Abstract
Objective:
This study aimed to find the key genes and miRNAs as potential biomarkers related to the progression of colorectal cancer (CRC) from Crohn’s disease (CD).
Background:
CD is widely accepted as one of the main risk factors leading to CRC. So, Identifying the novel molecular pathways involved in the development of CRC from CD can provide potential solutions for therapeutic interventions.
Methods:
By implementing a systematic approach, we have analyzed mRNA and miRNA datasets containing CRC and CD samples to determine differentially expressed genes (DEGs) and miRNAs (DEmiRNA). Then by selecting common genes involved in the progression from CD to CRC, different downstream analyses including mRNA-miRNA network, functional enrichment analysis, gene set enrichment analysis, and survival analysis were performed. Finally, quantitative real-time PCR (RT-PCR) analysis of tissue samples obtained from Normal/CRC samples was used to confirm the differential expression of selected genes and miRNA.
Results:
There were 10 DE miRNA and 181 genes DEGs common between progression from CD to CRC. The genes obtained for each of the 10 miRNAs were considered as the final target for downstream analyzes. In addition, analysis of RT-PCR indicated that miR-195-5p, PHLPP2, and LITAF were downregulated in the cancer group compared to the control group.
Conclusion:
This study showed that PHLPP2, LITAF, and miR-195-5p may have key roles in the tumorigenesis of CRC and they can be used as therapeutic targets and diagnostic biomarkers after further in-vitro and in-vivo evaluation.
Key Words: Colorectal cancer, Crohn’s disease, biomarker, miRNA
Introduction
Colorectal cancer (CRC) as one of the most common gastrointestinal tract malignancies is the second most cause of cancer-related death globally (Granados-Romero et al., 2017). There are numerous risk factors for CRC namely age, race, Family history of the CRC, Familial adenomatous polyposis (FAP), hereditary non-polyposis colorectal cancer (HNPCC), body mass index (BMI), Food habits, physical activity, cigarette smoking, and inflammatory bowel disease (IBD) (Amersi et al., 2005; Johnson et al., 2013).
IBD as a chronic inflammatory disease including Crohn’s disease (CD) and ulcerative colitis (UC) is considered a CRC risk factor. The CRC which initiates in IBD patients develops in a different pathway than sporadic CRC (Lucafò et al., 2021). These disorders are due to the dysregulation of T cells and imbalance of pro- and anti-inflammatory cytokines (Hnatyszyn et al., 2019a). The CD is characterized by a multifactorial etiology including environmental, genetic, and microbial factors (Larabi et al., 2020; Liu et al., 2015). Results of genome-wide association studies indicated that several genes have a role in inheritable vulnerability to CD. The protein encoded with these genes is associated with several homeostatic mechanisms including epithelial barrier integrity maintenance, Th17-lymphocyte differentiation, innate pattern recognition receptors, and the secondary immune response balance (Van Limbergen et al., 2009).
MiRNAs are small non-coding RNAs that control several biological functions such as proliferation, differentiation, apoptosis, metastasis, and invasion through suppressing the target mRNAs. (Afshar et al., 2020; Mahmoudi et al., 2022; Schee et al., 2010). MiRNAs play an important role in the pathogenesis of several human diseases, such as cancers and IBD. Based on the type and activity of the protein to which the miRNA binds to its target mRNA and changes its expression, they are called tumor-suppressor miRNAs or oncogenic miRNAs (oncomiRs) (DalalKwon, 2010; Zhang et al., 2015). In summary, miRNAs play essential roles in the pathogenesis of sporadic CRC, CRC which initiates in IBD, and IBDs, in particular CD (Grillo et al., 2021; Hnatyszyn et al., 2019b). In this regard, integrated analysis of omics data on miRNA-mRNA interactions has helped to identify key therapeutic targets in CRC and IBD diseases. Many studies are currently being conducted to extend our understanding of miRNA functions and regulatory mechanisms in the tumorigenic process of CRC (Vera et al., 2013).
