of the H-NOX signaling pathway, such as SoHnoD and SoHnoB, and promoting the SoHnoB PDE activity to reduce the cellular c-di-GMP levels (Fig. 6) (Plate and Marletta 2012). As a master regulator in the multicomponent signaling system, SoNosP can not only regulate the NosP/NaHK signaling pathway but also exert a regulatory effect upstream of the H-NOX/HaHK signaling pathway, enabling the two systems to establish an antagonistic relationship in a push–pull mechanism (Nisbett et al. 2019). Based on these results, the existing NO/H-NOX model of S. oneidensis was revised and a new NO/NosP/H-NOX pathway was established (Plate and Marletta 2012, Nisbett et al. 2019). However, considering that the reports of NosP regulating the H-NOX pathway are still limited, it could be only a single case. Besides S. oneidensis, other bacteria also have H-NOX, HnoB, HnoD, and HnoC homologs, but a complete NO signaling network has not been demonstrated in these bacteria, so we still retain the original regulatory model in Table 3.
Bioinformatic analysis of the distribution of H-NOX and NosP revealed that, in addition to S. oneidensis, many bacteria, especially Gram-positive ones, possess both H-NOX and NosP systems. Whether there is a regulatory relationship between the H-NOX and NosP systems in these bacteria as well, deserves further exploration. Beyond that, there are many questions to be answered. For example, the H-NOX of some bacteria can simultaneously act as a NO sensor and redox sensor. Does NosP affect the NO signaling pathway mediated by this type of H-NOX? Also, a subset of bacteria encodes neither H-NOX nor NosP. Do these bacteria sense NO, and, if so, how? In conclusion, the puzzle of the bacterial NO-sensing signaling pathways has not been fully resolved, and more research is needed in this field.
Regulation of c-di-GMP levels via light-sensing domains
Response to light was previously thought to be exclusive to photosynthetic bacteria, but recent studies have found that genes encoding photoreceptor proteins are also common in the genomes of nonphotosynthetic bacteria (van der Horst et al. 2007, Elías-Arnanz et al. 2011). Light is essential for photosynthetic bacteria to conserve energy but for other bacteria, light can also serve as a cue for optimal orientation and direction. In addition, light sensing is especially important for some pathogens. Because light can affect host immune responses and susceptibility by regulating circadian rhythms, pathogenic bacteria might benefit from the ability to adjust their behaviors in response to light signals in order to better infect the host (Verma et al. 2020).
A total of seven photoreceptor families have been found in bacteria (Table 4), namely phytochromes (Phys), light oxygen voltage (LOV) proteins, blue light sensing using flavin (BLUF) proteins, photoactive yellow proteins (PYPs), rhodopsins, cryptochromes, and orange carotenoid proteins (OCPs) (van der Horst and Hellingwerf 2004). Among them, the most studied families are PYPs, Phys, LOV, and BLUF proteins, which all participate in the signal transduction from light signals to c-di-GMP concentration change signals (Table 5).
Table 4.
Well-characterized bacterial photoreceptor families.
| Type | Chromophore | Spectral sensitivity | Reference |
|---|---|---|---|
| Phy | Linear tetrapyrrole bilin | Mainly red/far-red light photoreceptors (Cyanobacteriochromes have a broader spectrum) | Kraiselburd et al. (2017) |
| LOV | FMN/FAD/riboflavin | Blue light | Herrou and Crosson (2011) |
| BLUF | FAD | Blue light | Kraiselburd et al. (2017) |
| PYP | p-coumaric acid | Blue/UV-A light | Haker et al. (2003), Purcell and Crosson (2008) |
| Rhodopsin | Retinal | Visible region (400–700 nm) | Ernst et al. (2014) |
| Cryptochrome | FAD/pterin/flavin antenna | Blue/UV-A light | Geisselbrecht et al. (2012) |
| OCP | Carotenoid | Blue–green light | Muzzopappa and Kirilovsky (2020) |
Table 5.
Characteristics and distribution of selected photosensory DGC/PDEs from bacteria.
| Sensor type | Protein name | Species | Domains | Light dependence (in vitro) | Reference |
|---|---|---|---|---|---|
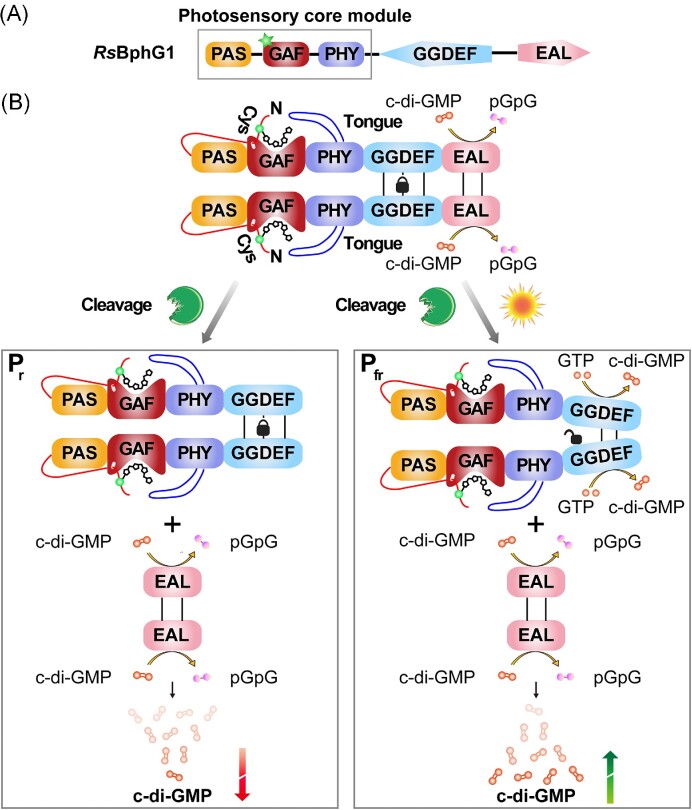
| Phy | RsBphG1 (Bph) | Rhodobacter sphaeroides | PAS-GAF-PHY-GGDEF-EAL | DGC activity is red light-dependent | Tarutina et al. (2006) |
| IdPadC (Bph) | Idiomarina sp. A28L | PAS-GAF-PHY-GGDEF | DGC activity is red light-dependent | Gourinchas et al. (2017) | |
| XoBphP (Bph) | Xanthomonas oryzae pv. oryzae | PAS-EAL-GAF-PHY-PAS | PDE activity is red light-dependent | Verma et al. (2020) | |
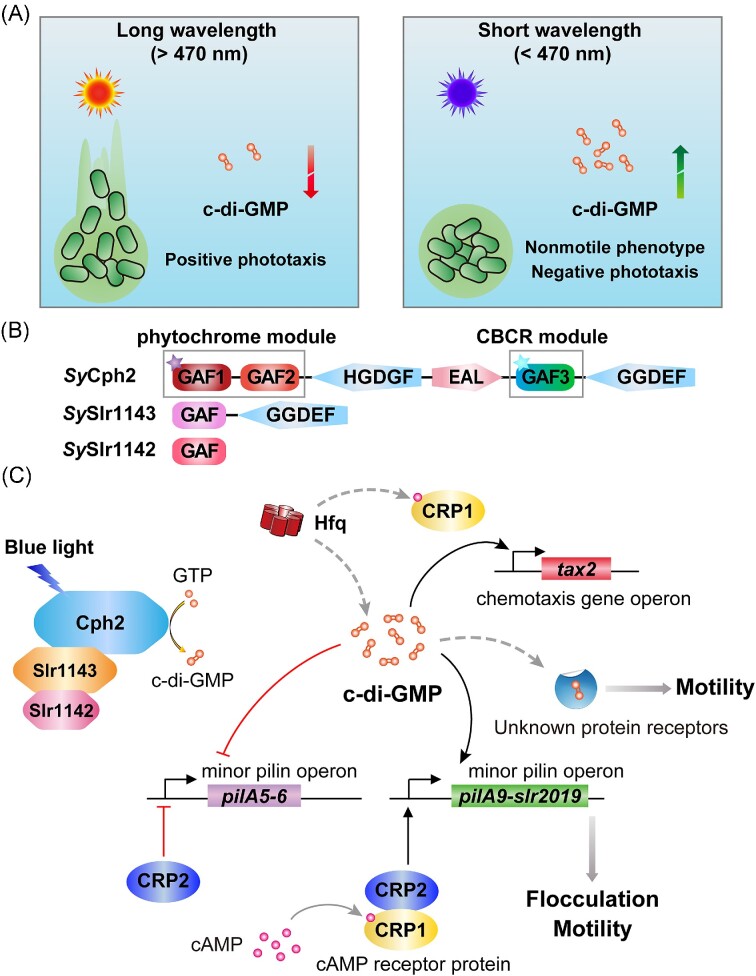
| SyCph2 (Cph) | Synechocystis sp. PCC 6803 | GAF‐GAF‐GGDEF‐EAL‐GAF‐GGDEF | PDE activity is red light-dependent; DGC activity is blue light-dependent |
Savakis et al. (2012) | |
| TeSesA/TeSesB/TeSesC (Cphs) | Thermosynechococcus elongatus | SesA: PAS-GAF‐GGDEF SesB: GAF-GGDEF‐EAL SesC:PAS-PAS-PAS-PAS-GAF-PAS-GGDEF‐EAL |
DGC activity of SesA/SesC is blue light-dependent; PDE activity of SesB is teal light-dependent; PDE activity of SesC is green light-dependent |
Enomoto et al. (2015) | |
| LOV | SeSL2 | Synechococcus elongatus | REC-PAS-PAC-LOV-GGDEF-EAL | PDE activity is blue light-dependent | Cao et al. (2010) |
| BLUF | KpBlrP1 | Klebsiella pneumoniae | BLUF-EAL | PDE activity is blue light-dependent | Barends et al. (2009) |
| MmBldP | Magnetococcus marinus | BLUF-EAL | PDE activity is blue light-dependent | Ryu et al. (2017a) | |
| RpPapA–RpPapB complex | Rhodopseudomonas palustris | PapA: EAL PapB: BLUF |
PapA interacts with PapB; PDE activity of PapA is blue light-dependent via PapB BLUF domain |
Kanazawa et al. (2010) |
Phys—more than simple red photoreceptors
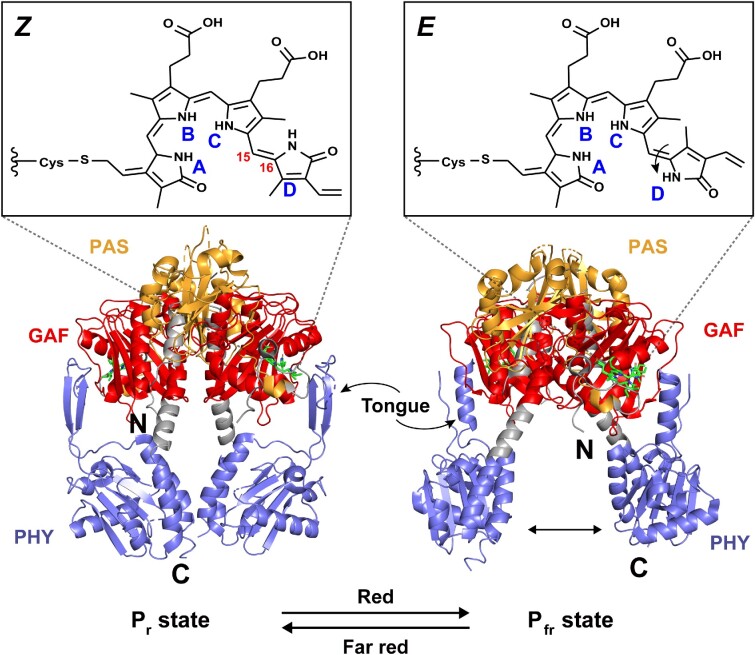
Phys are photoreceptors that utilize a linear bilin tetrapyrrole as a chromophore and are mainly present in plants, algae, fungi, and bacteria (Davis et al. 1999, Yu and Fischer 2019). The classic Phy photosensory core usually contains a conservative PAS-GAF-PHY light sensor module (the PHY domain is a phytochrome-specific domain likely belonging to the GAF family). Among them, GAF domain is the core, that binds chromophores to form biliary adducts (Aravind and Ponting 1997). The PAS and PHY domains are involved in biliary lyase activity and reversible photoconversion, respectively (Nagano 2016). In addition, some special structures are formed between the three domains, such as a figure-eight knot, a tongue-like structure, and a long centrally located α-helix, which further helps the transduction of light signals (Auldridge and Forest 2011, Fischer et al. 2020).
