Abstract
Objective:
Patients with triple-negative breast cancer (TNBC) frequently develop resistance to chemotherapy. Studies have shown that microRNAs (miRNAs) are often aberrantly expressed in TNBC and are associated with drug resistance. However, a prognostic strategy that correlates miRNAs with chemotherapy resistance remains largely unknown.
Methods:
To identify breast cancer chemoresistance-associated miRNAs, the miRNA microarray dataset GSE71142 was downloaded from the Gene Expression Omnibus database. Differentially expressed miRNAs (DE-miRNAs) in chemoresistant groups were identified using the LIMMA package in R. Potential target genes were predicted using the miRTarBase 9. Functional and pathway enrichment analyses was done using WebGestalt. A protein-protein interaction network was visualized using Cytoscape software. The top six hub genes regulated by DE-miRNAs were identified using the random forest model. The chemotherapy resistance index (CRI) in TNBC was defined as sum of the median expression levels of the top six hub genes. The association of CRI with distant relapse risk was evaluated using point-biserial correlation coefficient in the validation cohorts of patients with TNBC. The correlation between CRI and cumulative hazard rate was estimated using the Cox model, and the predicted rate of distant relapse was obtained from the Breslow-type estimator of the survival function. All statistical computations were performed using Origin2019b.
Results:
A total of 12 DE-miRNAs were screened, including six upregulated and six downregulated miRNAs in chemoresistant breast cancer tissues compared with chemosensitive tissues. Based on fold changes, miR-214-3p, miR-4758-3p, miR-200c-3p, miR-4254, miR-140-3p, and miR-24-3p were the top six most upregulated miRNAs, whereas miR-142-5p, miR-146-5p, miR-1268b, miR-1275, miR-4447, and miR-4472 were the top six most downregulated miRNAs. The top three hub genes for upregulated miRNAs were RAC1, MYC, and CCND1 and for downregulated miRNAs were IL-6, SOCS1, and PDGFRA. CRI was significantly associated with the risk of distant relapse.
Conclusion:
CRI predicted survival benefits with reduced hazard rate.
Key Words: Triple-Negative Breast Cancer, Chemotherapy resistance index, miRNAs, 5-Fluorouracil
Introduction
The absence of estrogen receptor, progesterone receptor, and human epidermal growth receptor 2 (HER2) is characteristic of TNBC. TNBC patients do not benefit from hormonal or trastuzumab-based therapies because of the loss of these receptors. Approximately 70-80% of patients with TNBC show chemosensitive behavior, with nearly 25-40% showing a pathologic complete response(Perez et al., 2010; Silver et al., 2010; Holanek et al., 2021; Li et al., 2022). However, ~30-50% of TNBC patients have residual disease burden (RCB) II/III after neoadjuvant chemotherapy and are at a high risk of relapse, with significantly worse survival, particularly in the first three years(Hamy et al., 2020).
The majority of TNBC patients are diagnosed late, with about 70% having advanced stages, resulting in a disproportionate number of breast cancer deaths compared to non-TNBC patients(Pratik K Jha and 2020). Despite their diagnosis at later stages, all TNBC patients (irrespective of their response to chemotherapy) were initially treated with standard four cycles of anthracycline/taxane-based chemotherapy, preferably within the neoadjuvant chemotherapy treatment (NACT) setting(Furlanetto and Loibl, 2020). Needless to say, it will be too late for the clinician to know about the non-responsive behavior of the tumor towards chemotherapy. However, this issue must be addressed. Therefore, there is a need to develop an assay that can predict the non-responsive behavior of TNBC tumors before or during the initial phases of chemotherapy.
MiRNAs regulate TNBC progression by targeting several genes. These regulatory mechanisms have been extended to include circulating miRNAs. MiRNAs are small non-coding RNAs that negatively regulate gene expression at the post-transcriptional level. The expression of several miRNAs, whether upregulated or downregulated, is associated with chemoresistance or poor survival in patients (Liang et al., 2016; Li et al., 2017; Piasecka et al., 2018; Song et al., 2018; Bao et al., 2019; Han et al., 2019; Liu et al., 2019; Sugita et al., 2019; Tormo et al., 2019). However, few studies have correlated the expression of miRNAs with the clinicopathological grade of patients with TNBC, with some limitations. For example, a study conducted by Gautam et al. (Bajaj et al., 2020) showed that overexpression of miR-182 and miR-18a correlates with worse clinical and pathological tumor characteristics in locally advanced TNBC, and hence could be used as a prognostic marker in these patients. However, the sensitivity and specificity of the assay was only 60 and 50%, respectively. Similarly, a study by Thakur et al. showed that miR-21 has 95% sensitivity and 81% specificity, and miR 195 has 78% specificity and 65% specificity (Thakur et al., 2016). These results suggest that studies conducted to correlate miRNAs with TNBC grade or chemoresistance should include a panel of miRNAs to enhance the accuracy of the assay. Furthermore, measurement of the expression levels of a panel of miRNAs in a patient’s tumor sample could overall predict the intrinsic sensitivity of TNBC to a particular type of chemotherapy. Therefore, a strategy that includes a panel of miRNAs for prognosing chemotherapy resistance needs to be developed.
