Abstract
Epigenetic changes such as histone deacetylation and DNA methylation play to regulate gene expression. DNA methylation plays a major role in cancer induction via transcriptional silencing of critical regulators such as tumor suppressor genes (TSGs). One approach to inhibit TSGs inactivation is to use chemical compounds, DNA methyltransferase inhibitors (DNMTIs). Previously, we investigated the effect of 5-aza-2’-deoxycytidine (5 AZA CdR or decitabine) on colon cancer and hepatocellular carcinoma cell lines. The present study aimed to investigate the effect of 5 AZA CdR on extrinsic (DR4, DR5, FAS, FAS-L, and TRAIL genes), intrinsic [pro- (Bax, Bak, and Bim) and anti- (Bcl-2, Bcl-xL, and Mcl-1) apoptotic genes], and JAK/STAT (SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B genes) pathways in neuroblastoma (IMR-32, SK-N-AS, UKF-NB-2, UKF-NB-3, and UKF-NB-4) and glioblastoma (SF-767, SF-763, A-172, U-87 MG, and U-251 MG) cell lines.
Materials and Methods:
The neuroblastoma and glioblastoma cells were cultured and treated with 5 AZA CdR. To determine cell viability, cell apoptosis, and the relative gene expression level, MTT assay, flow cytometry assay, and qRT-PCR were done respectively.
Results:
5 AZA CdR changed the expression level of the genes of the extrinsic, intrinsic, and JAK/STAT pathways by which induced cell apoptosis and inhibited cell growth in neuroblastoma and glioblastoma cell lines.
Conclusion:
5 AZA CdR can play its role through extrinsic, intrinsic, and JAK/STAT pathways to induce cell apoptosis.
Key Words: 5 AZA CdR, tumor suppressor gene, neoplasms
Introduction
Epigenetic changes such as histone deacetylation and DNA methylation play to regulate gene expression. In fact, epigenetic changes are susceptible to change and are excellent candidates to explain how certain factors may increase the risk of tumorigenesis and cancer induction. However, DNA methylation plays a major role in cancer via transcriptional silencing of critical regulators such as tumor suppressor genes (TSGs). Basically, tumorigenesis is directed by changes in two different groups of genes: TSGs that inhibit cell growth and oncogenes that promote this process. Meanwhile, chromatin modifications, such as DNA methylation, affect local chromatin structure without any changes in DNA sequences. The major step in tumorigenesis is gene inactivation by hypermethylation of CpG islands located in the promoter region. In mammals, DNA methylation occurs at the C5 position of cytosine, mostly within CpG dinucleotides (Grønbaek et al., 2007). Specific enzymes such as DNA methyltransferases (DNMTs) play a major role in DNA methylation and cause reduced expression of TSGs, resulting in cancer induction and progression. In mammals, DNA methylation is regulated by DNA methyltransferases (DNMTs), including DNMT1, DNMT3A, and DNMT3B (Zhang et al., 2017) The DNMTs are often overexpressed in various human cancers. DNMTs are important epigenetic targets for cancer treatment since DNA methylation is a reversible process. These enzymes are promising targets for the treatment of various types of cancers (Zhang et al., 2020; Yu et al., 2019). One approach to inhibit TSGs inactivation is to use chemical compounds, DNA methyltransferase inhibitors (DNMTIs), to reverse DNA hypermethylation. These compounds include 5-aza-2’-deoxycytidine (5 AZA CdR or decitabine), 5-aza-cytidine (5-aza-C), 5-fluoro-2’-deoxycytidine (FdCyd), 2-H pyrimidinone-1-β-D(2’-deoxyriboside) (zebularine), and pseudoisocytidine (Gowher et al., 2004). Previously, we investigated the effect of 5 AZA CdR on colon cancer (Sanaei et al., 2021; Sanaei et al., 2020; Sanaei et al., 2020) and hepatocellular carcinoma cell lines (Sanaei et al., 2020; Sanaei et al., 2020). This compound plays its role through multiple mechanisms comprising extrinsic, intrinsic, and JAK/STAT pathways. Recent experimental works have indicated that 5 AZA CdR induces apoptosis by re-activation of extrinsic pathway (FAS-ligand up-regulation) in neoplastic cells (Karlic et al., 2011). Further, this agent induces apoptosis via the intrinsic (mitochondrial) pathway (Mcl-1 cleavage; Bax, Puma, and Noxa up-regulation) (Kiziltepe et al., 2007). It has been reported that 5 AZA CdR suppresses cancer cell growth through JAK/STAT pathway, regulation of downstream targets of JAK2/STAT3/STAT5 signaling including Bcl-2, p16ink4a, p21waf1/cip1, and p27kip1 (Sanaei et al., 2021). The present study aimed to investigate the effect of 5 AZA CdR on extrinsic (DR4, DR5, FAS, FAS-L, and TRAIL genes), intrinsic [pro- (Bax, Bak, and Bim) and anti- (Bcl-2, Bcl-xL, and Mcl-1) apoptotic genes], and JAK/STAT (SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B genes) pathways in neuroblastoma (IMR-32, SK-N-AS, UKF-NB-2, UKF-NB-3, and UKF-NB-4) and glioblastoma (SF-767, SF-763, A-172, U-87 MG, and U-251 MG) cell lines.