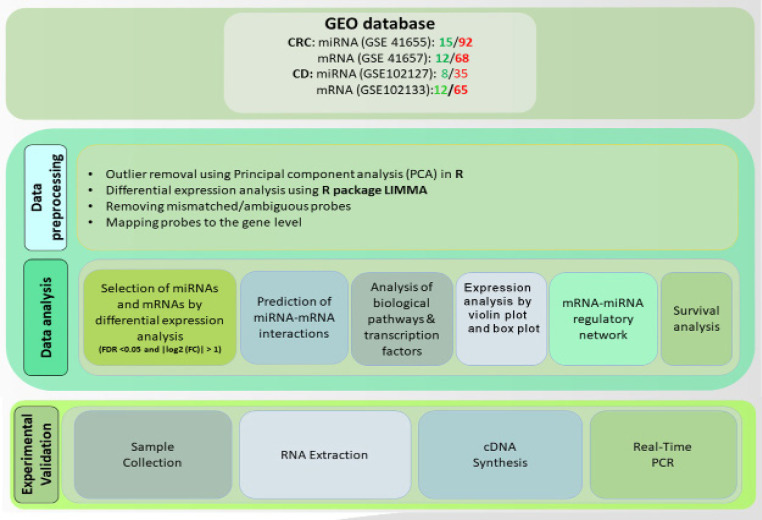
In this study, by evaluating the core miRNA-mRNA interactions related to CD and CRC, we search for the most important molecular factors related to the progression from CD to CRC. Moreover, with the identification of novel molecular factors, it will be possible to perform additional evaluations to prove the biomarker capability of these biomarkers and their importance as potential therapeutic targets. Finally, to confirm the results of systems biology approaches, the expression analysis of selected biomarkers was evaluated by the Real-Time PCR method on CRC tissue samples and adjacent normal tissue. An overview of the steps in this study is represented in Figure 1.
Figure 1.
Schematic Diagram Showing the Steps of This Study
Materials and Methods
Data collection and unsupervised analysis
Raw expression data of miRNA and mRNA of tissue samples from CRC patients were obtained from Gene Expression Omnibus (GEO) databases (Table 1). For pre-processing, we used R software and related Bioconductor packages.
Table1.
List of GEO Data Series Used in This Study
Differentially expressed miRNAs and mRNAs analysis
In order to identify miRNAs and mRNAs by differentiating between control and CRC samples, the LIMMA package under R software was used. In this study, significant DEGs were selected for subsequent analysis with criteria of adjusted P-value <0.05 and |log2 (FC)| > 1, while for DEmiRNAs, the cutoff criteria of adjusted P-value <0.05 and | log2 (FC) | > 0.5 was considered.
We also utilized the violin plot to demonstrate the expression patterns of DEGs and DE miRNAs in CD and CRC. The analysis was performed using the R package “ggplot2”. The violin plot was chosen as a useful visualization tool to show the distribution of expression values for each gene and miRNA, where the width of the plot indicates the frequency of expression values at different levels.
Prediction of miRNA-mRNA interactions
To identify miRNAs and their targets that show differential expression in control and disease samples, we used two miRNA target forecast software including miRTarBase (https://mirtarbase.cuhk.edu.cn) and miRWalk (http://mirwalk.umm.uni-heidelberg.de ). The mRNA-miRNA interactions present in both databases were considered as the final interactions for further analysis.
Analysis of KEGG pathway and Gene Ontology
To analyze the biological pathways in which common DEGs are involved, functional enrichment analysis for The Kyoto Encyclopedia Gene and Genomes (KEGG) pathway and tree gene ontology (GO) terms including molecular function (MF), biological process (BP), cellular component (CC) was performed using the R packages ClusterProfiler. KEGG pathways and GO terms with FDR <0.05 were considered statistically significant.
Analysis of mRNA-miRNA network
Finally, network analysis was performed to further investigate the hub genes involved in the pathogenesis of CD and CRC. The STRING online tool was used to construct The Protein-protein interaction (PPI) network (https://string-db.org/cgi/input.pl/) considering the required score = 0.4. The constructed network and bipartite miRNA- mRNA network was visualized and evaluated with Cytoscape software (V:3.7.2).
Survival analysis
The TCGA expression data was used to investigate the relationship between hub genes and the survival rate of CRC patients. The survival analysis and Kaplan-Meier plot depiction were performed with TCGA biolinks, and survminer packages in r software.
Investigation of gene expression in CRC samples
To validate the expression of selected miRNA and its target genes,12 CRC and 12 adjacent normal tissues were used (Table 3). All patients participating in this study accepted Informed consent. This study was approved by the Ethics Committee of the Hamadan University of Medical Sciences (IR.UMSHA.REC.1399.561.) In this regard, total RNA was isolated using the RNX-plus kit (Sinaclon, Iran) conferring to the manufacturer’s protocol. Afterward cDNA synthesis for miRNA and mRNA was performed with 1st strand cDNA Synthesis Kit (Takara, Japan) and miRNA stem-loop cDNA synthesis (Anacell, Iran), respectively. The expression level of miRNA-195-5p and U6 as an internal control was evaluated using specific primer pair (Anacell, Iran). The expression level of LITAF, PHLPP2, and GAPDH as an internal control was evaluated with specific primer pairs designed using AlleleId 6 (Table 2). Roche LightCycler® 96 system (Roche, Germany) was used for performing the real-time PCR analysis. Moreover, based on the microarray data of CD and CRC, stage plots were plotted using ggstatsplot package in R.