In this study, DE-miRNAs in chemoresistant and chemosensitive breast cancer tissues were screened using the miRNA expression profile of the breast cancer chemoresistance-associated miRNA microarray dataset, GSE71142. The genes targeted by DE-miRNAs were predicted, and their potential functions were analyzed using functional and pathway enrichment analyses. Furthermore, a protein-protein interaction (PPI) network of the predicted target genes was constructed. Next, we derived CRI using the target genes of the top six miRNAs in a large representative breast tumor cohort and tested the relationship between CRI and hazard rate. Next, the relationship between CRI and hazard rate was validated in the validation cohort. Through this comprehensive bioinformatic analysis, the present study aimed to explore the significance of CRI in identifying TNBC chemoresistance.
Materials and Methods
Patients and samples
Two datasets of TNBC patients (discovery and validation) who had undergone microarray analysis of their primary tumors were used in this study. The Discovery cohort (TCGA, Cell 2015) included a total of 55 TNBC patients. The median disease relapse-free survival (DRFS) was 29 months (range:0.33-113.18 months). The median age of the patients was 51 years (range:11.9-82 years). The baseline characteristics of the validation cohorts (TCGA, Firehose, and Legacy) are presented in Table 1.
Table 1.
Population Characteristics of Validation Cohort
| SN | Characteristics | Validation cohort |
|---|---|---|
| 1 | Number of TNBC patients | 55 |
| 2 | Platform | Agilent microarray |
| 3 | Age | |
| <50 | 24 | |
| ≥50 | 31 | |
| Mean | 51 | |
| SD | 11.9 | |
| 4 | T-Stage | |
| 1 | 11 | |
| 2 | 41 | |
| 3 | 2 | |
| 4 | 1 | |
| 5 | AJCC Stage | |
| I | 9 | |
| II | 36 | |
| III | 7 | |
| IV | 1 | |
| 6 | Node | |
| Positive | 36 | |
| Negative | 15 | |
| 7 | Menopausal status | |
| Pre | 21 | |
| Post | 29 | |
| Neither pre nor post | 2 | |
| Not available | 3 |
miRNA Microarray:
Breast cancer chemoresistance-associated miRNA microarray dataset GSE71142 was downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (http://www.ncbi. nlm.nih.gov/geo). The dataset GSE71142, based on the GPL20717 µParaflo ™ miRNA microarray platform (LC Sciences, Houston, TX, USA), included five cases of chemoresistant breast cancer tissues and five cases of chemosensitive tissues. The chemotherapy drugs used to treat patients with breast cancer were adriamycin, taxol, or a combination of two or more drugs (CTF: Cytoxan, pirarubicin, 5-Fluorouracil (5-FU)); CAF: Cytoxan, adriamycin, 5-FU, gemcitabine, and capecitabine; and CEF: Cytoxan, epirubicin, and 5-FU).
Screening for DE-miRNAs
Data were analyzed using linear models for microarray (LIMMA), which uses Bayesian statistics to minimize the type 1 false-positive error in R(Ritchie et al., 2015). To further reduce the possibility of a type 1 error, only genes present in all chemosensitive and chemoresistant tissues were used. miRNAs with fold change (FC) ≥ 2 and p-values ≤ 0.05 were selected for cluster analysis. The mean values of each group were used in cluster analysis using the ComplexHeatmap package in the R programming language.
Prediction of genes targeted by DE-miRNAs
In the present study, mirTarBase 9 was used to predict the target genes of the DE-miRNAs. In general, miRTarBase deals with experimentally validated targets using high-throughput approaches, such as reporter assays, western blots, and quantitative polymerase chain reaction (qPCR) (Chou et al., 2018). Only miRNAs with evidence in strong validation methods were chosen.
GO and pathway analysis
The WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) was used to perform functional enrichment analyses, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, to identify potential target genes of DE-miRNAs(Wang et al., 2017). Statistical significance was set at P < 0.05. The degree of connectivity in the networks was analyzed using high confidence (value=0.9) in Cytoscape software (version 3.9.1) to obtain significant nodes or hub proteins in the PPI networks(Shannon et al., 2003). The top ten hub genes were selected in each group (upregulated and downregulated) for further analysis (total hub genes =20).
Definition of Chemotherapy Resistance Index
The top genes out of selected 20 hub genes were identified using a random forest model(Yao et al., 2019). Random Forest in R programming builds and combines multiple decision trees to obtain more accurate predictions. It is a nonlinear classification algorithm. Genes were selected based on their importance level (identified by the random forest algorithm) and least variability in the z-value in both the discovery and validation cohorts. We defined a Chemotherapy Index (CI) based on the median expression levels of the genes regulated by the top six upregulated miRNAs (UmR) and the top six downregulated miRNAs (DmR). The median expression of these genes was further normalized using the Box-Cox power transformation to normalize their distribution. CI was calculated as CI = UmR + DmR. The CRI was calculated as CRI= CI + B, where B is a constant and the number determined to produce positive values for the CI.
Molecular docking
The RNAComposer system was used for the fully automated prediction of miRNA three-dimensional (3-D) structures. RNAComposer is a knowledge-based method for fully automated RNA 3D structure prediction from user-defined secondary structures (Biesiada et al., 2016). The secondary structure of miRNAs was determined using the mFold software (Zuker, 2003). The structures of the miRNAs were prepared and refined using AutoDock Vina (Trott and Olson, 2010). Charges were assigned, hydrogen atoms were added to the heavy atoms, and all the water molecules were deleted. For docking studies, 5-FU was retrieved from the PubChem database (compound identifier (CID):16213520). Molecular docking was performed using the PatchDock server (Schneidman-Duhovny et al., 2005).