Materials and Methods
Materials
Human neuroblastoma (IMR-32, SK-N-AS, UKF-NB-2, UKF-NB-3, and UKF-NB-4) and glioblastoma (SF-767, SF-763, A-172, U-87 MG, and U-251 MG) cell lines were purchased from the National Cell Bank of Iran-Pasteur Institute. 5 AZA CdR and Dulbecco’s modified Eagle’s medium (DMEM) were obtained from Sigma (St. Louis, MO, USA). The 5 AZA CdR was dissolved in dimethyl sulfoxide (DMSO) to make a work-stock solution. Further concentrations of 5 AZA CdR were obtained by diluting the provided stock solution. Other necessary materials and kits were purchased as provided for our previous works (Sanaei. et al., 2020; Sanaei et al., 2018). The cells were maintained in DMEM supplemented with fetal bovine serum of 10% and antibiotics in a humidified atmosphere of 5% CO2 in air at 37oC. This work was approved by the Ethics Committee of Jahrom University of Medical science with a code number of IR.JUMS.REC.1399.078.
Cell culture and cell viability
All cell lines were cultured in DMEM supplemented with 10% FBS and antibiotics at 37°C in 5% CO2 for 24 h. Subsequently, all cell lines were seeded into 96-well plates (3× 105 cells per well). After 24h, the culture medium was replaced with an experimental medium containing various concentrations of 5 AZA CdR. The human neuroblastoma and the glioblastoma cell lines were treated with 5 AZA CdR (0, 1, 2.5, 5, 7.5, 10, 15, and 20 μM) for 24h, the control cells were treated with the same amount of solvents, DMSO. After 24 h of treatment, all treated and untreated cells were investigated by MTT assay according to Standard protocols to determine cell viability, the MTT solution was added to each well for 4 h at 37oC and then it was changed by DMSO for 10 min to dissolve all of the crystals. Finally, the optical density was detected by a microplate reader at a wavelength of 570 nM. Each experiment was repeated three times (triplicates).
Cell apoptosis assay
To determine cell apoptosis, all cell lines were cultured at a density of 3 × 105 cells/well and incubated overnight. Then All of the cell lines were treated with 5 AZA CdR, based on IC50 values indicated in table 1, for 24h, the control cells were treated with the same amount of solvent, DMSO. Subsequently, the cells were harvested by trypsinization, washed with cold PBS, and resuspended in a Binding buffer (1x). Finally, Annexin-V-(FITC) and propidium iodide (PI) were used according to the protocol to determine the apoptotic cells by FACScan flow cytometry (Becton Dickinson, Heidelberg, Germany).
Table 1.
IC50 Values
| Cell line | Drug | Duration/Hour | IC50 | LogIC50 | R squared |
|---|---|---|---|---|---|
| Neuroblastoma IMR-32 | 5AZACdR | 24 | 2.178 | 0.3381 | 0.9358 |
| Neuroblastoma SK-N-AS | 5AZACdR | 24 | 3.154 | 0.4989 | 0.982 |
| Neuroblastoma UKF-NB-2 | 5AZACdR | 24 | 4.003 | 0.6024 | 0.9871 |
| Neuroblastoma UKF-NB-3 | 5AZACdR | 24 | 4.15 | 0.618 | 0.9218 |
| Neuroblastoma UKF-NB-4 | 5AZACdR | 24 | 5.259 | 0.7209 | 0.9723 |
| Glioblastoma SF-767 | 5AZACdR | 24 | 6.929 5AZACdR | 0.8407 | 0.9561 |
| Glioblastoma SF-763 | 5AZACdR | 24 | 5.306 | 0.7248 | 0.971 |
| Glioblastoma A-172 | 5AZACdR | 24 | 6.812 | 0.8333 | 0.9655 |
| Glioblastoma U-87 MG | 5AZACdR | 24 | 6.37 | 0.8041 | 0.9458 |
| Glioblastoma U-251 MG | 5AZACdR | 24 | 5.609 | 0.7489 | 0.9698 |
Real-time Quantitative Reverse Transcription Polymerase
Chain Reaction (qRT-PCR)
The qRT-PCR was done to determine the relative expression level of the extrinsic (DR4, DR5, FAS, FAS-L, and TRAIL), intrinsic [pro- (Bax, Bak, and Bim) and anti- (Bcl-2, Bcl-xL, and Mcl-1)], and JAK/STAT (SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B genes) genes expression. The cell lines were cultured at a density of 3 × 105 cells/well and treated with 5 AZA CdR, based on IC50 values indicated in table 1, for 24 h, except control groups which were treated with DMSO only. Then qRT-PCR was done as our previous works (Kavoosi et al., 2018; Sanaei et al., 2019). The primer sequences are addressed and shown in table 2.
Table 2.