Table 3.
Demographic Information of Patients Participated in This Study
| n | % | |
|---|---|---|
| Age | ||
| ≤63 | 6 | 50 |
| >63 | 6 | 50 |
| Gender | ||
| Male | 9 | 75 |
| Female | 3 | 25 |
| Stage | ||
| I | 6 | 50 |
| II | 5 | 41.7 |
| III | 1 | 8.3 |
| Grade | ||
| I | 7 | 58.3 |
| II | 4 | 33.3 |
| III | 1 | 8.3 |
Table 2.
The Specification of Primer Pairs Used to Evaluate the Expression Level of Target Genes
| Gene | Antisense strand | Sense strand | Gene ID | Product size |
|---|---|---|---|---|
| LITAF | GCAGTTACAGAATCTCAAAGCC | ACCATTATCTTATCCGTCTCTTCC | 9516 | 90 bp |
| PHLPP | GTCTACTTCCACCTCTACCTTGC | GCCTTCTTCTGCTACCTTCTC3 | 23035 | 115 bp |
| GAPDH | GCGTCAAAGGTGGAGGAGTGG | AAGGCTGTGGGCAAGGTCATC | 2597 | 248 bp |
Statistical analysis
Data are presented as the mean ± standard error mean. The t-test was used to evaluate the average values differences between groups using SPSS 16.0 and P-value less than 0.05 was considered statistically significant.
Results
Identification of DEGs and DE miRNAs
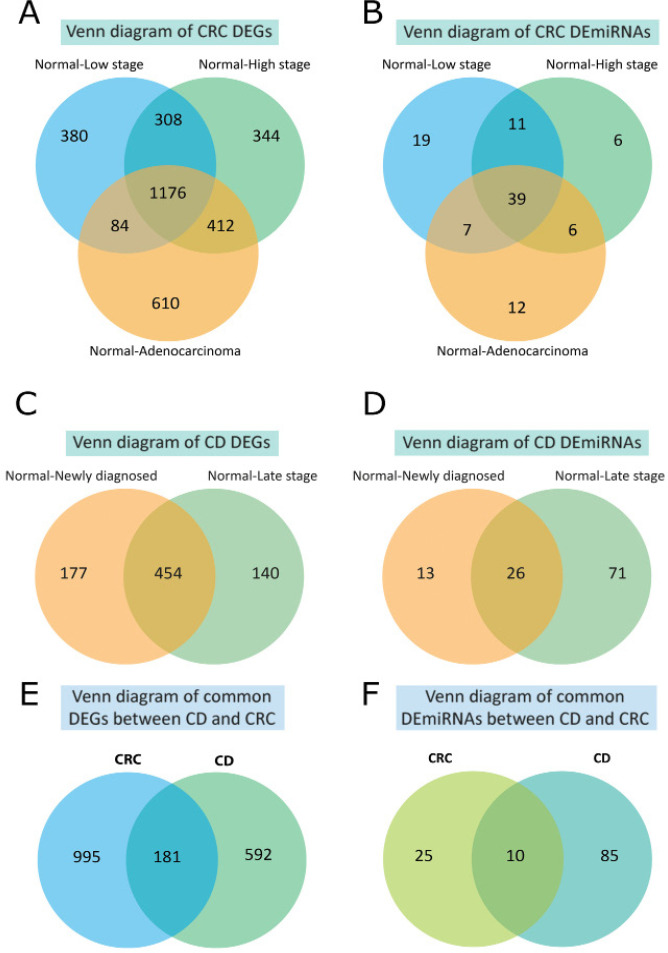
Evaluation of mRNA expression data indicated that in the low-grade adenomas, high-grade adenomas, and Adenocarcinoma samples 2,190, 2,772, and 2,677 DEGs were detected compared with healthy control respectively. For miRNA expression data 61, 48, and 55 DE miRNAs were detected in the low-grade adenomas, high-grade adenomas, and Adenocarcinoma samples respectively. The number of common DEGs and DE miRNAs between low-grade adenomas, high-grade adenomas, and Adenocarcinoma are 1,176 and 39 respectively. Regarding CD, the number of common DEGs and DE miRNAs between low and high-stage CD were 454 and 26. Finally, 181 DEGs and 10 DE miRNAs were found common between CRC and CD (Figure 2).
Figure 2.