Statistical Analysis
The correlation between CRI and cumulative hazard rate was estimated using the Cox model, and the predicted rate of distant relapse was obtained from the Breslow-type estimator of the survival function (OriginLab version 2019). CRI values were classified as low or high based on the cut-off values determined from the first discovery cohort using the Median ± 2SEM. The same thresholds were used for subsequent validation analyses and their correlations with hazard rate. All statistical computations were performed using OriginLab, version 2019.
Results
Identification of differentially expressed-miRNAs and their target genes
Based on FC (≥ 2) and p-values (≤ 0.05), a total of 12 DE-miRNAs were screened. Of these 12 DE-miRNAs, six were upregulated (miR-214-3p, miR-4758-3p, miR-200c-3p, miR-4254, miR-140-3p, and miR-24-3p) and six (miR-142-5p, miR-146-5p, miR-1268b, miR-1275, miR-4447, and miR-4472) were downregulated in chemoresistant breast cancer tissues compared to chemosensitive tissues (Figure 1). These miRNAs were used for further analyses. miRTarBase analysis generated 91 potential target genes, including 77 upregulated and 14 down-regulated miRNAs.
Figure 1.
Differentially Expressed miRNAs (DE-miRNAs) in Five Cases of Drug-Resistant (DR) Breast Cancer Tissues and Five Cases of Drug-Sensitive (DS) Tissues. Data are presented as a heat map. FC, fold change; CR, Chemoresistant; CS, Chemosensitive
Functional and pathway enrichment analyses
GO functional and KEGG pathway enrichment analyses were performed for the identified 91 potential target genes. The enriched GO functions for the target genes are presented in Tables 2A and 2B, including the positive regulation of cellular component movement, regulation of cell death, regulation of intracellular signal transduction, negative regulation of apoptotic process, regulation of cell proliferation, and positive regulation of molecular function in the biological process (BP) category; cyclin-dependent protein kinase holoenzyme complex, transcription factor complex, plasma membrane receptor complex and chromosome, telomeric region in the cellular component (CC) category; and cyclin-dependent protein kinase activity, transcription factor binding, proximal promoter sequence-specific DNA binding, Interleukin-6 receptor, platelet-derived growth factor and protein kinase binding, and phosphotransferase activity in the molecular function (MF) category. The enriched KEGG pathways for the target genes of the upregulated miRNAs (Supplementary Figure 1A) included miRNAs in cancer, pathways in cancer, PI3K-Akt signaling pathways, cellular senescence, and proteoglycans in cancer signaling pathways. For downregulated miRNAs, the enriched KEGG pathways included FoxO, Janus kinase/signal transducers and activators of transcription (JAK-STAT), epidermal growth factor receptor (EGFR), mitogen-activated protein kinase (MAPK), and toll-like receptor pathways (Supplementary Figure 1B).
Figure 2.
Corelation of Chemotherapy Resistance Index (CRI) with Hazard Rate for Distant Relapse in TNBC Patients from Validation Cohort. P values are from point-biserial corelation coefficient
Construction and analysis of PPI network and miRNA-target network
Data from the STRING database showed that many target genes interacted with each other. For better visualization, the top ten hub nodes with higher degrees were screened (Table 3). The hub genes for upregulated miRNAs were CTNNB1, CCND1, MYC, RHOA, MAPK8, BRCA1, RAC1, HIF1A, CCNA2, and KRAS. Among these genes, CTNNB1 had the highest node degree (degree=18). The hub genes for the downregulated miRNAs were IL-6, TRAF6, IRAK1, PDGFRA, SOCS1, ITGAV, TGFBR2, PTEN, SMAD3, and A2M. Among these genes, IL-6 showed the highest node degree (degree=06). A miRNA-hub gene network was constructed as shown in supplementary Figure 2A-B. The hub target genes of the upregulated miRNAs were potentially regulated by miR-24-3p, miR-200c-3p, and miR-214-3p. In particular, miR-24-3p and miR-200c-3p were predicted to target four hub genes (miR-24-3p: CCND1, MYC, BRCA1, CCNA2; miR-200c-3p: RHOA, RAC1, HIF-1A, and KRAS). The hub target genes of downregulated miRNAs were potentially regulated by miR-146b-5p (A2M, IL-6, IRAK1, PDGFRA, and TRAF6) and miR-142-5p (ITGAV, SMAD3, PTEN, TGFBR2, and SOCS1) (Table 3).
Table 3A.
