Highlights
-
•
The species L. aviarius, L. araffinosus and L. hohenheimius group in a cluster of species related to the intestinal environment of poultry.
-
•
L. aviarius is frequently found in poultry and highly prevalent across chicken faecal samples.
-
•
The pangenome of L. aviarius is closed having a core genome of 1179 gene clusters that codify to structural ribosomal, and biogenesis proteins and envelop biosynthesis.
-
•
The accessory genes are distributed within three clusters of orthologous genes, one of which contains genes of glycoside hydrolases and glycosyltransferases that mediate carbohydrate transport and interactions with the host cells.
-
•
Accessory features such as Crispr/Cas mechanism, mobilome and prophage inclusions are encoded at clusters 2 and 3 and variate amongst strains from different GIT sections.
Keywords: Ligilactobacillus aviarius, Pan-genome, Poultry, Chicken
Abstract
The genus Ligilactobacillus encompasses species adapted to vertebrate hosts and fermented food. Their genomes encode adaptations to the host lifestyle. Reports of gut microbiota from chicken and turkey gastrointestinal tract have shown a high persistence of Ligilactobacillus aviarius along the digestive system compared to other species found in the same host. However, its adaptations to poultry as a host has not yet been described. In this work, the pan-genome of Ligilactobacillus aviarius was explored to describe the functional adaptability to the gastrointestinal environment. The core genome is composed of 1179 gene clusters that are present at least in one copy that codifies to structural, ribosomal and biogenesis proteins. The rest of the identified regions were classified into three different functional clusters of orthologous groups (clusters) that codify carbohydrate metabolism, envelope biogenesis, viral defence mechanisms, and mobilome inclusions. The pan-genome of Ligilactobacillus aviarius is a closed pan-genome, frequently found in poultry and highly prevalent across chicken faecal samples. The genome of L. aviarius codifies different clusters of glycoside hydrolases and glycosyltransferases that mediate interactions with the host cells. Accessory features, such as antiviral mechanisms and prophage inclusions, variate amongst strains from different GIT sections. This information provides hints about the interaction of this species with viral particles and other bacterial species. This work highlights functional adaptability traits present in L. aviarius that make it a dominant key member of the poultry gut microbiota and enlightens the convergent ecological relation of this species to the poultry gut environment.
Graphical abstract

1. Introduction
There has been a growing interest on studying host specificity of the gut microbiome to understand to which extent factors such as diet, environment, host phylogeny and its evolutionary story impact the composition of the gut microbiome (Mallott and Amato 2021). Ecology and evolution play a long-term role shaping microbial dynamics and this idea has supported the emergence of “phylosymbiotic studies” defined as the observance of patterns in microbial communities that summarize and reflects the phylogeny of their host linking the microbiome to the host´s evolutionary story (Lim and Bordenstein 2020). In some mammals, the study of phylosymbiosis has related microbial dispersal to their phylogeny (Amato et al. 2019; Mazel et al. 2018). However, for non-mammalian vertebrate taxa, including birds and amphibians, studies still discuss the presence of phylosymbiosis (Capunitan et al. 2020; Song et al. 2020; Trevelline et al. 2020).
Chicken gastrointestinal colonisation has been described as a stochastic process driven by the contact with environmental microorganisms present in food and water (Diaz Carrasco et al. 2019). It is highly diverse and changes along the gastrointestinal tract (GIT) (Glendinning et al. 2020; Gong et al. 2007). Yet, previous studies typically convey on a constant presence of Enterobacteriaceae and Lactobacillaceae, suggesting that some microorganisms are more persistent to the gastrointestinal environment and are adpated to the physicochemical changes along poultry GIT environment, than others. Studies using 16S rRNA amplicon sequencing and metagenome-assembled genomes showed the dominance of Ligilactobacillus aviarius, Lactobacillus crispatus and Ligilactobacillus salivarius as host-associated commensal lactic acid bacteria in chickens representing 18% of the faecal clone sequences and present in over half of the chicken samples (Feng et al. 2021; Gong et al. 2007).
The genus Ligilactobacillus (formerly referred to as Lactobacillus salivarius group) encompasses 16 motile, homofermentative rod-shaped species with a G+C content of around 32.5 to 43.3 %. Most Ligilactobacillus species are adapted to vertebrate hosts and have been obtained from mammals, reptiles, and amphibians digestive samples (Zheng et al. 2020). Some other species of Ligilactobacillus are also present in fermented foods, silage, and soil (Tohno et al. 2019). Several strains of Ligilactobacillus express urease gene clusters dependant on low pH to mediate the resistance to gastric acids and buffer the environmental changes (Cotter and Hill 2003). L. aviarius has been reported to encode an alpha-glucanotransferase that converts amylose starch into isomalto-/malto-polysaccharides (IMMP) (Kralj et al. 2002; Meng et al. 2018). These highly branched α-glucans linked through α1–4 and α1–6 bonds are resistant to digestion by the host's α-amylase and therefore, represent a supply of soluble dietary fibre (Meng et al. 2018). Such digestibility resistance improves IMMP distribution to the large intestine and serves as a prebiotic source for the bacterial community to ferment and produce short-chain fatty acids (SCFAs) (Al-Khalaifa et al. 2019). Leading to the assumption that the production of IMMP from an active L. aviarius colonisation in the small intestine of birds, improves digestion and represents a potential supply of feed for fermentative bacteria in the lower part of the GIT. Experiments of probiotics co‑feeding mixtures of L. aviarius with L. salivarius and Ligilactobacillus agilis reported an increase of Lactobacilli, a decrease of Escherichia coli on the small intestine lumen of laying hens and an increase on the egg weight and laying performance, favoured by the increase of mucosal absorption and cytokine expression (Hong et al. 2021).
The frequent reports of L. aviarius to poultry highlights its persistence as a well-adapted coloniser of the avian GIT, and some studies agree on its presence as an indigenous Ligilactobacillus species along with L. agilis and L. salivarius (Hong et al. 2021; Lan et al. 2004; Liu et al. 2021). However, the identification approach must be taken cautiously due to the intra-species similarity and recent taxonomic re-assignations (Zheng et al. 2020). Additionally, many works focus on detecting and identifying poultry-associated species, but they lack a functional description (Qiao et al. 2019). Therefore, this work aims to describe the encoded functional adaptability of L. aviarius and the features that might make it host-specific adapted to the poultry gut environment. This is the first comparative genomic analysis for this species, which describes the genomic diversity and functional adaptability of Ligilactobacillus aviarius, not explored previously.