Considering the special taxonomic status of cyanobacteria, we further divided the bacterial Phys into bacteriophytochromes (Bphs) and cyanobacterial Phys (Cphs). The former are usually linked to biliverdin IXα as a chromophore through a conserved Cys residue upstream of the PAS domain, while the latter are mainly linked to phycocyanobilin (PCB) through a Cys residue in the GAF domain (Rockwell and Lagarias 2010).
Bphs use the PAS-GAF-PHY core structure to sense red light
Bphs typically act as photoswitches through reversible light conversion between the red-absorbing state (Pr) and far-red-absorbing state (Pfr) in cells (Fig. 7) (Takala et al. 2014). Their N-terminus conform to the classical Phy model, with complete PAS, GAF, and PHY domains. The C-termini of some Bphs also contain a domain related to c-di-GMP metabolism that helps bacteria regulate c-di-GMP levels in response to light signals. Such Bphs are present in both photosynthetic and nonphotosynthetic bacteria (Table 5).
Figure 7.
Light-driven changes in the structure and spectral properties of Deinococcus radiodurans Bph. The biliverdin chromophore contains four pyrrole rings, named A, B, C, and D rings. Under the irradiation by red and far-red light, biliverdin undergoes reversible Z/E isomerization around the C15/C16 double bond in the methine bridge between the C ring and the D ring, resulting in the rotation of the D ring and causing atomic rearrangements in the chromophore-binding pocket, which in turn leads to repositioning of the PHY domain and refolding of the tongue-like structure therein. In the Pr state, the tongue-like region appears as a β-sheet (PDB entry:4O0P) (Takala et al. 2014), while in the Pfr state, it transforms into an α-helix (PDB entry:4O01) (Kurokawa et al. 2004).
RsBphG1 was the first Bph with a nonkinase photoactivated enzymatic activity and also the first potentially bifunctional enzyme involved in c-di-GMP synthesis and hydrolysis. It is a photoreceptor protein from the photosynthetic bacterium Rhodobacter sphaeroides (recently renamed Cereibacter sphaeroides) that contains both GGDEF and EAL domains (Fig. 8A). When expressed at full length, the protein exhibited only light-independent PDE activity (Tarutina et al. 2006). In addition, RsBphG1 expressed in E. coli was found to be partially cleaved into two species; the smaller species was identified as an EAL domain with PDE activity, while the larger species lacking an EAL domain exhibited biliverdin-dependent and light-activated DGC activity. The authors speculated that the bifunctional CME RsBphG1 utilizes an “EAL lock” and a corresponding special unlocking mechanism to help bacteria decide whether DGC activity is required to be activated under specific conditions (Fig. 8B) (Tarutina et al. 2006). As previously mentioned, sensory domains are more widely distributed in GGDEF–EAL hybrid proteins, and this model can help us better understand the role of bifunctional GGDEF–EAL proteins in the dynamic regulation of the c-di-GMP levels.
Figure 8.
The photoreceptor protein RsBphG1 from Rhodobacter sphaeroides is involved in regulating c-di-GMP levels in bacteria. (A) Schematic representation of the domain composition of RsBphG1 that includes the classic PAS-GAF-PHY photosensory module and the GGDEF–EAL functional modules. Green stars represent the biliverdin chromophore. (B) An “EAL lock” model can be applied to explain how RsBphG1 modulates its catalytic activity in response to light signals (based on Tarutina et al. 2006). The EAL domain and the linker between GGDEF and EAL of RsBphG1 tend to homodimerize, although dimerization of the EAL domains is not required for PDE activity. Protein–protein interaction between the EAL domains can form an “EAL lock” to restrict the mobility of the upstream GGDEF domains. In this process, homodimer formation of GGDEF domains is necessary to receive signals from the sensory domain to undergo a conformational change and complete the transition from a nonproductive state with low or no enzymatic activity to a productive state with high activity. When “EAL lock” is present, the GGDEF domain cannot obtain enough mobility to change conformation, so DGC activity is inhibited; in other words, RsBphG1 gets locked in PDE mode. However, bacteria may use a cleavage mechanism to unlock this “EAL lock,” eventually splitting full-length RsBphG1 into two proteins. The smaller species was identified as the EAL domain and the linker with PDE activity, while the larger species was the rest of RsBphG1. When the unlocked RsBphG1 is not activated by light, its GGDEF domain is in a nonproductive state; when activated by light, the conformation of its GGDEF domain will shift to a productive mode, increasing the cellular c-di-GMP concentrations. RsBphG1 usually exists as a tetramer or a higher-order oligomer, but was drawn as a dimer in this figure for ease of presentation.
Cphs use simpler photoreceptor domains
Cyanobacteria, as photosynthetic prokaryotes, regulate their motility behaviors in response to light signals in order to grow under optimal conditions (Yang et al. 2018). This process may also involve the participation of c-di-GMP whose levels are regulated by cyanobacterial photoreceptor CMEs (Fig. 9A) (Savakis et al. 2012).
Figure 9.
c-di-GMP is involved in regulating Synechocystis motility. (A) Motility behaviors of Synechocystis sp. Its phototaxis is spectrally dependent, and the phototaxis behaviors have a transition point at a wavelength of about 470 nm. When the wavelength is higher than 470 nm, the cells show positive phototaxis, while when the wavelength is lower than 470 nm, the cells either do not move (e.g. under blue light) or exhibit negative phototaxis (e.g. from ultraviolet light) (Fiedler et al. 2005, Chau et al. 2017). This process may involve c-di-GMP, since artificial degradation of c-di-GMP in Synechocystis activates bacterial motility, while synthetic c-di-GMP inhibits phototaxis (Savakis et al. 2012, Wallner et al. 2020). (B) Schematic illustration of the domain composition of SyCph2 and its interaction partners that regulate phototaxis in Synechocystis. The PCB chromophore (purple star) and tetrapyrrole chromophore (cyan star) are covalently bound to the GAF1 and GAF3 domains of SyCph2, respectively, conferring SyCph2 light-sensing capabilities. (C) SyCph2-dependent model of Synechocystis blue light avoidance behaviors (based on Wallner et al. 2020). SyCph2, SySlr1143, and SySlr1142 may be able to form a protein complex that can flexibly adjust cellular c-di-GMP concentrations in response to different light signals. Under blue light irradiation, the C-terminal GGEDF domain of SyCph2 is activated to synthesize c-di-GMP. Elevated levels of c-di-GMP affect the expression of minor pilin operon (pilA5-pilA6 and pilA9-slr2019) and chemotaxis gene operon (tax2), in particular, up-regulated expression of pilA9-slr2019 significantly induces flocculation of Synechocystis, and this operon has also been shown to play an important role in the phototaxis motility of Synechocystis. Besides, the c-di-GMP receptor CdgR has been recently identified in cyanobacteria and demonstrated to control the cell size (Zeng et al. 2023). The possible existence of yet unknown c-di-GMP receptors that could regulate cyanobacterial motility remains to be investigated. Furthermore, this c-di-GMP-dependent signaling network also appears to crosstalk with the cAMP signal transduction system and the Hfq regulatory system due to the involvement of two CRP-like transcription factors, SyCRP1 (a cAMP receptor protein) and SyCRP2 (a potential c-di-GMP-dependent transcription factor lacking the key amino acids for binding cAMP) (Fu et al. 2021). The dotted lines represent mechanisms that have yet to be studied.
The photoreceptor domain composition of Cphs appears to be more diverse than that of Bphs. Canonical Cphs (e.g. Cph1), like Bphs, have a complete PAS-GAF-PHY architecture at the N-terminus, and in some cases, Cphs are knotless Phys that differ from the classical model. Knotless Phys do not have a typical figure-eight knot structure due to the lack of a PAS domain, but they retain the ability to sense light, such as Cph2-like Phys and cyanobacteriochromes (CBCRs) (Fushimi and Narikawa 2019).