The Common of Differentially Expressed Genes (DEGs) and Differentially Expressed microRNAs (DE-miRNAs) Across all CRC and CD Stages are Selected Using the VENN Diagram
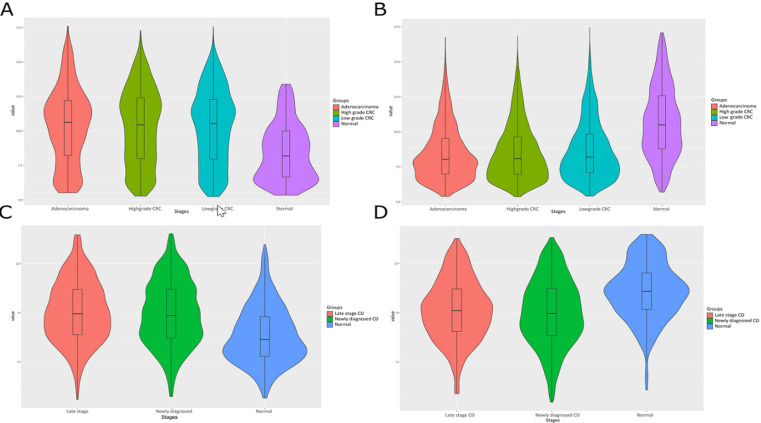
The violin plot analysis also revealed distinct expression patterns of DEGs and DE miRNAs in CD and CRC. Notably, DEGs and DE miRNAs showed significant up-regulation or down-regulation of each stage of CD and CRC in comparison to normal, indicating their potential as biomarkers and therapeutic targets for these diseases (Figure 3).
Figure 3.
The Violin Plot Analysis Shows Distinct Expression Patterns of Upregulated (A and C) and Downregulated (B and D) Differentially Expressed Genes (DEGs) in Colorectal Cancer (CRC) and Crohn’s Disease (CD)
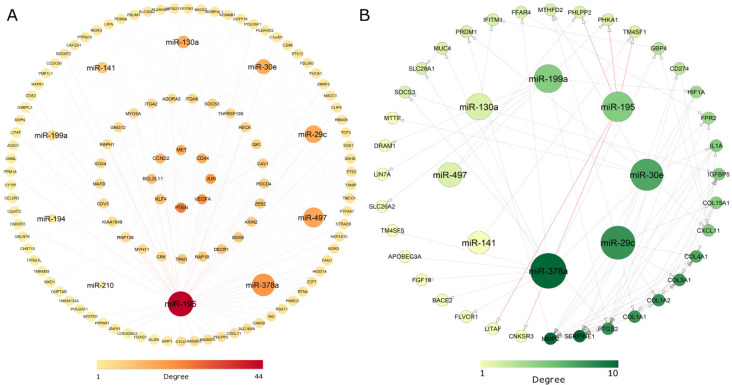
Bipartite miRNA-mRNA interaction analysis
The top 10 miRNA targets were searched in the miRTarBase and miRWalk databases. For CRC 111 DEGs and for CD 36 DEGs were found as a target of selected miRNAs respectively. Evaluation of the miRNA-mRNA network indicated that miR-195 as a hub miRNA due to the best scores in both CD and CRC networks. More ever, evaluation of the network indicated that LITAF, PHLPP2, and CNKSR3 were identified as common miR-195-5p target genes between colorectal and Crohn’s cancer (Figure 4). Due to the lack of correlation between the CNKSR3 gene and other nodes in the network, only the PHLPP2, LITAF, and miR-195 were selected for further experimental validation.
Figure 4.
The mRNA-miRNA Regulatory Network was Constructed for Significantly Differentially Expressed Genes (DEGs) and Differentially Expressed microRNAs (DE-miRNAs) in Colorectal Cancer (A) and Crohn's Disease (B). The inner circles in (A) are DEGs with higher degree. Significant interactions of miR-195-5p with its target genes are shown in red
KEGG pathway and Gene Ontology enrichment analysis
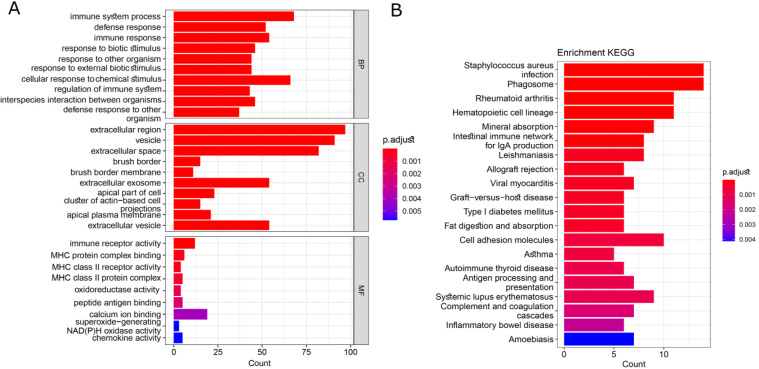
In this study, we analyzed the GO terms and KEGG pathways for the common differentially expressed genes (DEGs) using the clusterProfiler package in R. Our analysis revealed the top 10 enriched terms, which are illustrated in Figure 5. Notably, the largest terms identified were related to biological processes such as immune system process, and cellular components such as extracellular region and extracellular space. Additionally, immune receptor activity and calcium ion binding were also among the most prominent terms.
