Abstract
Chicken meat spoilage is a significant concern for food safety and quality, and this study aims to predict the spoilage point of chicken breast meat through various attributes and metabolites. Chicken meat was stored in anaerobic packaging at 4 °C for 13 days, and various meat quality attributes (pH, drip loss, color, volatile basic nitrogen [VBN], total aerobic bacteria [TAB], and metabolites) were examined. First, the spoiled point (VBN >20 mg/100 g and/or TAB >7 log CFU/g) of the chicken breast meat was determined. Using univariate and multivariate analyses, twenty-four candidate metabolites were identified. A receiver operating characteristic (ROC) analysis was used to validate the obtained binary logistic regression model using nine metabolites (proline, methionine, glutamate, threonine, acetate, uridine 5′-monophosphate, hypoxanthine, glycine, and glutamine). The results showed a high area under the ROC curve value (0.992). Thus, this study confirmed the predictability of spoilage points in chicken breast meat through these nine metabolites.
Keywords: Chicken breast, Meat quality, Spoilage point, Metabolomics, Prediction model, Pathway analysis
Graphical abstract
Highlights
-
•
NMR-based metabolites were used to detect the spoilage point of chicken breast meat.
-
•
Prediction model was created by data augmentation and binary logistic regression.
-
•
Pro, Met, Glu, Thr, Ace, UMP, Hyp, Gly, and Gln were selected to predict spoilage.
-
•
Prediction model has validated via ROC curve with AUC value of 0.992
1. Introduction
Chicken, as representative white meat, contains abundant amino acids, essential fatty acids, vitamins, and minerals (Mussa et al., 2022). It is a healthy food with a high percentage of unsaturated fatty acids (Sujiwo et al., 2018; Jung et al., 2022; Bae et al., 2014). However, because of its high percentage of unsaturated fatty acids, as well as sufficient water and dense nutrients, chicken can spoil quickly (Sujiwo et al., 2018). The use-by date of a food item refers to the period within which it must be eaten or thrown away to avoid spoilage (Kim et al., 2018). This period is calculated based on the spoilage point of each food type.
Indicators for the spoilage point of chicken meat include physicochemical, microbial, and metabolic measures such as torrymeter, total aerobic microbial counts (TAB), volatile basic nitrogen (VBN) content (Sujiwo et al., 2019). Particularly, VBN and TAB have been used as general spoilage standard because they highly correlated with sensory properties (Bekhit et al., 2021). The VBN value increase due to the breakdown of proteins by microbial growth, resulting in the production of ammonia and amino acids (Bekhit et al., 2021). However, some studies have reported that VBN values and TAB do not always accurately represent meat spoilage (Lee et al., 2018; Seleshe and Kang, 2021). Different microbial compositions can affect the production of VBN. For example, some microorganisms, such as lactic acid bacteria, do not always correlate with spoilage (Seleshe and Kang, 2021). Therefore, there is a need to verify various indicators for estimating the spoilage points of meat.
Metabolomics, which involves the study and classification of metabolites produced by biochemical reactions in cells, has been used in foodomics with respect to food quality and safety (Jung et al., 2022, Kim et al., 2021). Metabolites in meat can be changed by growth of spoilage microorganisms or endogenous enzymatic reactions (Kim et al., 2020; Wang et al., 2021; Hambrecht et al., 2005; Bae et al., 2014). One study used metabolomics to identify spoilage markers of chicken meat through multivariate analysis, which included amino acids, biogenic amines, and organic acids (Rukchon et al., 2014). In addition, metabolites can be used to distinguish between fresh chicken and frozen/thawing using selected biomarkers through multivariate analysis (Kim et al., 2021, Kim et al., 2022). Therefore, metabolomics can be an effective technique for identifying fingerprints related to biological and chemical changes related to meat spoilage.