Cph2-like Phys do not have a PAS domain but usually have multiple GAF domains. Some of these GAF domains contain conserved Cys residues that can covalently bind the chromophore, while others contain no such Cys residues and are homologous to the PHY domain. They utilize these domains to form a conserved GAF-PHY structure for Pr/Pfr photoconversion (Montgomery and Lagarias 2002). Compared to Cph2-like Phys, CBCRs, the main cyanobacterial photoreceptors, have simpler domain components for light sensing. They lack the PAS and PHY domains and only require the GAF domains to sense various light qualities covering the entire ultraviolet (UV)-to-visible spectrum (Fushimi and Narikawa 2019, Villafani et al. 2020).
Both classes of knotless Phys have been shown to convert light signals into c-di-GMP concentration change signals, which are involved in regulating cyanobacterial motility behaviors (Blain-Hartung et al. 2021, Nakane et al. 2022). In Thermosynechococcus elongatus, multiple CBCRs form a color-sensitive and highly specific c-di-GMP signaling network to synergistically induce cyanobacterial aggregation or dispersal (Enomoto et al. 2015). They can even appear as two modules in a single protein for different light signals to help cyanobacteria achieve flexible regulation of cellular c-di-GMP concentrations, such as SyCph2 from Synechocystis, which contains six domains in the order GAF-GAF-GGDEF-EAL-GAF-GGDEF. The first GGDEF domain is degenerated and loses its ability to synthesize c-di-GMP, but the remaining c-di-GMP metabolic domains can function accordingly (Fig. 9B). SyCph2 is a hybrid photoreceptor with an N-terminal module for receiving red/far-red light and a C-terminal module with CBCR functionality sufficient for photoconversion between green and blue absorbing states. When irradiated with blue light, the downstream GGDEF domain can be activated to promote c-di-GMP synthesis, resulting in hindered cyanobacterial motility (Fig. 9C) (Wallner et al. 2020); but when irradiated with white light, the long wavelength contribution of white light absorbed by the N-terminal GAF module abolishes blue light-induced motility inhibition, resulting in induced cyanobacterial motility (Savakis et al. 2012). Moreover, SySlr1143, an interaction partner of SyCph2, was also shown to regulate cyanobacterial motility. It is a highly active DGC containing a nonphotoactive GAF domain and a GGDEF domain, where the GGDEF domain can interact with the EAL domain of SyCph2 independently of light, and mutation of slr1143 affects the motility behavior of Synechocystis under red light. Since SySlr1143 itself is not photosensitive, it likely forms a photochemically active protein complex with SyCph2 and SySlr1142 (consisting of a single GAF domain encoded by a gene in the same operon as slr1143), to help cyanobacteria regulate c-di-GMP concentrations in response to different light signals. However, the exact mechanism behind it remains unknown and more research is required (Angerer et al. 2017a). These cases reflect the complexity of regulatory networks of cyanobacterial photoreceptor proteins, and it is because of them that cyanobacteria can improve their ability to adapt to different depths in the water.
Bacteria have more blue light than red light photoreceptors
In addition to red light-responsive systems, there are four major classes of blue-light photoreceptors that rely on flavins as chromophores in bacteria: LOV proteins, BLUF proteins, PYPs, and cryptochromes (Fig. 10). They can mediate bacterial responses to blue light, including photosynthesis (Metz et al. 2012), motility (Yang et al. 1995), DNA repair, stress response (Avila-Pérez et al. 2006), and regulation of development. Blue light usually inhibits the growth of bacteria. Therefore, exposure to blue light can trigger the dispersion of bacterial biofilms, helping bacteria migrate to a more favorable environment to survive (Halstead et al. 2016, Kahl et al. 2020). c-di-GMP has also been found to play an important role in the regulation of blue light signals in the first three types of photoreceptors.
Figure 10.
Structures of blue light photoreceptors in bacteria. (A) Crystal structure of the LOV domain of Bacillus subtilis YvtA (PDB entry: 2PR5) (Möglich and Moffat 2007). The YvtA core domain consists of a five-stranded antiparallel β-sheet (Aβ, Bβ, Gβ, Hβ, and Iβ) and four α-helices (Cα, Dα, Eα, and Fα), with a short linker helix at the C-terminus of the core domain. Critical Cys62 are highlighted and drawn in ball-and-stick. (B) Crystal structure of the BLUF domain of Rhodobacter sphaeroides AppA with FMN replacing the FAD cofactor of the native AppA (PDB entry: 1YRX) (Anderson et al. 2005). It contains two α-helices (Bα and Eα) and a five-stranded mixed β-sheet (Aβ, Cβ, Dβ, Gβ, and Fβ). (C) Crystal structure of Ectothiorhodospira halophila PYP (PDB entry: 2PHY) (Borgstahl et al. 1995). PYP consists of a central six-stranded β-sheet (Cβ, Dβ, Gβ, Jβ, Kβ, and Lβ) and six α-helices (Aα, Bα, Eα, Fα, Hα, and Iα). Key Cys69 covalently binding the chromophore are highlighted and drawn in ball-and-stick. (D) Protein fold of the Synechocystis sp. PCC6803 cryptochrome DASH (PDB entry: 1NP7) (Brudler et al. 2003). It contains several 310 helices (magenta) in addition to α-helices (red) and β-sheets (cyan). The chromophores in each domain are also indicated by arrows.
LOV domains
Proteins containing LOV domains are the most widely distributed blue light-sensitive proteins in bacteria, fungi, and plants (Losi et al. 2002). The LOV domain belongs to the PAS domain superfamily, and its core domain consists of a five-stranded antiparallel β-sheet (Aβ, Bβ, Gβ, Hβ, and Iβ) with helical linker elements (Cα, Dα, Eα, and Fα), which are flanked by variable and often helical N- and/or C-terminal extensions (Ncap or Ccap) (Fig. 10A) (Henry and Crosson 2011, Hart and Gardner 2021). The core domain of the LOV is capable of forming an internal pocket for noncovalent binding of flavin [most commonly flavin mononucleotide (FMN), occasionally FAD (He et al. 2002) or riboflavin (Rivera-Cancel et al. 2014)] as a chromophore. The form of flavin chromophore appears to depend on the expression system, expression conditions, and the characteristics of the domain itself (Dorn et al. 2013)). There is a conserved GXNCRFLQ motif that binds flavin on the Eα of LOV, and the Cys residue in this motif can form a covalent flavin-Cys thiol adduct with the flavin C4a atom when irradiated by blue light. This is the only criterion for the LOV domain to be photosensitive, and it is also the basis for participating in photocycling and signal transduction. At this point, the protein is in a “light state” that differs from the “dark state” in terms of spatial and electronic properties (strain, degrees of freedom, and intramolecular interactions), resulting in a rearrangement of the protein structure, accompanied by changes in protein dynamics, ultimately enabling regulation of downstream functional domain activity (Seifert and Brakmann 2018). Among them, antiparallel β-sheets and helical ends flanking the core domain play a major role in this process (Möglich and Moffat 2007).
Similar to the domain organization described in other bacterial signaling proteins, LOV proteins typically have a sensory domain at the N-terminus and a functional domain at the C-terminus. About half of the functional domains of LOV proteins are HK domains. In addition, the c-di-GMP metabolic domains occupy the second place, accounting for about 20%, which also means that some LOV proteins can sense blue light signals to regulate cellular c-di-GMP levels (Crosson et al. 2003, Herrou and Crosson 2011). One such LOV protein, SeSL2, was identified in Synechococcus elongatus. In addition to the LOV domain, SeSL2 contains a GGDEF and an EAL domain and exhibits blue light-induced PDE activity in vitro (Cao et al. 2010). It is worth emphasizing that the LOV–GGDEF–EAL construct is one of the most common domain organizations in bacterial LOV proteins, suggesting that regulation of cellular c-di-GMP concentrations by light may be a general regulatory mechanism in bacteria.
BLUF domain
Another class of blue-light sensing domains is the BLUF domain which utilizes FAD as a chromophore and is present in unicellular eukaryotes and about 10% of prokaryotes (Losi and Gärtner 2008). BLUF domains are small, compact photosensitive modules that form a ferredoxin-like protein fold with two α-helices aligned parallel to a five-stranded mixed β-sheet (Fig. 10B) (Anderson et al. 2005, Jung et al. 2006). Furthermore, a more variable helical C-cap is often stacked against the β-sheet on the side opposite the N-terminal helices (Conrad et al. 2014). In addition to the preferential noncovalent binding of the FAD chromophore, the BLUF domain, like LOV, can also bind other types of natural flavins under certain conditions (Laan et al. 2004). The uniqueness of the photochemical reaction of the BLUF domain is that its photoactivation process is not accompanied by a major structural change of the chromophore but merely rearranges the hydrogen bonds around the chromophore, resulting in a reversible red shift of the absorption spectrum of the BLUF domain (Kennis and Mathes 2013). In addition, among all blue light photoreceptors utilizing the flavin chromophore, BLUF proteins are also the only photoreceptor family showing photo-induced proton-coupled electron transfer (Park and Tame 2017).
BLUF domains can occur in single- or multidomain proteins, with 70% of BLUF proteins being represented by individual BLUF domains. BLUF–EAL fusions are the second most common construct (14%), such as KpBlrP1 from Klebsiella pneumoniae (Gomelsky and Hoff 2011). In solution, the isolated BLUF domain of KpBlrP1 is in stable monomeric form, while full-length KpBlrP1 is in dimeric form. Under blue light irradiation, the BLUF domain of one subunit of the antiparallel KpBlrP1 dimer absorbs photons, activating the EAL domain of the second subunit through allosteric communication transmitted through the conserved domain–domain interfaces, and hydrolyzes c-di-GMP (Barends et al. 2009).
However, most YcgFs in enteric bacteria, although containing a similar domain composition to KpBlrP1, have no enzymatic activity in the EAL domain (Gomelsky and Hoff 2011). EcYcgF is a direct antagonist of the MerR-like regulator EcYcgE, and the two encoding genes are located adjacent to each other. EcYcgF changes its conformation after blue light irradiation and then releases EcYcgE from bound DNA by interacting with EcYcgE, which ultimately affects the expression of bacterial biofilm formation-related proteins. Interestingly, not only the expression of ycgF and ycgE is strongly induced by low-temperature conditions, but the light-induced dimerization of EcYcgF also exhibits a significant temperature dependence (Tschowri et al. 2009, Nakasone et al. 2010). This indicates that the EcYcgE/EcYcgF signaling pathway involves not only the response to light but also the perception of temperature, which can help E. coli to survive better in aquatic environments other than the original host. In fungi, some Phys use the property that temperature can alter the rate of dark reversal to rely on light to sense temperature, acting as “temperature sensors” (Yu et al. 2019). It has been found that the optimal growth temperature of some bacteria varies under different light conditions (Mesquita et al. 2019), and the photosensitivity of some bacteria is temperature-dependent (Gomelsky and Hoff 2011, Dorey et al. 2019). These results imply that there is a cross-talk between the bacterial light signaling and the temperature signaling pathway.