The Primer Sequences of DR4, DR5, FAS, FAS-L, TRAIL, Bax, Bak, Bim, Bcl-xL, Mcl-1, SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B Genes were Used in the Current Study
| Primer | Primer sequences (5' to 3') | Product length | Reference |
|---|---|---|---|
| DR4 | 299 bp | Nakamoto et al., 2006 | |
| Forward | CAGAACATCCTGGAGCCTGTAAC | ||
| Reverse | ATGTCCATTGCCTGATTCTTTGTG | ||
| DR5 | 389 bp | Nakamoto et al., 2006 | |
| Forward | TGCAGCCGTAGTCTTGATTG | ||
| Reverse | GCACCAAG TCTGCAAAGTCA | ||
| FAS | Tao et al., 2012 | ||
| Forward | TTCTGCCATAAGCCCTGTCC | 103 bp | |
| Reverse | TGTACTCCTTCCCTTCTTGG | ||
| FAS-L | Tao et al., 2012 | ||
| Forward | GCCTGTGTCTCCTTGTGATG | 222 bp | |
| Reverse | TGGACTTGCCTGTTAAATGGG | ||
| TRAIL | 103 bp | Inoue et al., 213 | |
| Forward | GAAGCAACACATTGTCTTCTCCAA | ||
| Reverse | TTGCTCAGGAATGAATGCCC | ||
| Bax | 77 bp | Cao et al., 2002 | |
| Forward | AGTAACATGGAGCTGCAGAGGAT | ||
| Reverse | GCTGCCACTCGGAAAAAGAC | ||
| Bak | 82 bp | Ierano et al, 2013 | |
| Forward | CCTGCCCTCTGCTTCTGA | ||
| Reverse | CTGCTGATGGCGGTAAAAA | ||
| Bim | 101 bp | Zhang et al., 2017 | |
| Forward | ATTACCAAGCAGCCGAAGAC | ||
| Reverse | TCCGCAAAGAACCTGTCAAT | ||
| Bcl-2 | 147 bp | Xu et al., 2012 | |
| Forward | TGGCCAGGGTCAGAGTTAAA | ||
| Reverse | TGGCCTCTCTTGCGGAGTA | ||
| Bcl-xL | 62 bp | Zhang et al., 2008 | |
| Forward | TCCTTGTCTACGCTTTCCACG | ||
| Reverse | GGTCGCATTGTGGCCTTT | ||
| Mcl-1 | 198 bp | Wang et al., 2014 | |
| Forward | AAAGCCTGTCTGCCAAAT | ||
| Reverse | CCTATAAACCCACCACTC | ||
| SOCS1 | 119 bp | Masood et al., 2013 | |
| Forward | TTTTTCGCCCTTAGCGTGA | ||
| Reverse | AGCAGCTCGAAGAGGCAGTC | ||
| SOCS3 | 109 bp | Leon et al., 2009 | |
| Forward | GGCCACTCTTCAGCATCTC | ||
| Reverse | ATCGTACTGGTCCAGGAACTC | ||
| JAK1 | Chen et al., 2019 | ||
| Forward | CCACTACCGGATGAGGTTCTA | 213 | |
| Reverse | GGGTCTCGAATAGGAGCCAG | ||
| JAK2 | Xiong et al., 2009 | ||
| Forward | GATGAGAATAGCCAAAGAAAACG | 160 | |
| Reverse | TTGCTGAATAAATCTGCGAAAT | ||
| STAT3 | Xiong et al., 2009 | ||
| Forward | GCTTTTGTCAGCGATGGAGT | 174 | |
| Reverse | ATTTGTTGACGGGTCTGAAGTT | ||
| STAT5A | Xiong et al., 2009 | ||
| Forward | AATGAGAACACCCGCAACG | 101 | |
| Reverse | TTCCTGAAGTGGGCACTGAG | ||
| Primer | Primer sequences (5' to 3') | Product length | |
| STAT5B | |||
| Forward | ACTGCTAAAGCTGTTGATGGATAC | 174 | |
| Reverse | TGAGTCAGGGTTCTGTGGGTA | ||
| GAPDH | |||
| Forward | TGTGGGCATCAATGGATTTGG | 116 | |
| Reverse | ACACCATGTATTCCGGGTCAAT |
Statistical analysis
The database was set up with the SPSS 16.0 software package (SPSS Inc., Chicago, Illinois, USA) and Graph Pad Prism 8.0 for data analysis. Results are expressed as mean ± standard deviation (SD) for n=3 independent experiments. Statistical comparisons between groups were performed with ANOVA (oneway ANOVA). A significant difference was considered as P < 0.05.
Results
Result of cell viability by the MTT assay
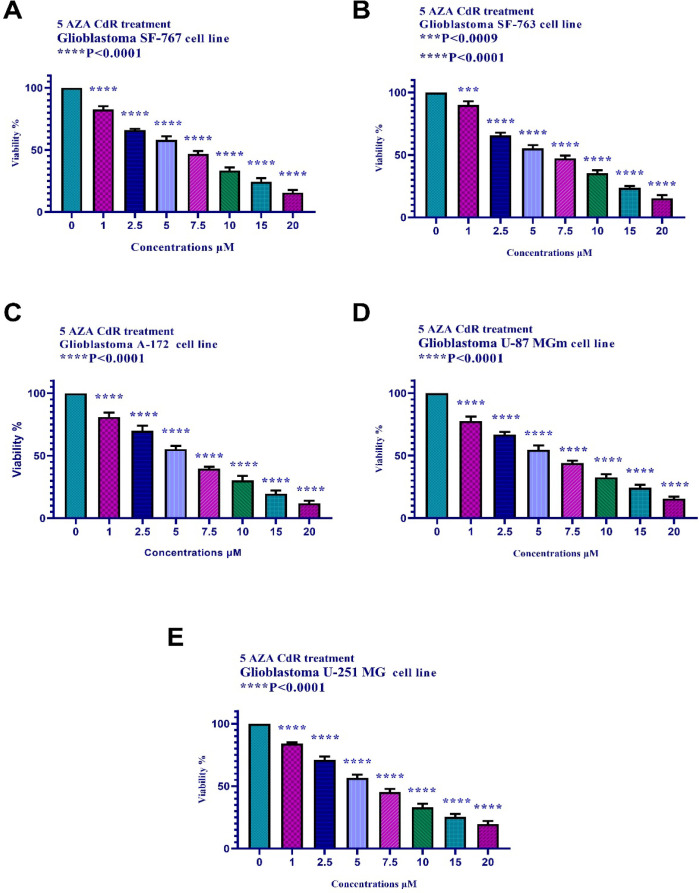
The cell viability of the neuroblastoma and glioblastoma cell lines treated with 5 AZA CdR (0, 1, 2.5, 5, 7.5, 10, 15, and 20 μM) for 24h was investigated by MTT assay thereby the activities of cellular enzymes produced a dark-blue formazan crystal by the tetrazolium salt MTT reduction. To determine the viable neuroblastoma and glioblastoma cells, the crystals were dissolvable in DMSO. As indicated in Figures 1 and 2, 5 AZA CdR induced significant cell growth inhibition in all treated groups in a dose-dependent manner.