Enriched Functions of the Target Genes of the Upregulated miRNAs
| GO terms: Upregulated genes | P-Value | FDR* | ER* | Genes overlap |
|---|---|---|---|---|
| Biological process (BP) | ||||
| GO:0051270: Regulation of cellular component movement | <2.2e-16 | <2.2e-16 | 6.44 | 35 |
| GO:2000145: Regulation of cell motility | 2.20E-16 | <2.2e-16 | 6.43 | 32 |
| GO:0040012: Regulation of locomotion | <2.2e-16 | <2.2e-16 | 5.4 | 34 |
| GO:0033993: Response to lipid | <2.2e-6 | <2.2e-16 | 5.17 | 32 |
| GO:0009628: Response to abiotic stimulus | <2.2e-6 | <2.2e-6 | 6.4 | 35 |
| GO:0010941: Regulation of cell death | <2.2e-16 | <2.2e-16 | 4.63 | 44 |
| GO:0051240: Positive regulation of multicellular organismal process | <2.2e-16 | <2.2e-16 | 4.38 | 42 |
| GO:1902531: Regulation of intracellular signal transduction | <2.2e-16 | <2.2e-16 | 4.37 | 46 |
| GO:0044093: Positive regulation of molecular function | <2.2e-16 | <2.2e-16 | 4.34 | 43 |
| GO:0006915: Apoptotic process | <2.2e-16 | <2.2e-16 | 4.26 | 47 |
| Cellular Component (CC) | ||||
| GO:0000307: Cyclin-dependent protein kinase holoenzyme complex | 3.36E-08 | 7.8974E-06 | 31.397 | 6 |
| GO:1902554: Serine/threonine protein kinase complex | 1.47E-07 | 0.000024689 | 17.463 | 7 |
| GO:1902911: Protein kinase complex | 5.80E-07 | 0.000072327 | 14.303 | 7 |
| GO:0000781: Chromosome, telomeric region | 3.36E-07 | 0.000049345 | 12.003 | 12 |
| GO:0005667: Transcription factor complex | 8.82E-08 | 0.000017281 | 7.2524 | 16 |
| GO:0044454: Nuclear chromosome part | 2.80E-09 | 0.000001096 | 6.4164 | 16 |
| GO:0000228: Nuclear chromosome | 9.37E-10 | 5.51E-07 | 6.3653 | 17 |
| GO:0000785: Chromatin | 6.16E-07 | 0.000072327 | 5.4796 | 13 |
| GO:0044427: Chromosomal part | 4.32E-10 | 5.08E-07 | 5.0852 | 21 |
| GO:0005694: Chromosome | 4.86E-09 | 1.4285E-06 | 4.4433 | 21 |
| Molecular Function (MF) | ||||
| GO:0097472: Cyclin-dependent protein kinase activity | 1.40E-09 | 4.69E-07 | 33.194 | 7 |
| GO:0004674: Protein serine/threonine kinase activity | 3.47E-08 | 7.2425E-06 | 5.8614 | 15 |
| GO:0004672: Protein kinase activity | 7.41E-11 | 6.95E-08 | 5.6772 | 21 |
| GO:0008134: Transcription factor binding | 3.94E-10 | 1.85E-07 | 5.5001 | 20 |
| GO:0019900: Kinase binding | 5.71E-11 | 6.95E-08 | 5.4289 | 22 |
| GO:0000987: Proximal promoter sequence-specific DNA binding | 3.93E-08 | 7.3845E-06 | 5.3676 | 16 |
| GO:0019901: Protein kinase binding | 2.25E-09 | 6.04E-07 | 5.283 | 19 |
| GO:0016773: Phosphotransferase activity, alcohol group as acceptor | 3.59E-10 | 1.85E-07 | 4.9297 | 22 |
| GO:0016301: Kinase activity | 1.50E-09 | 4.69E-07 | 4.568 | 22 |
| GO:0016772: Transferase activity, transferring phosphorus-containing groups | 5.53E-09 | 1.2984E-06 | 4.0639 | 23 |
*FDR, False discovery rate; ER, Enrichment ratio
Identification of top hub genes regulated by DE-miRNA
Using the random forest model, we identified the best model based on the tumor stage for 20 hub genes. Furthermore, these genes were screened based on their least variability of z-values in the discovery and validation cohorts. Genes with high importance in the random forest model and the least variability in the z-values were selected. This strategy led to the identification of the top three genes (RAC1, MYC, and CCND1) for upregulated miRNAs and the top three genes (IL-6, SOCS1, and PDGFRA) for downregulated miRNAs. The z-scores for these genes in both the discovery and validation cohorts showed similar trends. For example, in the discovery cohort, the z-scores for RAC1, MYC, CCND1, IL-6, SOCS1 and PDGFRA were − -0.16, 0.69, -0.89, 0.48, 0.46, and 0.18, respectively. In the validation cohort, the z-scores for these genes were -0.14, 0.75, -0.72, 0.37, 0.76, and 0.23, respectively. These data suggest that for upregulated miRNAs the expression of RAC1 and CCND1 is below the mean value and the expression of MYC is above the mean value. Similarly, for downregulated miRNAs, the expression of IL-6, SOCS1 and PDGFRA is above the mean value.
Relationship of CRI with distant relapse hazard rate
In the validation cohort, we observed a significant association between CRI and the risk (hazard rate) for distant relapse or death (P = 0.04, Figure 2), with a point-biserial correlation value of 0.26. However, no significant relationship was observed between box-cox transformed expression values of individual genes (RAC1, MYC, CCND1, IL-6, SOCS1, or PDGFRA) and hazard rate (Figure 3A-3F).
Figure 3.
