Abstract
Eukaryotic cells use chromatin marks to regulate the initiation of DNA replication. The origin recognition complex (ORC)‐associated protein ORCA plays a critical role in heterochromatin replication in mammalian cells by recruiting the initiator ORC, but the underlying mechanisms remain unclear. Here, we report crystal and cryo‐electron microscopy structures of ORCA in complex with ORC's Orc2 subunit and nucleosomes, establishing that ORCA orchestrates ternary complex assembly by simultaneously recognizing a highly conserved peptide sequence in Orc2, nucleosomal DNA, and repressive histone trimethylation marks through an aromatic cage. Unexpectedly, binding of ORCA to nucleosomes prevents chromatin array compaction in a manner that relies on H4K20 trimethylation, a histone modification critical for heterochromatin replication. We further show that ORCA is necessary and sufficient to specifically recruit ORC into chromatin condensates marked by H4K20 trimethylation, providing a paradigm for studying replication initiation in specific chromatin contexts. Collectively, our findings support a model in which ORCA not only serves as a platform for ORC recruitment to nucleosomes bearing specific histone marks but also helps establish a local chromatin environment conducive to subsequent MCM2‐7 loading.
Keywords: Chromatin, DNA replication initiation, MCM2‐7 loading, Origin licensing, Origin recognition complex
Subject Categories: Chromatin, Transcription & Genomics; DNA Replication, Recombination & Repair
Biochemical and structural studies suggest that ORCA not only recruits ORC to chromatin with specific histone marks but also reorganizes local chromatin structure in preparation for DNA replication initiation.
Introduction
Faithful transmission of genetic information requires accurate, once‐per‐cell‐cycle replication of chromosomal DNA prior to cell division. In mammalian cells, replication machineries are assembled at tens of thousands of genomic sites to complete replication in a timely manner and to sustain genome integrity (Ekundayo & Bleichert, 2019; Hu & Stillman, 2023). These replication origins are first licensed during the G1 phase of the cell cycle by the initiator, the origin recognition complex (ORC), which loads mini‐chromosome maintenance complexes (MCM2‐7) onto DNA with the help of the co‐loaders Cdc6 and Cdt1 (Bleichert, 2019; Costa & Diffley, 2022). When origins fire in S phase, a subset of these loaded MCM2‐7 double hexamers are activated by the action of Dbf4‐dependent and cell‐cycle‐dependent kinases (DDK and CDK, respectively) and converted into active CMG replicative helicases, which support bidirectional replisome assembly; concomitantly, CDK activity and metazoan geminin inhibit origin licensing in S phase to prevent re‐licensing of origins and DNA re‐replication (Parker et al, 2017; Costa & Diffley, 2022). Thus, cells rely on a sufficient number of MCM2‐7 double hexamers being loaded in G1 to complete genome replication in S phase. Since under‐licensing of origins in G1 poses a major threat to genome stability, it is critical to establish mechanistic models for how different genomic regions are efficiently licensed for DNA replication.
The natural substrate for eukaryotic genome replication is DNA, which is packaged to various degrees by histone proteins into nucleosomes and different types of chromatin. Generally, open chromatin or euchromatin is considered more permissive to replication initiation than condensed chromatin or heterochromatin. It is thus not surprising that chromatin context modulates various steps in DNA replication, including initiation factor recruitment, the locations and dynamics of MCM2‐7 loading in G1, and DNA replication timing in S phase (Prioleau & MacAlpine, 2016; Ekundayo & Bleichert, 2019; Ding & Koren, 2020; Nathanailidou et al, 2020; Mei et al, 2022). The metazoan initiator ORC, for example, directly binds the histone modification H4K20me2, associates with chromatin‐binding proteins (e.g., ORCA/LRWD1, HP1, HBO1, and several transcription factors), and is recruited to early mammalian replication origins that are enriched in H4K20me2 and the histone variant H2A.Z (Pak et al, 1997; Iizuka & Stillman, 1999; Bosco et al, 2001; Beall et al, 2002; Prasanth et al, 2004; Bartke et al, 2010; Shen et al, 2010; Kuo et al, 2012; Long et al, 2020); these properties contrast those of S. cerevisiae ORC, which recognizes specific DNA sequence elements at origins (Bell & Stillman, 1992). Timely replication of heterochromatic domains and the activity of late‐firing origins in mammalian cells, on the other hand, are in part regulated by trimethylation of H4K20 (Brustel et al, 2017). Altered H4K20 methylation levels cause DNA replication stress, DNA damage, defects in cell cycle progression, and have also been linked to cancer (Fraga et al, 2005; Tardat et al, 2007, 2010; Schotta et al, 2008; Oda et al, 2009; Beck et al, 2012). Despite the clear importance of chromatin factors in genome duplication, the molecular and structural mechanisms responsible for preparing chromatin as a substrate for replication initiation and for ensuring that origins are adequately distributed (to different chromatin domains) across the genome remain poorly defined and foremost problems in the field.
ORC‐associated protein (ORCA; also called LRWD1) has emerged as a central player in promoting origin licensing in chromatin. ORCA is a metazoan‐specific protein that binds the initiator ORC and has been suggested to constitute a bona fide member of the initiator complex in multicellular eukaryotes (Bartke et al, 2010; Shen et al, 2010; Vermeulen et al, 2010). Like ORC, ORCA associates with chromatin in a cell‐cycle‐dependent manner, and most ORCA‐binding sites co‐localize with replication origins (Shen et al, 2010; Wang et al, 2017). Depletion of ORCA reduces ORC recruitment to chromatin, origin licensing in primary cell lines, S‐phase progression, and impedes postnatal growth of ORCA knockout mice (Shen et al, 2010; Kang et al, 2022). Heterochromatin is particularly reliant on ORCA to maintain adequate MCM2‐7 loading rates and to sustain the activity of its origins and timely replication (Brustel et al, 2017; Mei et al, 2022); these functions appear mediated by ORCA's ability to bind repressive trimethylation marks on histones H3 and H4 (H3K9me3, H3K27me3, and H4K20me3) enriched in heterochromatin (Bartke et al, 2010; Vermeulen et al, 2010; Beck et al, 2012; Chan & Zhang, 2012; Giri et al, 2015). Yet, how ORCA binds these nucleosomes and ORC and whether these interactions are sufficient to target the initiator to specific chromatin contexts are not understood. Likewise, whether and how ORCA locally alters chromatin architecture to create a conducive environment for initiator recruitment and loading are unknown.
To resolve these questions, we determined structures of ORCA in complex with the ORCA‐binding region within ORC and with an H4K20me3‐modified nucleosome. These structures uncovered a conserved, 17‐amino‐acid peptide in Orc2 and an aromatic cage in ORCA that are responsible for ORCA's association with ORC and the recognition of repressive trimethyl‐lysine marks, respectively. While a WD40 domain in ORCA (ORCAWD40) is sufficient for these interactions, adjacent protein regions help stabilize ORCA on mononucleosomes and nucleosome arrays. Using confocal microscopy, we also show that ORCA is excluded from unmodified chromatin condensates but enriched in these structures when H4K20me3 is present, which in turn leads to H4K20me3‐dependent recruitment of ORC to chromatin. Surprisingly, H4K20me3‐chromatin condensates are disrupted upon ORCAWD40 binding. These findings suggest a model in which ORCA binding to H4K20me3‐chromatin not only serves as a recruitment platform for ORC but also reorganizes local chromatin architecture by diminishing nucleosome self‐association in preparation for ORC and MCM2‐7 loading in heterochromatin domains.
Results
Crystal structure of an ORCA•Orc2 complex
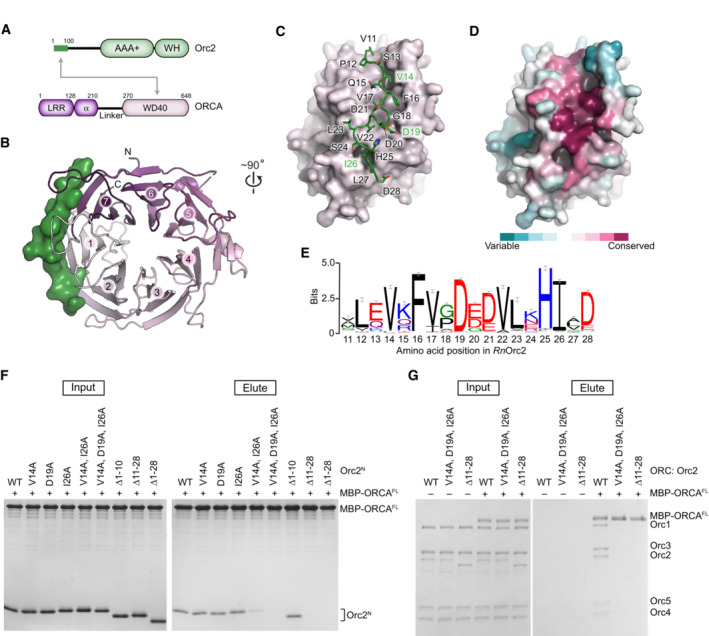
ORCA comprises several conserved protein folds, including N‐terminal LRR and α‐helical domains that are connected to a C‐terminal WD40 domain by a variable linker region (Fig 1A). Previous studies have mapped the ORC‐binding site to the WD40 domain in human ORCA (ORCAWD40), which is sufficient to associate with the first 100 amino acids in human Orc2 (Orc2N) (Bartke et al, 2010; Shen et al, 2010, 2012). We crystallized a complex containing the corresponding domains of the rat proteins (82.6 and 70% sequence identity to human ORCAWD40 and Orc2N, respectively) and determined its structure at 1.8 Å resolution (Table 1). The structure revealed that ORCAWD40 folds into a seven‐bladed β‐propeller (Fig 1B). Amino acid residues 11–28 of Orc2N bind to the circumference of the propeller and engage three of its blades, including the first and the last (Fig 1B and C). In this manner, Orc2N bridges N‐ and C‐terminal regions of ORCAWD40 and stabilizes the protein fold, rationalizing the dependence of ORCA on Orc2 for protein stability in cells (Bartke et al, 2010; Shen et al, 2012; Shen & Prasanth, 2012).
Figure 1. Orc2 binds a conserved region at the circumference of the ORCAWD40 β‐propeller.
-
ADomain architecture of ORCA and Orc2. The interaction between the N‐terminal region of Orc2 and ORCAWD40 is indicated by gray arrows. WH, winged helix domain; LRR, leucine‐rich repeat domain; α, alpha‐helical domain.
-
B–DCrystal structure of the ORCAWD40·Orc2N complex. (B)Top view of the ORCAWD40 β‐propeller shown in cartoon and Orc2N in surface representation. Blades are numbered from N to C terminus. (C) Side view of the ORCAWD40 β‐propeller (as surface) depicting the binding site for Orc2. Amino acid side chains for the interacting Orc2 region are shown as sticks. (D) ORCAWD40 surface colored by protein sequence conservation. This view is identical to that in C. Orc2N has been omitted for clarity.
-
ESequence logo of the ORCA‐binding region in Orc2 highlights conservation of this peptide sequence across metazoan Orc2 orthologs that have ORCA.
-
FMutations in or deletion of the Orc2 peptide region abrogate the association of Orc2N with ORCA. Coomassie‐stained SDS–PAGE gels of inputs and elutions from in vitro pulldown assays using MBP‐tagged ORCAFL as bait and Orc2N wild‐type (WT) or mutant proteins as prey. Corresponding control pulldowns with MBP instead of MBP‐ORCAFL as bait are included in Fig EV1E.
-
GMutations in or deletion of the Orc2 peptide region abrogate the association of ORCA with ORC. In vitro pulldown assay probing the interaction between MBP‐tagged ORCAFL (bait) and ORC assemblies containing wild‐type or mutant Orc2. Coomassie‐stained SDS–PAGE gels are shown. Rat ORCA and ORC proteins are used in this and all subsequent figures.
Source data are available online for this figure.
Table 1.
Summary of X‐ray diffraction data collection and refinement statistics.
| ORCAWD40·Orc2N (PDB 8SIU) | |
|---|---|
| Data collection | |
| Beamline | SLS‐Xo6DA‐PXIII |
| Wavelength (Å) | 0.979092 |
| Resolution range (Å) | 45.51–1.80 (1.83–1.80) |
| Space group | C2221 |
| Unit cell dimensions | |
| a, b, c (Å) | 91.02, 135.43, 78.57 |
| α, β, γ (°) | 90, 90, 90 |
| Reflections | |
| Total | 1,773,956 (90,294) |
| Unique | 45,458 (2,577) |
| Completeness (%) | 99.8 (97.4) |
| Multiplicity | 39.0 (35.0) |
| Mean I/σ | 14.8 (1.1) |
| CC1/2 | 0.999 (0.505) |
| Rmerge (all I+ & I‐) | 0.228 (4.364) |
| Rpim (all I+ & I‐) | 0.037 (0.716) |
| Wilson B factor | 31.41 |
| Phasing | |
| FOM | 0.33 |
| Refinement | |
| Resolution range (Å) | 45.51–1.80 |
| Rwork/Rfree | 0.1697/0.2032 |
| Model composition | |
| Non‐hydrogen atoms | 3,362 |
| Protein residues | 386 |
| Waters | 321 |
| Mean B factors (Å2) | |
| Protein | 30.99 |
| Waters | 39.39 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.091 |
| Ramachandran plot | |
| % favored | 97.09 |
| % allowed | 2.91 |
| % outliers | 0.0 |
| Rotamers | |
| % outliers | 0.0 |
| MolProbity | |
| Clashscore | 2.16 |
| MolProbity score | 1.15 |
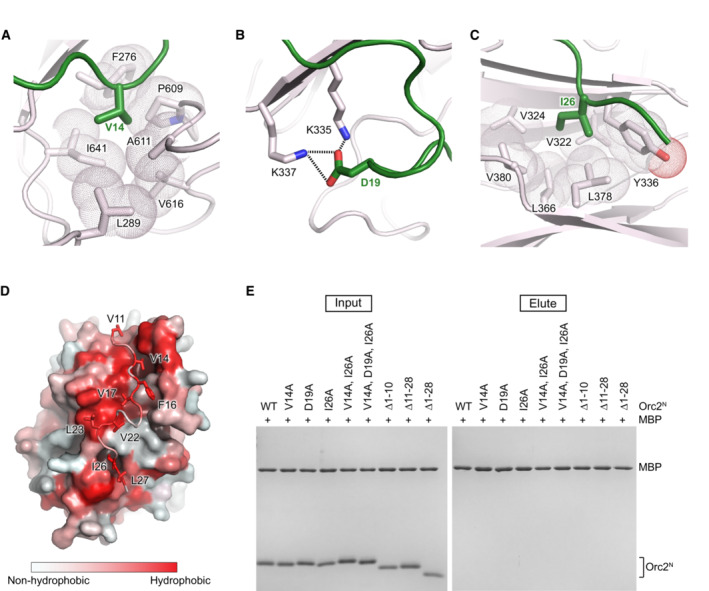
The ORCAWD40·Orc2N interface is composed of highly conserved residues and is predominantly hydrophobic in nature (Figs 1D and E and EV1A–D). V14 and I26 in Orc2, for example, are engulfed by hydrophobic pockets on the ORCA surface, although interactions are reinforced by electrostatic contacts between Orc2‐D19 and two ORCA lysines (K335 and K337; Fig EV1A–C). To test the significance of the observed crystallographic contacts, we asked whether purified single, double, and triple alanine mutants of V14, D19, and I26 in Orc2N, or deletion mutants of this conserved peptide sequence, can associate with full‐length ORCA (ORCAFL). In pulldown experiments, the Orc2N‐V14A/I26A double mutant showed reduced binding to ORCAFL, while the Orc2N‐V14A/D19A/I26A triple mutation and deletion of residues 11–28 (but not residues 1–10) abolished the interaction with ORCAFL (Figs 1F and EV1E). Incorporating these Orc2 mutations into the initiator ORC also abrogated the ability of ORC to bind to ORCA (Fig 1G). Thus, Orc2 residues 11–28 are essential for ORC·ORCA co‐association and constitute the main binding site for ORCA on ORC. We anticipate that these interactions flexibly tether ORCA to ORC since the N‐terminal Orc2 region is predicted to be disordered and not visible in prior ORC structures (Jaremko et al, 2020; Schmidt & Bleichert, 2020). Notably, ORCA orthologs are not omnipresent across the metazoan kingdom; the conservation of the ORCA‐binding peptide sequence in Orc2 correlates with the occurrence of an ORCA ortholog, indicating that ORCA binding is the major function of this N‐terminal Orc2 peptide sequence (Table 2; Appendix Fig S1).