Figure 1.
The Effect of 5AZACdR on the Viability of Neuroblastoma Cell Lines. The cells were treated without and with different doses of 5AZACdR for 24 and the cell viability was evaluated by MTT assay. Each experiment was achieved in triplicate. Mean values from the three experiments ± standard error of mean are indicated. Asterisks indicate significant differences between treated and untreated cells. **P <0.0015, ***P <0.0007, ***P <0.0004, and ****P< 0.0001
Figure 2.
The Effect of 5 AZA CdR on the Viability of Glioblastoma Cell Lines. The cells were treated without and with different doses of 5 AZA CdR for 24 and the cell viability was evaluated by MTT assay. Each experiment was achieved in triplicate. Mean values from the three experiments ± standard error of mean are indicated. Asterisks indicate significant differences between treated and untreated cells. ***P <0.0009, and ****P< 0.0001
Figure 5.
Part A. The apoptotic effect of 5AZACdR on glioblastoma cell lines versus control groups after 24h of treatment. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean. The results of the statistical analysis indicate significant differences between treated and untreated cells. *P< 0.0001. Part B. The apoptotic effect of 5AZACdR on neuroblastoma cell lines versus control groups after 24h of treatment. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean. The results of the statistical analysis indicate significant differences between treated and untreated cells. *P< 0.0001. Part C. Comparative analysis of the effect of 5AZACdR on neuroblastoma and glioblastoma cell lines. Maximal and minimal apoptosis was seen in SK-N-AS and U-87 MG cells treated with 5AZACdR (24 h) respectively
Result of cell apoptosis assay
To determine cell apoptosis, the neuroblastoma and glioblastoma cells were treated with 5 AZA CdR for 24h. Subsequently, the cells were stained using annexin-V-(FITC) and PI to determine apoptotic cells. As indicated in figures 3-5, this compound induced cell apoptosis significantly (P<0.0001). Based on statistical analysis, a significant difference was seen between treated and untreated cell groups. Maximal and minimal apoptosis was seen in SK-N-AS and U-87 MG cells treated with 5 AZA CdR (24 h) respectively.
Figure 3.
The Apoptosis-Inducing Effect of 5AZACdR was Investigated by Flow Cytometric Analysis of Neuroblastoma Cells Stained with Annexin V and PI. The result indicated that 5AZACdR induced cell apoptosis after 24 h of treatment significantly. P<0.0001
Result of determination of genes expression
Result of determination of genes expression in hepatocellular carcinoma SK-Hep 1
Neuroblastoma
5 AZA CdR and extrinsic pathway
To determine the expression level of the DR4, DR5, FAS, FAS-L, and TRAIL genes, neuroblastoma cell lines were treated with 5 AZA CdR, based on IC50 values demonstrated in table 2, was evaluated by quantitative real-time RT-PCR analysis. The result of the quantitative real-time RT-PCR indicated that treatment with 5 AZA CdR upregulated the expression level of DR4, DR5, FAS, FAS-L, and TRAIL genes significantly, Figure 6. *P< 0.0001.
Figure 6.
The Relative Expression Level of DR4, DR5, FAS, FAS-L, and TRAIL Genes Expression in Neuroblastoma Cell Lines Treated with 5AZACdR after 24h of Treatment. Quantitative reverse transcription-polymerase chain reaction analysis demonstrated that this compound up-regulated the expression of DR4, DR5, FAS, FAS-L, and TRAIL genes significantly. *P< 0.0001
5 AZA CdR and intrinsic pathway
To determine the expression level of the Bax, Bak, Bim, Bcl-2, Bcl-xL, and Mcl-1 genes, neuroblastoma cell lines were treated with 5 AZA CdR, based on IC50 values demonstrated in Table 2. The relative expression level was evaluated by quantitative real-time RT-PCR analysis. The result of the quantitative real-time RT-PCR indicated that treatment with 5 AZA CdR upregulated the expression level of Bax, Bak, and Bim genes and down-regulated the expression level of Bcl-2, Bcl-xL, and Mcl-1 genes significantly as indicated in Figure 7 *P< 0.0001.
Figure 7.
The Relative Expression Level of Bax, Bak, Bim, Bcl-2, Bcl-xL, and Mcl-1 Genes Expression in Neuroblastoma Cell Lines Treated with 5AZACdR at 24h. Quantitative reverse transcription-polymerase chain reaction analysis demonstrated that this compound upregulated the expression of Bax, Bak, and Bim and downregulated the expression of Bcl-2, Bcl-xL, and Mcl-1 genes significantly. *P< 0.0001
5 AZA CdR and JAK/STAT pathway
To determine the expression level of the SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B genes, neuroblastoma cell lines were treated with 5 AZA CdR, based on IC50 values demonstrated in Table 2. The relative expression level was evaluated by quantitative real-time RT-PCR analysis. The result of the quantitative real-time RT-PCR indicated that treatment with 5 AZA CdR upregulated the expression level of SOCS1 and SOCS3 genes and down-regulated the expression level of JAK1, JAK2, STAT3, STAT5A, and STAT5B genes significantly, Figure 8 *P< 0.0001.