As various metabolites can contribute to the meat spoilage system, a proper predictive model using them is necessary. It is established through biomarker selection, data augmentation, and a mathematical model application (Gandhi et al., 2022; Jang et al., 2020). Biomarker selection was performed using univariate and multivariate analyses (Gandhi et al., 2022; Lee et al., 2022; Jung et al., 2022). For example, univariate analysis has been used for t-test and multivariate analysis has been used for principal component analysis, partial least squares discriminant analysis, and random forest regression (Gandhi et al., 2022; Lee et al., 2022; Vinaixa et al., 2012; Jung et al., 2022). Overfitting may occur when establishing a predictive model using insufficient sample data (Deng et al., 2021). Thus, data augmentation is applied to selected biomarkers to prevent overfitting of the prediction model (Moreno-Barea et al., 2022). Then, the mathematical model can be applied for prediction, such as regression models (Deng et al., 2021; Jang et al., 2020; Moreno-Barea et al., 2022; Mundform et al., 2011), which contribute to predicting the spoilage of meat.
The main purpose of this study was to establish a prediction model for the spoilage point of chicken breast meat using changes in the metabolites and mathematical models. First, the quality characteristics of chicken meat during storage were analyzed to determine the spoilage points. Based on the determined spoilage point, metabolites were selected using univariate and multivariate analyses. Finally, a predictive equation was established using binary logistic regression equations with data augmentation.
2. Materials and methods
2.1. Sample preparation
Five packages (1 kg/package) of commercial broiler chicken (Ross strain) breast fillet (Pectoralis major) were purchased on the same day from the same slaughterhouse within 24 h of slaughter (Maniker Co., Ltd., Dongduchen, Korea). They were transported to the laboratory in an ice cooler. A total of 40 (N = 40) chicken breast fillets were individually vacuum packaged in polyethylene/nylon bags (oxygen permeability of 4.7 g/m2 for 24 h at 100% RH/25 °C). Each packaged fillet was weighed individually and stored at 4 °C for up to 13 d (0, 1, 3, 5, 7, 9, 11, and 13 d; n = 5). On the day of the analysis, the drip was removed from each packaged fillet and weighed for drip loss measurements. After drip loss and color were evaluated, the chicken breast fillet was minced using a chopper (CH180, Kenwood Appliances Co., Ltd., Dingguan, China). pH and total aerobic bacteria (TAB) assays were conducted on the same day. The remaining chopped samples were weighed, vacuum-packed, and stored at −70 °C until further analysis for volatile basic nitrogen (VBN), and nuclear magnetic resonance (NMR).
2.2. Physicochemical characteristics and total aerobic microbial counts of chicken breast meat during storage
2.2.1. Drip loss
Drip loss in chicken breast meat was determined as described by Bae et al. (2014). To determine the drip loss, each chicken breast fillet was weighed before packaging. After storage, the fillets were weighed again, and drip loss was calculated as follows:
2.2.2. Color
Color analysis of the surface color of chicken breast meat was performed using a chromameter (CR-310, Konica Minolta, NJ, USA), following the methodology outlined by Bae et al. (2014). A calibration plate was used for standardization prior to the analysis (Y = 92.8, x = 0.3134, y = 0.3193). The results were read three times per sample after a blooming period (30 min). Average lightness (L*), redness (a*), and yellowness (b*) were calculated from the readings.
2.2.3. pH
One gram of chicken breast meat was homogenized (T25 basic, Ika Co., Staufen, Germany) in 9 mL of distilled water and centrifuged (2265×g for 10 min at 4 °C). This procedure was slightly modified based on the methodology described by Lee et al. (2022). The supernatant was filtered using a Whatman No. 4 filter paper. The pH was measured using a pH meter (Seven2Go S2, Mettler-Toledo International Inc., Schwerzenbach, Switzerland).
2.2.4. TAB counts
TAB in chicken breast meat was determined as described by Bae et al. (2014). Minced chicken breast meat (3 g) was diluted with 27 mL of sterile saline (0.85% NaCl) for 2 min using a stomacher (BagMixer 400 P; Interscience Ind., St. Nom, France). Appropriate dilutions were spread on plate count agar (Difco Laboratories, USA), and agar plates were incubated at 37 °C for 48 h. Microorganisms were counted, and the results were expressed as log CFU/g.