Figure EV1. Conserved Orc2 residues mediate the interaction with ORCAWD40 .
-
A–CZoomed views of the Orc2·ORCA‐binding site highlighting contacts between conserved Orc2 residues (in green) V14 (in A), D19 (in B), and I26 (in C) with the ORCA β‐propeller circumference (in light pink). V14 and I26 in Orc2 bind hydrophobic pockets in ORCA, while Orc2‐D19 forms salt bridges with ORCA lysines.
-
DView of the ORCAWD40·Orc2N‐binding site colored according to the Eisenberg hydrophobicity scale (Eisenberg et al, 1984). ORCA is rendered as surface and Orc2 as cartoon with side chains in stick representation.
- E
Table 2.
Conservation of ORCA across the metazoan kingdom.
| Phylum | ORCA conservation | Comments |
|---|---|---|
| Chordata | + | |
| Mollusca | + | LRR is not present in Gastropoda and Cephalopoda |
| Echinodermata | + | |
| Cnidaria | + | |
| Platyhelminthes | + | ORCA protein sequences are very long (~ 1,500–2,250 aa) and in few cases contain other functional domains |
| Arthropoda | Variable | |
| Crustacea | ||
| Malacostraca | + | No LRR |
| Hexanauplia | + | No LRR |
| Branchiopoda | − | |
| Chelicerata | Variable | ORCA conserved in some of the orders in Arachnida |
| Hexapoda | Variable | ORCA (no LRR) conserved in some of the orders in Insecta |
| Nematoda | − | |
| Rotifera | − | |
| Annelida a | (+) | |
| Placozoa a | (+) | |
| Priapulida a | (+) | |
Only a limited number of sequences (≤3) is available from these phyla.
Cryo‐EM structure of an ORCA‐bound H4K20me3‐nucleosome
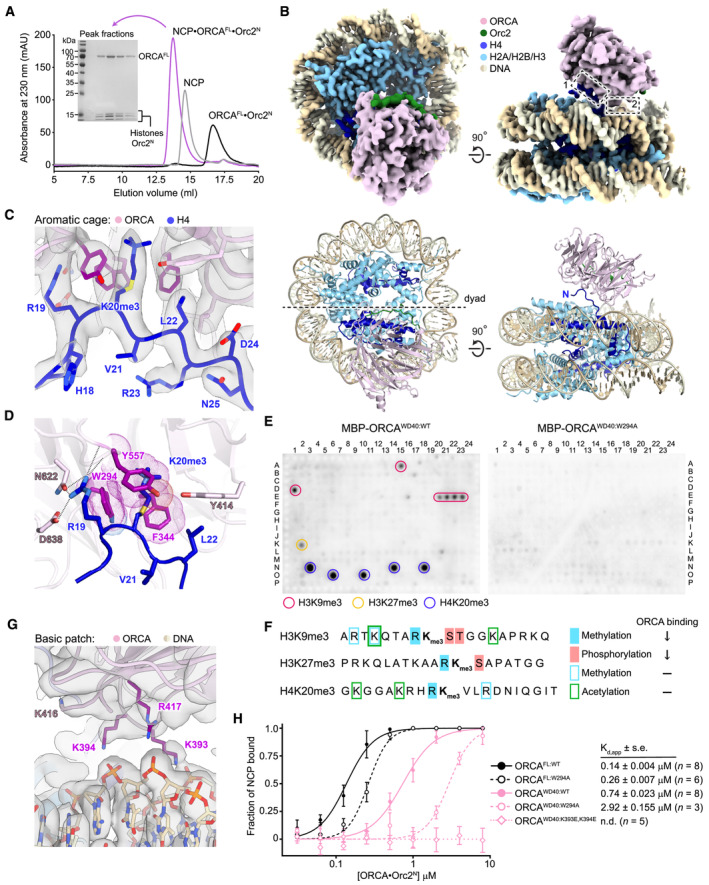
Despite the established importance of H4K20 trimethylation and ORCA in DNA replication, it is unknown how ORCA associates with nucleosomes and how it recognizes this and other repressive histone trimethylation marks. Therefore, we reconstituted a complex comprising full‐length ORCA, Orc2N, and an H4K20me3‐nucleosome (trimethylation at H4K20 was installed using methyl‐lysine analogs (Simon et al, 2007)) and determined its structure using cryo‐electron microscopy (cryo‐EM) at an overall resolution of 2.9 Å (Fig 2A and B and Table 3; Appendix Figs S2A and B, S3, and S4). The WD40 domains of either one or two ORCA molecules can be seen to engage the nucleosome disk, whereas the other ORCA regions (LRR, α‐helical fold, and linker) are not resolved, indicating flexibility of the N‐terminal half of the protein (Fig 2B; Appendix Figs S3C and S4D). Closer inspection of ORCA‐nucleosome contacts revealed that ORCAWD40 engages both the H4 tail and nucleosomal DNA. The H4 tail binds to a conserved surface on ORCA's β‐propeller, with H4K20 reaching into the central cavity, where it interacts with a cage of aromatic residues (W294, F344, and Y557) (Figs 2C and D and EV2A). This binding event is somewhat reminiscent of how the PRC2 component EED interacts with H3K27me3 peptides (Margueron et al, 2009; Xu et al, 2010), albeit the H4 tail approaches ORCAWD40 from a different direction compared to histone peptides in complex with EED (Fig EV2B and C). Disruption of the aromatic cage in ORCAWD40 by mutating tryptophan 294 to alanine abrogates the recognition of H3K9me3‐, H3K27me3‐, and H4K20me3‐containing peptides, which are bound by wild‐type ORCAWD40 and ORCAFL in histone peptide arrays (Figs 2E and EV2D). Interestingly, ORCA binding to histone peptides with repressive lysine trimethylations is inhibited when the arginine N‐terminal to the trimethylated lysine is itself methylated, or when the downstream serine and threonine in H3 are phosphorylated (Fig 2F). In our structure, the preceding arginine, H4R19, makes multiple electrostatic interactions with ORCA side chains (N622 and D638) and backbone carbonyls, which would be unfavorable upon H4R19 methylation (Fig 2D). These interactions also explain why ORCA only binds repressive trimethylation marks and not H3K4me3 or H3K36me3, which are not preceded by an arginine. Furthermore, H4binding positions ORCAWD40 near the nucleosome edge, facilitating contacts between the DNA backbone and conserved basic patch arginine and lysine residues on the β‐propeller surface (Figs 2G and EV2E and F).
Figure 2. ORCA binds nucleosomes using both an aromatic cage and a basic surface patch in the WD40 β‐propeller.
-
APurification of the ORCAFL•Orc2N•H4K20me3‐nucleosome complex for cryo‐EM. Size exclusion chromatography of ORCAFL·Orc2N, H4K20me3‐nucleosomes (NCP), and the ORCAFL•Orc2N•H4K20me3‐nucleosome complex. Coomassie‐stained SDS–PAGE gel demonstrates coelution of histones and ORCA in peak fractions upon complex formation.
-
BCryo‐EM structure of the ORCAFL•Orc2N•H4K20me3‐nucleosome complex. Sharpened cryo‐EM map (top) and model (as cartoon, bottom) are shown. Dashed boxes mark interaction sites between ORCA and the H4K20me3‐nucleosome.
-
C, DORCA binds the H4K20me3‐modified H4 tail using an aromatic cage. Zoomed views of the H4‐tail‐binding site (interaction site 1 in B) are shown. (C) Sharpened cryo‐EM map of the H4•ORCA interface with H4 tail residues and interacting ORCA side chains in stick representation. (D) A rotated view of the H4•ORCA interface, emphasizing recognition of H4K20me3 by ORCA's aromatic cage (in purple) and hydrogen bonds or salt bridges (dashed lines) between H4‐R19 and ORCA.
-
E, FORCA binding to histone peptides containing repressive trimethylation marks requires an intact aromatic cage and unmodified neighboring histone tail residues. (E) Histone peptide arrays of wild‐type (ORCAWD40:WT) and aromatic cage‐mutant ORCA (ORCAWD40:W294A). (F) Summary of histone peptide array results for wild‐type ORCA and effects of combinatorial histone modifications on ORCA binding (↓ reduced binding; − binding unaltered).
-
GA basic patch in ORCA engages nucleosomal DNA (interaction site 2 in B). Sharpened cryo‐EM map with basic patch side chains shown as sticks. ORCA residues contacting nucleosomal DNA are colored purple.
-
HMutations in ORCA's aromatic cage and basic patch decrease ORCA's affinity for H4K20me3‐mononucleosomes. Binding curves (mean and s.d.) for wild‐type and mutant ORCA constructs measured by electrophoretic mobility shift assays (EMSAs) are shown. Apparent dissociation constants (Kd,app), standard error of mean (s.e.), and number of replicate experiments are listed. Representative EMSA gels are included in Appendix Fig S5. N.d., not determined.
Source data are available online for this figure.
Table 3.
Summary of cryo‐EM data collection, refinement, and validation statistics.
| EM data collection and processing | |
| Microscope | Titan Krios |
| Camera | K3 |
| Voltage (kV) | 300 |
| Magnification | ×105,000 |
| Frames (no.) | 35 |
| Total electron dose (e−/Å2) | 51 |
| Calibrated pixel size (Å) | 0.832 |
| Defocus range (μm) | 1–2.2 |
| Movies | 3,845 |
| Initial picks (no.) | 3,271,302 |
| Refined particles (no.) | 31,420 |
| Symmetry imposed | C1 |
| Global resolution (Å) | |
| FSC 0.5 (unmasked/masked) | 7.0/3.2 |
| FSC 0.143 (unmasked/masked) | 3.8/2.9 |
| Local resolution range (Å) | 2.7–5.2 |
| Map sharpening B factor (Å2) | −80 |
| EMD accession number | EMD‐40522 |
| Model refinement and validation | |
| Initial models | 3LZ0 (nucleosome); Vasudevan et al, 2010 |
| 8SIU (ORCAWD40·Orc2N) | |
| Model composition | |
| Non‐hydrogen atoms | 14,844 |
| Protein residues | 1,118 |
| DNA residues | 294 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.854 |
| B factors (Å2) | |
| Protein | 62.78 |
| DNA | 55.46 |
| Ramachandran plot | |
| % favored | 97.24 |
| % allowed | 2.76 |
| % outliers | 0.0 |
| Rotamer outliers (%) | 1.38 |
| MolProbity | |
| Clashscore | 5.69 |
| MolProbity score | 1.56 |
| Model‐map comparison | |
| EM Ringer score | 3.65 |
| CCmask | 0.83 |
| FSCmodel/map 0.5 | 3.1 |
| PDB accession number | 8SIY |
Figure EV2. ORCA binds nucleosomes using two surfaces on the WD40 domain.
-
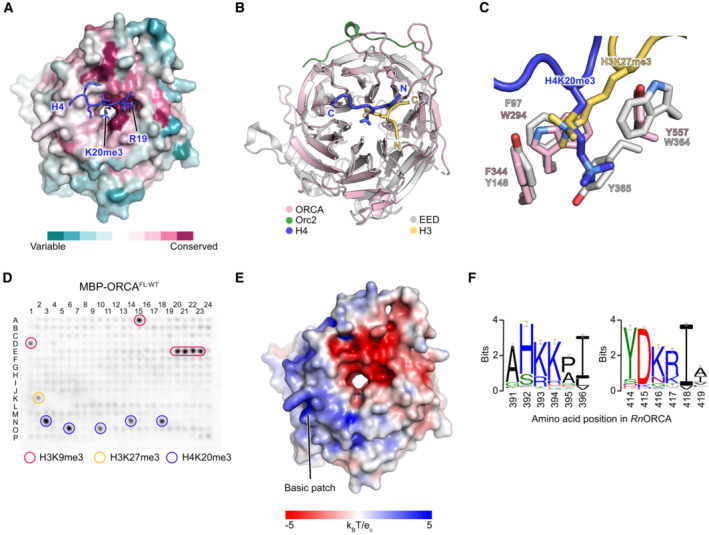
AThe H4‐binding site constitutes a conserved region of the ORCA β‐propeller. The ORCAWD40 surface is colored according to ConSurf conservation scores.
-
B, CORCA's WD40 domain is structurally similar to that of EED. (B) Structural superposition of the WD40 domains of ORCA (this study) and EED (PDB 3IIW; Margueron et al, 2009) with bound H4 and H3 peptides, respectively. (C) Zoomed view of the aromatic cages in ORCA and EED.
-
DHistone peptide array binding experiment for full‐length (FL) ORCA. A similar set of modified histone peptides is bound by the full‐length protein as by the WD40 domain alone (compared to Fig 2E).
-
E, FPositively charged residues form a basic patch on the surface of ORCA's WD40 domain. (E) View of the nucleosome‐facing surface of ORCA's WD40 domain colored by electrostatic potential. (F) Sequence logo of DNA‐binding residues (K393, K394, and R417) in ORCA's basic patch reveals conservation of chemical side chain properties among ORCA orthologs.
To test the importance of these interfaces for ORCA binding to mononucleosomes, we performed electrophoretic mobility shift assays (EMSAs) with H4K20me3‐nucleosomes and wild‐type ORCAFL, wild‐type ORCAWD40, as well as aromatic cage (ORCAFL:W294A and ORCAWD40:W294A) and basic patch mutant ORCA (ORCAWD40:K393E,K394E; Appendix Fig S2C). Mutation of the aromatic cage reduced the affinity of ORCAFL and ORCAWD40 for H4K20me3‐nucleosomes by two‐ and fourfold, respectively, while charge reversals in the DNA‐binding site abolished nucleosome binding of ORCAWD40 (Fig 2H; Appendix Fig S5A and B). Thus, both nucleosome‐binding sites in ORCAWD40 synergize to support co‐association of ORCA and H4K20me3‐mononucleosomes.
Multiple ORCA domains cooperate to stabilize ORCA on chromatin
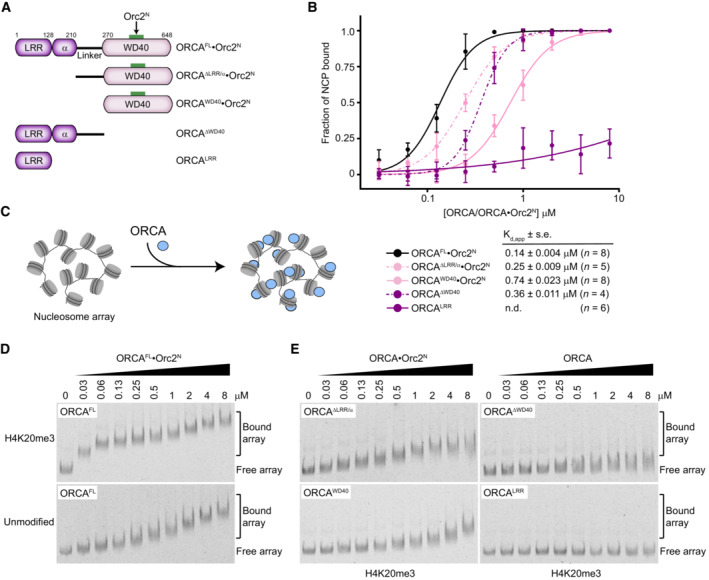
The WD40 domain has previously been reported to be sufficient and essential for ORCA localization on chromatin in human cells when ectopically expressed, while the roles of the conserved N‐terminal LRR, α‐helical, and linker domains for ORCA function are not understood (Shen et al, 2010). Moreover, only the WD40 domain is resolved in our cryo‐EM map, seen to bind the nucleosome (Fig 2B). Therefore, we were surprised that full‐length ORCA bound H4K20me3‐nucleosomes with fivefold higher affinity than the WD40 domain alone (Kd,app of 140 nM vs. 740 nM; Fig 2H). To clarify the contributions of different ORCA domains to nucleosome binding, we generated additional deletion constructs that harbored linker and WD40 domains (ORCAΔLRR/α), LRR, α‐helical and linker domains (ORCAΔWD40), or the LRR domain only (ORCALRR) and tested their ability to associate with H4K20me3‐nucleosomes (Fig 3A; Appendix Fig S2C). EMSA experiments showed that all ORCA variants except for the LRR domain associated with mononucleosomes more efficiently than ORCAWD40, including ORCA that lacked a WD40 domain (Fig 3B; Appendix Fig S5C). These results argue that multiple ORCA regions have an intrinsic affinity for binding mononucleosomes; we postulate that these non‐ORCAWD40‐mediated interactions likely involve heterogeneous associations with nucleosomal DNA, precluding the capture of these binding events in our cryo‐EM structure of the full‐length ORCA·nucleosome complex.