Pyp
PYP is a water-soluble protein and a class of small cytoplasmic blue light receptors. It was originally discovered in Halorhodospira halophila, and more than 100 bacteria are now known to encode PYPs (Schmidt 2017, Kim et al. 2020). PYP is the structural archetype of the PAS domain superfamily with an α/β folded structure consisting of a central six-stranded β-sheet packed by six α-helices (Borgstahl et al. 1995, Pellequer et al. 1998). PYP covalently binds the p-coumaric acid (pCA) chromophore to a conserved Cys residue, thereby conferring blue/UV light sensitivity to PYP. This chromophore is in a deprotonated trans form in the dark. After photoexcitation, the chromophore undergoes photo-isomerization from trans to cis, followed by protonation to achieve electrostatic neutrality. This results in a rearrangement of the hydrogen-bonding network, ultimately leading to a change in the overall conformation of the protein, which alters protein function (Imamoto and Kataoka 2007).
Due to the limited distribution of PYP, there is currently limited evidence for its association with c-di-GMP. In the genome of the gammaproteobacterium Idiomarina loihiensis, isolated from deep-sea samples and not expected to use photoreceptors, a gene encoding a PYP homolog is located next to a gene encoding the GGDEF domain. This protein has photochemical properties of the PYP family, and inhibition of bacterial biofilm formation was observed when bacteria were exposed to light (van der Horst et al. 2009). In addition, a protein combining PYP and GGDEF domains has been found in the thermophilic photosynthetic purple sulfur bacterium Thermochromatium tepidum, suggesting a possible functional link between PYPs and c-di-GMP (Kyndt et al. 2005).
Blue light photoreceptors may play different roles
Bacteria use abundant blue light photoreceptors to detect the blue light signals in the environment, regulating a wide range of biological responses. From the distribution of these blue photoreceptors, it is likely that their functions can be replaced by each other. For example, LOV homologs are generally absent in obligate anaerobes, obligate intracellular parasites, and extremophilic microbes that are infrequently stimulated by light. But there are also no LOV homologs in Enterobacterales and Vibrionales, which may have been exposed to light multiple times in their respective living environments. Instead, they encode BLUF proteins to take over blue light-sensing functions. At the same time, the results of bioinformatics analysis showed that the functional domains of BLUF proteins in bacteria were highly similar to those of LOV proteins, further confirming the previously mentioned opinion that the two blue photoreceptors may have overlapping functions (Krauss et al. 2009).
This conclusion seems to apply to other blue-light photoreceptor families as well. Compared with LOV- and BLUF-based blue photoreceptors, PYPs and cryptochromes have a very limited distribution. However, some halophilic bacteria that do not encode either LOV or BLUF, such as H. halophila and Halochromatium salexigens, have been found to encode PYP-containing proteins instead (Kumauchi et al. 2008). Therefore, the examples of c-di-GMP metabolic domains as the functional domains of blue-light photoreceptors may not be limited to LOV- and BLUF-containing proteins, and are likely to be experimentally confirmed in the other two classes of blue-light photoreceptors in the future. This also suggests that some bacteria with different blue light photoreceptors may share a common blue light avoidance mechanism.
At the same time, some bacteria encode more than one class of blue-light photoreceptors. For example, R. sphaeroides and Burkholderia phytofirmans possess both LOV-, BLUF-, and PYP-based photoreceptors (Kumauchi et al. 2008). From the available clues, although both LOV and BLUF domains are capable of coupling to c-di-GMP metabolic domains, LOV tends to be coupled to GGDEF–EAL (this coupling accounts for 20% of bacterial LOV proteins) (Herrou and Crosson 2011), while BLUF tends to couple to the EAL domain (this coupling constitutes 14% of bacterial BLUF proteins) (Gomelsky and Hoff 2011). Therefore, in these bacteria, although the blue light photoreceptors can all sense blue light to control c-di-GMP concentrations, they may play different roles in regulating the specific processes of the c-di-GMP signaling pathway.
Photosensitive CMEs have broad application prospects in synthetic biology
The light signals appear to be more controllable, safer and gentler than other inducers of bacterial behaviors and metabolism. Therefore, some researchers began to use photosensitive CMEs to explore the feasibility of regulating cellular c-di-GMP levels in bacteria and to develop tools for optogenetic applications (Ryu et al. 2017a,b, Angerer et al. 2017b). To a certain extent, light can help bacteria make important lifestyle-changing decisions. For example, it can help bacteria switch between the single-cell planktonic state and the attached biofilm state; in some pathogens, light can also affect bacterial virulence. However, a deeper understanding of the physiological significance of bacterial phototactic behaviors and the molecular mechanisms of light-dependent signal transduction is still required if we want to interfere with the lifestyle of bacteria to better control bacterial virulence and biofilms.
Summary and outlook
As the external environment changes, bacteria are challenged with fluctuations in various chemical and physical parameters, which pose a serious threat to the integrity and metabolic state of cells (Klinkert and Narberhaus 2009, Cornforth and Foster 2013). Therefore, the ability to sense and respond to environmental signals is a key factor for bacteria to adapt to an ever-changing environment to survive and reproduce. As an important nucleotide second messenger, c-di-GMP is a mediator that bridges signal perception with the cellular response and regulates the adaptive behaviors of bacteria. This process has been shown to benefit bacteria to improve their ability to infect hosts (Fu et al. 2018), fight environmental stress (Li et al. 2018), and establish symbiotic relationships (Rao et al. 2015), among others.
This review focused on those sensory domains that can sense gas and light to modulate CME activities, whether they achieve this function through direct sensing or indirect regulation. We summarized their signal transduction mechanisms and the corresponding phenotypic outputs and compared the differences between sensory domains responding to the same signals, which improved our understanding of bacterial adaptation behaviors. The existence of some unique domains (e.g. orphan H-NOX) and some bifunctional CMEs mentioned above illustrates the complexity of the bacterial signal transduction networks. Their physiological significance and related regulatory mechanisms are going to remain a key research direction in bacteriology for the foreseeable future.
In addition to the stimuli mentioned in our article, bacterial CMEs can sense and respond to other signals, such as nutrients, chemicals (Bernier et al. 2011, Heo et al. 2019), temperature (Almblad et al. 2021), and so on. In addition, the enzymatic properties of CMEs themselves can also determine whether they are affected by factors such as pH (Koestler and Waters 2014), ionic strength (Tamayo et al. 2005), and other signals. Due to the limitation of article length, they have not been discussed here.
In order to reduce the virulence of pathogens and solve the problems of antibiotic-resistant bacteria caused by biofilm formation in the medical field, researchers are trying to develop small molecule inhibitors as new antibacterial drugs targeting the c-di-GMP signaling pathway. These inhibitors mostly work by acting on the potential targets, like CMEs and c-di-GMP receptors, to block c-di-GMP signaling, but they often lack good selectivity in terms of biological activities and there is still a huge space for exploration in terms of chemical structure diversity (Xuan et al. 2021). We, therefore, advocate a new strategy, i.e. “understand what bacteria want and then use environmental signals to modulate c-di-GMP-mediated bacterial behaviors” (Galperin 2018). This strategy not only lifts the aforementioned limitations, but also helps prevent direct confrontation with bacteria during the use of traditional methods such as antibiotics, and avoiding damage to the human (animal) host (Galperin 2018). Thus, further understanding of the complete c-di-GMP-mediated signal transduction pathway and its roles in bacterial physiology and host–pathogen interactions may also provide good new targets for rational drug design.
More importantly, mastering the specific mechanisms by which bacteria sense and respond to signals, as well as the corresponding phenotypic outputs, is also beneficial to the development of new biosensors suitable for synthetic biology to a certain extent. These signaling CME-derived biosensors, especially related optogenetic tools, will enable rapid and reversible modulation of bacterial cellular c-di-GMP concentrations, thereby regulating bacterial physiological activities with a higher spatiotemporal resolution. Researchers can use these biosensors to regulate bacterial adaptation behaviors by exerting specific signals, so as to induce the formation of biofilms that can be used as living catalysts (Halan et al. 2012) and improve the colonization efficiency of intestinal probiotics (He et al. 2018).
However, there are still many problems in the practical application of the above strategy. First, different bacteria may respond differently to the same environmental signals, making it difficult to model the link between the initial signal and the final phenotypic output. Second, bacteria face a rapidly changing environment in the process of infecting the host, and not all the signals are controllable. Finally, and most importantly, the regulatory behavior of bacteria is inherently very complex. Notably, there are other enzymes (such as oligoribonucleases, which serve as degrading enzymes for the linear intermediate pGpG in c-di-GMP signaling) that can affect c-di-GMP concentrations, and their activities are also affected by environmental signals (Orr et al. 2018). In addition to regulating c-di-GMP levels, the same environmental signals can also regulate other cyclic dinucleotide messengers (e.g. c-di-AMP) and even other signal transduction pathways (Zhulin et al. 2003, Rao et al. 2010). Moreover, there is also a cross-talk between these second messenger networks and other signal transduction systems. For example, the DGC activity of Lcd1 from Leptospira interrogans and the PDE activity of Bd1971 from Bdellovibrio bacteriovorus can both be activated upon binding of cAMP to their sensory domains (Cadby et al. 2019, da Costa Vasconcelos et al. 2017); c-di-GMP and (p)ppGpp, the messenger molecule in the stringent response, appear to be antagonistic (Boehm et al. 2009). These problems all reduce the feasibility of the practical application of this strategy to control bacterial behavior. Therefore, there is still a long way to go before we can truly domesticate bacteria, and more research is needed in this field.
Supplementary Material
Acknowledgement
We are grateful to all researchers who have made a significant contribution to c-di-GMP research and apologize to those whose work could not be cited here because of space constraints.
Contributor Information
Zhaoqing Yu, National Key Laboratory of Agricultural Microbiology and Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, 1 Shizishan Street, Wuhan, Hubei 430070, PR China; Institute of Agro-Product Processing, Jiangsu Academy of Agricultural Sciences, 50 Zhongling Street, Nanjing, Jiangsu 210014, PR China.