Figure 5.
The Results of Enrichment Analysis for GO Terms and KEGG Pathways. The top 10 biological process, cellular component, molecular function, and top 20 KEGG pathways were shown in this graph
Furthermore, the top 20 significantly enriched KEGG pathways were revealed for the common DEGs. These included important signaling pathways such as Staphylococcus aureus infection, Rheumatoid arthritis, Phagosome, and Cell adhesion molecules. Importantly, the inflammatory bowel disease pathway was also significantly enriched and highlighted the role of our final set of DEGs in inflammation-related cancer progression. These results are depicted in Figure 5, underscoring the significance of our findings.
Find the prognostic index of hub genes by survival analysis
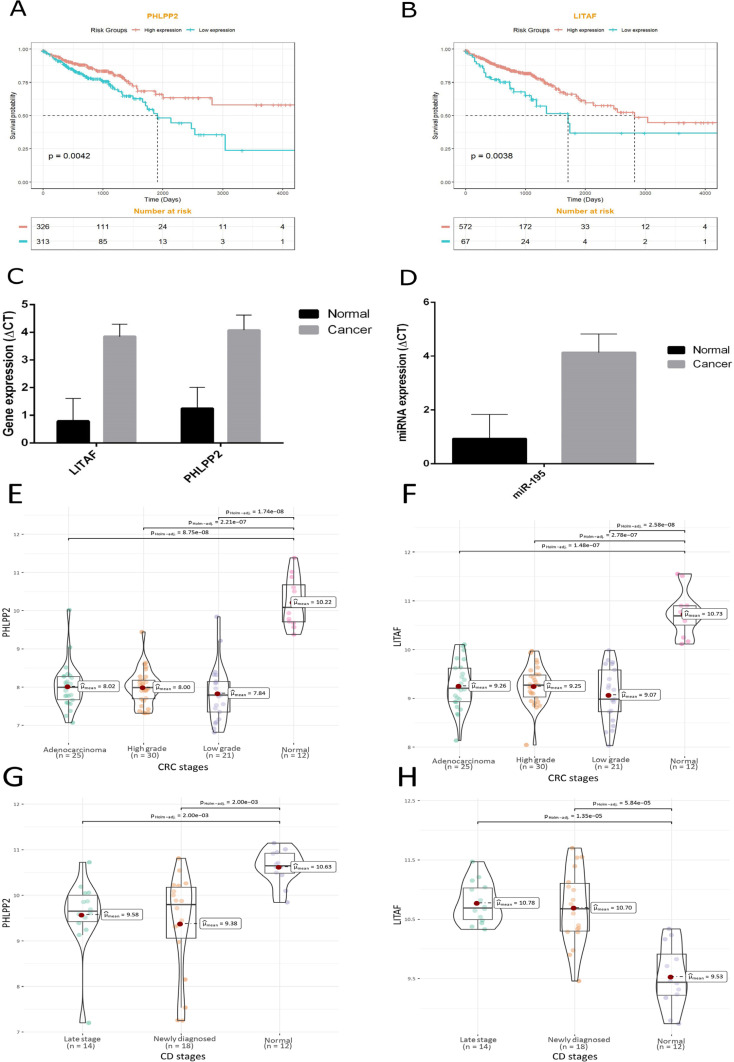
As shown in Figure 6A, B the expression level of PHLPP2 and LITAF was associated with the prognosis of patients. The results of the survival analysis indicated that the expression levels of PHLPP2 and LITAF were significantly associated with the overall survival of CRC patients (p-values of 0.0042 and 0.0038, respectively).
Figure 6.
Kaplan-Meier Curve for PHLPP2 (A) and LITAF (B). The expression level of a) PHLPP2 and b) LITAF was significantly associated with the patients' prognosis. The expression level of LITAF and PHLPP as genes (C), and miR-195-5p as the hub miRNA (D) in colorectal tumor samples and adjacent normal tissues. The ∆CT has a reverse correlation with the expression level of a gene, so PHLPP2 and LITAF were downregulated in CRC. Further validation of hub genes using microarray data of colorectal cancer (E, F) and Crohn's disease (G, H) via ggstatsplot package in R
Experimental validation of PHLPP2 and LITAF and miR-195
The results of the study of gene expression between the tumor sample groups and adjacent normal tissue groups in terms of ∆CT showed a significant difference for PHLPP2 LITAF and miR-195. The ∆CT has an inverse correlation with the expression level of a gene, so PHLPP2 LITAF and miR-195 are downregulated in CRC samples compared with adjacent normal tissues (Figure 6C, D). Moreover, using microarray expression datasets (Table 1), stage plots of final hub genes were also depicted for more validation of our experimental work (Figure 6E to 6H).