Figure 3. ORCA regions N‐terminal to the WD40 domain stabilize ORCA binding to nucleosomes and chromatin.
-
ASchematic of ORCA domain architecture and deletion constructs. Note that ORCA constructs harboring the WD40 repeat module were co‐expressed and copurified with Orc2N for increased stability.
-
BH4K20me3‐mononucleosome‐binding curves (means and standard deviations) for different ORCA deletion constructs. Apparent dissociation constants (Kd,app), standard error of mean (s.e.), and the number of replicate experiments are listed. Data for ORCAFL·Orc2N and ORCAWD40·Orc2N are replotted from Fig 2H for comparison. See Appendix Fig S5 for EMSA gels.
-
C–EFull‐length ORCA is required for high‐affinity binding to H4K20me3‐chromatin. (C) Chromatin arrays containing 12 nucleosomes (unmodified or H4K20me3‐modified) were used as a substrate for ORCA binding. (D) Full‐length ORCA has a higher affinity for H4K20me3‐nucleosome arrays than for unmodified arrays. EMSA gels are shown. (E) EMSA gels for binding of ORCA deletion constructs to H4K20me3‐nucleosome arrays.
Source data are available online for this figure.
ORCA promotes origin licensing in heterochromatin, a condensed form of chromatin with a high density of nucleosomes (Brustel et al, 2017; Imai et al, 2017; Mei et al, 2022); accordingly, mononucleosomes are imperfect substrates to comprehend mechanistic ORCA functions in chromatin. Therefore, we reconstituted chromatin arrays bearing 12 nucleosomes on Widom 601 sequences separated by 30 bp linker DNA (referred to as chromatin hereafter) as a more physiological substrate to ask (i) whether ORCA can discriminate unmodified from H4K20me3‐modified chromatin and (ii) which ORCA domains are required for efficient chromatin binding (Appendix Fig S6). Addition of full‐length ORCA to chromatin arrays at ORCA concentrations as low as 30–60 nM led to a marked reduction in the mobility of H4K20me3‐nucleosome arrays but not unmodified arrays in native agarose‐polyacrylamide gels, which is indicative of tight binding of ORCA to H4K20me3‐chromatin arrays (Fig 3C and D). ORCA's high affinity for H4K20me3‐arrays and ability to distinguish H4K20me3‐ and unmodified arrays was reliant on an intact aromatic cage (Appendix Fig S7). By contrast, truncated ORCA variants were unable to bind H4K20me3‐arrays (ORCALRR and ORCAΔWD40) or bound arrays less efficiently (ORCAWD40 and ORCAΔLRR/α) than ORCAFL (Fig 3E). Collectively, these results establish that multiple domains in ORCA cooperate to recruit and stabilize ORCA on polynucleosome substrates in a histone modification‐specific manner.
Binding of ORCA's WD40 domain to nucleosomes inhibits the self‐association of nucleosome arrays
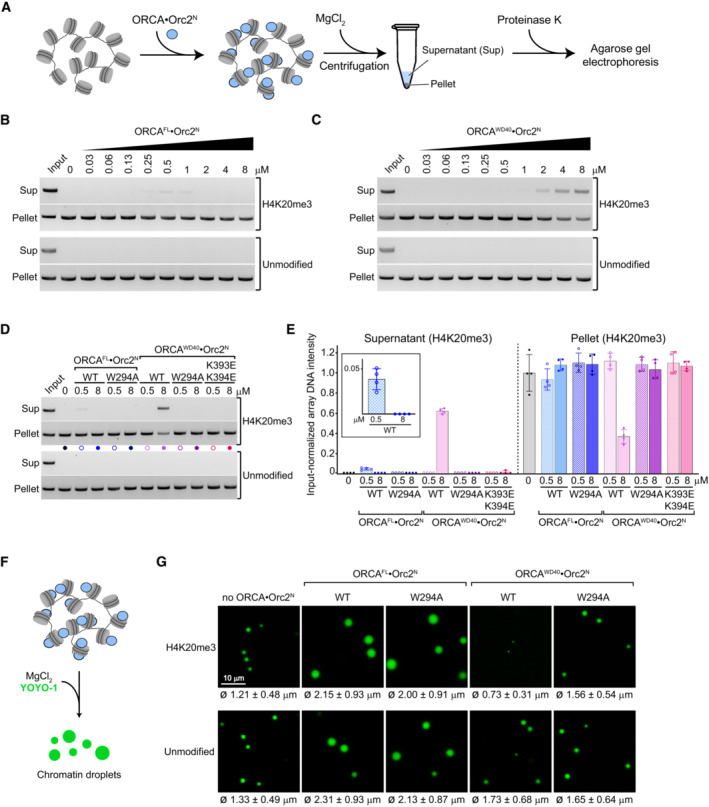
The supramolecular structure of chromatin regulates accessibility to DNA (Li & Reinberg, 2011; Luger et al, 2012). Chromatin compaction is contingent on multivalent inter‐nucleosomal interactions that are mediated by electrostatic forces between histones, as well as histones and DNA, and is regulated by chromatin‐binding proteins (Schwarz et al, 1996; Dorigo et al, 2003; Gordon et al, 2005; Zheng et al, 2005; Kan et al, 2009; Liu et al, 2011; Zhang & Kutateladze, 2019; Hansen et al, 2021). Classic examples include the linker histone H1 and heterochromatin protein HP1, which promote chromatin condensation (Carruthers et al, 1998; Fan et al, 2005; Maeshima et al, 2016; Gibson et al, 2019; Sanulli et al, 2019). Since ORCA is enriched in condensed chromatin in cells (Bartke et al, 2010; Shen et al, 2010; Chan & Zhang, 2012), we reasoned that ORCA recruitment may alter chromatin compaction. To test this premise, we examined the effect of ORCA on Mg2+‐induced oligomerization of nucleosome arrays by centrifugation and pelleting, an assay that has been used extensively to probe chromatin condensation and is considered a powerful approach to study chromatin structure in vitro (Schwarz et al, 1996; Gordon et al, 2005; Liu et al, 2011; Maeshima et al, 2016; Sanulli et al, 2019; Hansen et al, 2021). We incubated nucleosome arrays with purified ORCA and induced chromatin compaction by addition of MgCl2; array oligomers were then recovered from the pellet fraction after centrifugation (Fig 4A). In the absence of ORCA, both unmodified and H4K20me3‐nucleosome arrays pelleted efficiently. Surprisingly, addition of increasing amounts of full‐length ORCA led to a slight but reproducible increase in H4K20me3‐modified nucleosome arrays in the supernatant at ORCA concentrations of 0.25–1 μM (Fig 4B). The decrease in H4K20me3‐array pelleting was exacerbated when ORCA's WD40 domain was added instead of full‐length protein, especially at micromolar ORCAWD40 concentrations (Fig 4C). The higher concentration of ORCAWD40 needed to observe this effect is likely due to its lower affinity for binding H4K20me3‐arrays compared to full‐length ORCA (Fig 3D and E). Moreover, the inhibition of nucleosome array pelleting was specific to H4K20me3‐modified nucleosome arrays and also not seen with any of the other truncated ORCA variants (Figs 4B and C, and EV3A and B). Mutations in the aromatic cage or the basic patch restored sedimentation of H4K20me3‐nucleosome arrays in the presence of ORCAFL or ORCAWD40 (Fig 4D and E). These findings argue that the interaction of ORCA with H4 tails upon recognition of H4K20me3 inhibits inter‐array self‐association and chromatin compaction.
Figure 4. The WD40 domain of ORCA alters the ability of nucleosomes to self‐associate and chromatin compaction.
-
AExperimental outline of pelleting assay used to study the effect of ORCA on nucleosome array oligomerization.
-
B–EBinding of ORCA's WD40 domain to trimethylated H4 tails decreases the pelleting efficiency of nucleosome arrays. (B) Pelleting assays for H4K20me3‐modified and unmodified arrays in the absence or presence of increasing concentrations of full‐length (FL) ORCA. Input refers to proteinase K‐treated arrays without pelleting. (C) Pelleting assays for H4K20me3‐modified and unmodified arrays in the absence or presence of increasing concentrations of the ORCA WD40 domain. Input refers to proteinase K‐treated arrays without pelleting. (D) Pelleting assays for indicated wild‐type and mutant ORCA full‐length or WD40 domain constructs at 0.5 and 8 μM ORCA. (E) Quantification (mean and s.d.) of four independent experiments as done in D.
-
F, GBinding of ORCA's WD40 domain to trimethylated H4 tails inhibits condensation of nucleosome arrays into phase‐separated chromatin droplets. (F) Experimental setup for generating chromatin droplets. After ORCA binding, condensation was induced by MgCl2 addition and arrays were stained with YOYO‐1. (G) Confocal microscopy images of nucleosome array condensates in the absence and presence of wild‐type or aromatic cage‐mutant (W294A) ORCAFL and ORCAWD40 (at 4 μM ORCA concentration, chosen to be in a range that supports ORCAWD40 binding to H4K20me3‐nucleosome arrays (see Fig 3E) and that is also in line with previous studies examining the impact of proteins on chromatin condensation; Gibson et al, 2019). Mean and standard deviation of droplet diameters (n = 865, 1,428, 800, 78, and 773 for H4K20me3 and n = 674, 677, 742, 423, and 582 for unmodified chromatin droplets; order is as for images shown) are listed below each image. See Fig EV3C for cumulative frequency distributions. Note that ORCA·Orc2N complexes were used since Orc2 stabilizes the WD40 repeat fold.
Source data are available online for this figure.
Figure EV3. Binding of ORCA to nucleosome arrays alters inter‐nucleosomal interactions.
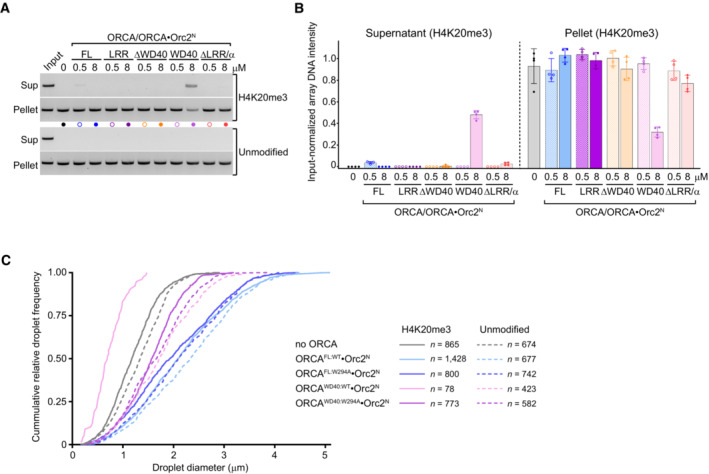
- Nucleosome array pelleting assays in the absence and presence of full‐length ORCA and indicated ORCA deletion constructs. Agarose gels of array DNA from supernatants and pellet fractions are shown. Input refers to nucleosome array without pelleting that is loaded for comparison.
- Quantification (mean and s.d.) of DNA band intensities from four independent experiments as done in A.
- Cumulative frequency distributions of chromatin droplet diameters in the absence and presence of wild‐type or mutant ORCA proteins (see Fig 4G). Number of droplets measured is listed.
Recent studies have shown that chromatin compaction is coupled to liquid–liquid phase separation (LLPS) (Gibson et al, 2019; Sanulli et al, 2019). Given our pelleting assay results, we predicted that binding of ORCAWD40 to nucleosomes in chromatin arrays would impede LLPS under physiological salt concentrations. Indeed, we observed a striking reduction in both size and number of Mg2+‐induced chromatin droplets when nucleosome arrays were incubated with wild‐type but not aromatic cage‐mutant ORCAWD40, and inhibition of LLPS was specific to H4K20me3‐nucleosome arrays (Figs 4F and G, and EV3C). Consistent with findings from pelleting assays, full‐length ORCA did not reduce the size of chromatin droplets at micromolar ORCA concentration, indicating ORCA regions outside the WD40 domain preserve oligomerization during ORCAWD40‐mediated chromatin decondensation (Fig 4B and G). Collectively, these results suggest that ORCA, and in the particular binding of ORCAWD40 to histone H4, alter the physiochemical properties of chromatin. We propose that recognition of H4K20me3 by ORCA's WD40 domain sequesters the H4 tail, preventing this histone tail from participating in inter‐nucleosome or nucleosome–DNA interactions that are critical for driving chromatin compaction and LLPS (Kan et al, 2009; Gibson et al, 2019); ORCA regions outside the WD40 repeat, on the other hand, compensate for this effect at least partially and stabilize nucleosome array oligomers.
The ORCA linker region stabilizes chromatin condensates
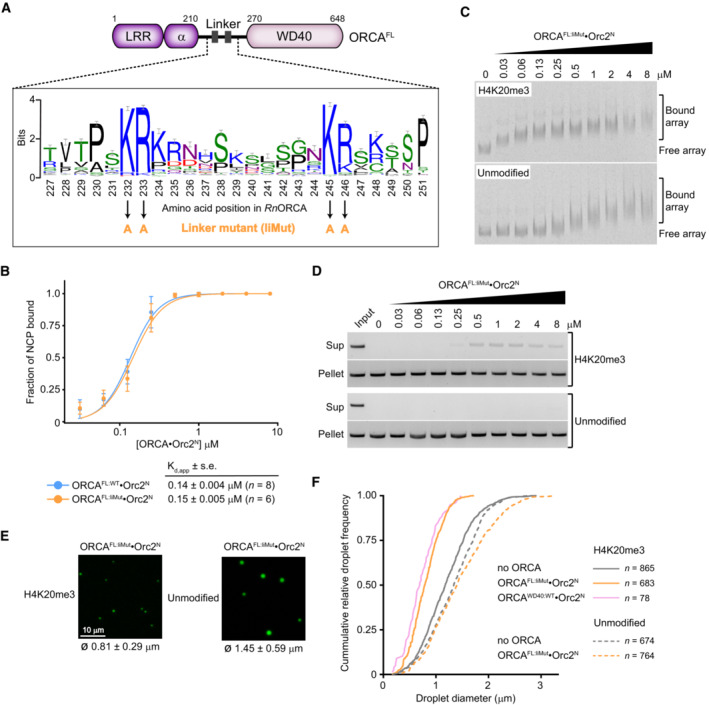
The opposing effects of the full‐length ORCA protein and the WD40 domain on nucleosome array oligomerization and chromatin condensate integrity indicate that ORCA regions located outside of the WD40 domain also modulate chromatin structure (Fig 4). Considering that truncated ORCA variants harboring the linker domain can bind mononucleosomes with sub‐micromolar affinity, even in the absence of a WD40 module, we hypothesized that the linker domain is involved in bridging different nucleosome arrays (Fig 3B). Phylogenetic analysis of the ORCA linker sequence uncovered four highly conserved arginine and lysine residues (K232, R233, K245, and R246 in rat ORCA) embedded in a region of high sequence variability (Fig 5A). Strikingly, mutation of these basic residues to alanine in ORCA (ORCALiMut) did not impede its binding to H4K20me3‐mononucleosomes or H4K20me3‐chromatin arrays (note that ORCALiMut binds more tightly to H4K20me3‐nucleosome arrays than unmodified ones as seen for ORCAWT; Fig 5B and C, compared with Fig 3B and D; Appendix Fig S5A); however, ORCA linker mutations compromised the pelleting efficiency of nucleosome arrays even at nanomolar concentrations as evinced by an increase in the amount of arrays present in the supernatant, and also reduced chromatin droplet size (Fig 5D–F). These effects of ORCALiMut were specific to H4K20me3‐modified chromatin arrays, phenocopying outcomes observed with ORCAWD40 (compare Figs 4 and 5). Thus, the ORCA linker domain helps maintain the integrity of H4K20me3‐chromatin condensates after sequestration of H4 tails by ORCA's WD40 domain, possibly by engaging in interactions that bridge different nucleosome arrays to stabilize H4K20me3‐chromatin oligomers.