2. Materials and methods
2.1. Phylogenomic analysis of the genus Ligilactobacillus
Reference genomes of the genus Ligilactobacillus (Table S1) were collected from public databases and previous studies. Briefly, 17 reference genomes were downloaded from the National centre for Biotechnology Information (NCBI) database (genomes obtained in August 2022). The genome of Ligilactobacillus hohenheimensis DSM 113,870 was used from our collection (Supplementary material S2) (Rios-Galicia et al. 2023). All reference genomes were phylogenetically placed based on 487 universal markers using Phylophlan 3.0 (Asnicar et al. 2020) and annotated using iTOL (v6.5.8) (Letunic and Bork 2019). Liquorilactobacillus vini (Rodas et al. 2006; Zheng et al. 2020) was used as an external group. A codon usage analysis was run in ATGme (http://atgme.org/) to detect the presence of less common codons.
2.2. Prevalence and abundance of poultry related Ligilactobacillus in chickens
To evaluate the prevalence and abundance of species of Ligilactobacillus related to poultry, 603 DNA sequences obtained from poultry samples were downloaded from the SRArun selector browser of the NCBI database. Samples of crop where obtained from the project PRJEB60928, (n= 48) obtained by our research group (internal communication, not yet public), samples from duodenum, jejunum, ileum, caeca and colorectum where obtained from the project PRJNA417359 (crop=38, duodenum=99, jejunum=98, ileum=97, caeca=99 and colorectum=98) (Huang et al. 2018). Finally, samples from faeces where obtained from the project PRJEB22062 “Gut microbiomes from 359 European pig and poultry herds” (n=159) (Duarte et al. 2021; Luiken et al. 2019; Munk et al. 2018). Details of each samples are listed at the Supplementary material file S1. The collected raw reads were mapped against the genomes of L. aviarius DSM 20,655 (GCA001436315), L. aviarius J01 (GCA947381835), L. hohenheimensis J14 DSM 113,870 (GCA947381805) and L. araffinosus DSM 20,653 (GCA001435375) using CoverM (v0.6.1) (https://github.com/wwood/CoverM).
2.3. Genomes collection and selection of Ligilactobacillus aviarius
Genome assembled sequences (n=21) of L. aviarius available at NCBI database as of July 2022 were collected from the projects PRJEB56193, PRJNA222257, PRJNA316009 and PRJNA377666 (Juricova et al. 2021; Rios-Galicia et al. 2023; Sun et al. 2015). Metagenome-Assembled Genomes (MAGs) with more than 90% completeness available at the Integrated Chicken Reference Genomes and Gene catalog (ICRGGC, https://nmdc.cn/icrggc/) (n = 12) were collected (Feng et al. 2021). All genomes were taxonomically analysed, screened and selected according to the index of completeness (>90%) and no contamination using Anvi'o v7.1 (Eren et al. 2021), and a pairwise genome comparison to discard clonal genomes (dereplication cluster >99% ANI) using dRep 3.2.2 (Olm et al. 2017) retaining 26 genomes.
2.4. Pan-genome analysis
The pan-genome of L. aviarius was calculated in Anvi'o v7.1 (Eren et al. 2021) using the set of 26 genomes. Genomes were pre-treated to discard contigs shorter than 2.5 kb. Gene prediction was made with HMMS included in the workflow of Anvi'o. The resulting pan-genome output was visualised through the interactive interface of Anvi'o. The core genome calculation was performed considering the presence of 90% of single-copy genes and an index of geometric homogeneity of 0.95, obtaining 1179 genes. The predicted annotation of ORFs of each genome was used to identify the number of total genes, core genes, and new genes present in the further pan-genome analysis and the resulting pan-genome output files were visualised using RStudio v1.1.463. Clusters were assigned following the microbial pangenomics workflow of anvi'o (https://merenlab.org/2016/11/08/pangenomics-v2/) that considers Euclidean distance of orthologous genes using linkage Ward method, defined by the gene tree in the centre of the pan-genome. The core genes shared amongst the 26 strains were concatenated and aligned using the workflow for homogeneity indices in pan-genomes of Anvi'o. The tree was visualised with iTOL (Letunic and Bork 2019).
2.5. Phenotype prediction and functional analysis
The annotation file of strains of Ligilactobacillus’ species isolated from small intestine of chicken (Rios-Galicia et al. 2023): L. aviarius J01 (GCA947381835), L. hohenheimensis J14 (GCA947381805), L. saerimneri (GCA947381505) and L. salivarius (GCA947381595) were compared to observe absence and presence of metabolic pathways encoded within the genomes to assess genome reduction within species related to chicken.
Functional analysis and gene absence/presence of the 26 genomes L. aviarius were performed using gene prediction in Anvi'o, to identify the protein domains present in each genome. Differences in presence and absence along the genome were subdivided into four regions (clusters) according to the protein domains present in that region. The identified regions were classified into different functional clusters of orthologous groups (COGs), and the differences amongst each strain were selected and highlighted in a heatmap.
Different functional features of the interaction of L. aviarius with the host were investigated using DRAM annotation (Distilled and Refined Annotation of Metabolism) (Shaffer et al. 2020). Briefly, DRAM annotates contigs using UniRef90 (Suzek et al. 2015), PFAM (Mistry et al. 2021), dbCAN (Yin et al. 2012), RefSeq viral (Brister et al. 2015), VOGDB (https://vogdb.org/), and the MEROPS peptidase database (Rawlings et al. 2018) and curates these annotations into useful functional categories. The results were hand curated to show the positive presence of metabolic pathways and subunits completion.
2.6. Prophage identification
Prediction of prophage genes and regions insertions in the 26 genomes of L. aviarius was performed using PHASTER (Arndt et al. 2019) (www.phaster.ca). The prophage regions were compared against a bacterial and phage/prophage database available within PHASTER by October 2022. The insertion was considered complete when the region contained known phage sequences and when more than >90% of the proteins in the detected regions were associated with known phage sequences. Proteins present within the operon insertion and its distribution were directly exported from PHASTER's website.
2.7. Antiviral defence systems detection
Antiviral systems presence was identified utilising Prokaryotic Antiviral Defence LOCator (PADLOC) (Payne et al. 2022) using HMM-based homologue searches and gene presence/absence/synteny criteria. CRISPR–Cas systems were detected using CRISPR-Cas++ 1.1.2 using the genome assemblies as input (Grissa et al. 2007). The occurrence of defence systems in strains from different origins was compared.
3. Results and discussion
3.1. Phylogenomic analysis of Ligilactobacillus strains related to poultry
The genus Ligilactobacillus encompasses homofermentative organisms frequently associated with the gut environment of different hosts or fermented substrates such as silage, food, or soil. The size of genomes used for the phylogenomic analysis (reference genomes) ranges between 1.4 and 2.3 Mb with a G+C content of 32.5 to 43.3%. The species of L. aviarius, L. araffinosus and L. hohenheimensis (Fig. 1) have been reported on poultry and show a smaller genome size (1.46 Mb) than the average of the genus (1.87 Mb).
Fig. 1.