Discussion
In recent years, the discovery of molecular identifiers which are involved in cancer progression has created an interesting challenging research field. Nevertheless, there are some issues such as the absence of in-depth studies to compare healthy tissues, different stages of cancer, and precancerous stages. For instance, in a comprehensive integrated analysis, Samadi et al. have demonstrated a signature comprising DEGs, DE-miRNAs, and DE-lncRNAs (long non-coding RNAs) biomarkers related to the diagnosis, prognosis, and therapy of CRC (Samadi et al., 2022). Therefore, finding biomarkers based on diseases that promote cancer risk, such as inflammatory diseases, can be more effective in the treatment of cancer in the early stages and improve the patients’ prognosis(Liu et al., 2021; Sheng et al., 2020).
In the present study, in order to detect mRNA and miRNA biomarkers involved in the progression of Crohn’s disease to various stages of CRC, separate miRNA and mRNA expression data were evaluated. Subsequently, the most significant mRNAs and miRNAs that are involved in the progression from inflammation to adenocarcinoma were used for further evaluation. Furthermore, the results of the current study indicated that miR-195-5p was significantly downregulated in CRC tissues. Recent studies have examined the central role of this miRNA in controlling CRC growth and proliferation. Various other studies have also shown that miR-195 through targeting Bcl-2 (Liu et al., 2010), FGF2, Wnt / β-catenin (Zhang et al., 2016), and GDPD5 pathways (Feng et al., 2018) can induce apoptosis and suppress the growth and proliferation of CRC cells and. It has also been shown that miR-195-5p can play an important role in response to chemotherapy and radiotherapy (Jin et al., 2018; Zheng et al., 2017). Results of recent studies indicated that miR-195 -5p has an essential role in the pathogenesis of IBD by regulating inflammation and intestinal tight junctions(Scalavino et al., 2022; Tian et al., 2020). Chen et al. in their study showed that miR-195 has a role in regulation of UC development and response to glucocorticoid in UC by targeting the SMAD7 and regulating the TGFβ signaling pathway (Chen et al., 2015).
Results of the current study indicated that PHLPP and LITAF upregulated in colorectal tumor tissues. PHLPP2 which was a tumor-suppressor gene in several cancers regulates tumor cell apoptosis and survival. More ever, this gene has a regulatory role in metastasis, invasion, and chemoresistance of CRC through the stemness regulation of colorectal tumor cells (Yongfu et al., 2022). Downregulation of PHLPP2 in CRC patients was common and its expression level was associated with CRC patients’ prognosis (Li et al., 2021). Li et al., (2022) in their study indicated that PHLPP2 was downregulated in UC and associated with immune cell infiltration. PHLPP2 has an essential role in intestinal epithelial cell pyroptosis via the NF-κB signaling pathway (Li et al., 2021). LITAF acts as a tumor suppressor and reduces properties in various types of cancer and its expression is controlled by p53. LITAF which is dysregulated in IBD may play a role in inflammatory disease (Zou et al., 2015). Zhou et al., (2017) in their study showed that protein and mRNA expression levels of LITAF were downregulated in pancreatic cancer and associated with patients’ prognosis. Stucchi et al., (2006) in their study showed that the expression level of LITAF was significantly upregulated in CD compared with healthy control and inflammatory colon area compared with the normal colorectal area. Similarly, in UC patients, the expression level of LITAF was significantly increased compared with healthy control.
Results of the current study indicated that the expression level of mir-195 had not a reverse correlation with LITAF and PHLPP2 which were bioinformatically determined as target genes of miR-195. Usually, miRNAs suppress the expression level of target genes and miRNA expression level has a reverse correlation with its target genes (Afshar et al., 2020). Ritchie et al., (2009) in their study showed that the expression level of miR-92 and miR-32 has a positive correlation with PCNA as a target gene. The positive correlation between PCNA and miR-92 and miR-32 is due to the intermediated regulation. More ever, miR-92 and miR-32 repress the RFX1 and this intermediate gene represses the PCNA. So we can hypothesize that miR-195 may have a positive regulator role for LITAF and PHLPP2 through the intermediate gene.
Taken together, miR-195, LITAF, and PHLPP2 may have critical roles in the tumorigenesis of CRC and the progression of Crohn’s disease to adenocarcinoma. More ever, miR-195, LITAF, and PHLPP2 could be used as therapeutic targets and diagnostic biomarkers after further in-vitro and in-vivo evaluation. The positive correlation between miR-195 and bioinformatically determined target genes also should be evaluated through more precise studies.