Figure 5. Conserved basic residues in the ORCA linker are required to maintain higher‐order nucleosome assemblies upon ORCA binding.
-
ASequence logo of the ORCA linker region highlighting highly conserved arginine and lysine residues.
-
BMutation of conserved basic residues in the linker region does not interfere with binding of ORCA to H4K20me3‐mononucleosomes. Binding curves (mean and s.d.) for wild‐type and linker mutant ORCA measured by electrophoretic mobility shift assays (EMSAs) are shown. Apparent dissociation constants (Kd,app), standard error of mean (s.e.), and number of replicate experiments are listed. Representative EMSA gels are included in Appendix Fig S5A. Data for wild‐type ORCAFL·Orc2N are replotted from Fig 2H for comparison.
-
CMutation of conserved basic residues in the linker region does not interfere with binding of ORCA to nucleosome arrays. EMSA gels are shown.
-
D–FLinker mutations in ORCA reduce the pelleting of H4K20me3‐arrays and chromatin droplet formation. (D) Pelleting assays of H4K20me3‐modified and unmodified arrays in the absence or presence of increasing concentrations of full‐length (FL) ORCA‐containing mutations in the linker region. Input refers to proteinase K‐treated arrays without pelleting. (E) Confocal microscopy images of nucleosome array condensates in the presence of full‐length ORCA‐containing mutations in the linker domain. Mean and standard deviation of droplet diameters are listed. (F) Cumulative frequency distribution of chromatin droplet diameter in the presence of the ORCA linker mutant. Number of droplets measured is indicated. Distributions for other experimental conditions are replotted from Fig EV3C for comparison.
Source data are available online for this figure.
ORCA recruits ORC to chromatin condensates in an H4K20me3‐dependent manner
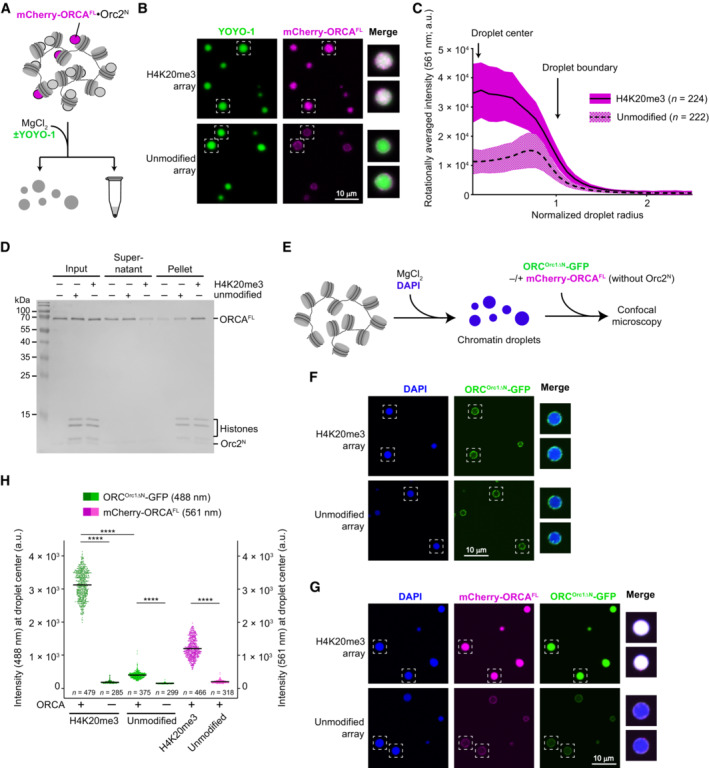
In cells, ORCA and ORC co‐localize to heterochromatin regions, and ORCA is able to recruit ORC when tethered to a specific chromosome locus (Bartke et al, 2010; Shen et al, 2010). We thus asked whether ORCA is located in chromatin condensates and whether ORCA in turn regulates the partitioning of ORC to chromatin phases in a histone modification‐dependent manner. To answer the first question, we purified mCherry‐tagged, full‐length ORCA and analyzed its distribution pattern in unmodified and H4K20me3‐chromatin condensates by confocal microscopy (Fig 6A). The data show that ORCA was uniformly distributed within H4K20me3‐chromatin droplets but accumulated at the phase boundary of unmodified chromatin condensates (Fig 6B). Quantification of fluorescence intensities revealed a three‐ to fourfold enrichment of ORCA inside H4K20me3‐chromatin droplets over unmodified ones (Fig 6C). We confirmed these results biochemically by demonstrating that ORCA co‐sediments more efficiently with H4K20me3‐modified than unmodified nucleosome array oligomers (or condensates) upon Mg2+‐induced compaction (Fig 6D). Moreover, ORCA enrichment in H4K20me3 chromatin was also evident when ORCA was added after MgCl2‐induced chromatin compaction (Fig EV4A–C). Taken together, these data indicate that ORCA is recruited into the chromatin phase by recognition of H4K20me3 (by the WD40 domain) and that the linker region (which helps maintain H4K20me3‐chromatin oligomers upon binding of full‐length ORCA) is unable by itself to promote efficient partitioning of ORCA into unmodified nucleosome array oligomers. Accumulation of ORCA at the droplet boundary, however, likely alters the physicochemical surface properties of condensates, which may promote droplet fusion and possibly explain the larger size of chromatin droplets observed in the presence of full‐length ORCA (Figs 4G and EV3C).
Figure 6. ORCA and ORC are recruited to chromatin condensates in a histone modification‐specific manner.
-
A–DORCA is distributed inside H4K20me3‐nucleosome array condensates but not unmodified chromatin condensates. (A)Experimental setup for examining ORCA enrichment in H4K20me3‐chromatin. The endpoint is either confocal imaging of chromatin droplets or pelleting of nucleosome arrays by centrifugation. (B) Confocal microscopy images of unmodified and H4K20me3‐modified chromatin droplets in the presence of mCherry‐labeled ORCA. Merged images for YOYO‐1 and mCherry channels are shown for boxed droplets. (C) Radially averaged distribution profiles for mCherry‐ORCA intensities in unmodified and H4K20me3‐chromatin droplets. The means are represented as solid and dashed lines, while colored areas mark standard deviations. (D) Coomassie‐stained SDS‐PAGE gel of histones and ORCA partitioning into supernatants and pellet fractions in nucleosome array pelleting assays.
-
E–HORCA‐mediated recruitment of ORC into preformed H4K20me3‐chromatin droplets. (E) Schematic of experimental setup. DAPI is used to stain array DNA. (F) Confocal microscopy images of chromatin droplets in the presence of GFP‐labeled ORC (harboring a deletion of the N‐terminal region in Orc1) and the absence of mCherry‐labeled ORCA. (G) Confocal microscopy images of chromatin droplets in the presence of GFP‐labeled ORC (harboring a deletion of the N‐terminal region in Orc1) and mCherry‐labeled ORCA. (H) Quantification of ORC‐GFP and mCherry‐ORCA intensities at droplet centers. Statistical significance according to the Kruskal–Wallis test and Dunn's multiple comparisons test is indicated by asterisks (****P < 0.0001). See also Fig EV4.
Source data are available online for this figure.
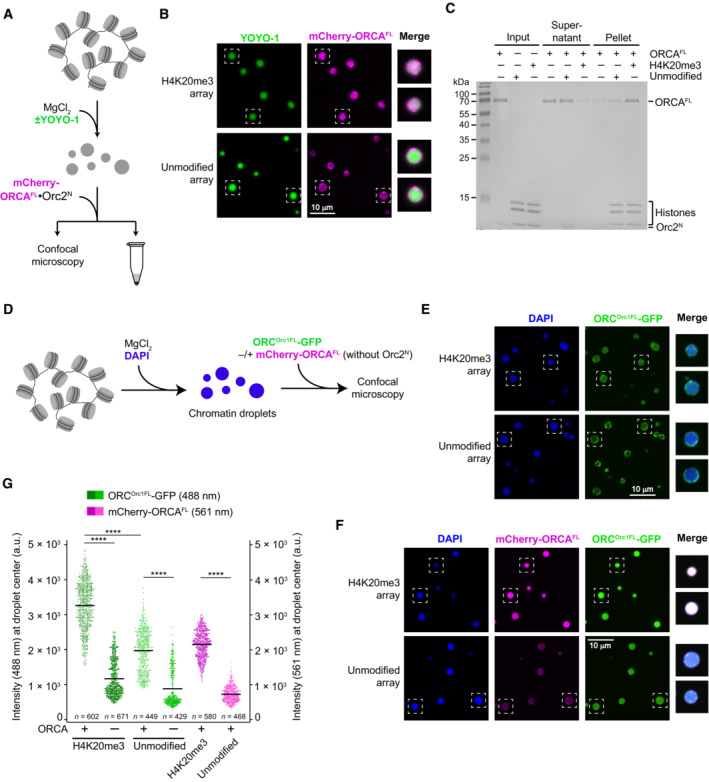
Figure EV4. ORCA and ORC are recruited to nucleosome arrays in a histone modification‐specific manner.
-
A–CORCA is recruited into preformed H4K20me3‐chromatin but not unmodified chromatin condensates. (A) Experimental setup. The endpoint is either confocal imaging of chromatin droplets or pelleting of nucleosome arrays by centrifugation. (B) Confocal microscopy images of unmodified and H4K20me3‐modified chromatin droplets after addition of mCherry‐labeled ORCA. Merged images for YOYO‐1 and mCherry channels are shown for boxed droplets. (C) Coomassie‐stained SDS–PAGE gel of histones and ORCA partitioning into supernatants and pellet fractions in nucleosome array pelleting assays. The difference between A‐C in this figure and Fig 6A–D is that here nucleosome arrays were first allowed to oligomerize prior to addition of ORCA.
-
D–GORCA enriches full‐length ORC in condensed nucleosome arrays in a histone modification‐dependent manner. (D) Schematic of experimental setup. ORCA and ORC are added to preformed chromatin droplets. (E) Confocal microscopy images of chromatin droplets with GFP‐labeled full‐length ORC in the absence of mCherry‐labeled ORCA. (F) Confocal microscopy images of chromatin droplets with GFP‐labeled full‐length ORC in the presence of mCherry‐labeled ORCA. (G) Quantification of ORC‐GFP and mCherry‐ORCA intensities at droplet centers. Statistical significance according to the Kruskal–Wallis test and Dunn's multiple comparisons test is indicated by asterisks (****P < 0.0001). The higher background recruitment of ORC to unmodified arrays as compared to Fig 6H is likely caused by interactions of the Orc1‐IDR with nucleosome array DNA.
To understand how ORCA modulates recruitment of the initiator ORC to chromatin, we next examined the distribution pattern of ORC in chromatin condensates as a dependency of ORCA and H4K20 trimethylation. Here, we used preformed chromatin condensates (by MgCl2 addition) to mimic a compacted chromatin state (Fig 6E). Initially, we focused on ORC that contained an N‐terminally truncated Orc1 subunit (Orc1ΔN) to avoid confounding effects of the intrinsically disordered region (IDR), which can drive phase separation of metazoan ORC in the presence of DNA (Parker et al, 2019). Addition of GFP‐tagged ORCOrc1ΔN alone to chromatin droplets did not lead to any substantial accumulation of ORC inside H4K20me3‐ or unmodified condensates but only at the phase boundary (Fig 6F and H). By contrast, in the presence of ORCA, ORCOrc1ΔN (like ORCA) strongly accumulated inside droplets formed with H4K20me3‐modified nucleosome arrays but not unmodified ones (Fig 6G and H). Similar results were obtained when full‐length ORC was used in these experiments, with ORC being enriched inside condensed chromatin droplets in an H4K20me3‐ and ORCA‐dependent manner (Fig EV4D–G). The higher background partitioning of full‐length ORC into chromatin condensates that is seen without H4K20me3 or ORCA is likely attributable to the Orc1‐IDR participating in multivalent interactions with DNA regions in nucleosome arrays. On account of these findings, we conclude that ORCA is essential and sufficient to specifically target ORC to condensed chromatin in a histone modification‐specific manner in this reconstituted system.
Discussion
Licensing of replication origins is a prerequisite for subsequent DNA replication and must be adequately distributed across different chromatin regions to ensure that no part of the genome is left unreplicated. How precisely this task is achieved in tightly packed and less accessible chromatin environments remains an open question. In this study, we provide a structural and mechanistic framework for how the heterochromatin‐binding protein ORCA interfaces with the MCM2‐7‐loading factor ORC and nucleosomes to alter the physicochemical properties of chromatin and to recruit the initiator complex to condensed chromatin in a histone modification‐specific manner.
Our structural and biochemical findings support the role of the ORCA‐WD40 repeat domain as a major hub in coordinating the interactions between the initiator ORC and nucleosomes. ORC and ORCA co‐associate into a stoichiometric complex, in which the initiator core is flexibly tethered to ORCA's WD40 domain through a conserved sequence fragment near the N‐terminus of Orc2 (Fig 7A). The coevolution of the Orc2 peptide sequence with the presence of an ORCA ortholog among a subset of metazoan phyla lends credence to the importance of this interaction (Table 2; Appendix Fig S1). Flexibility of the ORCA·ORC linkage could help ensure that ORCA binding does not interfere with the binding of core initiation factors (Cdc6, Cdt1, and MCM2‐7) to ORC during MCM2‐7 loading. Collectively, our findings rationalize at a structural level how ORCA, through its WD40 module, can recruit ORC (and in turn, MCM2‐7) to a chromosomal locus when artificially tethered to this region, allowing the creation of a replication origin (Takeda et al, 2005; Shen et al, 2010).
Figure 7. Model for ORCA's dual role in targeting ORC to chromatin.
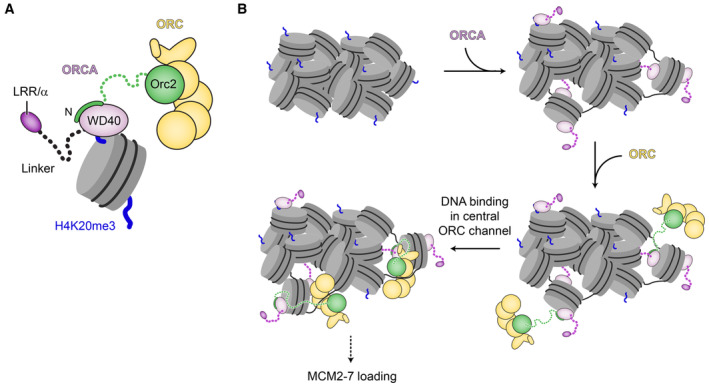
- Summary of ORC·ORCA·nucleosome (H4K20me3) interactions.
- Model for ORCA‐mediated chromatin reorganization and ORC recruitment to chromatin for origin licensing. ORCA and ORC may be recruited individually or as a preformed complex.