Wei Zhang, National Key Laboratory of Agricultural Microbiology and Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, 1 Shizishan Street, Wuhan, Hubei 430070, PR China.
He Yang, National Key Laboratory of Agricultural Microbiology and Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, 1 Shizishan Street, Wuhan, Hubei 430070, PR China.
Shan-Ho Chou, National Key Laboratory of Agricultural Microbiology and Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, 1 Shizishan Street, Wuhan, Hubei 430070, PR China.
Michael Y Galperin, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jin He, National Key Laboratory of Agricultural Microbiology and Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, 1 Shizishan Street, Wuhan, Hubei 430070, PR China.
Conflicts of interest statement
None declared.
Funding
This paper is supported by the National Natural Science Foundation of China (grants 31970074, 31770087, and 32171424). M.Y.G. is supported by the NIH Intramural Research Program at the U.S. National Library of Medicine.
References
- Abreu IA, Lourenço AI, Xavier AV et al. A novel iron centre in the split-soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774. J Biol Inorg Chem. 2003;8:360–70. [DOI] [PubMed] [Google Scholar]
- Agari Y, Kashihara A, Yokoyama S et al. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol Microbiol. 2008;70:60–75. [DOI] [PubMed] [Google Scholar]
- Ali-Ahmad A, Fadel F, Sebban-Kreuzer C et al. Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa. Sci Rep. 2017;7:11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almblad H, Randall TE, Liu F et al. Bacterial cyclic diguanylate signaling networks sense temperature. Nat Commun. 2021;12:1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Dragnea V, Masuda S et al. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44:7998–8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer V, Essen LO, Wilde A. Analysis of c-di-GMP levels synthesized by a photoreceptor protein in response to different light qualities using an in vitro enzymatic assay. Methods Mol Biol. 2017b;1657:187–204. [DOI] [PubMed] [Google Scholar]
- Angerer V, Schwenk P, Wallner T et al. The protein Slr1143 is an active diguanylate cyclase in Synechocystis sp. PCC 6803 and interacts with the photoreceptor Cph2. Microbiology. 2017a;163:920–30. [DOI] [PubMed] [Google Scholar]
- Aono S. Metal-containing sensor proteins sensing diatomic gas molecules. Dalton Trans. 2008;24:3137–46. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–9. [DOI] [PubMed] [Google Scholar]
- Arora DP, Boon EM. Nitric oxide regulated two-component signaling in Pseudoalteromonas atlantica. Biochem Biophys Res Commun. 2012;421:521–6. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Forest KT. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol. 2011;46:67–88. [DOI] [PubMed] [Google Scholar]
- Avila-Pérez M, Hellingwerf KJ, Kort R. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188:6411–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon B, Nisbett LM, Boon E. Bacterial haemoprotein sensors of NO: H-NOX and NosP. Adv Microb Physiol. 2017;70:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon BA, Liu Y, Kincaid JR et al. Spectral characterization of a novel NO sensing protein in bacteria: nosP. Biochemistry. 2018;57:6187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban M, Shcherbakova DM, Pletnev S et al. Designing brighter near-infrared fluorescent proteins: insights from structural and biochemical studies. Chem Sci. 2017;8:4546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends TR, Hartmann E, Griese JJ et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–8. [DOI] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang SH et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale J, Lee SY, Iwata S et al. Structure of the aliphatic sulfonate-binding protein SsuA from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrunka P, Olbrisch F, Rüger M et al. Nitric oxide controls c-di-GMP turnover in Dinoroseobacter shibae. Microbiology. 2018;164:1405–15. [DOI] [PubMed] [Google Scholar]
- Bell CE, Barry J, Matthews KS et al. Structure of a variant of lac repressor with increased thermostability and decreased affinity for operator. J Mol Biol. 2001;313:99–109. [DOI] [PubMed] [Google Scholar]
- Bernier SP, Ha DG, Khan W et al. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res Microbiol. 2011;162:680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson C, Adams NBP, Stevenson B et al. The molecular basis of phosphite and hypophosphite recognition by ABC-transporters. Nat Commun. 2017;8:1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain-Hartung M, Rockwell NC, Lagarias JC. Natural diversity provides a broad spectrum of cyanobacteriochrome-based diguanylate cyclases. Plant Physiol. 2021;187:632–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F et al. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol. 2009;72:1500–16. [DOI] [PubMed] [Google Scholar]
- Boon EM, Davis JH, Tran R et al. Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase β1 H-NOX domain. J Biol Chem. 2006;281:21892–902. [DOI] [PubMed] [Google Scholar]
- Boon EM, Huang SH, Marletta MA. A molecular basis for NO selectivity in soluble guanylate cyclase. Nat Chem Biol. 2005;1:53–9. [DOI] [PubMed] [Google Scholar]
- Boon EM, Marletta MA. Ligand specificity of H-NOX domains: from sGC to bacterial NO sensors. J Inorg Biochem. 2005;99:892–902. [DOI] [PubMed] [Google Scholar]
- Boraston AB, Nurizzo D, Notenboom V et al. Differential oligosaccharide recognition by evolutionarily-related β-1,4 and β-1,3 glucan-binding modules. J Mol Biol. 2002;319:1143–56. [DOI] [PubMed] [Google Scholar]
- Borgstahl GE, Williams DR, Getzoff ED. 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–87. [DOI] [PubMed] [Google Scholar]
- Borziak K, Zhulin IB. FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics. 2007;23:2518–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudes M, Lazar N, Graille M et al. The structure of the NasR transcription antiterminator reveals a one-component system with a NIT nitrate receptor coupled to an ANTAR RNA-binding effector. Mol Microbiol. 2012;85:431–44. [DOI] [PubMed] [Google Scholar]
- Bradshaw WJ, Bruxelle JF, Kovacs-Simon A et al. Molecular features of lipoprotein CD0873: a potential vaccine against the human pathogen Clostridioides difficile. J Biol Chem. 2019;294:15850–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster JL, McKellar JL, Finn TJ et al. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci Rep. 2016;6:35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H et al. Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell. 2003;11:59–67. [DOI] [PubMed] [Google Scholar]
- Burns JL, Jariwala PB, Rivera S et al. Oxygen-dependent globin coupled sensor signaling modulates motility and virulence of the plant pathogen Pectobacterium carotovorum. ACS Chem Biol. 2017;12:2070–7. [DOI] [PubMed] [Google Scholar]
- Burns JL, Rivera S, Deer DD et al. Oxygen and bis(3',5')-cyclic dimeric guanosine monophosphate binding control oligomerization state equilibria of diguanylate cyclase-containing globin coupled sensors. Biochemistry. 2016;55:6642–51. [DOI] [PubMed] [Google Scholar]
- Cadby IT, Basford SM, Nottingham R et al. Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase. EMBO J. 2019;38:e100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Livoti E, Losi A et al. A blue light-inducible phosphodiesterase activity in the cyanobacterium Synechococcus elongatus. Photochem Photobiol. 2010;86:606–11. [DOI] [PubMed] [Google Scholar]
- Carlson HK, Vance RE, Marletta MA. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol Microbiol. 2010;77:930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary SP, Winger JA, Derbyshire ER et al. Nitric oxide signaling: no longer simply on or off. Trends Biochem Sci. 2006;31:231–9. [DOI] [PubMed] [Google Scholar]
- Chang AL, Tuckerman JR, Gonzalez G et al. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry. 2001;40:3420–6. [DOI] [PubMed] [Google Scholar]
- Chau RM, Bhaya D, Huang KC. Emergent phototactic responses of cyanobacteria under complex light regimes. Mbio. 2017;8:e02330–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Chuankhayan P, Wu HH et al. The bacteriohemerythrin from Methylococcus capsulatus (Bath): crystal structures reveal that Leu114 regulates a water tunnel. J Inorg Biochem. 2015;150:81–9. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nat Chem Biol. 2014;10:801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–93. [DOI] [PubMed] [Google Scholar]
- Crane BR, Sudhamsu J, Patel BA. Bacterial nitric oxide synthases. Annu Rev Biochem. 2010;79:445–70. [DOI] [PubMed] [Google Scholar]
- Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. [DOI] [PubMed] [Google Scholar]
- Cutruzzolà F, Frankenberg-Dinkel N. Origin and impact of nitric oxide in Pseudomonas aeruginosa biofilms. J Bacteriol. 2016;198:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Vasconcelos FN, Maciel NK, Favaro DC et al. Structural and enzymatic characterization of a cAMP-dependent diguanylate cyclase from pathogenic Leptospira species. J Mol Biol. 2017;429:2337–52. [DOI] [PubMed] [Google Scholar]
- Dahlstrom KM, O'Toole GA. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol. 2017;71:179–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vener AV, Vierstra RD. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 1999;286:2517–20. [DOI] [PubMed] [Google Scholar]
- Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–91. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR et al. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–7. [DOI] [PubMed] [Google Scholar]
- Dorey AL, Lee BH, Rotter B et al. Blue light sensing in Listeria monocytogenes is temperature-dependent and the transcriptional response to it is predominantly SigB-dependent. Front Microbiol. 2019;10:2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn M, Jurk M, Wartenberg A et al. LOV takes a pick: thermodynamic and structural aspects of the flavin-LOV-interaction of the blue-light sensitive photoreceptor YtvA from Bacillus subtilis. PLoS ONE. 2013;8:e81268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D et al. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–82. [DOI] [PubMed] [Google Scholar]
- Elías-Arnanz M, Padmanabhan S, Murillo FJ. Light-dependent gene regulation in nonphototrophic bacteria. Curr Opin Microbiol. 2011;14:128–35. [DOI] [PubMed] [Google Scholar]
- Enomoto G, Ni-Ni-Win, Narikawa R et al. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc Natl Acad Sci USA. 2015;112:8082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Lodowski DT, Elstner M et al. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA. 2008;105:14709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhana A, Saini V, Kumar A et al. Environmental heme-based sensor proteins: implications for understanding bacterial pathogenesis. Antioxid Redox Signal. 2012;17:1232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler B, Börner T, Wilde A. Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: role of different photoreceptors. Photochem Photobiol. 2005;81:1481–8. [DOI] [PubMed] [Google Scholar]