Phylogeny based on universal marker genes of species belonging to the genus Ligilactobacillus (reference genomes). Reconstruction of a maximum-likelihood tree based on 497 single-copy core protein concatenated sequences. The tree is rooted in Liquorilactobacillus vini DSM 20,605. Bootstrap percentages (1000 replicates) are shown at the branch points. Bar depicts 0.1 substitutions per nucleotide position. Bar plots represent genome size and GC% content: The last column represents the source of isolation of each species described in more detail in table S1 and the asterisks marks a specie that has been reported in chicken.
Such reduction might indicate a reductive adaptation to the gut environment of poultry (genome reduction) (Nayfach et al. 2019). Other species found in poultry like L. saerimneri= 1525,592 bp (GCA944326145) (Plaza Onate et al. 2022) (assembly not considered in the phylogenomic analysis) have a relatively similar genome size, while some other species such as L. agilis = 2168,186 bp (GCA025311455) (PRJNA880302-no publication related), L. salivarius = 1807,598 bp (GCA002159345) (Juricova et al. 2021) and L. animalis = 1940,664 bp (GCA001705475) (PRJNA337943 no publication related), also reported in poultry do not share such size. L. aviarius and L. hohenheimensis lack a complete operon of repair system genes and peptides transport (Supplementary Fig. S1) compared to other Ligilactobacilli, which might partly explain the differences in genome size. The loss of repair DNA genes might be explained by the Proteomic Constraint theory, that propose that in order to minimize mutations (errors), organisms reduce its proteome size leading to a reduction of selection pressure (Garcia-Gonzalez et al. 2012). Amongst the group of poultry-related species, L. hohenheimensis has a relatively high content of G+C (50.1%) compared to the rest of Ligilactobacillus species, with an average of 40%. The particular traits of this species contradict the general understanding that larger bacterial genomes tend to have higher G+C contents (Almpanis et al. 2018). Other factors should be considered to explain the higher G+C content, such as the organism's normal optimal temperature range, a restriction of the genetic code (where encoding certain amino acids requires at least some usage of A/T or G/C), or the presence of rare codons in genes. Although some codons resulted less frequent such as CGG instead of AGA (15.6‰) or CGU (13.2‰), or the stop codon UAG instead of UAA (2.3‰), the presence of rare codons must be taken with care since the codon usage table used to compare was based on the codon usage of L. salivarius available at the databases, a relatively distant member of the genus Ligilactobacillus (Supplementary material S2). Moreover, the genomic GC content of L. hohenheimensis (50%) is less common within the bimodal GC content distribution of bacteria, with peak values either below 45% or above 60%, and 10% higher than the average GC content observed in Firmicutes (Teng et al. 2023). Although all species share the same ancestor and similar fermentation pathways, the clade of L. salitolerans, L. acidipiscis and L. pobuzihii show a bigger genome size (∼2.3 Mb) and have been reported in associations with fermented food and mushroom substrates, which is a different lifestyle from the host associated environment.
3.2. Presence of Ligilactobacillus-related species on poultry
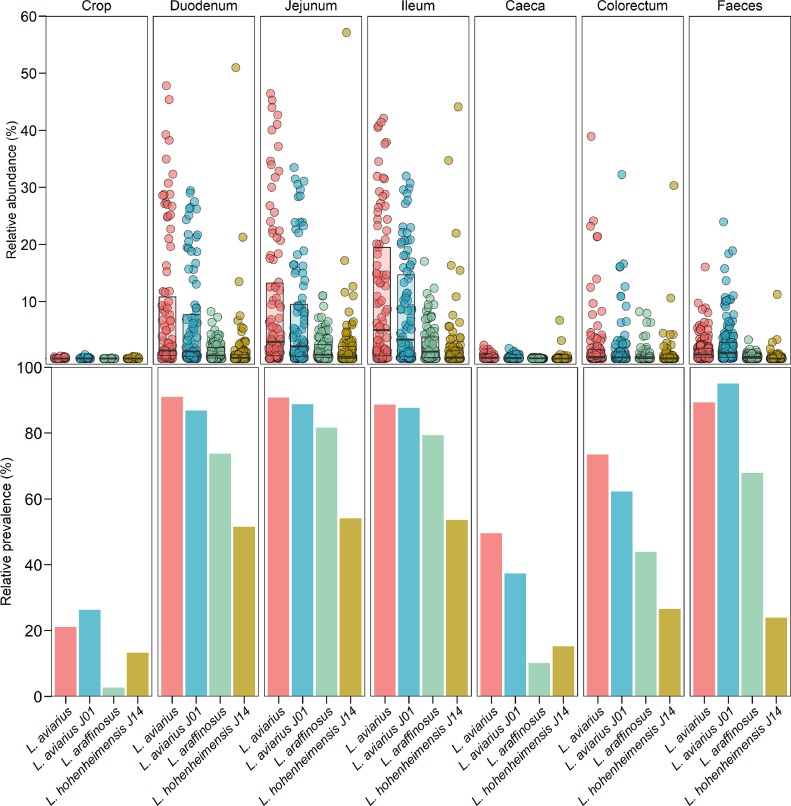
To observe the prevalence and abundance of L. aviarius, L. araffinosus, and L. hohenheimensis as relevant community members in the digestive tract of poultry, its presence was estimated using a large-scale comparison of the genomes against samples of chicken digesta available at public databases. Both strains of L. aviarius (GCA947381835 and GCA001436315) that were detected abundant above 0.1%, depicted a prevalence of at least 80% on duodenum, jejunum, ileum and faecal samples. Its abundance ranged between 0.5 to 40% (Fig. 2). In both cases, our isolate of L. aviarius J01 and the reference genome DSM 20,655, maintained a prevalence between 20 and 60% in crop, colorectum and caeca, where its abundance reduces, suggesting to be a persistant member of the intestinal community with a dominance in small intestine. The presence of L. araffinosus follows the same pattern as L. aviarius along the chicken intestine. However, its presence in crop and caeca depicts lower percentage of prevalence and abundance than the rest of the strains considered in the analysis. This recently separated species from L. aviarius (Zheng et al. 2020) appears to have a bigger sensitivity to the environmental conditions of crop and caeca, dropping its abundance to 0.1%, which highlights its individual environmental adaptations from those of L. aviarius. The lowest prevalence and abundance was detected for L. hohenheimensis DSM 113,870, which presented the highest prevalence at small intestine (50%) and the lowest (10%) in crop and caeca, above L. araffinosus. Previous works have reported similar abundance proportions of L. aviarius and L. hohenheimensis on the upper GIT sections, crop, and ileum (Rios-Galicia et al. 2023). The high occurence of L. aviarius in poultry samples (high abundance and dominance along the GIT) and the availability of genomic/metagenomic assemblies in the databases further supported the functional analyses to understand the adaptability of this species to the chicken's gut environment.
Fig. 2.