Corelation of Median Expression Level of A) MYC, B) RAC1, C) CCND1, D) IL-6, 5) SOCS1, and 6) PDGFRA genes with hazard rate for distant relapse in TNBC patients from validation cohort. P values are from point-biserial corelation coefficient
Two groups of chemotherapy resistance defined by CRI
Having validated the concept that a higher CRI is associated with a higher risk of distant relapse (hazard rate), we sought to establish clinically useful categories. Two cut-off points (corresponding to index values of 0.83 and 1.34) were chosen to maximize the association of CRI with distant relapse events or death. Patients were divided into high- and low-risk groups based on the cut-off points of the CRI (≥1.3 and ≤0.8). All patients with relapsed events showed a high CRI value (≥1.3), and none of the patients in the low-risk group showed a relapse event (≤0.8). However, no significant association was observed between the risk of distant relapse and CRI (P=0.09; Figure 4A-B). This could be because of the small number of patients with disease recurrence. A high CRI index and node-positive status were independent predictors of the risk of distant relapse in a univariate Cox model (Table 4).
Figure 4.
Kaplan-Meier Estimates of A) relapse-free survival and B) hazard rate in high and low risk groups of TNBC patients from validation cohort. Patients were divided into high (CRI ≥1.3) and low-risk (CRI ≤ 0.8) groups based on the cut-off points of the CRI
Table 4.
Univariate Cox Regression Analysis of Association with Hazard Rate
| Factor | Hazard ratio | p-value |
|---|---|---|
| Age | 1.25 | 0.7500 |
| Node (Positive vs. Negative) | 7.91 | 0.0152 |
| Menopausal status | 1.26 | 0.4861 |
Molecular models of miRNAs involved in chemoresistance
RNAComposer was used to produce a molecular model of the six miRNAs expressed in chemoresistance through in silico projection processing of their molecular structures (Figure 5A-F). Using the SM2miR tool, three and five drugs were identified that upregulated miR-142-5p and miR-146b-5p, respectively. Similarly, 6, 15, 6, and 4 drugs were identified that downregulated the expression of miR-214-3p, miR-24-3p, miR-140-3p and miR200c-3p respectively. Our data suggest that 5-FU is a common drug that can downregulate miR-200c-3p and miR-24-3p and up-regulate the expression of miR-146b-5p (Supplementary Table 1). Therefore, 5-FU can enhance sensitivity to personalized chemotherapy when the CRI of patients is high. Further investigations are necessary to validate this hypothesis.
Figure 5.
Predictive Models of miRNAs by RNAComposer A) miR-24-3p, B) miR-200c-3p, C) miR-140-3p, D) miR-214-3p, E) miR-146b-5p, and F) miR-142-5p
Molecular docking of 5-FU with miRNAs
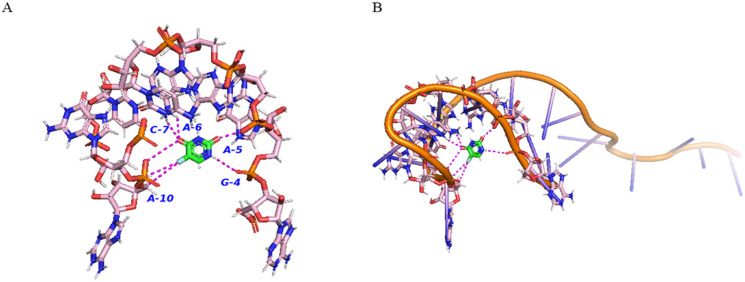
Visualization of 5-FU with miR146-5p showed 7 hydrogen bonds with 5-FU [Adenine10 (A10)-3.5 & 4Aº, Adenine5 (A5)-2.2 Aº, Adenine6 (A6)-2.3 Aº, Guanine4 (G4)-3.1 Aº and Cytosine7 (C7)-3.3 Aº] (Figure 6). However, three and two hydrogen bonds were observed for miR24-3p [Guanine16 (G16)-2 Aº, Guanine 17(G17)-2.9 Aº, and Adenine12 (A12)- 3.2 Aº] and miR200c-3p [Adenine16 (A16)-3.3 Aº and cytosine15 (C15)-2.7 Aº)], respectively (Figure 7-8). These results indicate the strongest interaction between 5-FU and miR146-5p in comparison to the other two miRNAs. These sites represent the active sites of miRNAs with 5-FU. According to the docking analysis, the atomic contact energy (ACE) for miR-146-5p and 5-FU was − -157 Kcal/mol which indicated a strong binding with miR-146-5p. The ACE for miR-24-3p with 5-FU was − -83-Kcal/mol and for miR-200c-3p with 5-FU was −64 kcal/mol. These data further support that the interaction of miR-146-5p is strongest with 5-FU; however, the other two miRNAs also showed strong interactions with 5-FU.
Figure 6.
Molecular Docking of miR-146b-5p with 5-FU A) Interactions between residues of miR-146b-5p with 5-FU (Residues of miR-146b-5p-Pink, 5-FU-Green). Magenta colored lines are showing hydrogen bonding. Amino acids were labelled with a one-letter code. Residues shown in the figures are within the range of 5Aº from the 5-FU. B) Complete three-dimensional (3D) structure of miR-146b-5p showing interactions with 5-FU. Predictive 3D structure of mir-146b-5p shown in orange. Residues within the range of 5Aº from 5-FU are shown in pink and 5-FU is shown in green
Figure 7.