- Fischer JT, Hossain S, Boon EM. NosP modulates cyclic-di-GMP signaling in Legionella pneumophila. Biochemistry. 2019;58:4325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Xu Q, Zhao KH et al. Effect of the PHY domain on the photoisomerization step of the forward Pr →Pfr conversion of a knotless phytochrome. Chem. 2020;26:17261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yu Z, Liu S et al. c-di-GMP regulates various phenotypes and insecticidal activity of gram-positive Bacillus thuringiensis. Front Microbiol. 2018;9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yu Z, Zhu L et al. The multiple regulatory relationship between RNA-chaperone hfq and the second messenger c-di-GMP. Front Microbiol. 2021;12:689619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Narikawa R. Cyanobacteriochromes: photoreceptors covering the entire UV-to-visible spectrum. Curr Opin Struct Biol. 2019;57:39–46. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Chou SH. Sequence conservation, domain architectures, and phylogenetic distribution of the HD-GYP type c-di-GMP phosphodiesterases. J Bacteriol. 2022;204:e0056121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. What bacteria want. Environ Microbiol. 2018;20:4221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisselbrecht Y, Frühwirth S, Schroeder C et al. CryB from Rhodobacter sphaeroides: a unique class of cryptochromes with new cofactors. EMBO Rep. 2012;13:223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacalone D, Smith TJ, Collins AJ et al. Ligand-mediated biofilm formation via enhanced physical interaction between a diguanylate cyclase and its receptor. Mbio. 2018;9:e01254–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G. Signal transduction by heme-containing PAS-domain proteins. J Appl Physiol. 2004;96:774–83. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Sousa EHS. Escherichia coli DosC and DosP: a role of c-di-GMP in compartmentalized sensing by degradosomes. Adv Microb Physiol. 2019;75:53–67. [DOI] [PubMed] [Google Scholar]
- Goers Sweeney E, Henderson JN, Goers J et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20:1177–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M, Hoff WD. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 2011;19:441–8. [DOI] [PubMed] [Google Scholar]
- Gourinchas G, Etzl S, Göbl C et al. Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Sci Adv. 2017;3:e1602498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cooper MM, Bromberg R et al. A dual-H-NOX signaling system in Saccharophagus degradans. Biochemistry. 2018;57:6570–80. [DOI] [PubMed] [Google Scholar]
- Guo Y, Marletta MA. Structural insight into H-NOX gas sensing and cognate signaling protein regulation. ChemBioChem. 2019;20:7–19. [DOI] [PubMed] [Google Scholar]
- Ha DG, O’Toole GA. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol Spectr. 2015;3:MB–0003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habazettl J, Allan MG, Jenal U et al. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J Biol Chem. 2011;286:14304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haker A, Hendriks J, van Stokkum IH et al. The two photocycles of photoactive yellow protein from Rhodobacter sphaeroides. J Biol Chem. 2003;278:8442–51. [DOI] [PubMed] [Google Scholar]
- Halan B, Buehler K, Schmid A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012;30:453–65. [DOI] [PubMed] [Google Scholar]
- Hall CL, Lee VT. Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip Rev RNA. 2018;9. doi: 10.1002/wrna.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead FD, Thwaite JE, Burt R et al. Antibacterial activity of blue light against nosocomial wound pathogens growing planktonically and as mature biofilms. Appl Environ Microbiol. 2016;82:4006–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart JE, Gardner KH. Lighting the way: recent insights into the structure and regulation of phototropin blue light receptors. J Biol Chem. 2021;296:100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ruan W, Sun J et al. Functional characterization of c-di-GMP signaling-related genes in the probiotic Lactobacillus acidophilus. Front Microbiol. 2018;9:1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yin W, Galperin MY et al. Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 2020;48:2807–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y et al. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–3. [DOI] [PubMed] [Google Scholar]
- Henares BM, Higgins KE, Boon EM. Discovery of a nitric oxide responsive quorum sensing circuit in Vibrio harveyi. ACS Chem Biol. 2012;7:1331–6. [DOI] [PubMed] [Google Scholar]
- Hengge R, Galperin MY, Ghigo JM et al. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J Bacteriol. 2015;198:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K, Park YH, Lee KA et al. Sugar-mediated regulation of a c-di-GMP phosphodiesterase in Vibrio cholerae. Nat Commun. 2019;10:5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst S, Lorkowski M, Sarenko O et al. Transmembrane redox control and proteolysis of PdeC, a novel type of c-di-GMP phosphodiesterase. EMBO J. 2018;37:e97825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzik MA Jr, Jonnalagadda R, Kuriyan J et al. Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins. Proc Natl Acad Sci USA. 2014;111:E4156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespen CW, Bruegger JJ, Phillips-Piro CM et al. Structural and functional evidence indicates selective oxygen signaling in Caldanaerobacter subterraneus H-NOX. ACS Chem Biol. 2016;11:2337–46. [DOI] [PubMed] [Google Scholar]
- Hill S, Austin S, Eydmann T et al. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc Natl Acad Sci USA. 1996;93:2143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Huang Z, Guo L et al. The ligand-binding domain of a chemoreceptor from Comamonas testosteroni has a previously unknown homotrimeric structure. Mol Microbiol. 2019;112:906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Boon EM. Discovery of a novel nitric oxide binding protein and nitric-oxide-responsive signaling pathway in Pseudomonas aeruginosa. ACS Infect Dis. 2017;3:454–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Heckler I, Boon EM. Discovery of a nitric oxide responsive quorum sensing circuit in Vibrio cholerae. ACS Chem Biol. 2018;13:1964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Nisbett LM, Boon EM. Discovery of two bacterial nitric oxide-responsive proteins and their roles in bacterial biofilm regulation. Acc Chem Res. 2017;50:1633–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Dabi T, Chory J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat Chem Biol. 2011;7:766–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Larsen RW, Boudko D et al. Myoglobin-like aerotaxis transducers in archaea and bacteria. Nature. 2000;403:540–4. [DOI] [PubMed] [Google Scholar]
- Hu X, Zhao J, DeGrado WF et al. Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc Natl Acad Sci USA. 2013;110:678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature. 1993;364:259–62. [DOI] [PubMed] [Google Scholar]
- Hughes HQ, Floyd KA, Hossain S et al. Nitric oxide stimulates type IV MSHA pilus retraction in Vibrio cholerae via activation of the phosphodiesterase CdpA. Proc Natl Acad Sci USA. 2022;119:e2108349119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. GAF domains: cyclic nucleotides come full circle. Sci STKE. 2003;2003:PE1. [DOI] [PubMed] [Google Scholar]
- Ilgü H, Jeckelmann JM, Gapsys V et al. Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC. Proc Natl Acad Sci USA. 2016;113:10358–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto Y, Kataoka M. Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochem Photobiol. 2007;83:40–9. [DOI] [PubMed] [Google Scholar]
- Jain R, Chan MK. Mechanisms of ligand discrimination by heme proteins. J Biol Inorg Chem. 2003;8:1–11. [DOI] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–84. [DOI] [PubMed] [Google Scholar]
- Jing X, Jaw J, Robinson HH et al. Crystal structure and oligomeric state of the RetS signaling kinase sensory domain. Proteins. 2010;78:1631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A, Domratcheva T, Tarutina M et al. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc Natl Acad Sci USA. 2005;102:12350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A, Reinstein J, Domratcheva T et al. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J Mol Biol. 2006;362:717–32. [DOI] [PubMed] [Google Scholar]
- Junkermeier EH, Hengge R. A novel locally c-di-GMP-controlled exopolysaccharide synthase required for bacteriophage N4 infection of Escherichia coli. Mbio. 2021;12:e0324921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarczyk A, Hempel AM, von Arx C et al. Precise timing of transcription by c-di-GMP coordinates cell cycle and morphogenesis in Caulobacter. Nat Commun. 2020;11:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl LJ, Price-Whelan A, Dietrich LEP. Light-mediated decreases in cyclic di-GMP levels inhibit structure formation in Pseudomonas aeruginosa biofilms. J Bacteriol. 2020;202:e00117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander T, Sachs JN, Goldman A et al. Electrostatic interactions of hsp-organizing protein tetratricopeptide domains with Hsp70 and Hsp90: computational analysis and protein engineering. J Biol Chem. 2009;284:25364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Ren S, Maekawa M et al. Biochemical and physiological characterization of a BLUF protein-EAL protein complex involved in blue light-dependent degradation of cyclic diguanylate in the purple bacterium Rhodopseudomonas palustris. Biochemistry. 2010;49:10647–55. [DOI] [PubMed] [Google Scholar]
- Kennis JT, Mathes T. Molecular eyes: proteins that transform light into biological information. Interface Focus. 2013;3:20130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppner A, Maric D, Correia M et al. Lessons from the post-genomic era: globin diversity beyond oxygen binding and transport. Redox Biol. 2020;37:101687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kwak GH, Lee K et al. Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from thermoplasma acidophilum. Arch Biochem Biophys. 2014;560:10–9. [DOI] [PubMed] [Google Scholar]
- Kim S, Nakasone Y, Takakado A et al. Wavelength-dependent photoreaction of PYP from Rhodobacter capsulatus. Biochemistry. 2020;59:4810–21. [DOI] [PubMed] [Google Scholar]
- Kim T, Choi J, Lee S et al. Structural studies on the extracellular domain of sensor histidine kinase YycG from Staphylococcus aureus and its functional implications. J Mol Biol. 2016;428:3074–89. [DOI] [PubMed] [Google Scholar]
- Kitanishi K, Igarashi J, Matsuoka A et al. Identification and characterization of a redox sensor phosphodiesterase from Ferrovum sp. PN-J185 containing bacterial hemerythrin and HD-GYP domains. Biochemistry. 2020;59:983–91. [DOI] [PubMed] [Google Scholar]
- Kitanishi K, Kobayashi K, Kawamura Y et al. Important roles of Tyr43 at the putative heme distal side in the oxygen recognition and stability of the Fe(II)-O2 complex of YddV, a globin-coupled heme-based oxygen sensor diguanylate cyclase. Biochemistry. 2010;49:10381–93. [DOI] [PubMed] [Google Scholar]
- Kitanishi K. Bacterial hemerythrin domain-containing oxygen and redox sensors: versatile roles for oxygen and redox signaling. Front Mol Biosci. 2022;9:967059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B, Narberhaus F. Microbial thermosensors. Cell Mol Life Sci. 2009;66:2661–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler BJ, Waters CM. Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect Immun. 2014;82:3002–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiselburd I, Moyano L, Carrau A et al. Bacterial photosensory proteins and their role in plant-pathogen interactions. Photochem Photobiol. 2017;93:666–74. [DOI] [PubMed] [Google Scholar]