How does ORCA localize to heterochromatin? Previous reports have shown that ORCA associates with repressive histone trimethylation marks, an activity that has also been attributed to its WD40 domain (Bartke et al, 2010; Vermeulen et al, 2010; Beck et al, 2012; Chan & Zhang, 2012; Giri et al, 2015). Our cryo‐EM structure of ORCA in complex with a nucleosome, in conjunction with biochemical experiments, uncovered an aromatic cage at the center of ORCA's WD40 propeller that is responsible for recognition of H3K9me3, H3K27me3, and H4K20me3 (Fig 2). While aromatic cages are common in chromatin readers, to our knowledge, this example is only the second one in a WD40 domain to recognize histone lysine trimethylation, the other being EED (Margueron et al, 2009; Xu et al, 2010). Histone H3 and H4 residues surrounding the trimethylated lysine are likely bound by ORCA in an analogous manner, which is corroborated by an inhibitory effect of arginine methylation prior to Kme3 in all three peptide contexts (Fig 2). When H4K20 is trimethylated, binding of ORCA to the H4 tail positions a basic patch in the WD40 domain near nucleosomal DNA between superhelical locations 1 and 2, promoting electrostatic interactions and stabilizing ORCA on the nucleosome. We predict that ORCA will approach nucleosomes with K9 and K27 trimethylation in the H3 tail in a distinct manner than those with H4K20me3 due to different positions and restraints of the histone tails, allowing the formation of alternate contacts with nucleosomal DNA. It is also feasible that more than two ORCA proteins associate with a single nucleosome containing multiple repressive trimethylations in different histone tails. Of note, nucleosome binding by ORCA has been suggested to be modulated by CpG methylation, possibly by recognizing 5‐methylcytosine directly in DNA (Bartke et al, 2010; Wang et al, 2017). When bound to H4K20me3‐nucleosomes, we observe no contacts between ORCA and the DNA major groove (the location of the methyl group of 5‐methylcytosine in double‐stranded DNA), indicating that DNA methylation may be read out either by flexible, N‐terminal ORCA domains or indirectly by an ORCA‐interacting protein.
Our experimental observations demonstrate that ORCA is capable of reorganizing nucleosome self‐associations in a histone modification‐specific manner and provide a molecular mechanism for how this outcome is accomplished. Using nucleosome arrays, we uncovered that ORCA binding to H4 tails containing K20me3 attenuates the self‐association of nucleosome arrays and chromatin aggregation even at nanomolar ORCA concentrations (Figs 4B and 5D). This outcome is driven by the WD40 domain, while other ORCA protein regions, including the linker domain, stabilize chromatin condensates. Histone tails, especially that of H4, have established roles in promoting higher‐order chromatin organization through multivalent interactions with nearby nucleosomes and DNA, and through charge neutralization during chromatin compaction (Kan et al, 2009; Gibson et al, 2019). For example, mutations in the H4 tail that abolish positive charges inhibit the formation of chromatin condensates at physiological salt concentrations (Gibson et al, 2019). The same H4 tail region is sequestered by ORCA's WD40 domain upon binding to H4K20me3, preventing H4 tail interactions that drive nucleosome array oligomerization, chromatin compaction, and LLPS. In this regard, ORCA engagement of H4 tails would mimic effects on inter‐nucleosomal interactions seen in the presence of certain H4 tail modifications, such as acetylation and sumoylation (but not H4K20 trimethylation; Shogren‐Knaak et al, 2006; Liu et al, 2011; Dhall et al, 2014). ORCA regions outside the WD40 domain are capable of counteracting the inhibitory effect of ORCAWD40 on H4K20me3‐nucleosome self‐association to some extent, likely by bridging different nucleosomes to help maintain chromatin oligomers that are structurally distinct from those without ORCA (and those composed of unmodified nucleosomes). ORCA interactions with the nucleosome acidic patch may contribute to nucleosome bridging since mutation of this negatively charged surface diminished ORCA association in a recent nucleosome interactome screen (Skrajna et al, 2020). Besides H4 tails, H3 tails are also involved in nucleosome–nucleosome and nucleosome–DNA interactions (Zheng et al, 2005); while ORCA binds H3K9me3‐ and H3K27me3‐peptides and nucleosomes (Bartke et al, 2010; Vermeulen et al, 2010; Fig 2), whether sequestration of the H3 tail by ORCA elicits similar consequences on chromatin compaction as that of the H4 tail is currently unknown but will be an important focus of future studies.
H4K20 trimethylation, a histone modification enriched in heterochromatin, plays a pivotal role in ensuring timely replication of this chromatin domain in mammalian cells (Schotta et al, 2004; Brustel et al, 2017). At a molecular level, this outcome is thought to be mediated by ORCA (Shen et al, 2010; Beck et al, 2012; Brustel et al, 2017; Mei et al, 2022). Collectively, our structural and biochemical findings afford the integration of the multiple ORCA activities into a mechanistic model of how ORCA could promote replication of condensed chromatin regions (Fig 7B). In cells, ORCA associates with a subset of ORC assemblies in G1 (Shen et al, 2012) and recruits these to chromatin with repressive histone trimethylation marks. Binding of ORCA to H4K20me3 will compete with nucleosome self‐associations and locally decondense chromatin, thereby increasing accessibility to DNA. ORC, tethered to ORCA, will then form productive interactions (in the center of the ORC ring; Li et al, 2018; Schmidt & Bleichert, 2020) with DNA needed for MCM2‐7 loading. This model does not rely on high nuclear ORCA concentrations since only a few sites need to serve as origins. We envision that this dual functionality of ORCA (i.e., serving as a recruitment platform for ORC and locally remodeling chromatin) is particularly suited to overcome specific challenges faced during MCM2‐7 loading in condensed chromatin regions due to reduced DNA accessibility. This model provides a molecular rationale for the H4K20me3‐dependent enrichment of ORCA, ORC, and MCM2‐7 at genomic sites, in particular, at late‐replicating origins, and the ORCA‐dependent acceleration of origin licensing in heterochromatin observed in cells (Beck et al, 2012; Brustel et al, 2017; Mei et al, 2022). We note that such a scenario does not prohibit contributions of other proteins (e.g., HP1; Pak et al, 1997; Prasanth et al, 2004) and of other ORC‐chromatin interactions (e.g., H4K20me2 binding by the bromo‐adjacent homology domain in Orc1 or DNA associations mediated by the IDR in Orc1; Oda et al, 2010; Beck et al, 2012; Kuo et al, 2012; Parker et al, 2019) to initiator recruitment, nor does it preclude a role for ORCA in MCM2‐7 loading at H4K20me3‐independent origins. Indeed, ORCA also associates with H4K20me3‐independent origins, as well as those marked by H3K9me3 (Brustel et al, 2017; Wang et al, 2017). Future efforts will define how the different repressive histone marks cooperate to drive recruitment of ORCA·ORC to chromatin, whether these associations can enhance MCM2‐7 loading in condensed chromatin in our reconstituted system, and whether and how specific heterochromatin components such as HP1 and histone H1 modulate origin licensing efficiency.
Our phylogenetic analysis of ORCA helps reconcile some disparate experimental observations pertaining to the role of H4K20 methylation in DNA replication in metazoa. In mammalian systems, altered expression levels of the enzymes that catalyze H4K20 methylation interfere with proper control of replication origin activity, inducing under‐ or re‐replication of DNA caused by deregulated origin licensing (Tardat et al, 2007, 2010; Beck et al, 2012). By contrast, origin function in flies is not sensitive to aberrant H4K20 methylation (McKay et al, 2015; Li et al, 2016). Conspicuously, Drosophila species and various other arthropods do not encode an ORCA ortholog, while ORCA is universally conserved across vertebrates. The lack of ORCA in flies, in conjunction with the dispensability of the Orc1‐BAH domain (which binds H4K20me2 in mammals) for the recruitment of Drosophila ORC to chromatin (Parker et al, 2019), can explain why origins in flies are not subject to regulation by H4K20 methylation. The absence of ORCA is shared by some other invertebrates; future research will be needed to test whether the role of H4K20 methylation in origin licensing is correlated with the presence of ORCA more broadly.
ORCA and ORC have established functions in heterochromatin organization. While ORC interacts with HP1, ORCA has been found in complex with several histone methyl transferases and helps sustain H3K9me2/3 and H4K20me3 marks (Pak et al, 1997; Prasanth et al, 2004, 2010; Giri et al, 2015; Giri & Prasanth, 2015; Brustel et al, 2017; Wang et al, 2017). Both initiation factors remain associated with heterochromatin in post‐G1 cells after completion of origin licensing. How these distinct functions of ORCA and ORC are regulated throughout the cell cycle is not immediately apparent. In this regard, it is interesting that ORCA contains several annotated phosphorylation sites that cluster in its linker domain (some near the conserved basic linker residues), a region that we show stabilizes ORCA on nucleosomes and antagonizes the chromatin decondensation activity of the WD40 repeat domain in vitro. Since phosphorylation of other proteins, including HP1, can influence their ability to engage in multivalent interactions (Larson et al, 2017; Yamazaki et al, 2022), it is tempting to speculate that ORCA's functions in origin licensing and heterochromatin organization may be in part regulated by differential phosphorylation throughout the cell cycle to control local chromatin compaction. Our reconstituted system provides a powerful tool for exploring these exciting possibilities in the future and for studying the coordination of both origin licensing and heterochromatin organization functions of ORCA.
Materials and Methods
Cloning, expression, and purification of rat ORCA and ORCA·Orc2
Wild‐type full‐length (FL) Rattus norvegicus (Rn) ORCA and ORCA truncations ORCAWD40 (aa 269–648) and ORCAΔLRR/α (aa 211–648) were cloned into modified pFastBac baculovirus expression vectors by ligation‐independent cloning (LIC), while ORCA truncations ORCALRR (aa 1–128) and ORCAΔWD40 (aa 1–270) were cloned into pET28a‐derived E. coli expression vectors. A dual affinity tag containing 6xHis and maltose‐binding protein (MBP) followed by a tobacco etch virus (TEV) cleavage site was added in frame 5′ to the ORCA coding sequences. mCherry‐ORCA fusions were generated by adding the mCherry coding sequence in between the 6xHis‐MBP‐TEV tag and the ORCA open reading frame. Orc2N encoding amino acid residues 1–100 was cloned as an N‐terminal 6xHis‐TEV fusion into baculovirus expression vector 4B (QB3 MacroLab, UC Berkeley). Expression vectors encoding ORCA mutants (ORCAFL:W294A, ORCAWD40:W294A, ORCAWD40:K393E,K394E, and ORCAFL:K232A,R233A,K245A,R246A or also referred to as ORCAFL:liMut) were generated by site‐directed mutagenesis and verified by DNA sequencing.
ORCALRR and ORCAΔWD40 were expressed in BL21 RIL E. coli cells. Four liters of shaker culture in 2xYT media with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol were grown at 37°C to an OD600 nm of 0.6 and expression was induced with 0.5 mM IPTG for ~ 18 h at 18°C. Cells were harvested by centrifugation at 3,500 g in a Sorvall Evolution RC centrifuge and lysed by sonication in lysis buffer containing 50 mM Tris–HCl pH 7.8, 1 M NaCl, 10% glycerol, 30 mM imidazole, 1 mM β‐mercaptoethanol, 200 μM PMSF, and 1 μg/ml leupeptin. The lysate was clarified by centrifugation at 23,426 g for 30 min and loaded onto a 5 ml HisTrap HP column (Cytiva). After a wash with 250 ml lysis buffer, ORCALRR and ORCAΔWD40 were eluted with elution buffer (50 mM Tris–HCl pH 7.8, 300 mM KCl, 250 mM imidazole, 10% glycerol, 1 mM ß‐mercaptoethanol) and further purified by affinity chromatography using 5–8 ml amylose resin (New England Biolabs). Proteins were eluted with 50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, 20 mM maltose, and 1 mM ß‐mercaptoethanol. Affinity tags were cleaved off by digestion with 4–6 mg His‐tagged TEV protease. Uncleaved protein and TEV were removed by passing the protein solution through a 5 ml HisTrap HP column (Cytiva). Finally, cleaved ORCALRR and ORCAΔWD40 were subjected to gel filtration chromatography on a HiLoad 16/600 Superdex 75 pg column (Cytiva) equilibrated in 50 mM Tris pH 7.8, 300 mM KCl, 10% glycerol, 1 mM DTT, concentrated in 10 K Amicon Ultra‐15 concentrators (Millipore), and flash frozen in liquid nitrogen for storage at −80°C.
Full‐length ORCA, ORCAWD40, ORCAΔLRR/α, and ORCA mutants were expressed in insect cells by infecting 2–4 l High5 cell cultures with ORCA baculoviruses (generated according to the Bac‐to‐Bac protocol, Thermo Fisher Scientific) either alone or by co‐infection with Orc2N baculovirus for ~ 48 h. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, 50 mM imidazole, 1 mM β‐mercaptoethanol, 200 μM PMSF, and 1 μg/ml leupeptin), and lysed by sonication. Soluble proteins were recovered after two rounds of ultracentrifugation at 38,724 g for 45 min and a 30 min precipitation step with 5–10% ammonium sulfate, and loaded onto a 5 ml HisTrap HP column (Cytiva). The HisTrap HP column was washed with 60–100 ml lysis buffer and bound proteins eluted with a 50–250 mM imidazole gradient in 50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, and 1 mM β‐mercaptoethanol. ORCA or ORCA·Orc2N‐containing fractions were subsequently loaded onto an amylose column prepacked with 3.5–5 ml resin. After washing the column with 50 ml wash buffer (50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, and 1 mM β‐mercaptoethanol), bound proteins were eluted in wash buffer supplemented with 20 mM maltose and incubated with 6xHis‐tagged TEV protease for 18–36 h. Subsequently, uncleaved protein, His‐TEV, and affinity tags were removed by nickel affinity chromatography on a 5 ml HisTrap HP column (Cytiva) in 50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, 50 mM imidazole, and 1 mM β‐mercaptoethanol. ORCA and ORCA·Orc2N were then further purified by gel filtration chromatography using a 16/600 HiLoad Superdex 200 pg column (Cytiva) equilibrated in 50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, and 1 mM DTT. Peak fractions for ORCA and ORCA·Orc2N were pooled, concentrated, flash‐frozen in liquid nitrogen, and stored at −80°C. 6x‐His‐MBP‐tagged ORCA was purified in an analogous manner, but TEV digestion and the second nickel affinity chromatography step were omitted. Note that rat ORCA and ORC proteins were used throughout this study because human ORCA is very poorly expressed in several heterologous expression systems.
Except for ORCALRR, all protein concentrations were determined according to the Beer–Lambert equation with NanodropOne (Thermo Fisher Scientific) absorbance readings at 280 nm. Extinction coefficients for each protein construct were calculated based on protein sequence. Due to the lack of tryptophans, ORCALRR protein concentration was measured using the Qubit protein assay kit (Thermo Fisher Scientific).
Expression and purification of recombinant rat Orc2N
Wild‐type and mutant N‐terminal fragments (amino acids 1–100) of RnOrc2 (Orc2N) were cloned into a LIC‐converted, pET‐derived E. coli expression vector (vector 1B, QB3 Macrolab, UC Berkeley) as N‐terminal 6xHis‐TEV fusions. Orc2N constructs were expressed in BL21 RIL E. coli cells cultured in 2xYT medium with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol upon induction with 0.5 mM IPTG for 3 h at 37°C. Two liters of culture per construct were harvested by centrifugation, resuspended in 60 ml lysis buffer (50 mM Tris–HCl pH 7.6, 600 mM KCl, 50 mM imidazole, 10% glycerol, 1 mM β‐mercaptoethanol, 200 μM PMSF, and 1 μg/ml leupeptin), and lysed by sonication. The clarified lysate (by 30 min centrifugation at 23,426 g) was loaded onto a 5 ml HisTrap HP affinity chromatography column (Cytiva), which was washed with 150 ml wash buffer (50 mM Tris–HCl 7.6, 600 mM KCl, 50 mM imidazole, 10% glycerol, and 1 mM β‐mercaptoethanol). Bound proteins were eluted with 250 mM imidazole in wash buffer and further purified by gel filtration on a HiLoad 16/600 Superdex 75 pg column (Cytiva) equilibrated in gel filtration buffer (50 mM Tris–HCl pH 7.6, 600 mM KCl, 10% glycerol, and 1 mM β‐mercaptoethanol). Peak fractions containing 6xHis‐Orc2N were pooled, concentrated, flash‐frozen, and stored at −80°C.