Molecular Docking of miR-24-3p with 5-FU A) Interactions between residues of miR-24-3p with 5-FU (Residues of miR-24-3p-Pink, 5-FU-Green). Magenta coloured lines are showing hydrogen bonding. Amino acids were labelled with a one-letter code. Residues shown in the figures are within the range of 5Aº from the 5-FU. B) Complete 3D of miR-24-3p showing interactions with 5-FU. Predictive 3D structure of mir-24-3p shown in orange. Residues within the range of 5Aº from 5-FU are shown in pink and 5-FU is shown in green
Figure 8.
Molecular Docking of miR-200c-3p with 5-FU A) Interactions between residues of miR-200c-3p with 5-FU (Residues of miR-200c-3p-Pink, 5-FU-Green). Magenta coloured lines are showing hydrogen bonding. Amino acids were labelled with a one-letter code. Residues shown in the figures are within the range of 5Aº from the 5-FU. B) Complete 3D structure of miR-200c-3p showing interactions with 5-FU. Predictive 3D structure of mir-200c-3p shown in orange. Residues within the range of 5Aº from 5-FU are shown in pink and 5-FU is shown in green
Table 2.
Hub Genes Identified in PPI Network
| Gene Symbol | Degree | Gene Symbol |
|---|---|---|
| CTNNB1 | 18 | IL6 |
| CCND1 | 15 | TRAF6 |
| MYC | 15 | IRAK1 |
| RHOA | 15 | PDGFRA |
| MAPK8 | 14 | SOCS1 |
| BRCA1 | 14 | ITGAV |
| RAC1 | 14 | TGFBR2 |
| HIF1A | 13 | PTEN |
| CCNA2 | 12 | SMAD3 |
| KRAS | 10 | A2M |
Table 3B.
Enriched Functions of the Target Genes of the Downregulated miRNAs
| GO terms: Downregulated genes | P-Value | FDR* | ER* | Genes overlap |
|---|---|---|---|---|
| Biological process (BP) | ||||
| GO:0045785: Positive regulation of cell adhesion | 3.11E-09 | 0.000003821 | 18.893 | 8 |
| GO:0030155: Regulation of cell adhesion | 7.40E-09 | 6.7306E-06 | 12.74 | 9 |
| GO:0043066: Negative regulation of apoptotic process | 2.47E-10 | 6.62E-07 | 11.29 | 11 |
| GO:0043069: Negative regulation of programmed cell death | 2.91E-10 | 6.62E-07 | 11.117 | 11 |
| GO:0060548: Negative regulation of cell death | 7.19E-10 | 1.3074E-06 | 10.214 | 11 |
| GO:0042127: Regulation of cell proliferation | 2.29E-10 | 6.62E-07 | 7.6951 | 13 |
| GO:0042981: Regulation of apoptotic process | 3.40E-09 | 0.000003821 | 7.3281 | 12 |
| GO:0043067: Regulation of programmed cell death | 3.78E-09 | 0.000003821 | 7.261 | 12 |
| GO:0008283: Cell proliferation | 2.15E-10 | 6.62E-07 | 6.5261 | 14 |
| GO:0006915: Apoptotic process | 2.80E-09 | 0.000003821 | 6.2978 | 13 |
| Cellular Component (CC) | ||||
| GO:0098802: Plasma membrane receptor complex | 9.09E-07 | 0.00053419 | 26.559 | 5 |
| GO:0005811: Lipid droplet | 0.0027644 | 0.32482 | 25.387 | 2 |
| GO:0043235: Receptor complex | 2.21E-09 | 2.5929E-06 | 19.745 | 8 |
| GO:0009897: External side of plasma membrane | 0.00018196 | 0.032174 | 13.528 | 4 |
| GO:0098552: Side of membrane | 0.00000568 | 0.0022255 | 12.192 | 6 |
| GO:0098797: Plasma membrane protein complex | 0.00019167 | 0.032174 | 8.758 | 5 |
| GO:0009986: Cell surface | 0.00090472 | 0.11812 | 6.2493 | 5 |
| GO:0098796: Membrane protein complex | 0.00073894 | 0.10853 | 5.0773 | 6 |
| GO:0005887: Integral component of plasma membrane | 0.00008592 | 0.02524 | 4.8992 | 8 |
| GO:0031226: Intrinsic component of plasma membrane | 0.00012021 | 0.02825 | 4.6737 | 8 |
| Molecular Function (MF) | ||||
| GO:0005138: Interleukin-6 receptor binding | 0.0000205 | 0.0064129 | 280.13 | 2 |
| GO:0005161: Platelet-derived growth factor receptor binding | 0.00010201 | 0.021274 | 130.73 | 2 |
| Gene set: GO:0031435: Mitogen-activated protein kinase kinase kinase binding | 0.00016573 | 0.031107 | 103.21 | 2 |
| GO:0019838: Growth factor binding | 2.07E-07 | 0.00019393 | 35.524 | 5 |
| GO:0019955: Cytokine binding | 0.00000708 | 0.0030997 | 30.881 | 4 |
| GO:0070851: Growth factor receptor binding | 0.00000825 | 0.0030997 | 29.711 | 4 |
| GO:0005126: Cytokine receptor binding | 0.00000609 | 0.0030997 | 17.892 | 5 |
| GO:0019901: Protein kinase binding | 0.00002488 | 0.006672 | 9.323 | 6 |
| GO:0019900: Kinase binding | 0.00004874 | 0.011436 | 8.274 | 6 |
| GO:0005102: Signaling receptor binding | 2.92E-08 | 0.000054871 | 7.0125 | 11 |
*FDR, False discovery rate; ER, Enrichment ratio
Discussion