Relative abundance and prevalence of species of Ligilactobacillus related to poultry across chicken samples obtained from public databases. Dots represent the relative abundance of each sample (crop=38, duodenum=99, jejunum=98, ileum=97, caeca=99 colorectum=98, and faeces=159) obtained from the projects PRJEB60928, PRJNA417359 and PRJEB22062. Boxplots depict the mean relative abundance of each species across all samples. Barplots represent the relative prevalence of genomes abundant above 0.1%.
3.3. Pan-genome determination of Ligilactobacillus aviarius
The pan-genome of 26 dereplicated genomes of L. aviarius was determined, including 17 publicly available genomes from the NCBI (July 2022) and nine newly sequenced genomes from our bacterial collection (PRJEB56193). From this dataset, three genomes correspond to isolates from the ileum of turkeys, and the others originate from the different intestinal sections of the chicken: six from the jejunum, five from the ileum, three from caeca, and nine isolated from faeces (Table S2). The average genome size was 1.52 Mb with a standard deviation of ± 0.88 considering the size of metagenomic assemblies (MAGs) and of ± 0.48 considering only isolated genomic assemblies. Such difference was considered in the pan-genome analysis lowering the core gene threshold and predicting genes in metagenome mode (included in Anvi'o´s protocol) to attenuate the loss of precision in the analysis (Li and Yin 2022). The number of genes detected in each genome varies by 5.36% (1456 ± 78.18) (mean ± SD), with no correlation observed between the number of genes and the number of contigs nor the length of the contigs, showing that the quality of the genome did not influence the genome annotation and the pan-genome analysis. The pan-genome of L. aviarius cannot be considered closed since the number of pan-genome genes represented against the number of genomes does not reach the plateau. In this analysis, the addition of the last two genomes decreased the number of total genes, and reduced the standard deviation (Fig. 3A). Despite the number of new genes did not increase, and the number of core genes was close to reach a constant with the addition of genomes (Fig. 3B, C), a future addition of novel genomes will be necessary to reach a possible plateau and enrich the pan-genome analysis.
Fig. 3.
Number of core genes and pan-genome of 26 genomes L. aviarius. (A) The number of genes of the pan-genome increases and flattens as a function of the number of genomes included in the analysis while (B) the core genome and (C) number of new genes decreases. Bars depict standard deviation.
The core genome threshold in this analysis was lowered to a presence of 90% of single copy genes shared throughout the dataset. 90% of the genes resulted in 1179 genes (cluster named core90) present at least in one copy in 23 out of 26 of the genomes (Fig. 4). The analysis of strict core genes shared amongst 100% of the genomes reduced the number to 857 core genes (cluster named core). The functional annotation displayed that most core genes are related to basic biological functions such as translation, ribosomal structure, replication, cell wall/envelope biosynthesis, carbohydrate transport, and metabolism, as expected. The rest of the identified regions were classified into three different functional clusters of orthologous groups (COGS) that varies according to the type of genome assembly and source of isolation. The first cluster detected (cluster 1, 58 genes) included genes encoding carbohydrate metabolism and transporters. Carbohydrate metabolism was represented by genes for glycoside hydrolases and sugar transporters of arabinose, lactose, melibiose, maltose, sucrose and starch, together with some glycosyltransferases that are predicted to mediate interactions with the host cell mucosa layer (glucans, dextrins, galactans) and digestive cell receptors. In addition, genes assigned to peptidases were detected in core and cluster 1, which might serve as possible indicators for host-specific bacterial interactions. Further points are discussed in the paragraph 3.5. The distribution of genes assigned to carbohydrate metabolism and transport showed that these genes are constitutive and present in all genomes (core). However, some genes codifying for cellulose and maltodextrin utilization and transportation were exclusively found in cluster 1, representing an individual adaptation. Within these two clusters (core and cluster 1), some peptidase genes such as insulin protease (ptrA2/M16B), bleomycin hydrolase (C01B) and endothelin protease (M13), related to the maturation and degradation of peptide hormones were detected. The presence of these genes might indicate a host-lifestyle adaptation of L. aviarius whose peptidase activity might cross-talk the signalisation of the host. However, no evidence has been demonstrated in chicken. Cluster 2 encompass 41 genes encoding viral defence and replication mechanisms such as the abortive infection strategy (Abi), the Cispr/Cas type IIA system, the defence reverse transcriptase (DRTs) mechanism and the restriction-modification (RM) systems that recognize and cleaves foreign DNA which will be discussed in detail at the accessory features section. This information hints at the species's constant interaction with phages and foreign DNA. Finally, cluster 3 comprehended 16 genes encoding mainly mobilome inclusions (prophages/transposons), viral defence mechanisms, and cell wall envelope biogenesis/transcriptional genes. In general, the repertoire of genes included in all clusters might illustrate the importance of such housekeeping genes to persist in the chicken gut environment.
Fig. 4.
Pan-genome analysis of L. aviarius. Comparative genomic analysis of the 26 non-clonal strains of L. aviarius available in the databases. The inner layers represent individual genomes obtained from chicken (blue) or turkey (orange), arranged according to their phylogenetic relation. Genomes are depicted, organised by clusters of orthologous genes where each colour shadow indicates a cluster: Core genes in orange, Cluster 1 genes in green, Cluster 2 genes in blue, and Cluster 3 genes in yellow. An absence of colour depicts an absence of genes. The origin of each genome (either jejunum, ileum, caeca or faeces)is displayed next to the genome label. The barplots below represent the relative abundance of genes found on each cluster.
3.4. Metabolism of L. aviarius
The species of L. aviarius is a heterotrophic bacteria able to obtain energy and carbon by oxidating glucose and arabinose (pentose) via the Embden-Meyerhof glycolysis and the pentose phosphate cycle. ATP is hydrolysed to generate ion gradients across the membranes by the F-type ATPase unit that couples with H+ transport across a membrane to obtain energy (Fig. 5). Although relevant genes were only partially found, Entner-Doudoroff pathway (glucose-6-phosphate to pyruvate) and citrate cycle might be also important to oxidise carbon and reduce cofactors. Across all genomes, whole clusters of arabinose and chitin metabolism were detected codified, while genes for carbohydrate active enzymes (CAZymes) for cellulose and maltodextrin utilisation (bcsZ and yvdJ) in cluster 1, and mannose, galactose and xylose utilisation (nucleotide sugar metabolism) in cluster 2 were individually found in some genomes. Finally, only three genomes of strains isolated from ileum and one of caeca presented carbohydrate active enzyme genes for the hydrolysis of the plant polysaccharides galactan and mannan. The presence of genes related to carbohydrate transport and metabolism, exclusively found in some genomes (genes clustered in cluster 1) might be an indication of an adaptive nutrient utilisation strategy of L. aviarius within the intestine environment. Their potential expression would provide the strain a metabolic advantage on the use of these sugars at a specific GIT section. However, a clear separation regarding carbohydrate utilisation amongst strains originated from different GIT sections was not observed (Figs. 5 and 6).
Fig. 5.