- Krauss U, Minh BQ, Losi A et al. Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. J Bacteriol. 2009;191:7234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumauchi M, Hara MT, Stalcup P et al. Identification of six new photoactive yellow proteins–diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochem Photobiol. 2008;84:956–69. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Lee DS, Watanabe M et al. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–93. [DOI] [PubMed] [Google Scholar]
- Kyndt JA, Fitch JC, Meyer TE et al. Thermochromatium tepidum photoactive yellow protein/bacteriophytochrome/diguanylate cyclase: characterization of the PYP domain. Biochemistry. 2005;44:4755–64. [DOI] [PubMed] [Google Scholar]
- Laan W, Bednarz T, Heberle J et al. Chromophore composition of a heterologously expressed BLUF-domain. Photochem Photobiol Sci. 2004;3:1011–6. [DOI] [PubMed] [Google Scholar]
- Lau RK, Ye Q, Birkholz EA et al. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol Cell. 2020;77:723–733.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li M, Hu L et al. HpoR, a novel c-di-GMP effective transcription factor, links the second messenger’s regulatory function to the mycobacterial antioxidant defense. Nucleic Acids Res. 2018;46:3595–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Heine S, Entian M et al. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol. 2013;195:3531–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Xu Y, Hossain S et al. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry. 2012;51:2087–99. [DOI] [PubMed] [Google Scholar]
- Lori C, Ozaki S, Steiner S et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–9. [DOI] [PubMed] [Google Scholar]
- Losi A, Gärtner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–78. [DOI] [PubMed] [Google Scholar]
- Losi A, Polverini E, Quest B et al. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J. 2002;82:2627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe EC, Baslé A, Czjzek M et al. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci USA. 2012;109:7298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey B, Whiteley AT, Keszei AFA et al. CBASS immunity uses CARF-related effectors to sense 3'-5'- and 2'-5'-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell. 2020;182:38–49.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínková M, Kitanishi K, Shimizu T. Heme-based globin-coupled oxygen sensors: linking oxygen binding to functional regulation of diguanylate cyclase, histidine kinase, and methyl-accepting chemotaxis. J Biol Chem. 2013;288:27702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Mora D, Ortega Á, Matilla MA et al. The molecular mechanism of nitrate chemotaxis via direct ligand binding to the PilJ domain of McpN. Mbio. 2019;10:e02334–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Rodríguez AJ, Higdon SM, Thorell K et al. Comparative genomics of cyclic di-GMP metabolism and chemosensory pathways in Shewanella algae strains: novel bacterial sensory domains and functional insights into lifestyle regulation. Msystems. 2022;7:e0151821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr Opin Microbiol. 2013;16:148–55. [DOI] [PubMed] [Google Scholar]
- Mashruwala AA, Guchte AV, Boyd JM. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife. 2017;6:e23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslennikov I, Klammt C, Hwang E et al. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc Natl Acad Sci USA. 2010;107:10902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Velando F, Martín-Mora D et al. A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol Rev. 2022;46:fuab043. [DOI] [PubMed] [Google Scholar]
- Mesquita MCB, Lürling M, Dorr F et al. Combined effect of light and temperature on the production of saxitoxins in Cylindrospermopsis raciborskii strains. Toxins. 2019;11:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz S, Haberzettl K, Frühwirth S et al. Interaction of two photoreceptors in the regulation of bacterial photosynthesis genes. Nucleic Acids Res. 2012;40:5901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Privé GG, Milligan DL et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–7. [DOI] [PubMed] [Google Scholar]
- Miller LD, Russell MH, Alexandre G. Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol. 2009;66:53–75. [DOI] [PubMed] [Google Scholar]
- Minasov G, Padavattan S, Shuvalova L et al. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem. 2009;284:13174–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocellin S, Bronte V, Nitti D. Nitric oxide, a double edged sword in cancer biology: searching for therapeutic opportunities. Med Res Rev. 2007;27:317–52. [DOI] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of per-ARNT-Sim domains. Structure. 2009;17:1282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BL, Lagarias JC. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 2002;7:357–66. [DOI] [PubMed] [Google Scholar]
- Morgan R, Kohn S, Hwang SH et al. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura T, Suga M, Hienerwadel R et al. Crystal structure and redox properties of a novel cyanobacterial heme protein with a His/Cys heme axial ligation and a Per-Arnt-Sim (PAS)-like domain. J Biol Chem. 2017;292:9599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Chacón KN, Jarvis JM et al. Structural insights into the mechanism of oxidative activation of heme-free H-NOX from Vibrio cholerae. Biochem J. 2020;477:1123–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadyay R, Sudasinghe N, Schaub T et al. Heme-independent redox sensing by the heme-nitric oxide/oxygen-binding protein (H-NOX) from Vibrio cholerae. J Biol Chem. 2016;291:17547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muok AR, Deng Y, Gumerov VM et al. A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species. Proc Natl Acad Sci USA. 2019;116:14955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzopappa F, Kirilovsky D. Changing color for photoprotection: the orange carotenoid protein. Trends Plant Sci. 2020;25:92–104. [DOI] [PubMed] [Google Scholar]
- Nagano S. From photon to signal in phytochromes: similarities and differences between prokaryotic and plant phytochromes. J Plant Res. 2016;129:123–35. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Kitanishi K, Kobayashi K et al. Leu65 in the heme distal side is critical for the stability of the Fe(II)-O2 complex of YddV, a globin-coupled oxygen sensor diguanylate cyclase. J Inorg Biochem. 2012;108:163–70. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mine S, Hagihara Y et al. Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus. J Mol Biol. 2008;381:670–80. [DOI] [PubMed] [Google Scholar]
- Nakane D, Enomoto G, Bähre H et al. Thermosynechococcus switches the direction of phototaxis by a c-di-GMP-dependent process with high spatial resolution. Elife. 2022;11:e73405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasone Y, Ono TA, Ishii A et al. Temperature-sensitive reaction of a photosensor protein YcgF: possibility of a role of temperature sensor. Biochemistry. 2010;49:2288–96. [DOI] [PubMed] [Google Scholar]
- Navarro MV, Newell PD, Krasteva PV et al. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 2011;9:e1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, Bootman MD, Scott JD. Second messengers. Cold Spring Harb Perspect Biol. 2016;8:a005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya AN, Mulkidjanian AY, Beech IB et al. MASE1 and MASE2: two novel integral membrane sensory domains. J Mol Microbiol Biotechnol. 2003;5:11–6. [DOI] [PubMed] [Google Scholar]
- Nioche P, Berka V, Vipond J et al. Femtomolar sensitivity of a NO sensor from Clostridium botulinum. Science. 2004;306:1550–3. [DOI] [PubMed] [Google Scholar]
- Nisbett LM, Binnenkade L, Bacon B et al. NosP signaling modulates the NO/H-NOX-mediated multicomponent c-di-GMP network and biofilm formation in Shewanella oneidensis. Biochemistry. 2019;58:4827–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BH, Pandit J, Kang CH et al. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem. 1993;268:11348–55. [PubMed] [Google Scholar]
- Olea C, Herzik MA Jr, Kuriyan J et al. Structural insights into the molecular mechanism of H-NOX activation. Protein Sci. 2010;19:881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Galperin MY, Lee VT. Sustained sensing as an emerging principle in second messenger signaling systems. Curr Opin Microbiol. 2016;34:119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Weiss CA, Severin GB et al. A subset of exoribonucleases serve as degradative enzymes for pGpG in c-di-GMP signaling. J Bacteriol. 2018;200:e00300–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Á, Zhulin IB, Krell T. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev. 2017;81:e00033–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Suquet C, Satterlee JD et al. Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH). Biochemistry. 2004;43:2738–46. [DOI] [PubMed] [Google Scholar]
- Park SY, Tame JRH. Seeing the light with BLUF proteins. Biophys Rev. 2017;9:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DC, Ruiz MP, Yoon H et al. Differential ligand-selective control of opposing enzymatic activities within a bifunctional c-di-GMP enzyme. Proc Natl Acad Sci USA. 2021;118:e2100657118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L, O’Hara B, Drew R et al. Crystal structure of AmiC: the controller of transcription antitermination in the amidase operon of Pseudomonas aeruginosa. EMBO J. 1994;13:5810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellequer JL, Wager-Smith KA, Kay SA et al. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA. 1998;95:5884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicena P, Karow DS, Boon EM et al. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci USA. 2004;101:12854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Cherny KE, Sauer K. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol. 2015;197:174–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfiffer V, Sarenko O, Possling A et al. Genetic dissection of Escherichia coli’s master diguanylate cyclase DgcE: role of the N-terminal MASE1 domain and direct signal input from a GTPase partner system. PLoS Genet. 2019;15:e1008059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflüger T, Hernández CF, Lewe P et al. Signaling ammonium across membranes through an ammonium sensor histidine kinase. Nat Commun. 2018;9:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate L, Marletta MA. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell. 2012;46:449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate L, Marletta MA. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci. 2013a;38:566–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate L, Marletta MA. Phosphorylation-dependent derepression by the response regulator HnoC in the Shewanella oneidensis nitric oxide signaling network. Proc Natl Acad Sci USA. 2013b;110:E4648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, Crosson S. Photoregulation in prokaryotes. Curr Opin Microbiol. 2008;11:168–78. [DOI] [PubMed] [Google Scholar]
- Qi Y, Rao F, Luo Z et al. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry. 2009;48:10275–85. [DOI] [PubMed] [Google Scholar]
- Randall TE, Eckartt K, Kakumanu S et al. Sensory perception in bacterial cyclic diguanylate signal transduction. J Bacteriol. 2022;204:e0043321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, See RY, Zhang D et al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Herzik MA Jr, Iavarone AT et al. Nitric oxide-induced conformational changes govern H-NOX and histidine knase interaction and regulation in Shewanella oneidensis. Biochemistry. 2017;56:1274–84. [DOI] [PubMed] [Google Scholar]