Figure 8.
The Relative Expression Level of SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B Expression in Neuroblastoma Cell Lines Treated with 5AZACdR at 24h. Quantitative reverse transcription-polymerase chain reaction analysis demonstrated that this compound up-regulated the expression of SOCS1 and SOCS3 and downregulated JAK1, JAK2, STAT3, STAT5A, and STAT5B genes expression significantly. *P< 0.0001
Glioblastoma
5 AZA CdR and extrinsic pathway
To determine the expression level of the DR4, DR5, FAS, FAS-L, and TRAIL genes, glioblastoma cell lines were treated with 5 AZA CdR, based on IC50 values demonstrated in Table 2, was evaluated by quantitative real-time RT-PCR analysis. The result of the quantitative real-time RT-PCR indicated that treatment with 5 AZA CdR upregulated the expression level of DR4, DR5, FAS, FAS-L, and TRAIL genes significantly, Figure 9. *P< 0.0001.
Figure 9.
The Relative Expression Level of DR4, DR5, FAS, FAS-L, and TRAIL Genes Expression in Glioblastoma Cell Lines Treated with 5AZACdR after 24h of Treatment. Quantitative reverse transcription-polymerase chain reaction analysis demonstrated that this compound up-regulated the expression of DR4, DR5, FAS, FAS-L, and TRAIL genes significantly. *P< 0.0001
5 AZA CdR and intrinsic pathway
To determine the expression level of the Bax, Bak, Bim, Bcl-2, Bcl-xL, and Mcl-1 genes, glioblastoma cell lines were treated with 5 AZA CdR, based on IC50 values demonstrated in Table 2. The relative expression level was evaluated by quantitative real-time RT-PCR analysis. The result of the quantitative real-time RT-PCR indicated that treatment with 5 AZA CdR upregulated the expression level of Bax, Bak, and Bim genes and down-regulated the expression level of Bcl-2, Bcl-xL, and Mcl-1 genes significantly as indicated in figure 10. *P< 0.0001.
Figure 10.
The Relative Expression Level of Bax, Bak, Bim, Bcl-2, Bcl-xL, and Mcl-1 Genes Expression in Glioblastoma Cell Lines Treated with 5AZACdR at 24h. Quantitative reverse transcription-polymerase chain reaction analysis demonstrated that this compound upregulated the expression of Bax, Bak, and Bim and downregulated the expression of Bcl-2, Bcl-xL, and Mcl-1 genes significantly. *P< 0.0001
5 AZA CdR and JAK/STAT pathway
To determine the expression level of the SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B genes, glioblastoma cell lines were treated with 5 AZA CdR, based on IC50 values demonstrated in Table 2. The relative expression level was evaluated by quantitative real-time RT-PCR analysis. The result of the quantitative real-time RT-PCR indicated that treatment with 5 AZA CdR upregulated the expression level of SOCS1 and SOCS3 genes and down-regulated the expression level of JAK1, JAK2, STAT3, STAT5A, and STAT5B genes significantly, Figure 11 *P< 0.0001.
Figure 11.
The Relative Expression Level of SOCS1, SOCS3, JAK1, JAK2, STAT3, STAT5A, and STAT5B Expression in Glioblastoma Cell Lines Treated with 5AZACdR at 24h. Quantitative reverse transcription-polymerase chain reaction analysis demonstrated that this compound up-regulated the expression of SOCS1 and SOCS3 and downregulated JAK1, JAK2, STAT3, STAT5A, and STAT5B genes expression significantly. *P< 0.0001
Figure 4.
The Apoptosis-Inducing Effect of 5AZACdR was Investigated by Flow Cytometric Analysis of Glioblastoma Cells Stained with Annexin V and PI. The result indicated that 5AZACdR induced cell apoptosis after 24 h of treatment significantly. P<0.0001
Discussion
It has been reported that DNMTIs induce apoptosis throw extrinsic (Zhao et al., 2013), intrinsic (Ruiz-Magaña et al., 2012) and JAK/STAT (Calvisi et al., 2006) apoptotic pathways. In the present study, we indicated that 5 AZA CdR induced apoptosis via extrinsic (up-regulation of DR4, DR5, FAS, FAS-L, and TRAIL genes), intrinsic (up-regulation of Bax, Bak, and Bim, and down-regulation of Bcl-2, Bcl-xL, and Mcl-1 genes), and JAK/STAT (up-regulation of SOCS1 and SOCS3 and down-regulation of JAK1, JAK2, STAT3, STAT5A, and STAT5B genes) pathways in neuroblastoma (IMR-32, SK-N-AS, UKF-NB-2, UKF-NB-3, and UKF-NB-4) and glioblastoma (SF-767, SF-763, A-172, U-87 MG, and U-251 MG) cell lines. Similarly, in vitro studies have shown that 5 AZA CdR can induce apoptosis via an extrinsic pathway (re-expression of FAS and DR4 gene) in colon cancer and AML cell lines (Torres et al., 201; Zhang et al., 2018). Further, it up-regulates DR4 mRNA in choriocarcinoma (BeWo, JEG 3, JAR) and HTR-8/SVneo trophoblast cell lines (Wu et al., 2016). As we reported in this work, it has been shown that 5 AZA CdR and MS-275 play their roles by intrinsic pathway (up-regulation of p21waf1 and down-regulation of Mcl-1) in MV4-11 TP53 R248W cell line (Nishioka et al., 2011). Besides, 5 AZA CdR inhibits cell growth and induces apoptosis by MCL-1 down-regulation in the AML cell line (Tsao et al., 2012). Other researchers have shown that this compound up-regulates the Bax gene and leads to apoptosis induction in the human pancreatic cancer cell line (PANC-1) (Dastjerdi et al., 2018). 