Abbreviations
CRC, colorectal cancer; DEGs, differentially expressed genes; DEmiRNA, differentially expressed miRNAs; RT-PCR, quantitative real-time PCR; HNPCC, hereditary non-polyposis colorectal cancer; FAP, Familial adenomatous polyposis; IBD, Inflammatory bowel disease; BMI, Body mass index; UC, ulcerative colitis; CD, Crohn’s disease; oncomiRs, oncogenic miRNAs; GEO, Gene Expression Omnibus; KEGG, Kyoto Encyclopedia Gene and Genomes; GO, gene ontology; BP, biological process; MF, Molecular function; CC, Cellular component; DAVID, Database for Annotation Visualization and Integrated Discovery.
Author Contribution Statement
SA, PS conceived and designed the analysis. NS, SA PS collected the data. NS, SA PS, MP contributed to analysis tools. NS, SA PS preformed the analysis. NS, SA PS, MP, AJ, and RD contributed to the interpretation of the results. SA wrote the manuscript in consultation with all authors.
Acknowledgements
In memory of Prof. Massoud Saidijam, who is no longer with us, but continues to inspire his students over the course of his career.
Funding
This work is supported by a grant from Hamadan University of Medical Sciences, Hamadan, Iran (No. 9907295200).
Ethical approval
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors. The ethical protocol of this study was approved by the Ethics Committee of Hamadan University of Medical Sciences. (Ethical code: IR.UMSHA.REC.1399.561.) and written informed consent was obtained from all patients to participate in the study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the [https://www.ncbi.nlm.nih.gov/geo/database]. The original contributions presented in the study are included in the article/Supplementary Material, further inquiries are available from the corresponding author on reasonable request.
Conflict of Interest
The authors declare no conflict of interest.
References
- Afshar S, Sedighi Pashaki A, Najafi R, et al. Cross-Resistance of Acquired Radioresistant Colorectal Cancer Cell Line to gefitinib and regorafenib. Iran J Med Sci. 2020;45:50–8. doi: 10.30476/ijms.2019.44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amersi F, Agustin M, Ko CY. Colorectal cancer: epidemiology, risk factors, and health services. Clin Colon Rectal Surg. 2005;18:133–40. doi: 10.1055/s-2005-916274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Cao S, Liu F, Liu Y. miR-195 plays a role in steroid resistance of ulcerative colitis by targeting Smad7. Biochem J. 2015;471:357–67. doi: 10.1042/BJ20150095. [DOI] [PubMed] [Google Scholar]
- Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol. 2010;6:714–22. [PMC free article] [PubMed] [Google Scholar]
- Feng C, Zhang L, Sun Y, et al. GDPD5, a target of miR-195-5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed Pharmacother. 2018;101:945–52. doi: 10.1016/j.biopha.2018.03.028. [DOI] [PubMed] [Google Scholar]
- Granados-Romero J, Valderrama-Treviño A, Contreras Flores E, et al. Colorectal cancer: a review. Int J Res Med Sci. 2017;5:4667. [Google Scholar]
- Grillo TG, Quaglio AEV, Beraldo RF, et al. MicroRNA expression in inflammatory bowel disease-associated colorectal cancer. World J Gastrointest Oncol. 2021;13:995–1016. doi: 10.4251/wjgo.v13.i9.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnatyszyn A, Hryhorowicz S, Kaczmarek-Ryś M, et al. Colorectal carcinoma in the course of inflammatory bowel diseases. Hered Cancer Clin Pract. 2019a;17:18. doi: 10.1186/s13053-019-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnatyszyn A, Hryhorowicz S, Kaczmarek-Ryś M, et al. Colorectal carcinoma in the course of inflammatory bowel diseases. Hered Cancer Clin Pract. 2019b;17:1–9. doi: 10.1186/s13053-019-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang M, Hu H, et al. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol. 2018;117:445–53. doi: 10.1016/j.ijbiomac.2018.05.151. [DOI] [PubMed] [Google Scholar]
- Johnson CM, Wei C, Ensor JE, et al. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control. 2013;24:1207–22. doi: 10.1007/s10552-013-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larabi A, Salesse L, Cordonnier C, et al. Differential miRNA-Gene Expression in M Cells in Response to Crohn’s Disease-Associated AIEC. Microorganisms. 2020;8:1205. doi: 10.3390/microorganisms8081205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DF, Chang X, Zhao JL, et al. Colonic Epithelial PHLPP2 Deficiency Promotes Colonic Epithelial Pyroptosis by Activating the NF-κB Signaling Pathway. Oxid Med Cell Longev. 2021;2021:5570731. doi: 10.1155/2021/5570731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Han T, Wang X, Wang Y, Yang Q. Identification of novel survival-related lncRNA-miRNA-mRNA competing endogenous RNA network associated with immune infiltration in colorectal cancer. Am J Trans Res. 2021;13:5815. [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu W, Hui W, et al. Integrated Analysis of Ulcerative Colitis Revealed an Association between <i>PHLPP2</i> and Immune Infiltration. Dis Markers. 