Metabolic pathways detected along the chicken GIT and across the pan-genome input genomes of L.aviarius. The figure is separated in two parts to highlight pathway completion (superior heatmap) and genes presence/absence (inferior heatmap). Tiles are coloured by pathway coverage according to the scale in the superior right corner. Presence/absence in the second heatmap represent the presence of genes necessary to express a particular process. An absence of colour indicate absence genes.
Fig. 6.
Distribution of carbohydrate metabolism and transport genes codified at the core cluster, cluster 1 or both across all input genomes of L. aviarius originating from four different origins along the chicken GIT.
Genes for acetate production including pta and ackA (Fig. 5) were also detected, these genes convert acetyl-CoA into acetate forming ATP, and present a reversible reaction that depends on the environmental conditions. In nutrient-rich conditions, acetyl-CoA is converted to acetate, while under starving conditions acetyl-CoA is generated allowing the cell to dump the excess of acetylation potential in exchange for ATP formation (Campos‐Bermudez et al. 2010). These genes were constitutively detected in all genomes. Both, lactate and acetate production can contribute to the production of butyrate and propionate when coupled with butyrate cycle via a NAD-independent d-lactate dehydrogenase (d-iLDH) also present within all genomes of L. aviarius (ldh) (Sheridan et al. 2022).
A cysteine biosynthesis gene was present in all genomes, meaning that the species is auxotroph to the rest of amino acids. A dihydrolipoamide dehydrogenase gene to hydrolyse leucine, lysin and methionine was constitutive in all genomes as well as many catalytic peptidases including aspartic peptidases (A08, A24A), cysteine peptidases (C01, C10, C26, C40, C56, C59, C60 C69 and C82), metallo-peptidases (M01, M03, M13, M16, M20, M24, M29, M38, M41, M48, M50, M60, M78, M79), serine peptidases (S01, S08, S09, S11, S12, S14, S15, S16, S24, S26, S33, S54) and threonine peptidase (T01B). The presence of the majority of these genes within the core cluster reflects the essential adaptations that L. aviarius gathers to efficiently colonise and adapt the chicken intestinal environment abundant on organic matter. The additional genes found individually represent a signature element of this strain that might help during niche occupation.
3.5. Functional adaptations of Ligilactobacillus aviarius to the host
Intestinal bacteria have been described to hold some adaptations that help them to survive in the intestine, such as transforming primary bile salts into secondary bile salts, degrading urea as a survival mechanism in acidic conditions, or producing large protein domains involved in extracellular matrix binding (Frese et al. 2011). The pan-genome of Ligilactobacillus aviarius did not encode an urease cluster, and no biliary salt dihydroxylation gene was detected. Its strategy to survive the acidic conditions of the GIT lays on an adaptive presence of an FoF1-type ATP synthase (atp ABCDEFGH) (Fig. 6) present in the core cluster, generating a constant gradient between extracellular and cytoplasmic pH due to protons exclusion. These adaptations have been described on the homofermentative species of Lacticaseibacillus rhamnosus, a well-known human probiotic (Corcoran et al. 2005). Across the pan-genome, different genes related to surface structures that interact with the host epithelia were detected, such as the pilus assembly protein pulE and the ATPase pilB protein domains that provide motility to the cell and might mediate interaction and adherence to the chicken gut epithelia (Castelain et al. 2016). Glycosyltransferases genes related to EPS (galM) and glucan binding domain (yg repeat) (Fig. 6) were detected in all genomic assembled genomes and partially in the metagenomic assemblies, meaning that completeness has to be taken into account in downstream analysis (T. Li and Yin 2022). Expression of genes of mucin hydrolase (peptidase M60) that targets complex host glycoproteins, such as mucins (Bardoel et al. 2012), or peptidase M24 related to collagen recycling, might play a role in the host cross-talk interaction between bacteria and the glycolipids of the host cell surface.
Some of the key genes detected along the genomes that might mediate the interaction of L. aviarius were detected at the core cluster and cluster 1 and encode peptidases that can interact with host molecules such as endothelins (neprilysin/neutral endopeptidases, pepO), insulin (insulinase, ptrA2), collagen (Xaa-Pro-aminopeptidase), mucin (enhancing-like peptidase), anti-inflammatory glycopeptides (bleomycin hydrolases), and peptide hormones maturation (prolyl oligopeptidase) (Fig. 7), enzymes described to be involved in the maturation and degradation of peptide hormones and neuropeptides such as alpha-melanocyte-stimulating hormone, luteinizing hormone-releasing hormone (LH-RH) or insulin (Wei et al. 2020; H. Zhang et al. 2016). Although the sole presence of those genes do not ensure an interaction, a possible expression might be related to a host lifestyle adaptation where the formed molecules, if absorbed, can interact with the host signalling. Being present along all the genomes, these possible interactions would affect the host's biological response and might be the key to the successful colonisation of L. aviarius across the intestine of poultry.
Fig. 7.
Distribution of peptidases with a potential to interact with the host structures and signalisation across all input genomes of L. aviarius originating from four different origins along the chicken GIT. The scale represents the number of gene-copies detected in each genome.
The potential to hydrolyse and assimilate carbohydrates with different chemical natures suggests a dynamic and planktonic lifestyle even in distal regions where absorption and competence would limit carbon intake. Additionally, the dominance and presence across the gut epithelium require fast multiplication rates that seem to be supported by the number of genes involved in replication, envelope biogenesis, and transcription, which represent 30% of total genes.
Studies on the pan-genome of L. salivarius have shown two ways of adaptation within the intestinal environment, either by gaining adhesion abilities or by developing efficient utilization of nutrients (Lee et al. 2017). In the case of L. aviarius, the homogeneous presence of genes for carbon utilisation, enzymes that potentially interact with the host, and its persistence along the whole GIT indicate that despite its narrow genome size, the gene repertoire of L. aviarius provides the necessary tools for efficiently colonise and adapt to the chicken intestinal environment. However, studies of gene expression and interaction are still needed.
3.6. Accessory features of Ligilactobacillus aviarius
The detected accessory genes differed from genome to genome and were mainly found in clusters 2 and 3 of orthologous genes. amongst these inclusions, several genes encoded antiviral defence, such as the abortive system (Abi) that prevents phage maturation by autolyzing the infected cell. In this case, the genes for abiD and abiL were more abundant amongst most of the genomes. Both proteins provide resistance by avoiding the phage's replication cycle completion and interfering with the phage endonuclease (Lopatina et al. 2020). Additional to a suicidal strategy, two enzymatic mechanisms that involve the detection and modification of foreign DNA were detected: the restriction-modification system (RM) that signals endonucleases to degrade double-strand DNA, and the defence reverse transcriptase system (DRTs) that signal retro transcriptase enzymes that suppress the expression of phage genes. Some of these mechanisms have a non-specific activity against specific phages, and might be active against a broad range of foreign DNA at an early stage of the infection after the intrusion of foreign DNA into the cell, before transcription (Gao et al. 2020).