Cloning, expression, and purification of recombinant rat ORC
Full‐length, wild‐type RnORC subunits Orc1‐5 were cloned into a single multibaculovirus expression vector (series 11 vector, QB3 Macrolab, UC Berkeley) by BioBrick assembly. A 6xHis‐tag and a TEV cleavage site were added to the N‐terminus of RnOrc1, while an MBP‐TEV tag was added to the N‐terminus of RnOrc4. Multibaculovirus vectors containing Orc2 with V14A, D19A, and I26A mutations (Orc2V14A,D19A,I26A), Orc2 lacking amino acids 11–28 (Orc2Δ11–28), or Orc1 lacking amino acids 1–425 encompassing the BAH domain and the intrinsically disordered region (Orc1ΔN) were also generated. For LLPS experiments, Orc1FL or Orc1ΔN425 fused to a C‐terminal msfGFP tag were used for multibaculovirus construction.
Bacmids were generated in DH10Bac cells and used to transfect Sf9 cells for baculovirus production according to the Bac‐to‐Bac protocol (Thermo Scientific Fisher, catalog number 12659017). Two to four liters of High5 cell cultures (Thermo Scientific Fisher, catalog number B85502) were infected for 48 h with high‐titer multibaculovirus and harvested by centrifugation at 500 g for 20–30 min. 35 ml lysis buffer (50 mM Tris–HCl pH 7.8, 300 mM KCl, 50 mM imidazole, 10% glycerol, 200 μM PMSF, 1 μg/ml leupeptin, and 1 mM β‐mercaptoethanol) per liter High5 cells were used to resuspend cells that were then lysed by sonication. The lysate was clarified by a combination of ultracentrifugation at 38,724 g for 45 min, precipitation with 20% ammonium sulfate, and a second round of ultracentrifugation at 38,724 g for 45 min. RnORC was purified by nickel and amylose affinity chromatography using a 5 ml HisTrap HP column (Cytiva) and 2.5–4 ml amylose column (New England Biolabs), respectively. Nickel and amylose columns were washed with 50–60 ml lysis buffer or amylose wash buffer (50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, and 1 mM β‐mercaptoethanol), and proteins were eluted with a 30 ml 50–250 mM imidazole gradient (from HisTrap HP column) or 20 ml of 20 mM maltose (from amylose column), both in 50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, and 1 mM β‐mercaptoethanol buffer. Affinity tags (except for msfGFP) were cleaved off by incubation of RnORC with 2–4 mg TEV protease overnight, followed by another nickel affinity step using a 5 ml HisTrap HP column (Cytiva) to remove TEV. Concentrated RnORC was subsequently loaded onto a Superose 6 Increase 10/300 GL, HiPrep 16/60 Sephacryl S‐400 HR, or HiLoad 16/600 Superdex 200 pg gel filtration column equilibrated in 50 mM Tris–HCl pH 7.8, 300 mM KCl, 10% glycerol, and 1 mM DTT or in 25 mM HEPES‐KOH pH 7.6, 500 mM potassium glutamate, 10% glycerol, and 1 mM DTT. RnORC peak fractions were concentrated, flash‐frozen in liquid nitrogen, and stored at −80°C.
Expression and purification of Xenopus histones
Wild‐type recombinant X. laevis histones H2A, H2B, H3, and H4, as well as H4K20C mutant histone, were expressed in BL21 RIL or BL21 (DE3) pLysS cells and purified from inclusion bodies under denaturing conditions as described previously (Luger et al, 1999a, 1999b). Unfolded histone proteins (in 7 M guanidinium hydrochloride, 20 mM Tris–HCl pH 7.5, 10 mM DTT) were loaded onto a HiPrep 26/10 desalting column (Cytiva) equilibrated in purification buffer (7 M urea, 20 mM Tris–HCl pH 7.8, 150 mM NaCl, 1 mM Na‐EDTA, 5 mM β‐mercaptoethanol). Pooled histone fractions were then added to HiPrep 16/10 Q FF and Hiprep 16/10 SP FF columns (Cytiva) connected in series. Columns were washed with purification buffer, after which the HiPrep 16/10 Q FF column was removed and the protein eluted from the Hiprep 16/10 SP FF column by gradually increasing the NaCl concentration from 150 mM to 1 M. Purified histones were dialyzed three times against 4 liters of MilliQ water with 2 mM β‐mercaptoethanol and lyophilized for storage at −80°C. H4K20C histone was purified using a similar protocol except that the NaCl concentration in the purification buffer was reduced to 50 mM.
MLA installation on H4K20C
The methyl‐lysine analog (MLA) approach described previously (Simon et al, 2007; Simon & Shokat, 2012) was used to generate histone H4 trimethylated on lysine 20 (H4K20cme3, referred to as H4K20me3 throughout the manuscript). Ten milligrams of lyophilized histone H4K20C were dissolved in alkylation buffer (1 M HEPES‐NaOH pH 7.8, 4 M guanidinium chloride, 10 mM d/l‐methionine, and 20 mM DTT) and 100 mg of (2‐bromoethyl) trimethylammonium bromide (Sigma‐Aldrich) were added to the reaction. After 2 to 2.5 h incubation at 50°C, DTT was added to a final concentration of 10 mM with further incubation at 50°C for another 2–2.5 h. Subsequently, the 1 ml reaction was quenched by the addition of 50 μl 14.2 M β‐mercaptoethanol (Sigma‐Aldrich) and purified on a PD‐10 column (Cytiva) pre‐equilibrated with 3 mM β‐mercaptoethanol in MilliQ water. H4Kc20me3 was lyophilized and stored at −80°C.
Successful installation of trimethyl‐lysine analogs on H4K20C was confirmed by intact protein mass spectrometry as a shift of approximately +86 Da. Lyophilized H4K20C and H4Kc20me3 were dissolved in LCMS‐grade water with 0.1% formic acid and analyzed using a 6550 iFunnel Q‐TOF LC/MS system (Agilent Technologies) with a Dual AJS electrospray ion source with direct injection at a flow rate of 0.150 ml/min in 90% water and 10% acetonitrile mobile phase. Prior to data acquisition, the system was tuned to the 3,200 m/z mass range with positive‐ion polarity in extended dynamic range (2GHz) instrument mode. Data analysis was performed with the Bioconfirm module of the Agilent MassHunter Qualitative Analysis B.07.00 software (Agilent Technologies).
Reconstitution of histone octamers
Unmodified and H4K20me3‐histone octamers were reconstituted and purified as described previously (Luger et al, 1999a, 1999b). Lyophilized histones were dissolved in unfolding buffer (20 mM Tris–HCl pH 7.5, 6 M guanidinium hydrochloride, and 10 mM DTT), mixed at a final concentration of 1 mg/ml with a 1.1‐ to 1.2‐fold excess of H2A and H2B compared to H3 and H4, and dialyzed three times against 2 l of refolding buffer (10 mM Tris–HCl pH 7.5, 2 M NaCl, 1 mM EDTA pH 8, 5 mM β‐mercaptoethanol, or 5 mM DTT). The histone octamer was further purified by gel filtration chromatography on a HiLoad 16/600 Superdex 200 pg gel filtration column in refolding buffer. Peak fractions containing equimolar ratios of all four histones were pooled, concentrated, and stored at 4°C.
Nucleosomal DNA purification
Widom 601 DNA for reconstitution of mononucleosomes was obtained by large‐scale purification of a plasmid containing 32 copies of Widom 601 DNA (Lowary & Widom, 1998). Six liters of NovaBlue E. coli cells transformed with plasmid DNA were grown in 2xYT media with 50 μg/ml kanamycin for 24 h. Plasmid DNA was released by alkaline lysis and further purified on a 600 ml Sepharose 6 column (Cytiva) in TES2000 (50 mM Tris–HCl pH 8.0, 2 M KCl, 1 mM EDTA pH 8). Fractions containing plasmid DNA were pooled, precipitated with 0.5 volumes isopropanol for 30 min at 22°C, and digested overnight at 37°C with 5,000 units EcoRV in Cutsmart buffer to release Widom 601 DNA inserts. Subsequently, Widom 601 DNA was separated from the vector backbone by precipitation with PEG 6000 and concentrated by ethanol precipitation. Pellets were resuspended in 10 mM Tris–HCl pH 7.5 and 1 mM EDTA pH 8, and stored at 4°C.
Nucleosome array DNA containing 12 repeats of Widom 601 DNA separated by a 30 bp linker was obtained by amplifying plasmid pWM‐12x601‐30bplinker (a kind gift from Mike Rosen) in HB101 E. coli cells grown in 2xYT with 100 μg/ml ampicillin. Cells were harvested from 5 l of culture grown for 23 h and plasmid DNA was isolated with the PureLink HiPure Expi Gigaprep Kit (Thermo Fisher Scientific) according to the manufacturer's instruction. The 12x601 DNA array was released by digestion with 2,000 units EcoRV for 18 h at 37°C and further purified by precipitation with 5–6% PEG 6000. Array DNA was stored in 10 mM Tris–HCl pH 8.0, and 0.1 mM EDTA pH 8.
Reconstitution of mononucleosomes and nucleosome arrays
Nucleosome core particles were assembled by mixing histone octamers and DNA at a 1.1:1 molar ratio at 4 μM DNA concentration in 2 M KCl, followed by stepwise dialysis against 200 ml high‐salt buffer (20 mM Tris pH 7.5, 1.4 mM KCl, 0.1 mM EDTA pH 8, 1 mM DTT). The salt concentration was lowered in a stepwise manner by the addition of low‐salt buffer (20 mM Tris pH 7.5, 10 mM KCl, 0.1 mM EDTA pH 8, 1 mM DTT) to 1.2 M KCl, 1.0 mM KCl, 0.8 M KCl, and 0.6 mM KCl over a duration of ~ 20 h. Afterward, the reconstitution was dialyzed twice against 200 ml low‐salt buffer. The nucleosome concentrations were determined by measuring DNA absorbance at 260 nm.
Reconstitution of nucleosome arrays was initiated by mixing histone octamer with 12x601 array DNA at 3 μM of 601 DNA repeats, equimolar amounts of histone octamer, and 1 μM MMTV buffer DNA in 2 M KCl. Excess H2A/H2B dimer was also added to reconstitution reactions to a final concentration of 0.6–1.5 μM, respectively, to avoid hexasome incorporation into nucleosome arrays. Reconstitution reactions were dialyzed against 1 l high‐salt buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 2 M KCl, 1 mM DTT) for 1 h, after which the KCl concentration was gradually lowered during salt gradient dialysis by adding ~ 1.8 l low‐salt buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 200 mM KCl, 1 mM DTT) at a flow rate of 0.9 ml/min. Subsequently, the dialysis buffer volume was reduced to 500 ml and the KCl concentration was further lowered by gradually adding ~ 800 ml no‐salt buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM DTT). In a final dialysis step, nucleosome arrays were dialyzed for 4–5 h against 500 ml buffer containing 10 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, and 1 mM DTT and then purified by two rounds of precipitation with 4 mM MgCl2 and stored in 10 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, and 1 mM DTT. Successful reconstitution of nucleosome arrays was confirmed by restriction digest with BsiWI (cuts in Widom 601 repeat) or NlaIII (cuts in 30 bp linker DNA) in Cutsmart buffer and analysis of products by agarose gel electrophoresis (after deproteinization with proteinase K) or native 1% agarose/2% polyacrylamide gel electrophoresis. Concentrations of nucleosome arrays were determined by measuring the absorbance of DNA at 260 nm. Throughout the manuscript, array concentrations are given as concentrations of individual nucleosomes or DNA repeats within the array.
Crystallization and structure determination of rat ORCAWD40 ·Orc2N
For crystallization and structure determination of rat ORCAWD40·Orc2N, a selenomethionine (SeMet)‐derivatized protein complex was expressed and purified from High5 cells grown in media lacking methionine but supplemented with 2 mM l‐cysteine (Sigma‐Aldrich). High5 cells were infected with baculoviruses encoding 6xHis‐TEV‐tagged RnOrc2N (amino acids 1–100) and 6xHis‐MBP‐TEV‐tagged RnORCAWD40 for 48 h before cell harvesting. Twenty‐four hours after infection, selenomethionine (Sigma‐Aldrich) was added to a final concentration of 100 μg/ml. Purification of selenomethionine‐derivatized ORCAWD40·Orc2N was performed as described for other ORCA·Orc2N assemblies above, except that the Superdex 200 gel filtration chromatography was performed in buffer composed of 20 mM HEPES‐NaOH pH 7.6, 150 mM NaCl, and 0.5 mM TCEP and that 0.5 mM TCEP was added to all purification buffers. We note that the ORCAWD40 crystallization construct contains point mutations K646R/T647R at the C‐terminus that escaped detection during cloning and sequencing analysis but that do not interfere with Orc2 binding.
The rat ORCAWD40·Orc2N complex was crystallized by vapor diffusion in sitting drop format. Crystals grew as clusters of small plates within 12–14 days at 19–22°C upon combining 1.5 μl of ORCAWD40·Orc2N complex at a concentration of 8 mg/ml (in 20 mM HEPES‐NaOH pH 7.6, 150 mM NaCl, and 0.5 mM TCEP) with an equal volume of reservoir solution containing 1.2 M ammonium sulfate and 100 mM MES pH 5.3. These crystals were used for streak seeding into fresh 3 μl drops containing 5 mg/ml protein complex and reservoir solution (1.2 M ammonium sulfate and 100 mM MES pH 5.3) to obtain larger crystals, which grew to their maximum size within 5–7 days. Crystals were cryoprotected by exchanging the mother liquor to 0.1 M ammonium sulfate, 40% ethylene glycol, and 0.1 M MES pH 5.3, harvested by looping, and flash‐frozen in liquid nitrogen.
Crystal screening and data collection were performed at the Xo6DA‐PXIII beamline at the Swiss Light Source (Villigen PSI, Switzerland), which is equipped with a PILATUS 2 M‐F detector and multi‐axis PRIGo goniometer. SeMet single‐wavelength anomalous dispersion (SAD) datasets were collected from a crystal that diffracted to greater than 2 Å at a wavelength of λ = 0.979092 Å with an oscillation range of 0.2 degrees and Χ‐angles ranging from 0 to 90° at 5° increments. Diffraction data were processed with XDS, and reflection intensities merged with the CCP4 program AIMLESS (Kabsch, 2010; Winn et al, 2011). Three SeMet datasets collected from the same crystal at different Χ‐angles on the PRIGo goniometer were merged for further processing. ORCAWD40·Orc2N crystals belonged to space group C 2 2 21, with unit cell dimensions of a = 91.02 Å, b = 135.43 Å, and c = 78.57 Å.
Phases were determined experimentally by single‐wavelength anomalous dispersion. Five SeMet sites were identified with HYSS in PHENIX and used for phase calculation (Adams et al, 2010; Liebschner et al, 2019). Density modification resulted in an electron density map that was of sufficient quality for automated de novo model building using PHENIX AUTOBUILD (Adams et al, 2010; Liebschner et al, 2019). The model was improved by iterative rounds of refinement in PHENIX (real space, individual xyz, individual B‐factors, occupancies, and TLS) with subsequent manual model rebuilding in COOT (Emsley et al, 2010). The final model has excellent geometry (MOLPROBITY; Chen et al, 2010; Williams et al, 2018, score of 1.15, Table 1) and contains ORCAWD40 and residues 11–28 of Orc2.