2.2.5. Volatile basic nitrogen (VBN)
The VBN content of chicken breast meat was measured according to Lee et al. (2022) using the Conway micro-diffusion technique. Chicken breast meat (5 g) was homogenized in 20 mL distilled water for 30 s. The homogenate was filtered through Whatman No. 1 filter paper. The filtrate (1 mL) and 1 mL of K2CO3 were added to the outer space of the Conway tool (Sibata Ltd., Sitama, Japan), and 1 mL of 0.01 N H3BO3 was added to the inner space with Conway's reagent (0.066% methyl red: 0.0066% bromocresol green, 1:1; v/v) to the inner space, and then sealed the Conway tool with grease. The sealed Conway was incubated at 25 °C for 1 h. Following incubation, the samples were titrated with 0.01 N NaOH after incubation. The VBN values were expressed as follows:
where a is the titration volume of 0.01 N HCl (mL) in the sample and b is the titration volume of 0.01 N HCl (mL) in the blank. F is the standardization index of 0.01 N NaOH, and S is the sample weight (g).
2.3. Metabolites of chicken breast extract, identification, and quantification using nuclear magnetic resonance (NMR)
2.3.1. Extract metabolites of chicken breast meat for NMR analysis
Metabolite extraction was performed using the method described by Kim et al. (2022), with slight modifications. Chicken breast meat (5 g) was thawed at 4 °C for 24 h before analysis. After thawing, the samples were homogenized at 1720×g for 30 s with 20 mL of 0.6 M perchloric acid. The homogenate was centrifuged (Continent 512R; Hanil Co., Ltd., Incheon, Korea) at 3000×g for 20 min at 4 °C. After centrifugation, the supernatant was transferred to a fresh test tube. Each supernatant sample was neutralized with potassium hydroxide. The neutralized samples were centrifuged (3000×g for 20 min at 4 °C) and filtered using Whatman No. 1 filter paper. The filtrate samples were lyophilized (Freezer dryer 18, Labco Corp., Mo, USA) and reconstituted using 1 mM 3-(trimethylsilyl)propionic acid-2,2,3,3-d4 (internal standard, TSP) in D2O (pD 7.4, 20 mM phosphate buffered). The samples were vortexed and stored in a water bath at 37 °C for 10 min. Thereafter, the solution was centrifuged (3000×g, 20 min, 4 °C) to eliminate insoluble substances. The supernatant was loaded into a 5 mm NMR tube for NMR acquisition.
2.3.2. NMR acquisition and metabolites concentration
According to Kim et al. (2019), the NMR data were recorded at 298 K on a Bruker 850 MHz NMR spectrometer in D2O (Bruker Biospin GmbH, Baden-Wuttemberg, Germany). The standard zg30 pulse sequence was used to analyze 1D 1H NMR in Topspin 4.1.1 (Bruker). Pulse sequences were obtained using 64 K data points, sweep width of 17,007.803 Hz, and 128 scans. TSP resonance was used for the chemical shifts (δ). Baseline corrections were performed manually. The Chenomx deconvolution program and heteronuclear single quantum coherence (HSQC) were used to qualify metabolites. Qualification of metabolites was performed by mixing all samples (quality control samples) and measuring the 2D 1H–13C HSQC spectra. The measured 2D spectra of the peaks were identified based on the biological magnetic resonance bank BMRB (bmrb.wise.edu) and human metabolome database (HMDB; hmdb.ca). HSQC was performed with 2 K data points in the t2 domain and 512 increments in t1, each with 8 and 32 scans, respectively. The spectral widths were 12.0016 ppm for the f2 dimension and 180.0045 ppm for the f1 dimension. Coupling constant values of 145 Hz were employed to set the delay duration for the short-range correlations. The peaks identified by 2D HSQC NMR were quantified using 1H NMR spectroscopy. The quantified dataset from the 1H NMR spectrum of each metabolite was processed using the Topspin 4.1.1. The internal standard for metabolite quantification was 1 mM TSP. Quantification of the samples was performed with five replicates. Metabolite concentrations were quantified using the following equation:
2.3.3. Multivariate analysis and statistical analysis by storage period
Meat quality values and quantified metabolites were statistically analyzed using analysis of variance (ANOVA) in SAS software (Version 9.4, SAS Institute Inc., Cary, NC, USA). Differences among the means by storage date were assessed using Tukey's multiple comparison test. Statistical significance was set at P < 0.05. The results of quantified metabolites were analyzed by multivariate analysis (principal component analysis [PCA] and correlation analysis) using MetaboAnalyst 5.0 (www.metaboanalyst.ca). Before the PCA and correlation analysis, the integrated data were log10 transformed and auto scaled. PCA analysis of metabolite samples was performed to distinguish the differences according to storage days. A loading plot of the PCA was used to confirm the contribution to the distinction between storage days. Correlation analysis of the metabolite samples was performed using Pearson's correlation coefficient, and a heat map was used to visualize the relationships between each metabolite. To interpret the correlation, a rule of thumb was applied to the Pearson correlation coefficient (r); for |r|, 0–0.5 a weak correlation, 0.5–0.7 a high correlation and 0.7–1.0 a very high correlation (Jung et al., 2019).