2022;2022:4983471. doi: 10.1155/2022/4983471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–40. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- Liu Z, Lu T, Li J, et al. Clinical Significance and Inflammatory Landscape of aNovel Recurrence-Associated Immune Signature in Stage II/III Colorectal Cancer. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.702594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucafò M, Curci D, Franzin M, Decorti G, Stocco G. Inflammatory Bowel Disease and Risk of Colorectal Cancer: An Overview From Pathophysiology to Pharmacological Prevention. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.772101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi R, Saidijam M, Nikzad S, et al. Human exposure to low dose ionizing radiation affects miR-21 and miR-625 expression levels. Mol Biol Rep. 2022;49:1321–7. doi: 10.1007/s11033-021-06960-3. [DOI] [PubMed] [Google Scholar]
- Ritchie W, Rajasekhar M, Flamant S, Rasko J. Conserved Expression Patterns Predict microRNA Targets. PLoS Comput Biol. 2009;5:e1000513. doi: 10.1371/journal.pcbi.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadi P, Soleimani M, Nouri F, et al. An integrative transcriptome analysis reveals potential predictive, prognostic biomarkers and therapeutic targets in colorectal cancer. BMC Cancer. 2022;22:1–22. doi: 10.1186/s12885-022-09931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalavino V, Piccinno E, Lacalamita A, et al. miR-195-5p Regulates Tight Junctions Expression via Claudin-2 Downregulation in Ulcerative Colitis. Biomedicines. 2022;10:919. doi: 10.3390/biomedicines10040919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schee K, Fodstad Ø, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. Am J Pathol. 2010;177:1592–9. doi: 10.2353/ajpath.2010.100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng KL, Kang L, Pridham KJ, et al. An integrated approach to biomarker discovery reveals gene signatures highly predictive of cancer progression. Sci Rep. 2020;10:21246. doi: 10.1038/s41598-020-78126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucchi A, Reed K, O’Brien M, et al. A new transcription factor that regulates TNF-α gene expression, LITAF, is increased in intestinal tissues from patients with CD and UC. Inflamm Bowel Dis. 2006;12:581–7. doi: 10.1097/01.MIB.0000225338.14356.d5. [DOI] [PubMed] [Google Scholar]
- Tian Y, Cui L, Lin C, et al. LncRNA CDKN2B-AS1 relieved inflammation of ulcerative colitis via sponging miR-16 and miR-195. Int Immunopharmacol. 2020;88:106970. doi: 10.1016/j.intimp.2020.106970. [DOI] [PubMed] [Google Scholar]
- Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn’s disease. Annu Rev Genomics Hum Genet. 2009;10:89–116. doi: 10.1146/annurev-genom-082908-150013. [DOI] [PubMed] [Google Scholar]
- Vera J, Lai X, Schmitz U, Wolkenhauer O. MicroRNA-regulated networks: the perfect storm for classical molecular biology, the ideal scenario for systems biology. MicroRNA Cancer Regulation. 2013;2013:55–76. doi: 10.1007/978-94-007-5590-1_4. [DOI] [PubMed] [Google Scholar]
- Yongfu X, He Z, Xujian H, et al. PLHPP2 inhibits the stemness of colorectal cancer by inactivating the Nrf2 signaling pathway. J Cancer. 2022;13:1313–23. doi: 10.7150/jca.65444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu J, Jiang T, et al. MicroRNA-195 suppresses colorectal cancer cells proliferation via targeting FGF2 and regulating Wnt/β-catenin pathway. Am J Cancer Res. 2016;6:2631. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yu J, Liu H, et al. Novel epigenetic CREB-miR-630 signaling axis regulates radiosensitivity in colorectal cancer. PLoS One. 2015;10:e0133870. doi: 10.1371/journal.pone.0133870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Chen J, Zhou Z, He Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. Oncotargets Ther. 2017;10:1027. doi: 10.2147/OTT.S125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Huang J, Yu X, et al. LITAF is a potential tumor suppressor in pancreatic cancer. Oncotarget. 2017;9:3131–42. doi: 10.18632/oncotarget.23220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo P, Lv N, Huang D. Lipopolysaccharide-induced tumor necrosis factor-α factor enhances inflammation and is associated with cancer. Mol Med Rep. 2015;12:6399–404. doi: 10.3892/mmr.2015.4243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the [https://www.ncbi.nlm.nih.gov/geo/database]. The original contributions presented in the study are included in the article/Supplementary Material, further inquiries are available from the corresponding author on reasonable request.