5 AZA CdR-induced apoptosis in the human leukemia cell lines U937 and HL60 is correlated with the downregulation of anti-apoptotic Bcl-2, cIAP-1, XIAP, and cIAP-2 protein levels, the cleavage of Bid proteins, the activation of caspases resulting in cell apoptosis (Shin et al., 2012). Another apoptotic molecular mechanism of 5 AZA CdR includes JAK/STAT pathway. This agent reactivates the suppressor of the cytokine signaling-1 (SOCS-1) gene and affected the Janus kinase/signal transducers and activators of the transcription (JAK/STAT) pathway in pancreatic cancer cells (Fukushima et al., 2003). In multiple myeloma, this compound plays its role via STAT3 down-regulation (Chim et al., 2004). Further, in hepatocellular carcinoma and endometrial cell lines, it up-regulates SOCS-3 expression (Niwa et al., 2005; Chen et al., 2015). Meanwhile, it induces apoptosis through both JAK/STAT (SOCS3 re-expression) and intrinsic (down-regulation of BCL2 and BCL-XL) apoptosis pathways in mantle cell lymphoma (MCL) (Molavi et al., 2013). It should be noted that the mentioned pathways are not the only pathway of 5 AZA CdR. Previously, we reported that 5 AZA CdR can induce apoptosis by DNMT 1 inhibition, and CIP/KIP family (p21, p27, and p57) genes up-regulation in colon cancer SW480 cell line (Sanaei et al., 2019). Besides, recent studies have demonstrated that 5 AZA CdR reactivates the expression of BRCA1, and p53 and increases p15, p16, and BRCA2 genes in Breast cancer stem cells (BCSCs) (Phan et al., 2016). Furthermore, treatment with 5 AZA CdR induces upregulation of p16, FHIT, CRBP1, WWOX, and DLC-1 in the gastric cancer cell line (He et al., 2015). Finally, 5 AZA CdR induces apoptosis by various molecular mechanisms, we investigated only three ways comprising extrinsic, intrinsic, and JAK/STAT apoptotic pathways in neuroblastoma (IMR-32, SK-N-AS, UKF-NB-2, UKF-NB-3, and UKF-NB-4) and glioblastoma (SF-767, SF-763, A-172, U-87 MG, and U-251 MG) cell lines. Therefore, the investigation of other mechanisms is recommended.
In conclusion, our results indicated that 5 AZA CdR can induce its apoptotic effect through several molecular mechanisms including intrinsic, extrinsic, and JAK/STAT pathways in neuroblastoma and glioblastoma cell lines. Besides, this compound changes the relative expression level of the pro-and anti-apoptotic genes by which inhibits cell growth inhibition and induces apoptosis induction. Our findings will form the basis for further studies on the effects of 5 AZA CdR in neuroblastoma and glioblastoma.
Author Contribution Statement
All authors generated the ideas and contributed to the writing of the manuscript, acquisition of data, analysis, interpretation of data, critical revision of the manuscript for important intellectual content, statistical analysis, and technical or material support. All authors approved the final revision..
Acknowledgements
This article was supported by the adjutancy of research of Jahrom University of Medical Sciences, Iran.
Conflict Of Interest
The authors report no conflict of interest.
References
- Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–28. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Cao XX, Mohuiddin I, Chada S, et al. Adenoviral transfer of mda-7 leads to BAX up-regulation and apoptosis in mesothelioma cells, and is abrogated by over-expression of BCL-XL. Mol Med. 2002;8:869–76. [PMC free article] [PubMed] [Google Scholar]
- Chen B, Lai J, Dai D, et al. JAK1 as a prognostic marker and its correlation with immune infiltrates in breast cancer. Aging (Albany NY) 2019;11:11124–35. doi: 10.18632/aging.102514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang C, Sheng Y, et al. Frequent SOCS3 and 3OST2 promoter methylation and their epigenetic regulation in endometrial carcinoma. Am J Cancer Res. 2004;5:180–90. [PMC free article] [PubMed] [Google Scholar]
- Chim CS, Fung TK, Cheung WC, et al. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood. 2004;103:4630–35. doi: 10.1182/blood-2003-06-2007. [DOI] [PubMed] [Google Scholar]
- Dastjerdi MN, Azarnezhad A, Hashemibeni B, et al. An effective concentration of 5-Aza-CdR to induce cell death and apoptosis in human pancreatic cancer cell line through reactivating RASSF1A and up-regulation of bax genes. Iran J Med Sci. 2018;43:533–39. [PMC free article] [PubMed] [Google Scholar]
- Fukushima N, Sato N, Sahin F, et al. Aberrant methylation of suppressor of cytokine signalling-1 (SOCS-1) gene in pancreatic ductal neoplasms. Br J Cancer. 2003;89:338–43. doi: 10.1038/sj.bjc.6601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønbaek K, Hother C, Jones PA. Epigenetic changes in cancer. Apmis. 2007;115:1039–59. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- He D, Zhang YW, Zhang NN, et al. Aberrant gene promoter methylation of p16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein–Barr virus-associated gastric carcinomas. Med Oncol. 2015;32:92–8. doi: 10.1007/s12032-015-0525-y. [DOI] [PubMed] [Google Scholar]