Genes for the adaptive immune system CRISPR CAS type II and the ATPases/protease were detected amongst all strains while antiviral system GAO 19 was found codified only at strains obtained from jejunum and ileum. In this case, the defence locus herA-SIR2 of the protease-helicase system GAO 19 (Gao et al. 2020), might play a protective role on strains that colonise jejunum and ileum (Fig. 8). In general, the diversity of antiviral systems detected in this single species present across the GIT, emphasizes the adaptation plasticity of L. aviarius and the importance of counting on multiple defence mechanisms to succeed within this environment.
Fig. 8.
Distribution of antiviral systems across all input genomes of L. aviarius originating from four different origins along the chicken GIT. Tiles are coloured to indicate the antiviral system to which the protein belongs.
Finally, four genomes were detected to carry complete prophage inclusions (tail, head, capsid, protease, portal, and terminase). All the detected inclusions are detailed in Table S3. Despite the taxonomic distance, these prophages have been found in genomes of Ligilactobacillus equi (Li et al. 2022), a member of the genus Ligilactobacillus that colonises the intestine of horses. Previous studies have shown that the number of prophages (phages in a lysogenic state) might be larger in a healthy gut, which allows for inferring the physiological state of the host by observing the composition and number of prophages (Bakhshinejad and Ghiasvand 2017). Moreover, studies on humans have reported that people with similar diets converge to a similar viral community (Minot et al. 2011). In this sense, the presence of similar prophages in different species of the same genus might result from the availability of similar nutrients and an adaptation of the genus Ligilactobacillus to the intestinal environment. Such inclusions play a key role in the exchange of genetic material and serve as a repository of mobile elements to maintain balance in the GIT.
This work highlights functional adaptability traits in L. aviarius making it a persistent key member of the poultry gut microbiota. The metabolic potential predicted on L. aviarius genomes reflect a constitutive utilization of glucose, arabinose, chitin, acetate and lactate, with the potential to hydrolyse additional plant polysaccharides and nucleotide sugars. The description of some genome signature elements of L. aviarius can improve the optimization of diets for the host, fermentation conditions, implementation of health biomarkers or enhance the production of specific metabolites, specially for a persistent specie along the digestive tract.
The comparison of genomes and identification of orthologous genes of commensal organisms such as L. aviarius, improves search and description of genes and pathways at unexplored microbial communities with similar environmental conditions. This approach facilitates the discovery of novel enzymes, biosynthetic gene clusters, or metabolic capabilities. Additionally, these genes can be used as markers to infer the relatedness of species and their evolutionary divergence, especially to understand dynamics of colonisation at a highly domesticated animal like chicken. Finally, a deep characterisation of proteins that interact with the host such as peptidases that potentially interact with hormone peptides can be utilized in various biotechnological applications, including bioregulation of communities or heterologous expression in other species of interest. In addition, the study and characterization of accessory genes provide information on the environmental dynamics and evidence the adaptation plasticity of L. aviarius to the GIT of poultry. Such characterization and functional descriptions broaden the understanding of the close host-interactions and provide information for further biotechnological applications.
Funding
This work was in part funded by the Rehovot-Hohenheim partnership program.
CRediT authorship contribution statement
Bibiana Rios Galicia: Conceptualization, Methodology, Investigation, Data curation, Validation, Formal analysis, Visualization, Writing – original draft. Johan Sebastian Sáenz: Methodology, Software, Validation, Data curation, Visualization, Writing – review & editing. Timur Yergaliyev: Methodology, Software, Formal analysis, Data curation, Visualization. Amélia Camarinha-Silva: Resources, Validation, Supervision, Writing – review & editing, Funding acquisition. Jana Seifert: Methodology, Investigation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2023.100199.
Contributor Information
Amélia Camarinha-Silva, Email: amelia.silva@uni-hohenheim.de.
Jana Seifert, Email: jana.seifert@uni-hohenheim.de.
Appendix. Supplementary materials
Data availability
Data are available in public databases as given in the main text.
References
- Al-Khalaifa H., Al-Nasser A., Al-Surayee T., Al-Kandari S., Al-Enzi N., Al-Sharrah T., Ragheb G., Al-Qalaf S., Mohammed A. Effect of dietary probiotics and prebiotics on the performance of broiler chickens. Poult. Sci. 2019;98(10):4465–4479. doi: 10.3382/ps/pez282. [DOI] [PubMed] [Google Scholar]
- Almpanis A., Swain M., Gatherer D., McEwan N. Correlation between bacterial G+C content, genome size and the G+C content of associated plasmids and bacteriophages. Microb Genom. 2018;4(4) doi: 10.1099/mgen.0.000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato K.R., J G.S., Song S.J., Nute M., Metcalf J.L., Thompson L.R., Morton J.T., Amir A., V J.M., Humphrey G., Gogul G., Gaffney J., A L.B., G A.O.B., F P.C., Di Fiore A., N J.D., T L.G., Gomez A., Kowalewski M.M., R J.L., Link A., M L.S., Tecot S., B A.W., K E.N., R M.S., Knight R., S R.L. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019;13(3):576–587. doi: 10.1038/s41396-018-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D., Marcu A., Liang Y., Wishart D.S. PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief Bioinform. 2019;20(4):1560–1567. doi: 10.1093/bib/bbx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnicar F., Thomas A.M., Beghini F., Mengoni C., Manara S., Manghi P., Zhu Q., Bolzan M., Cumbo F., May U., Sanders J.G., Zolfo M., Kopylova E., Pasolli E., Knight R., Mirarab S., Huttenhower C., Segata N. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020;11(1):2500. doi: 10.1038/s41467-020-16366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshinejad B., Ghiasvand S. Bacteriophages in the human gut: our fellow travelers throughout life and potential biomarkers of heath or disease. Virus Res. 2017;240:47–55. doi: 10.1016/j.virusres.2017.07.013. [DOI] [PubMed] [Google Scholar]
- Bardoel B.W., Hartsink D., Vughs M.M., de Haas C.J., van Strijp J.A., van Kessel K.P. Identification of an immunomodulating metalloprotease of Pseudomonas aeruginosa (IMPa) Cell. Microbiol. 2012;14(6):902–913. doi: 10.1111/j.1462-5822.2012.01765. [DOI] [PubMed] [Google Scholar]