Reconstitution and cryo‐EM structure determination of an ORCA·H4K20me3‐nucleosome complex
A complex containing full‐length RnORCA, the N‐terminal 100 residues of RnOrc2 (co‐purified with ORCA as described above), and an H4K20me3‐mononucleosome was obtained by mixing ORCAFL·Orc2N and nucleosomes at a molar ratio of 2.5 to 1 (final 10 μM ORCAFL·Orc2N and 4 μM nucleosome) in 20 mM HEPES‐KOH pH 7.5, 50 mM KCl, and 1 mM DTT. 300 μl of the reconstitution reaction were incubated on ice for 30 min and subsequently crosslinked for 10 min by adding an equal volume of crosslinking buffer (20 mM HEPES‐KOH pH 7.5, 50 mM KCl, 1 mM DTT, and 0.1% glutaraldehyde). Crosslinking was stopped by the addition of 60 μl quenching solution with 900 mM Tris pH 7.5, 9 mM lysine, and 9 mM aspartate. 300 μl of crosslinked complex were purified by gel filtration chromatography on a Superose 6 Increase 10/300 GL column (Cytiva) equilibrated in reconstitution buffer. Peak fractions were concentrated to an absorbance of 10 at 280 nm for cryo‐EM grid preparation.
Cryo‐EM grids were prepared by applying 3.5 μl of concentrated ORCAFL·Orc2N·H4K20me3‐nucleosome complex on a 300‐mesh R1.2/1.3 UltrAuFoil grid (Quantifoil Micro Tools GmbH), which had been glow discharged in a PELCO easiGlow system (Ted Pella) for 25 s at 25 mA. The sample was allowed to adsorb to grids for 30 s and then plunge frozen at 4°C in liquid ethane using a Vitrobot Mark IV (Thermo Fischer Scientific). Cryo‐EM data were collected using Serial EM (Mastronarde, 2005) on a 300 kV Titan Krios G2 transmission electron microscope (Thermo Fischer Scientific) equipped with a post‐GIF K3 direct detector in super‐resolution mode at a nominal magnification of 105,000× and a physical pixel size of 0.832 Å. A multi‐hole exposure scheme was applied with 3 × 3 holes imaged per stage position. Movies (35 frames) were recorded at a defocus range of 1–2.2 μm using a total electron dose of 51 electrons per Å2.
Super‐resolution cryo‐EM movies were binned twofold and motion corrected with MotionCor2, followed by estimation of contrast transfer function (CTF) parameters with GCTF (Zhang, 2016; Zheng et al, 2017). Particles were auto‐picked in RELION in a template‐free manner and imported into CryoSPARC, which was used for all subsequent processing steps (Fernandez‐Leiro & Scheres, 2017; Punjani et al, 2017, 2020; Zivanov et al, 2018; Punjani & Fleet, 2021). Particles were subjected to 2D classification and those in poorly defined classes were discarded. Cleaned particles were used for ab initio reconstruction and subsequent heterogeneous refinement against 10 initial volumes. Two classes resembling nucleosomes were kept for further processing by several rounds of homogeneous 3D refinement and 3D classification using a focus mask encompassing weak ORCA density to increase ORCA occupancy and resolve ORCA flexibility. The final set of particles was subjected to CTF refinement and non‐uniform refinement, and the resulting 3D volume was post‐processed using DeepEMhancer (Sanchez‐Garcia et al, 2021).
Model building was initiated by docking crystal structures of ORCAWD40·Orc2N (this study) and of the nucleosome (PDB 3LZ0; Vasudevan et al, 2010) into the cryo‐EM map, followed by several rounds of manual rebuilding in COOT and real‐space refinement in PHENIX (using secondary structure and Ramachandran restraints for protein, and base pair and base stacking restraints for DNA; Adams et al, 2010; Afonine et al, 2018). The final model has excellent geometry as indicated by a MOLPROBITY score of 1.56 (Table 3; Williams et al, 2018).
Structure analysis
Structure analysis was performed with UCSF Chimera, UCSF Chimera X, and PyMOL (Pettersen et al, 2004; Goddard et al, 2007, 2018; The PyMOL Molecular Graphics System, Version 1.8.2.0 Schrödinger, LLC). Multiple sequence alignments were performed with MAFFT and conservation scores were calculated using CONSURF (Katoh et al, 2005; Katoh & Toh, 2008; Ashkenazy et al, 2010; Yariv et al, 2023). Sequence logos were calculated with WebLogo3 (Crooks et al, 2004). Figures were rendered in UCSF Chimera X and PyMOL (Pettersen et al, 2004; Goddard et al, 2007, 2018; The PyMOL Molecular Graphics System, Version 1.8.2.0 Schrödinger, LLC).
Identification of Orc2 and ORCA orthologs and multiple sequence alignments
Orc2 and ORCA orthologs across the metazoan kingdom were identified using the human and rat Orc2 and ORCA protein sequences (UniProt identifiers Q13416, Q75PQ8, Q9UFC0, and A0A140UHX1) as a query in NCBI blastp searches. In the case of ORCA, candidate ORCA orthologs outside the phylum chordata, particularly those that lack an LRR domain, were tested in reverse blast searches against the human proteome database to ensure they are true homologs of ORCA. In addition, blastp searches with human EED (UniProt identifier O75530), which is similar in structure and protein sequence to ORCA, were done to avoid misassignment of EED orthologs as ORCA. Protein hits from initial blastp searches that retrieved proteins other than ORCA with high confidence or that were more similar to EED than ORCA are unlikely true ORCA orthologs and were excluded from further analysis. We note that several ORCA orthologs identified in Mollusca (Gastropoda and Cephalopoda) and most in Arthropoda appear to lack an LRR domain, while ORCA orthologs in Platyhelminthes are embedded in very long protein sequences (1,800–2,500 aa) that in few cases contain other functional domains. Using the above criteria, no orthologs were identified in Nematoda or Rotifera, and many classes or subclasses within the phylum Arthropoda.
In vitro pulldown assays
The interaction of RnORCA with wild‐type or mutant RnOrc2N was examined by affinity pulldowns. Reactions were set up at 18.5 μM Orc2N and 10 μM MBP‐tagged ORCAFL or MBP alone in 50 mM Tris–HCl pH 7.6, 300 mM KCl, 10% glycerol, and 1 mM β‐mercaptoethanol. After incubation on ice for 30 min, 35 μl of each binding reaction were added to 30 μl amylose bead slurry (New England Biolabs). Amylose beads were washed three times with 1 ml binding buffer and bound proteins were eluted with 20 μl binding buffer supplemented with 20 mM maltose. Input and eluted proteins were analyzed by SDS–PAGE and stained with Coomassie brilliant blue.
RnORCA binding to RnORC containing wild‐type or mutant Orc2 was analyzed in pulldown assays using MBP‐tagged ORCA as bait. MBP‐ORCA and ORC were mixed at 0.5 μM each in 25 mM HEPES‐KOH pH 7.5, 300 mM potassium glutamate, 10% glycerol, 10 mM magnesium acetate, and 1 mM DTT. 45 μl of binding reactions were incubated with 25 μl amylose bead slurry for 20 min on ice. Beads were then washed twice with 1 ml binding buffer and proteins eluted in 30 μl 25 mM HEPES‐KOH pH 7.5, 300 mM potassium glutamate, 10% glycerol, 10 mM magnesium acetate, 20 mM maltose, and 1 mM DTT and detected by SDS–PAGE and Coomassie staining.
Peptide array binding assays
Modified histone peptide arrays (Active Motif) containing 384 unique histone modification combinations were used to examine binding of RnORCA to histone peptides. Arrays were blocked at room temperature for 1 h in a solution of 5% non‐fat dry milk in TBST (10 mM Tris–HCl pH 7.5, 150 mM NaCl, and 0.05% Tween20). After three 5‐min washes in TBST, arrays were equilibrated in binding buffer (50 mM Tris–HCl pH 7.6, 300 mM KCl, 10% glycerol, 1 mM β‐mercaptoethanol, and 0.4 mg/ml BSA) and incubated overnight at 4°C with MBP‐tagged ORCAFL, ORCAWD40, or ORCAWD40:W294A at a final concentration of 2–4 μM in binding buffer. Arrays were washed three times with binding buffer and incubated with an HRP‐conjugated anti‐MBP monoclonal antibody (New England Biolabs, RRID: AB_1559738) at a dilution of 1:4,000 in wash buffer (50 mM Tris–HCl pH 7.6, 300 mM KCl, 10% glycerol, 1 mM β‐mercaptoethanol, 0.05% Tween 20) for 8 h at 4°C. Arrays were then washed three times in wash buffer for 5 min, incubated with ECL reagent (BioRad), and imaged in an Azure C400 chemiluminescence reader. Data were analyzed with Active Motif's Array Analyze Software.
Electrophoretic mobility shift assays (EMSAs)
Binding affinities of different RnORCA or RnORCA·Orc2N proteins to H4K20me3‐nucleosomes were determined using electrophoretic mobility shift assays (EMSAs). EMSA experiments were performed in 20 μl reactions containing twofold serially diluted ORCA or ORCA·Orc2N ranging from 8 μM to 31.25 nM and 50 nM mononucleosome in binding buffer (20 mM HEPES‐KOH pH 7.5, 70 mM KCl, 1 mM DTT, and 6.25% sucrose). After 1‐h incubation on ice, 5 μl of each reaction were loaded onto a 5% native polyacrylamide gel and bound and unbound nucleosomes separated at 200 V for 2.5 h in 0.5 × Tris‐Borate‐EDTA buffer (TBE) at 4°C. Gels were stained with ethidium bromide and imaged in a Gel Doc EZ Imager (BioRad). Band intensities of unbound nucleosomes were quantified with ImageJ and used to calculate the percentage of ORCA‐bound nucleosomes. Data from at least three independent experiments were plotted as a function of ORCA concentration and fit to the Hill equation in GraphPad Prism 9 to determine apparent dissociation constants (Kd,app).
Binding of ORCA or ORCA·Orc2N to nucleosome arrays was determined similarly as described for mononucleosomes except that the nucleosome concentration (or 601 DNA repeat concentration) was reduced to 20 nM and ORCA‐bound and unbound nucleosome arrays were separated by electrophoresis in native 1% agarose/1.5% polyacrylamide gels (APAGE) run for 1.5 h at 100 V at 4°C. APAGE gels were stained with 1:50,000 dilution of SYBR Gold and imaged in a Typhoon 5 laser scanner (Cytiva) using LD488 laser and 525BP20 filter settings.
Note that ORCA proteins harboring a WD40 repeat domain were copurified with Orc2N for increased expression and stability.
Pelleting assays of nucleosome arrays
Unmodified or H4K20me3‐nucleosome arrays were incubated at 100 nM repeat concentration with increasing amounts of ORCA or ORCA·Orc2N complexes (increasing twofold from 31.25 nM to 8 μM, or at 0.5 and 8 μM) in 20 mM HEPES‐KOH pH 7.5, 70 mM KCl, and 1 mM DTT for 1 h on ice. Magnesium chloride was then added to each 15 μl reaction to a final concentration of 4 mM. Reactions were incubated at 22°C for 30 min, followed by 10 min centrifugation at 20°C at 21,300 g. Supernatants were transferred to a fresh Eppendorf tube, and proteins in both supernatants and pellets were digested with proteinase K (0.1 mg/ml proteinase K/0.2% SDS final concentrations) for 30 min at 37°C. Nucleosome array DNA in supernatants and pellets was separated in 1% agarose gels containing ethidium bromide and visualized using a Gel Doc EZ Imager (BioRad). Band intensities in supernatants and pellets from at least three independent experiments were quantified with ImageJ and normalized to input array DNA run on each gel for comparison. Again, we note that ORCA proteins harboring a WD40 repeat domain were copurified with Orc2N for increased expression and stability.
For examining co‐pelleting of ORCA with nucleosome arrays, unmodified and H4K20me3‐nucleosome arrays were incubated at 0.5 μM (refers to each nucleosome repeat) with 1 μM ORCAFL·Orc2N in 20 mM HEPES pH 7.5, 100 mM KCl, and 1 mM DTT for 1 h on ice. Afterwards, MgCl2 was added to 4 mM and reactions kept at 22°C for 30 min and then centrifuged at 21,300 g for 10 min. To analyze ORCAFL·Orc2N recruitment to preformed chromatin oligomers, nucleosome arrays were first incubated at 1 μM in 20 mM HEPES pH 7.5, 100 mM KCl, 1 mM DTT, and 4 mM MgCl2, for 30 min at 22°C prior to adding an equal volume of ORCAFL·Orc2N to 1 μM concentration, or buffer only. Reactions were incubated for 1 h at 22°C before centrifugation. Proteins in supernatants and pellets were analyzed by SDS–PAGE and stained with Coomassie brilliant blue.
Confocal microscopy of ORCA‐chromatin droplets
The effect of RnORCA·Orc2N complexes (ORCAFL:WT·Orc2N, ORCAFL:W294A· Orc2N, ORCAWD40:WT·Orc2N, ORCAWD40:W294A·Orc2N, and ORCAFL:liMut· Orc2N) on condensation of nucleosome arrays was examined by confocal fluorescence microscopy of chromatin droplets stained with YOYO‐1. Unmodified and H4K20me3‐nucleosome arrays were mixed at 100 nM per repeat with 4 μM ORCA·Orc2N in 20 mM HEPES pH 7.5, 100 mM KCl, and 1 mM DTT for 1 h on ice to allow protein binding. An ORCA concentration of 4 μM was chosen to be in a range that supports ORCAWD40 binding to H4K20me3‐nucleosome arrays (see Fig 3E) and that is in line with prior experiments studying chromatin condensation (Gibson et al, 2019). Subsequently, MgCl2 and YOYO‐1 were added to 4 mM and 45 nM, respectively, to induce oligomerization of nucleosome arrays. 20 μl of each sample were transferred to a passivated 384‐well plate with glass bottom and imaged after 60 min incubation at 22°C. For localization of ORCA, an N‐terminal mCherry fusion of full‐length ORCA bound to His‐tagged Orc2N was added to 10% of the final ORCA concentration, which was incubated with nucleosome arrays either before (Fig 6A–C) or after Mg2+‐induced chromatin oligomerization (Fig EV4A and B).
Passivation of 384‐well plates was performed as described in Gibson et al (2019). The 384‐well glass bottom plates (Cellvis) were washed five times with MilliQ water and treated with 2% Hellmanex solution (Hellma) for 2–3 h at room temperature. Wells were then rinsed three times with MilliQ water, treated with 0.5 M NaOH for 30 min, and washed again with MilliQ water three times. mPEG‐silane (Creative PEGworks) dissolved in 95% ethanol at 20 mg/ml was added to each well and incubated overnight (> 18 h) in a humidified container protected from light. mPEG‐silane solution was removed by 10 washes with 95% ethanol and 3–5 washes with MilliQ water. Afterward, plates were air‐dried, sealed, and stored for future use. Immediately prior to adding samples, wells were incubated with 60 μl 100 mg/ml BSA per well for at least 30 min, followed by three washes with MilliQ water.
Chromatin condensates formed in the presence of various ORCA·Orc2N assemblies were imaged in an inverted laser scanning confocal Zeiss LSM880 microscope with a 100 × oil objective (1.46 numerical objective aperture) using the ZEN v2.1 software (Zeiss). Image acquisition was performed with 488 nm (to excite YOYO‐1) and 561 nm (to excite mCherry, if applicable) lasers. Images were saved as czi file format with metadata.
Confocal fluorescence microscopy of ORC recruitment to chromatin droplets
To analyze the ability of RnORCA to recruit RnORC into condensed chromatin, nucleosomal arrays were first allowed to oligomerize into chromatin condensates at 400 nM repeat concentration in 20 mM HEPES‐KOH pH 7.5, 150 mM KCl, 1 mM DTT, and 4 mM MgCl2. DAPI was added to a concentration of 25 nM for subsequent visualization of arrays. After 30 min incubation at room temperature, an equal volume of ORCOrc1FL, ORCOrc1ΔN425, ORCAFL (without Orc2N), or premixed ORC and ORCA were added to achieve final concentrations of 100 nM for ORC (50% unlabeled and 50% GFP‐labeled Orc1) and 400 nM for ORCA (75% unlabeled and 25% mCherry‐labeled ORCA). ORC and/or ORCA were incubated with nucleosomal arrays for 1 h at 22°C in a passivated 384‐well plate with glass bottom prior to imaging.