- Ierano C, Chakraborty A, Nicolae A, et al. Loss of the proteins Bak and Bax prevents apoptosis mediated by histone deacetylase inhibitors. Cell Cycle. 2013;12:2829–38. doi: 10.4161/cc.25914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Anai S, Onishi S, et al. Inhibition of COX-2 expression by topical diclofenac enhanced radiation sensitivity via enhancement of TRAIL in human prostate adenocarcinoma xenograft model. BMC Urol. 2013;13:1–9. doi: 10.1186/1471-2490-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlic H, Reitermaier R, Ghanim V, et al. 5-azacytidine and decitabine induce demethylation and re-expression of FAS (CD95) and promote apoptosis in neoplastic cells in acute myeloid leukemia (AML) Blood. 2011;118:3463–71. [Google Scholar]
- Kavoosi F. Effect of curcumin and trichostatin a on the expression of DNA methyltransfrase 1 in hepatocellular carcinoma cell line hepa 1-6. Iran J Pediatr Hematol Oncol. 2018;8:193–201. [Google Scholar]
- Kiziltepe T, Hideshima T, Catley L, et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol Cancer Ther. 2007;6:1718–27. doi: 10.1158/1535-7163.MCT-07-0010. [DOI] [PubMed] [Google Scholar]
- Leon AJ, Gomez E, Garrote JA, et al. High levels of proinflammatory cytokines, but not markers of tissue injury, in unaffected intestinal areas from patients with IBD. Mediators Inflamm. 2009;5:1–10. doi: 10.1155/2009/580450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masood KI, Rottenberg ME, Salahuddin N, et al. Expression of M tuberculosis-induced suppressor of cytokine signaling (SOCS) 1 SOCS3 FoxP3 and secretion of IL-6 associates with differing clinical severity of tuberculosis. BMC Infect Dis. 2013;13:13–22. doi: 10.1186/1471-2334-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molavi O, Wang P, Zak Z, et al. Gene methylation and silencing of SOCS 3 in mantle cell lymphoma. Br J Haematol. 2013;161:348–56. doi: 10.1111/bjh.12262. [DOI] [PubMed] [Google Scholar]
- Nakamoto N, Higuchi H, Kanamori H, et al. Cyclooxygenase-2 inhibitor and interferon-β synergistically induce apoptosis in human hepatoma cells in vitro and in vivo. Iran J Ped Hematol Oncol. 2019;9:219–28. [PubMed] [Google Scholar]
- Nishioka C, Ikezoe T, Yang J, et al. Simultaneous inhibition of DNA methyltransferase and histone deacetylase induces p53-independent apoptosis via down-regulation of Mcl-1 in acute myelogenous leukemia cells. Leuk Res. 2011;35:932–39. doi: 10.1016/j.leukres.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Kanda H, Shikauchi Y, et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406–17. doi: 10.1038/sj.onc.1208788. [DOI] [PubMed] [Google Scholar]
- Phan NLC, Van Trinh N, Van Pham P. Low concentrations of 5-aza-2′-deoxycytidine induce breast cancer stem cell differentiation by triggering tumor suppressor gene expression. Onco Targets Ther. 2016;9:49–59. doi: 10.2147/OTT.S96291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Magaña MJ, Rodríguez-Vargas JM, Morales JC, et al. The DNA methyltransferase inhibitors zebularine and decitabine induce mitochondria mediated apoptosis and DNA damage in p53 mutant leukemic T cells. Int J Cancer. 2012;130:1195–207. doi: 10.1002/ijc.26107. [DOI] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Pourahmadi M. Effect of Decitabine (5-aza-2ˈ-deoxycytidine, 5-aza-CdR) in Comparison with Vorinostat (Suberoylanilide Hydroxamic Acid, SAHA) on DNMT1, DNMT3a and DNMT3b, HDAC 1-3, SOCS 1, SOCS 3, JAK2, and STAT3 Gene Expression in Hepatocellular Carcinoma HLE and LCL-PI 11 Cell Lines. Asian Pac J Cancer Prev. 2021;22:2089–98. doi: 10.31557/APJCP.2021.22.7.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Sahraeian H. The effects of 5-aza-2′-deoxycytidine and valproic acid on apoptosis induction and cell growth inhibition in colon cancer HT 29 cell line. Int J Prev Med. 2021;12:33. doi: 10.4103/ijpvm.IJPVM_410_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Nasiri S. Effect of 5-aza-2ˈ-deoxycytidine on p27Kip1, p21Cip1/Waf1/Sdi1, p57Kip2, and DNA methyltransferase 1 Genes Expression, Cell Growth Inhibition and Apoptosis Induction in Colon Cancer SW 480 and SW 948 Cell Lines. Galen Med J. 2020;9:1899–908. doi: 10.31661/gmj.v9i0.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F. Effect of 5-aza-2′-deoxycytidine on estrogen receptor alpha/beta and DNA methyltransferase 1 genes expression, apoptosis induction, and cell growth prevention of the colon cancer HT 29 cell line. Int J Prev Med. 2020;11:147–53. doi: 10.4103/ijpvm.IJPVM_140_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Ghasemi A. Investigation of the Effect of 5-Aza-2ʹ-Deoxycytidine on p15INK4, p16INK4, p18INK4, and p19INK4 Genes Expression, Cell Growth Inhibition, and Apoptosis Induction in Hepatocellular Carcinoma PLC/PRF/5 Cell Line. Adv Biomed Res. 2020;9:33–8. doi: 10.4103/abr.abr_68_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Esmi Z. The effect of 5-aza-2′-deoxycytidine in combination to and in comparison with vorinostat on DNA methyltransferases, histone deacetylase 1, glutathione S-transferase 1 and suppressor of cytokine signaling 1 genes expression, cell growth inhibition and apoptotic induction in hepatocellular LCL-PI 11 cell line. Int J Hematol Oncol Stem Cell Res. 2020;14:45–55. [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Esmi