The treatment of TNBC remains challenging because of the heterogeneity of the disease, drug resistance, tumor relapse, and lack of single targetable mutations. The effects of miRNAs on multiple targets may improve the response rates in the context of this genetically and biologically heterogeneous disease. In this scenario, miRNAs-based therapeutics offer a very attractive area to treat this difficult to treat cancer. The differential expression of miRNAs based on disease severity is an essential consideration when assessing the utility of miRNA biomarkers for clinical use. In this study, we identified the differential expression of the top 12 miRNAs in chemoresistant TNBC compared to that in chemosensitive TNBC. Among the dysregulated miRNAs, miR-214-3p (upregulated) and miR-142-5p (downregulated) showed the greatest fold-change in expression between chemoresistant and chemosensitive tissues. miR-214-3p promotes carcinogenesis in TNBC, and its inhibition attenuates the migration, invasion, and viability of TNBC cells (Zhang et al., 2019). Similarly, stable re-expression of miR-142-5p robustly and significantly increased the sensitivity of TNBC cells embedded in collagen to doxorubicin (Qattan, 2020). The delivery of this miRNA to drug-resistant TNBC patients in combination with chemotherapy may be a promising therapeutic approach. These two miRNAs (miR-214-3p and miR-142-5p) have not been systemically investigated in breast cancer. This is the first report to link these two miRNAs with TNBC chemotherapy resistance. Other studies that correlate miRNAs with chemoresistance in TNBC have less diagnostic testing accuracy and, thus, have less clinically significant value. These results highlight the importance of including a panel of miRNAs to increase the sensitivity of the diagnostic/prognostic assay.
The miRNAs identified in this study can be used to calculate and validate the CRI if a validation cohort describing detailed patient data, miRNA signatures, and their correlation with chemoresistance is available. However, these data were not available. Therefore, we utilized the target hub genes of miRNAs to calculate CRI, which was used to define the hazard rate or DRFS in TNBC patients. The target genes were trained using a random forest model. This study showed that the gene expression-based random forest classification model accurately predicts and distinguishes patients according to risk. Top six genes identified in this study played an important role in regulation of TNBC (Hartman et al., 2013; Klauber-DeMore et al., 2018; Qian et al., 2018; Ganapathy-Kanniappan, 2020; Zhang et al., 2020).
The CRI and lymph node status were significantly correlated with the hazard rate or DRFS in TNBC. Lymph node status was an independent prognostic factor for hazard rate in this study. Furthermore, an elevated CRI was more strongly associated with an increased hazard rate and reduced survival. This supports our interpretation that CRI is a chemoresistance predictor and predicts a lower response to chemotherapy in TNBC tumors. These data further suggest that patients with node-negative TNBC status and low CRI could reasonably select standard chemotherapy alone, but other patients at high risk might benefit from additional treatment, such as immunotherapy, to have better DFRS. Validation cohort data showed that approximately 33% of patients with clinical stage II or III disease had a high CRI and may not benefit from chemotherapy. Therefore, it is important to consider whether chemotherapy should be encouraged for patients with node-negative and low CRI groups, or whether a predictive test for chemosensitivity could identify node-negative patients with excellent survival from standard chemotherapy. We expect that additional studies that evaluate the predictive performance of the CRI will further highlight its clinical interpretation. The performance of the CRI-based predictive model in both discovery and validation cohorts was highly positive. This suggests that this model has the potential to assist clinicians in making decisions for patients whose tumors are chemoresistant due to the differential expression of the top six genes regulated by miRNAs.
The clinical relevance of the independent prediction of chemoresistance should not dissuade the use of standard chemotherapy as a standard treatment for eligible patients. Rather, CRI results should be better interpreted in the context of nodal status and combined to predict the chemoresistant nature of TNBC. The CRI was calculated based on differential gene expression profiling. In this context, CRI could be considered an important piece of a prognostic puzzle for personalized breast cancer treatment. For example, our findings from the molecular docking studies suggest that 5-FU has a potential chemical affinity and ability to alter the expression of the three miRNAs. These results suggest that 5-FU, in combination with a potent small-molecule miRNA targeting drugs, could be used as a remedial approach for chemoresistant TNBC.
Author Contribution Statement
RG and MS was responsible for the conceptualization of the manuscript and manuscript drafting. ASC, MS, DN, and RG performed all the analyses.
Acknowledgements
Ethics approval
This bioinformatics study was waived from ethics approval.
Availability of data
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
Conflict of interest
The authors declare no conflict of interest.