Chromatin droplets in the presence of ORC and ORC/ORCA were collected with a 100 × oil immersion objective (1.49 numerical objective aperture) using a spinning disk confocal Nikon Ti2 inverted microscope (Yokogawa CSU‐W1) equipped with a Hamamatsu ORCA‐Fusion Gen‐III sCMOS camera. Fluorophores were excited using 405 nm (for DAPI), 488 nm (for GFP), and 561 nm (for mCherry) lasers. Images were acquired using the Nikon Elements AR software (Nikon), saved in nd2 file format with metadata, and further processed using ImageJ/FIJI and MATLAB (Schindelin et al, 2012). Exposure times and gain settings were kept constant for each channel between equivalent experiments.
Chromatin droplet image analysis
Acquired raw images (~ 20) were combined in ImageJ/FIJI into a .tiff stack for further processing using custom‐written MATLAB scripts for measuring droplet size and intensity distributions. Briefly, all stack images were first pre‐processed by applying a 4 × 4 median filter to downweigh noise in the image, normalization, and binarization into black and white using Otsu's global thresholding method (Otsu, 1979). The built‐in MATLAB functions “imclearborder”, “bwareafilt”, and “regionprops” were used for removing boundary objects, eliminating small clusters close to the diffraction limit or noise, and extracting image features for measuring area, centroid, as well as major and minor axes lengths. For images with very small clusters close to the optical diffraction limit (~ 300 nm), manual intensity thresholding was used instead of Otsu's global threshold. Each cluster in the binary image after pre‐processing represented a condensate.
For droplet size measurement, major and minor axes lengths determined by MATLAB “regionprops” were averaged to compute the diameter of each droplet. Droplet diameter measurements from all image stacks were stored in an array to plot the size distribution. Size distributions of droplets were plotted and descriptive statistical parameters were calculated using GraphPad Prism 9.
Fluorescence intensity distributions in condensates were used to describe the localization of ORC and ORCA and calculated in MATLAB. Fluorescence intensities for ORC‐GFP and ORCA‐mCherry in the respective channels were radially averaged, and 1D radial profiles were plotted. Briefly, droplets between 2.5 μm and 4 μm diameter were selected for analysis to avoid boundary effects in the intensity distribution for smaller droplets. A polar coordinate binary mask (r, θ) was defined for intensity averaging along the θ‐direction from the center r = 0 to the exterior of the condensate. The radial coordinate (r) was normalized by the radius of the condensate to rnorm. The intensity versus rnorm profiles were binned into equal‐width bins, and mean and standard deviation of the profiles were calculated in MATLAB.
Author contributions
Sumon Sahu: Software; formal analysis; investigation; methodology; writing – review and editing. Babatunde Ekundayo: Resources; formal analysis; investigation; methodology; writing – review and editing. Ashish Kumar: Resources; formal analysis; investigation; writing – review and editing. Franziska Bleichert: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interests.
Supporting information
Appendix
Expanded View Figures PDF
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We are grateful to Kelly Long for help in compiling protein sequences of ORCA and Orc2 orthologs. We thank Kaifeng Zhou, Shenping Wu, and Terence Wu for assistance with cryo‐EM grid screening, data collection, and mass spectrometry, respectively. We also like to thank Lilian Kabeche and lab members for providing access to their spinning disk confocal microscope, and members of the Bleichert lab and Yannick Jacob for critical reading of the manuscript. The plasmid containing the 12x601 Widom sequence array with 30 bp linker was a kind gift from Michael Rosen. This study utilized the following Yale core facilities: Yale CryoEM Resource, West Campus Analytical Core, and the MCDB Imaging Facility on Science Hill. This work was supported by the European Research Council under the European Union's Horizon 2020 research and innovation program (ERC‐STG‐757909 to F.B.), the National Institutes of Health (5R01GM141313 to F.B.), and an Endowed Rudolph Anderson Fellowship from Yale University. B.E.E. was supported by a Swiss National Science Foundation Postdoc.Mobility fellowship (191050). A.K. was supported by a Leslie H. Warner Postdoctoral Fellowship from the Yale Cancer Center. F.B. is a member of the Yale Cancer Center.
The EMBO Journal (2023) 42: e114654
Data availability
The PDB coordinates, structure factors, and cryo‐EM maps have been deposited into the Protein Data Bank and Electron Microscopy Data Bank under the following accession numbers: PDB 8SIU (http://www.rcsb.org/pdb/explore/explore.do?structureId=8SIU) for the RnORCAWD40·Orc2N crystal structure, and PDB 8SIY (http://www.rcsb.org/pdb/explore/explore.do?structureId=8SIY) and EMD‐40522 (http://www.ebi.ac.uk/pdbe/entry/EMD‐40522) for the RnORCAFL·Orc2N·H4K20me3‐nucleosome complex cryo‐EM structure. The code for analyzing LLPS data has been deposited in the Zenodo repository and is available at https://doi.org/10.5281/zenodo.8147618.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD (2018) Real‐space refinement in PHENIX for cryo‐EM and crystallography. Acta Crystallogr D Struct Biol 74: 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben‐Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38: W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T (2010) Nucleosome‐interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb‐containing protein complex in site‐specific DNA replication. Nature 420: 833–837 [DOI] [PubMed] [Google Scholar]
- Beck DB, Burton A, Oda H, Ziegler‐Birling C, Torres‐Padilla ME, Reinberg D (2012) The role of PR‐Set7 in replication licensing depends on Suv4‐20h. Genes Dev 26: 2580–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B (1992) ATP‐dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Bleichert F (2019) Mechanisms of replication origin licensing: a structural perspective. Curr Opin Struct Biol 59: 195–204 [DOI] [PubMed] [Google Scholar]
- Bosco G, Du W, Orr‐Weaver TL (2001) DNA replication control through interaction of E2F‐RB and the origin recognition complex. Nat Cell Biol 3: 289–295 [DOI] [PubMed] [Google Scholar]
- Brustel J, Kirstein N, Izard F, Grimaud C, Prorok P, Cayrou C, Schotta G, Abdelsamie AF, Dejardin J, Mechali M et al (2017) Histone H4K20 tri‐methylation at late‐firing origins ensures timely heterochromatin replication. EMBO J 36: 2726–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers LM, Bednar J, Woodcock CL, Hansen JC (1998) Linker histones stabilize the intrinsic salt‐dependent folding of nucleosomal arrays: mechanistic ramifications for higher‐order chromatin folding. Biochemistry 37: 14776–14787 [DOI] [PubMed] [Google Scholar]
- Chan KM, Zhang Z (2012) Leucine‐rich repeat and WD repeat‐containing protein 1 is recruited to pericentric heterochromatin by trimethylated lysine 9 of histone H3 and maintains heterochromatin silencing. J Biol Chem 287: 15024–15033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Diffley JFX (2022) The initiation of eukaryotic DNA replication. Annu Rev Biochem 91: 107–131 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhall A, Wei S, Fierz B, Woodcock CL, Lee TH, Chatterjee C (2014) Sumoylated human histone H4 prevents chromatin compaction by inhibiting long‐range internucleosomal interactions. J Biol Chem 289: 33827–33837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Koren A (2020) Positive and negative regulation of DNA replication initiation. Trends Genet 36: 868–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N‐terminal tail. J Mol Biol 327: 85–96 [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Schwarz E, Komaromy M, Wall R (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 179: 125–142 [DOI] [PubMed] [Google Scholar]
- Ekundayo B, Bleichert F (2019) Origins of DNA replication. PLoS Genet 15: e1008320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Fernandez‐Leiro R, Scheres SHW (2017) A pipeline approach to single‐particle processing in RELION. Acta Crystallogr D Struct Biol 73: 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar‐Garea A, Boix‐Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K et al (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK (2019) Organization of chromatin by intrinsic and regulated phase separation. Cell 179: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri S, Prasanth SG (2015) Association of ORCA/LRWD1 with repressive histone methyl transferases mediates heterochromatin organization. Nucleus 6: 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri S, Aggarwal V, Pontis J, Shen Z, Chakraborty A, Khan A, Mizzen C, Prasanth KV, Ait‐Si‐Ali S, Ha T et al (2015) The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. Elife 4: e06496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157: 281–287 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon F, Luger K, Hansen JC (2005) The core histone N‐terminal tail domains function independently and additively during salt‐dependent oligomerization of nucleosomal arrays. J Biol Chem 280: 33701–33706 [DOI] [PubMed] [Google Scholar]
- Hansen JC, Maeshima K, Hendzel MJ (2021) The solid and liquid states of chromatin. Epigenetics Chromatin 14: 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Stillman B (2023) Origins of DNA replication in eukaryotes. Mol Cell 83: 352–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Stillman B (1999) Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem 274: 23027–23034 [DOI] [PubMed] [Google Scholar]
- Imai R, Nozaki T, Tani T, Kaizu K, Hibino K, Ide S, Tamura S, Takahashi K, Shribak M, Maeshima K (2017) Density imaging of heterochromatin in live cells using orientation‐independent‐DIC microscopy. Mol Biol Cell 28: 3349–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaremko MJ, On KF, Thomas DR, Stillman B, Joshua‐Tor L (2020) The dynamic nature of the human origin recognition complex revealed through five cryoEM structures. Elife 9: e58622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ (2009) The H4 tail domain participates in intra‐ and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol 29: 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TZE, Wan YCE, Zhang Z, Chan KM (2022) Lrwd1 impacts cell proliferation and the silencing of repetitive DNA elements. Genesis 60: e23475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe‐Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O (2012) The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier‐Gorlin syndrome. Nature 484: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ (2017) Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547: 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Reinberg D (2011) Chromatin higher‐order structures and gene regulation. Curr Opin Genet Dev 21: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Armstrong RL, Duronio RJ, MacAlpine DM (2016) Methylation of histone H4 lysine 20 by PR‐Set7 ensures the integrity of late replicating sequence domains in Drosophila . Nucleic Acids Res 44: 7204–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lam WH, Zhai Y, Cheng J, Cheng E, Zhao Y, Gao N, Tye BK (2018) Structure of the origin recognition complex bound to DNA replication origin. Nature 559: 217–222 [DOI] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ et al (2019) Macromolecular structure determination using X‐rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lu C, Yang Y, Fan Y, Yang R, Liu CF, Korolev N, Nordenskiold L (2011) Influence of histone tails and H4 tail acetylations on nucleosome‐nucleosome interactions. J Mol Biol 414: 749–764 [DOI] [PubMed] [Google Scholar]
- Long H, Zhang L, Lv M, Wen Z, Zhang W, Chen X, Zhang P, Li T, Chang L, Jin C et al (2020) H2A.Z facilitates licensing and activation of early replication origins. Nature 577: 576–581 [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence‐directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999a) Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119: 1–16 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999b) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ (2012) New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Rogge R, Tamura S, Joti Y, Hikima T, Szerlong H, Krause C, Herman J, Seidel E, DeLuca J et al (2016) Nucleosomal arrays self‐assemble into supramolecular globular structures lacking 30‐nm fibers. EMBO J 35: 1115–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V et al (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51 [DOI] [PubMed] [Google Scholar]
- McKay DJ, Klusza S, Penke TJ, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD et al (2015) Interrogating the function of metazoan histones using engineered gene clusters. Dev Cell 32: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Kedziora KM, Song EA, Purvis JE, Cook JG (2022) The consequences of differential origin licensing dynamics in distinct chromatin environments. Nucleic Acids Res 50: 9601–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanailidou P, Taraviras S, Lygerou Z (2020) Chromatin and nuclear architecture: shaping DNA replication in 3D. Trends Genet 36: 967–980 [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres‐Padilla ME, Heard E, Reinberg D (2009) Monomethylation of histone H4‐lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D (2010) Regulation of the histone H4 monomethylase PR‐Set7 by CRL4(Cdt2)‐mediated PCNA‐dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N (1979) A threshold selection method from gray‐level histograms. IEEE Trans Syst Man Cybern 9: 62–66 [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Parker MW, Botchan MR, Berger JM (2017) Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol 52: 107–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Bell M, Mir M, Kao JA, Darzacq X, Botchan MR, Berger JM (2019) A new class of disordered elements controls DNA replication through initiator self‐assembly. Elife 8: e48562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Shen Z, Prasanth KV, Stillman B (2010) Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci USA 107: 15093–15098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, MacAlpine DM (2016) DNA replication origins‐where do we begin? Genes Dev 30: 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Fleet DJ (2021) 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo‐EM. J Struct Biol 213: 107702 [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA (2017) cryoSPARC: algorithms for rapid unsupervised cryo‐EM structure determination. Nat Methods 14: 290–296 [DOI] [PubMed] [Google Scholar]
- Punjani A, Zhang H, Fleet DJ (2020) Non‐uniform refinement: adaptive regularization improves single‐particle cryo‐EM reconstruction. Nat Methods 17: 1214–1221 [DOI] [PubMed] [Google Scholar]
- Sanchez‐Garcia R, Gomez‐Blanco J, Cuervo A, Carazo JM, Sorzano COS, Vargas J (2021) DeepEMhancer: a deep learning solution for cryo‐EM volume post‐processing. Commun Biol 4: 874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ (2019) HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Bleichert F (2020) Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co‐loader Cdc6. Nat Commun 11: 4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3‐K9 and H4‐K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa‐Velazquez IA et al (2008) A chromatin‐wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22: 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC (1996) Reversible oligonucleosome self‐association: dependence on divalent cations and core histone tail domains. Biochemistry 35: 4009–4015 [DOI] [PubMed] [Google Scholar]
- Shen Z, Prasanth SG (2012) Orc2 protects ORCA from ubiquitin‐mediated degradation. Cell Cycle 11: 3578–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG (2010) A WD‐repeat protein stabilizes ORC binding to chromatin. Mol Cell 40: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Chakraborty A, Jain A, Giri S, Ha T, Prasanth KV, Prasanth SG (2012) Dynamic association of ORCA with prereplicative complex components regulates DNA replication initiation. Mol Cell Biol 32: 3107–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren‐Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4‐K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Simon MD, Shokat KM (2012) A method to site‐specifically incorporate methyl‐lysine analogues into recombinant proteins. Methods Enzymol 512: 57–69 [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM (2007) The site‐specific installation of methyl‐lysine analogs into recombinant histones. Cell 128: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrajna A, Goldfarb D, Kedziora KM, Cousins EM, Grant GD, Spangler CJ, Barbour EH, Yan X, Hathaway NA, Brown NG et al (2020) Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res 48: 9415–9432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A (2005) Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev 19: 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E (2007) PR‐Set7‐dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR‐Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Vasudevan D, Chua EYD, Davey CA (2010) Crystal structures of nucleosome core particles containing the '601′ strong positioning sequence. J Mol Biol 403: 1–10 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG et al (2010) Quantitative interaction proteomics and genome‐wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Wang Y, Khan A, Marks AB, Smith OK, Giri S, Lin YC, Creager R, MacAlpine DM, Prasanth KV, Aladjem MI et al (2017) Temporal association of ORCA/LRWD1 to late‐firing origins during G1 dictates heterochromatin replication and organization. Nucleic Acids Res 45: 2490–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB et al (2018) MolProbity: More and better reference data for improved all‐atom structure validation. Protein Sci 27: 293–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A et al (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F et al (2010) Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci USA 107: 19266–19271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Takagi M, Kosako H, Hirano T, Yoshimura SH (2022) Cell cycle‐specific phase separation regulated by protein charge blockiness. Nat Cell Biol 24: 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv B, Yariv E, Kessel A, Masrati G, Chorin AB, Martz E, Mayrose I, Pupko T, Ben‐Tal N (2023) Using evolutionary data to make sense of macromolecules with a “face‐lifted” ConSurf. Protein Sci 32: e4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016) Gctf: real‐time CTF determination and correction. J Struct Biol 193: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kutateladze TG (2019) Liquid‐liquid phase separation is an intrinsic physicochemical property of chromatin. Nat Struct Mol Biol 26: 1085–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Lu X, Hansen JC, Hayes JJ (2005) Salt‐dependent intra‐ and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem 280: 33552–33557 [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. Elife 7: e42166 [DOI] [PMC free article] [PubMed] [Google Scholar]