- Brister J.R., Ako-Adjei D., Bao Y., Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43(Database issue):D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Bermudez V.A., Bologna F.P., Andreo C.S., Drincovich M.F. Functional dissection of Escherichia coli phosphotransacetylase structural domains and analysis of key compounds involved in activity regulation. FEBS J. 2010;277(8):1957–1966. doi: 10.1111/j.1742-4658.2010.07617.x. [DOI] [PubMed] [Google Scholar]
- Capunitan D.C., Johnson O., Terrill R.S., Hird S.M. Evolutionary signal in the gut microbiomes of 74 bird species from Equatorial Guinea. Mol. Ecol. 2020;29(4):829–847. doi: 10.1111/mec.15354. [DOI] [PubMed] [Google Scholar]
- Castelain M., Duviau M.P., Canette A., Schmitz P., Loubiere P., Cocaign-Bousquet M., Piard J.C., Mercier-Bonin M. The nanomechanical properties of Lactococcus lactis pili are conditioned by the polymerized backbone pilin. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0152053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran B.M., Stanton C., Fitzgerald G.F., Ross R.P. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl. Environ. Microb. 2005;71(6):3060–3067. doi: 10.1128/AEM.71.6.3060-3067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Hill C. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003;67(3):429–453. doi: 10.1128/MMBR.67.3.429-453.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Carrasco J.M., Casanova N.A., Fernandez Miyakawa M.E. Microbiota, gut health and chicken productivity: what is the connection? Microorganisms. 2019;7(10) doi: 10.3390/microorganisms7100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A.S.R., Roder T., Van Gompel L., Petersen T.N., Hansen R.B., Hansen I.M., Bossers A., Aarestrup F.M., Wagenaar J.A., Hald T. Metagenomics-based approach to source-attribution of antimicrobial resistance determinants - identification of reservoir resistome signatures. Front. Microbiol. 2021;11 doi: 10.3389/fmicb.2020.601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren A.M., Kiefl E., Shaiber A., Veseli I., Miller S.E., Schechter M.S., Fink I., Pan J.N., Yousef M., Fogarty E.C., Trigodet F., Watson A.R., Esen O.C., Moore R.M., Clayssen Q., Lee M.D., Kivenson V., Graham E.D., Merrill B.D., Karkman A., Blankenberg D., Eppley J.M., Sjodin A., Scott J.J., Vazquez-Campos X., McKay L.J., McDaniel E.A., Stevens S.L.R., Anderson R.E., Fuessel J., Fernandez-Guerra A., Maignien L., Delmont T.O., Willis A.D. Community-led, integrated, reproducible multi-omics with anvi'o. Nat. Microbiol. 2021;6(1):3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Wang Y., Zhu B., Gao G.F., Guo Y., Hu Y. Metagenome-assembled genomes and gene catalog from the chicken gut microbiome aid in deciphering antibiotic resistomes. Commun. Biol. 2021;(1):4. doi: 10.1038/s42003-021-02827-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese S.A., Benson A.K., Tannock G.W., Loach D.M., Kim J., Zhang M., Oh P.L., Heng N.C., Patil P.B., Juge N., Mackenzie D.A., Pearson B.M., Lapidus A., Dalin E., Tice H., Goltsman E., Land M., Hauser L., Ivanova N., Kyrpides N.C., Walter J. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLos Genet. 2011;7(2) doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Altae-Tran H., Bohning F., Makarova K.S., Segel M., Schmid-Burgk J.L., Koob J., Wolf Y.I., Koonin E.V., Zhang F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science. 2020;369(6507):1077–1084. doi: 10.1126/science.aba0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez A., Rivera-Rivera R.J., Massey S.E. The presence of the DNA repair genes mutM, mutY, mutL, and mutS is related to proteome size in bacterial genomes. Front. Genet. 2012;3:3. doi: 10.3389/fgene.2012.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning L., Stewart R.D., Pallen M.J., Watson K.A., Watson M. Assembly of hundreds of novel bacterial genomes from the chicken caecum. Genome Biol. 2020;21(1):34. doi: 10.1186/s13059-020-1947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Si W., Forster R.J., Huang R., Yu H., Yin Y., Yang C., Han Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 2007;59(1):147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhou Z., Yu L., Jiang K., Xia J., Mi Y., Zhang C., Li J. Lactobacillus salivarius and Lactobacillus agilis feeding regulates intestinal stem cells activity by modulating crypt niche in hens. Appl. Microbiol. Biotechnol. 2021;105(23):8823–8835. doi: 10.1007/s00253-021-11606-2. [DOI] [PubMed] [Google Scholar]
- Huang P., Zhang Y., Xiao K., Jiang F., Wang H., Tang D., Liu D., Liu B., Liu Y., He X., Liu H., Liu X., Qing Z., Liu C., Huang J., Ren Y., Yun L., Yin L., Lin Q., Zeng C., Su X., Yuan J., Lin L., Hu N., Cao H., Huang S., Guo Y., Fan W., Zeng J. The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome. 2018;6(1):211. doi: 10.1186/s40168-018-0590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juricova H., Matiasovicova J., Kubasova T., Cejkova D., Rychlik I. The distribution of antibiotic resistance genes in chicken gut microbiota commensals. Sci. Rep. 2021;11(1):3290. doi: 10.1038/s41598-021-82640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj S., van Geel-Schutten G., Rahaoui H., Leer R., Faber E., Van Der Maarel M., Dijkhuizen L. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with α-(1→ 4) and α-(1→ 6) glucosidic bonds. Appl. Environ. Microb. 2002;68(9):4283–4291. doi: 10.1128/AEM.68.9.4283-4291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P.T., Sakamoto M., Benno Y. Effects of two probiotic Lactobacillus strains on jejunal and cecal microbiota of broiler chicken under acute heat stress condition as revealed by molecular analysis of 16S rRNA genes. Microbiol. Immunol. 2004;48(12):917–929. doi: 10.1111/j.1348-0421.2004.tb03620.x. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Han G.G., Kim E.B., Choi Y.J. Comparative genomics of Lactobacillus salivarius strains focusing on their host adaptation. Microbiol. Res. 2017;205:48–58. doi: 10.1016/j.micres.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Yin Y. Critical assessment of pan-genomic analysis of metagenome-assembled genomes. Brief Bioinform. 2022;23(6):bbac413. doi: 10.1093/bib/bbac413. doi:org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu C., Liu Q., Liu W. Comparative genomic analysis reveals intestinal habitat adaptation of Ligilactobacillus equi rich in prophage and degrading cellulase. Molecules. 2022;(6):27. doi: 10.3390/molecules27061867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.J., Bordenstein S.R. An introduction to phylosymbiosis. Proc. R. Soc. B. 2020;287(1922) doi: 10.1098/rspb.2019.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhou Z., Hong Y., Jiang K., Yu L., Xie X., Mi Y., Zhu S.J., Zhang C., Li J. Transplantion of predominant Lactobacilli from native hens to commercial hens could indirectly regulate their ISC activity by improving intestinal microbiota. Microb. Biotechnol. 2021;15(4):1235–1252. doi: 10.1111/1751-7915.13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina A., Tal N., Sorek R. Abortive Infection: bacterial suicide as an antiviral immune strategy. Annu. Rev. Virol. 2020;7(1):371–384. doi: 10.1146/annurev-virology-011620-040628. [DOI] [PubMed] [Google Scholar]