References
- Bajaj R, Tripathi R, Sridhar TS, et al. Prognostic role of microRNA 182 and microRNA 18a in locally advanced triple negative breast cancer. PLoS One. 2020;15:e0242190. doi: 10.1371/journal.pone.0242190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao C, Lu Y, Chen J, et al. Exploring specific prognostic biomarkers in triple-negative breast cancer. Cell Death Dis. 2019;10:807. doi: 10.1038/s41419-019-2043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiada M, Purzycka KJ, Szachniuk M, et al. Automated RNA 3D Structure Prediction with RNAComposer. Methods Mol Biol. 2016;1490:199–215. doi: 10.1007/978-1-4939-6433-8_13. [DOI] [PubMed] [Google Scholar]
- Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:296–302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlanetto J, Loibl S. Optimal Systemic Treatment for Early Triple-Negative Breast Cancer. Breast Care (Basel) 2020;15:217–26. doi: 10.1159/000508759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S. Rac1 repression reverses chemoresistance by targeting tumor metabolism. Cancer Biol Ther. 2020;21:888–90. doi: 10.1080/15384047.2020.1809923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamy AS, Darrigues L, Laas E, et al. Prognostic value of the Residual Cancer Burden index according to breast cancer subtype: Validation on a cohort of BC patients treated by neoadjuvant chemotherapy. PLoS One. 2020;15:e0234191. doi: 10.1371/journal.pone.0234191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lim W, You D, et al. Chemoresistance in the Human Triple-Negative Breast Cancer Cell Line MDA-MB-231 Induced by Doxorubicin Gradient Is Associated with Epigenetic Alterations in Histone Deacetylase. J Oncol. 2019;2019:1345026. doi: 10.1155/2019/1345026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman ZC, Poage GM, den Hollander P, et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013;73:3470–80. doi: 10.1158/0008-5472.CAN-12-4524-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holanek M, Selingerova I, Bilek O, et al. Neoadjuvant Chemotherapy of Triple-Negative Breast Cancer: Evaluation of Early Clinical Response, Pathological Complete Response Rates, and Addition of Platinum Salts Benefit Based on Real-World Evidence. Cancers (Basel) 2021:13. doi: 10.3390/cancers13071586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauber-DeMore N, Schulte BA, Wang GY. Targeting MYC for triple-negative breast cancer treatment. Oncoscience. 2018;5:120–1. doi: 10.18632/oncoscience.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lai Y, Ma J, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer. 2017;17:745. doi: 10.1186/s12885-017-3674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang H, Merkher Y, et al. Recent advances in therapeutic strategies for triple-negative breast cancer. J Hematol Oncol. 2022;15:121. doi: 10.1186/s13045-022-01341-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Bian X, Shim H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer. Biochem Biophys Res Commun. 2016;477:461–6. doi: 10.1016/j.bbrc.2016.06.076. [DOI] [PubMed] [Google Scholar]
- Liu M, Gong C, Xu R, et al. MicroRNA-5195-3p enhances the chemosensitivity of triple-negative breast cancer to paclitaxel by downregulating EIF4A2. Cell Mol Biol Lett. 2019;24:47. doi: 10.1186/s11658-019-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EA, Patel T, Moreno-Aspitia A. Efficacy of ixabepilone in ER/PR/HER2-negative (triple-negative) breast cancer. Breast Cancer Res Treat. 2010;121:261–71. doi: 10.1007/s10549-010-0824-0. [DOI] [PubMed] [Google Scholar]
- Piasecka D, Braun M, Kordek R, et al. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol. 2018;144:1401–11. doi: 10.1007/s00432-018-2689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratik K Jha MAA, Vivek Srivastava, Mohit Mangla. Triple Negative Breast Cancer: Alarming Burden and Future Challenges in Indian Perspective. J Sci Res. 2020;64:126–30. [Google Scholar]
- Qattan A. Novel miRNA Targets and Therapies in the Triple-Negative Breast Cancer Microenvironment: An Emerging Hope for a Challenging Disease. Int J Mol Sci. 2020:21. doi: 10.3390/ijms21238905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Lv Y, Li P. SOCS1 is associated with clinical progression and acts as an oncogenic role in triple-negative breast cancer. IUBMB Life. 2018;70:320–7. doi: 10.1002/iub.1728. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, et al. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Richardson AL, Eklund AC, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–53. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Li D, Wu T, et al. MicroRNA-301b promotes cell proliferation and apoptosis resistance in triple-negative breast cancer by targeting CYLD. BMB Rep. 2018;51:602–7. doi: 10.5483/BMBRep.2018.51.11.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita BM, Pereira SR, de Almeida RC, et al. Integrated copy number and miRNA expression analysis in triple negative breast cancer of Latin American patients. Oncotarget. 2019;10:6184–203. doi: 10.18632/oncotarget.27250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur S, Grover RK, Gupta S, et al. Identification of Specific miRNA Signature in Paired Sera and Tissue Samples of Indian Women with Triple Negative Breast Cancer. PLoS One. 2016;11:e0158946. doi: 10.1371/journal.pone.0158946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo E, Ballester S, Adam-Artigues A, et al. The miRNA-449 family mediates doxorubicin resistance in triple-negative breast cancer by regulating cell cycle factors. Sci Rep. 2019;9:5316. doi: 10.1038/s41598-019-41472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, et al. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:130–7. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Zhan X, Kwoh CK. An improved random forest-based computational model for predicting novel miRNA-disease associations. BMC Bioinformatics. 2019;20:624. doi: 10.1186/s12859-019-3290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yuan C, Peng J, et al. SHP-2-Mediated Upregulation of ZEB1 Is Important for PDGF-B-Induced Cell Proliferation and Metastatic Phenotype in Triple Negative Breast Cancer. Front Oncol. 2020;10:1230. doi: 10.3389/fonc.2020.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao Z, Li S, et al. Inhibition of miR214 attenuates the migration and invasion of triplenegative breast cancer cells. Mol Med Rep. 2019;19:4035–42. doi: 10.3892/mmr.2019.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.