- Rao M, Smith BC, Marletta MA. Nitric oxide mediates biofilm formation and symbiosis in silicibacter sp. strain TrichCH4B. Mbio. 2015;6:e00206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo S, Giardina G, Mantoni F et al. Beyond nitrogen metabolism: nitric oxide, cyclic-di-GMP and bacterial biofilms. FEMS Microbiol Lett. 2018;365:29401255. [DOI] [PubMed] [Google Scholar]
- Rivera-Cancel G, Ko WH, Tomchick DR et al. Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc Natl Acad Sci USA. 2014;111:17839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. A brief history of phytochromes. ChemPhysChem. 2010;11:1172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AB, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194:2904–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Fomicheva A, Moskvin OV et al. Optogenetic module for dichromatic control of c-di-GMP signaling. J Bacteriol. 2017a;199:e00014–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Fomicheva A, O'Neal L et al. Using light-activated enzymes for modulating intracellular c-di-GMP levels in bacteria. Methods Mol Biol. 2017b;1657:169–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarenko O, Klauck G, Wilke FM et al. More than enzymes that make or break cyclic di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. Mbio. 2017;8:e01639–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savakis P, De Causmaecker S, Angerer V et al. Light-induced alteration of c-di-GMP level controls motility of synechocystis sp. PCC 6803. Mol Microbiol. 2012;85:239–51. [DOI] [PubMed] [Google Scholar]
- Sawai H, Yoshioka S, Uchida T et al. Molecular oxygen regulates the enzymatic activity of a heme-containing diguanylate cyclase (HemDGC) for the synthesis of cyclic di-GMP. Biochim Biophys Acta. 2010;1804:166–72. [DOI] [PubMed] [Google Scholar]
- Schaller RA, Ali SK, Klose KE et al. A bacterial hemerythrin domain regulates the activity of a Vibrio cholerae diguanylate cyclase. Biochemistry. 2012;51:8563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Sauthof L, Szczepek M et al. Structural snapshot of a bacterial phytochrome in its functional intermediate state. Nat Commun. 2018;9:4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. A short history of structure based research on the photocycle of photoactive yellow protein. Struct Dyn. 2017;4:032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert S, Brakmann S. LOV domains in the design of photoresponsive enzymes. ACS Chem Biol. 2018;13:1914–20. [DOI] [PubMed] [Google Scholar]
- Sellner B, Prakapaitė R, van Berkum M et al. A new sugar for an old phage: a c-di-GMP-dependent polysaccharide pathway sensitizes Escherichia coli for bacteriophage infection. Mbio. 2021;12:e0324621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshasayee AS, Fraser GM, Luscombe NM. Comparative genomics of cyclic-di-GMP signalling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res. 2010;38:5970–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Potts M, Kennelly PJ. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol Rev. 1998;22:229–53. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Huang D, Yan F et al. Gaseous O2, NO, and CO in signal transduction: structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem Rev. 2015;115:6491–533. [DOI] [PubMed] [Google Scholar]
- Shimizu T. The heme-based oxygen-sensor phosphodiesterase ec DOS (DosP): structure-function relationships. Biosensors. 2013;3:211–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisinni L, Cendron L, Favaro G et al. Helicobacter pylori acidic stress response factor HP1286 is a YceI homolog with new binding specificity. FEBS J. 2010;277:1896–905. [DOI] [PubMed] [Google Scholar]
- Spiro S. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev. 2007;31:193–211. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. [DOI] [PubMed] [Google Scholar]
- Stone JR, Marletta MA. Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry. 1994;33:5636–40. [DOI] [PubMed] [Google Scholar]
- Stülke J, Krüger L. Cyclic di-AMP signaling in bacteria. Annu Rev Microbiol. 2020;74:159–79. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu Y, Liu X et al. Azorhizobium caulinodans c-di-GMP phosphodiesterase Chp1 involved in motility, EPS production, and nodulation of the host plant. Appl Microbiol Biotechnol. 2020;104:2715–29. [DOI] [PubMed] [Google Scholar]
- Takala H, Björling A, Berntsson O et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem. 2005;280:33324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Shimizu T. Ligand binding to the Fe(III)-protoporphyrin IX complex of phosphodiesterase from Escherichia coli (Ec DOS) markedly enhances catalysis of cyclic di-GMP: roles of Met95, Arg97, and Phe113 of the putative heme distal side in catalytic regulation and ligand binding. Biochemistry. 2008;47:13438–46. [DOI] [PubMed] [Google Scholar]
- Tarnawski M, Barends TR, Schlichting I. Structural analysis of an oxygen-regulated diguanylate cyclase. Acta Crystallogr D Biol Crystallogr. 2015;71:2158–77. [DOI] [PubMed] [Google Scholar]
- Tarutina M, Ryjenkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem. 2006;281:34751–8. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB, Johnson MS. Aerotaxis and other energy-sensing behavior in bacteria. Annu Rev Microbiol. 1999;53:103–28. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol Microbiol. 2007;65:1415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschler JK, Nadell CD, Drescher K et al. Mechanisms underlying Vibrio cholerae biofilm formation and dispersion. Annu Rev Microbiol. 2022;76:503–32. [DOI] [PubMed] [Google Scholar]
- Tomita T, Gonzalez G, Chang AL et al. A comparative resonance Raman analysis of heme-binding PAS domains: heme iron coordination structures of the BjFixL, AxPDEA1, EcDos, and MtDos proteins. Biochemistry. 2002;41:4819–26. [DOI] [PubMed] [Google Scholar]
- Trakhanov S, Vyas NK, Luecke H et al. Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity. Biochemistry. 2005;44:6597–608. [DOI] [PubMed] [Google Scholar]
- Tschowri N, Busse S, Hengge R. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 2009;23:522–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol. 2011;407:633–9. [DOI] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Sousa EH et al. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry. 2009;48:9764–74. [DOI] [PubMed] [Google Scholar]
- Ueno T, Fischer JT, Boon EM. Nitric oxide enters quorum sensing via the H-NOX signaling pathway in Vibrio parahaemolyticus. Front Microbiol. 2019;10:2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay AA, Fleetwood AD, Adebali O et al. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol. 2016;12:e1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano R, Karlinsey JE, Libby SJ et al. Host nitric oxide disrupts microbial cell-to-cell communication to inhibit staphylococcal virulence. Cell Host Microbe. 2018;23:594–606.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem. 2016;291:12547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Filloux A. Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu Rev Microbiol. 2019;73:387–406. [DOI] [PubMed] [Google Scholar]
- van Aalten DM, Crielaard W, Hellingwerf KJ et al. Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution. Acta Crystallogr D Biol Crystallogr. 2002;58:585–90. [DOI] [PubMed] [Google Scholar]
- van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004;37:13–20. [DOI] [PubMed] [Google Scholar]
- van der Horst MA, Key J, Hellingwerf KJ. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol. 2007;15:554–62. [DOI] [PubMed] [Google Scholar]
- van der Horst MA, Stalcup TP, Kaledhonkar S et al. Locked chromophore analogs reveal that photoactive yellow protein regulates biofilm formation in the deep sea bacterium Idiomarina loihiensis. J Am Chem Soc. 2009;131:17443–51. [DOI] [PubMed] [Google Scholar]
- Verma RK, Biswas A, Kakkar A et al. A bacteriophytochrome mediates interplay between light sensing and the second messenger cyclic di-GMP to control social behavior and virulence. Cell Rep. 2020;32:108202. [DOI] [PubMed] [Google Scholar]
- Villafani Y, Yang HW, Park YI. Color sensing and signal transmission diversity of cyanobacterial phytochromes and cyanobacteriochromes. Mol Cells. 2020;43:509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov SN, Hoogewijs D, Bailly X et al. Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc Natl Acad Sci USA. 2005;102:11385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Rivera S, Weinert EE. Mechanism and role of globin-coupled sensor signalling. Adv Microb Physiol. 2017;71:133–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Wu Y, Potter JR et al. π-helix controls activity of oxygen-sensing diguanylate cyclases. Biosci Rep. 2020;40:BSR20193602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner T, Pedroza L, Voigt K et al. The cyanobacterial phytochrome 2 regulates the expression of motility-related genes through the second messenger cyclic di-GMP. Photochem Photobiol Sci. 2020;19:631–43. [DOI] [PubMed] [Google Scholar]
- Wan X, Saito JA, Newhouse JS et al. The importance of conserved amino acids in heme-based globin-coupled diguanylate cyclases. PLoS ONE. 2017;12:e0182782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Tuckerman JR, Saito JA et al. Globins synthesize the second messenger bis-(3'-5')-cyclic diguanosine monophosphate in bacteria. J Mol Biol. 2009;388:262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dufour YS, Carlson HK et al. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci USA. 2010;107:8375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Chin KH, Tu ZL et al. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun. 2016;7:12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DE, Boon EM. Towards understanding the molecular basis of nitric oxide-regulated group behaviors in pathogenic bacteria. J Innate Immun. 2019;11:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DE, Nisbett LM, Bacon B et al. Bacterial heme-based sensors of nitric oxide. Antioxid Redox Signal. 2018;29:1872–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Su X, Potluri N et al. NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. Elife. 2016;5:e18790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan TF, Wang ZQ, Liu J et al. Design and synthesis of novel c-di-GMP G-quadruplex inducers as bacterial biofilm inhibitors. J Med Chem. 2021;64:11074–89. [DOI] [PubMed] [Google Scholar]
- Yang H, Inokuchi H, Adler J. Phototaxis away from blue light by an Escherichia coli mutant accumulating protoporphyrin IX. Proc Natl Acad Sci USA. 1995;92:7332–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lam V, Adomako M et al. Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2018;115:E12378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Ali A, Igbalajobi OA et al. Two hybrid histidine kinases, TcsB and the phytochrome FphA, are involved in temperature sensing in Aspergillus nidulans. Mol Microbiol. 2019;112:1814–30. [DOI] [PubMed] [Google Scholar]
- Yu Z, Fischer R. Light sensing and responses in fungi. Nat Rev Microbiol. 2019;17:25–36. [DOI] [PubMed] [Google Scholar]
- Zeng X, Huang M, Sun QX et al. A c-di-GMP binding effector controls cell size in a cyanobacterium. Proc Natl Acad Sci USA. 2023;120:e2221874120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Cronin S, Lefèvre CT et al. MamA as a model protein for structure-based insight into the evolutionary origins of magnetotactic bacteria. PLoS ONE. 2015;10:e0130394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J Bacteriol. 2003;185:285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R, Wong HC, Calhoon R et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.