- Luiken R.E.C., Van Gompel L., Munk P., Sarrazin S., Joosten P., Dorado-Garcia A., Borup Hansen R., Knudsen B.E., Bossers A., Wagenaar J.A., Aarestrup F.M., Dewulf J., Mevius D.J., Heederik D.J.J., Smit L.A.M., Schmitt H., consortium E. Associations between antimicrobial use and the faecal resistome on broiler farms from nine European countries. J. Antimicrob. Chemother. 2019;74(9):2596–2604. doi: 10.1093/jac/dkz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallott E.K., Amato K.R. Host specificity of the gut microbiome. Nat. Rev. 2021;19(10):639–653. doi: 10.1038/s41579-021-00562-3. [DOI] [PubMed] [Google Scholar]
- Mazel F., Davis K.M., Loudon A., Kwong W.K., Groussin M., Parfrey L.W. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems. 2018;3(5) doi: 10.1128/mSystems.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Gangoiti J., de Kok N., van Leeuwen S.S., Pijning T., Dijkhuizen L. Biochemical characterization of two GH70 family 4,6-alpha-glucanotransferases with distinct product specificity from Lactobacillus aviarius subsp. aviarius DSM 20655. Food Chem. 2018;253:236–246. doi: 10.1016/j.foodchem.2018.01.154. [DOI] [PubMed] [Google Scholar]
- Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D., Lewis J.D., Bushman F.D. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk P., Knudsen B.E., Lukjancenko O., Duarte A.S.R., Van Gompel L., Luiken R.E., Smit L.A., Schmitt H., Garcia A.D., Hansen R.B. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 2018;3(8):898–908. doi: 10.1038/s41564-018-0192-9. [DOI] [PubMed] [Google Scholar]
- Nayfach S., Shi Z.J., Seshadri R., Pollard K.S., Kyrpides N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568(7753):505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olm M.R., Brown C.T., Brooks B., Banfield J.F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11(12):2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne L.J., Meaden S., Mestre M.R., Palmer C., Toro N., Fineran P.C., Jackson S.A. PADLOC: a web server for the identification of antiviral defence systems in microbial genomes. Nucleic Acids Res. 2022;50(W1):W541–W550. doi: 10.1093/nar/gkac400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza Onate, F., Jeammet, M., Pons, N., Ehrlich, S.D., Estelle, J., & Calenge, F. (2022). MetaChick: characterization of the chicken caecal metagenome by deep shotgun sequencing. Retrieved from: https://doi.org/10.15454/FHPJH5 </Dataset>.
- Qiao H., Shi H., Zhang L., Song Y., Zhang X., Bian C. Effect of Lactobacillus plantarum supplementation on production performance and fecal microbial composition in laying hens. Open Life Sci. 2019;14:69–79. doi: 10.1515/biol-2019-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46(D1):D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Galicia B., Saenz J.S., Yergaliyev T., Roth C., Camarinha-Silva A., Seifert J. Novel taxonomic and functional diversity of bacteria from the upper digestive tract of chicken including eight novel species. Biorxiv. 2023 doi: 10.1101/2023.05.10.540237. [DOI] [PubMed] [Google Scholar]
- Rodas A.M., Chenoll E., Macian M.C., Ferrer S., Pardo I., Aznar R. Lactobacillus vini sp. nov., a wine lactic acid bacterium homofermentative for pentoses. Int. J. Syst. Evol. Microbiol. 2006;56(Pt 3):513–517. doi: 10.1099/ijs.0.63877-0. [DOI] [PubMed] [Google Scholar]
- Shaffer M., Borton M.A., McGivern B.B., Zayed A.A., Rosa La, Sabina L., Solden L.M., Liu P., Narrowe A.B., Rodríguez-Ramos J., Bolduc B., Gazitúa M.C., Daly R.A., Smith G.J., Vik D.R., Pope P.B., Sullivan M.B., Roux S., Wrighton K.C. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48(16):8883–8900. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan P.O., Louis P., Tsompanidou E., Shaw S., Harmsen H.J., Duncan S.H., Flint H.J., Walker A.W. Distribution, organization and expression of genes concerned with anaerobic lactate utilization in human intestinal bacteria. Microb Genom. 2022;8(1) doi: 10.1099/mgen.0.000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.J., Sanders J.G., Delsuc F., Metcalf J., Amato K., Taylor M.W., Mazel F., Lutz H.L., Winker K., Graves G.R. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. mBio. 2020;11(1) doi: 10.1128/mbio.02901-19. 10.1128/mbio. 02901-02919. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Harris H.M., McCann A., Guo C., Argimon S., Zhang W., Yang X., Jeffery I.B., Cooney J.C., Kagawa T.F., Liu W., Song Y., Salvetti E., Wrobel A., Rasinkangas P., Parkhill J., Rea M.C., O'Sullivan O., Ritari J., Douillard F.P., Paul Ross R., Yang R., Briner A.E., Felis G.E., de Vos W.M., Barrangou R., Klaenhammer T.R., Caufield P.W., Cui Y., Zhang H., O'Toole P.W. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015;6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt C. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31(6):926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng W., Liao B., Chen M., Shu W. Genomic legacies of ancient adaptation illuminate GC-content evolution in bacteria. Microbiol. Spectr. 2023;11(1) doi: 10.1128/spectrum.02145-22. e02145-02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohno M., Tanizawa Y., Kojima Y., Sakamoto M., Nakamura Y., Ohkuma M., Kobayashi H. Lactobacillus salitolerans sp. nov., a novel lactic acid bacterium isolated from spent mushroom substrates. Int. J. Syst. Evol. Microbiol. 2019;69(4):964–969. doi: 10.1099/ijsem.0.003224. [DOI] [PubMed] [Google Scholar]
- Trevelline B.K., Sosa J., Hartup B.K., Kohl K.D. A bird's-eye view of phylosymbiosis: weak signatures of phylosymbiosis among all 15 species of cranes. Proc. R. Soc. B. 2020;287(1923) doi: 10.1098/rspb.2019.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Keller C., Li L. Neuropeptides in gut-brain axis and their influence on host immunity and stress. Comput. Struct. Biotechnol. J. 2020;18:843–851. doi: 10.1016/j.csbj.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang L., Yao H., He Y., Wang X., Xu W., Song Z., Yin Y., Zhang X. Streptococcus pneumoniae endopeptidase O (PepO) elicits a strong innate immune response in mice via TLR2 and TLR4 signaling pathways. Front. Cell. Infect. Microbiol. 2016;6:23. doi: 10.3389/fcimb.2016.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wittouck S., Salvetti E., Franz C., Harris H.M.B., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., Watanabe K., Wuyts S., Felis G.E., Gänzle M.G., Lebeer S. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70(4):2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in public databases as given in the main text.