Z. The effect of 5-aza-2′-deoxycytidine in combination to and in comparison with vorinostat on DNA methyltransferases, histone deacetylase 1, glutathione S-transferase 1 and suppressor of cytokine signaling 1 genes expression, cell growth inhibition and apoptotic induction in hepatocellular LCL-PI 11 cell line. Int J Hematol Oncol Stem Cell Res. 2020;14:45–53. [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F. Histone deacetylases and histone deacetylase inhibitors: molecular mechanisms of action in various cancers. Adv Biomed Res. 2019;8:63–75. doi: 10.4103/abr.abr_142_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei M, Kavoosi F, Mohammadi M, et al. Effect of 5-aza-2′-deoxycytidine on p16INK4a, p14ARF, p15INK4b Genes Expression, Cell Viability, and Apoptosis in PLC/PRF5 and MIA Paca-2 Cell Lines. Iran J Pediatr Hematol Oncol. 2019;9:219–28. [Google Scholar]
- Sanaei M, Kavoosi F. Effect of 5-aza-2’-deoxycytidine in comparison to valproic acid and trichostatin A on histone deacetylase 1, DNA methyltransferase 1, and CIP/KIP family (p21, p27, and p57) genes expression, cell growth inhibition, and apoptosis induction in colon cancer SW480 cell line. Adv Biomed Res. 2019;8:52–60. doi: 10.4103/abr.abr_91_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DY, Park Y-S, Yang K, et al. Decitabine, a DNA methyltransferase inhibitor, induces apoptosis in human leukemia cells through intracellular reactive oxygen species generation. Int J Oncol. 2012;41:910–8. doi: 10.3892/ijo.2012.1546. [DOI] [PubMed] [Google Scholar]
- Soncini M, Santoro F, Gutierrez A, et al. The DNA demethylating agent decitabine activates the TRAIL pathway and induces apoptosis in acute myeloid leukemia. Biochimica et Biophys Acta Mol Basis Dis. 2013;1832:114–20. doi: 10.1016/j.bbadis.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Tao J, Qiu B, Zhang D, et al. Expression levels of Fas/Fas-L mRNA in human brain glioma stem cells. Mol Med Rep. 2012;5:1202–6. doi: 10.3892/mmr.2012.791. [DOI] [PubMed] [Google Scholar]
- Torres CM, Yang D, Zimmerman M, et al. Decitabine and vorinostat cooperate to sensitize metastatic human colon carcinoma cells to Fas-mediated apoptosis. AACR. 2012;8:4888–96. doi: 10.4049/jimmunol.1103035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao T, Shi Y, Kornblau S, et al. Concomitant inhibition of DNA methyltransferase and BCL-2 protein function synergistically induce mitochondrial apoptosis in acute myelogenous leukemia cells. Ann Hematol. 2012;91:1861–70. doi: 10.1007/s00277-012-1537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Ni Z, Dai X, et al. The Bcl-2/xL inhibitor ABT-263 increases the stability of Mcl-1 mRNA and protein in hepatocellular carcinoma cells. Mol Cancer. 2014;13:98–109. doi: 10.1186/1476-4598-13-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PH, Chen XM, Liu XQ, et al. Activation of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor gene expression following DNA demethylation in placental choriocarcinoma and transformed cell lines. Reprod Fertil Dev. 2016;28:1844–53. doi: 10.1071/RD14408. [DOI] [PubMed] [Google Scholar]
- Xiong H, Chen ZF, Liang QC, et al. Inhibition of DNA methyltransferase induces G2 cell cycle arrest and apoptosis in human colorectal cancer cells via inhibition of JAK2/STAT3/STAT5 signalling. J Cell Mol Med. 2009;13:3668–79. doi: 10.1111/j.1582-4934.2009.00661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu L, Qiu X, et al. CCL21/CCR7 prevents apoptosis via the ERK pathway in human non-small cell lung cancer cells. PLoS One. 2012;7:17–25. doi: 10.1371/journal.pone.0033262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Xie T, Wang Z, et al. DNA methyltransferases: emerging targets for the discovery of inhibitors as potent anticancer drugs. Drug Discov Today. 2019;24:2323–31. doi: 10.1016/j.drudis.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang C, Wu C, et al. DNA methyltransferases in cancer: Biology, paradox, aberrations, and targeted therapy. Cancers. 2020;12:2123–45. doi: 10.3390/cancers12082123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen Y, Pei X, et al. Effects of Decitabine on the proliferation of K562 cells and the expression of DR4 gene. Saudi J Biol Sci. 2018;25:242–47. doi: 10.1016/j.sjbs.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xu J. DNA methyltransferases and their roles in tumorigenesis. Biomark Res. 2017;5:1–8. doi: 10.1186/s40364-017-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Pang LQ, Wu Y, et al. Significance of Bcl-xL in human colon carcinoma. World J Gastroenterol. 2008;14:3069–73. doi: 10.3748/wjg.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Dong JJ, Cai T, et al. High glucose induces apoptosis via upregulation of Bim expression in proximal tubule epithelial cells. Oncotarget. 2017;8:24119–29. doi: 10.18632/oncotarget.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Guo J, Zhao Y, et al. Chidamide, a novel histone deacetylase inhibitor, inhibits the viability of MDS and AML cells by suppressing JAK2/STAT3 signaling. Am J Transl Res. 2016;8:3169–78. [PMC free article] [PubMed] [Google Scholar]