2.4. Selection of biomarker and establishment of predictive model for spoilage point
Metabolite results from each sample were divided into fresh group = 0 (n = 20) and spoiled group = 1 (n = 20) based on the freshness standard, which was VBN 20 mg/100 g or TAB 7.0 log CFU/g (Sujiwo et al., 2018; Kim et al., 2018). Univariate analysis was performed using Student's t-test with Benjamini–Hochberg False Discovery Rates (FDR) correction (set to 0.05) to identify differentially regulated metabolites between the fresh and spoiled groups (Benjamini & Hochberg, 1995). After cut-off, metabolites were normalized by log10 transformation and auto-scaling for subsequent multivariate analysis and receiver operating characteristic (ROC) curve (Moreno-Barea et al., 2022; Deng et al., 2021). Various multivariate analyses (PLS-DA, partial least squares-discriminant analysis; RF, random forest) were conducted to select preliminary biomarkers using MetaboAnalyst 5.0. By calculating the weighted sum of the PLS regression coefficients, we identified significant buckets with the most outstanding contribution to cluster segregation in PLS-DA, and assigned their metabolites (Moreno-Barea et al., 2022). The RF technique combines many decision trees constructed by classifying each tree and voting for popular classes by bootstrap sampling (Percival et al., 2021). The number of trees selected was 5000 (Gandhi et al., 2022). One-third of the samples were excluded from bootstrapping during the tree construction. Small clusters of input information were randomly used as nodes to construct a simple RF with a random function. A classification and regression approach was used to grow each tree.
Before establishing the prediction formula, data augmentation was performed to prevent the overfitting of the model. Data augmentation was performed using the Monte Carlo simulation technique, and 1000 samples were randomly generated on each storage day based on a normal distribution (Jang et al., 2020). One thousand repetitions are commonly used in Monte Carlo analyses to produce stable results (Mundform et al., 2011). Afterward, 70% of the data were randomly selected for the binary logistic regression (BLR) model training data and the remaining 30% were used for validation. Finally, BLR was applied to select preliminary biomarkers using SPSS software (SPSS Inc., USA). A step-wise algorithm was used in the Binary Logistic Regression (BLR) model to iteratively select the most predictive variables based on statistical criteria. Predicted regression models were validated using ROC curves with area under the curve (AUC) values.
2.5. Pathway analysis
To identify metabolite pathways related to spoilage points of chicken breast meat, the selected metabolites as biomarkers were assessed by pathway analysis using MetaboAnalyst 5.0, based on the estimated spoilage point on day 7. Pathway analysis was conducted using the metabolite results on days 0 and 7 to determine whether the types of pathways were changed during the initial 7 days of storage. Afterward, metabolite results on days 7 and 13 were used to confirm the affected metabolite pathway after spoilage. The results of the metabolomic pathway were sorted by important values (-log P value > 1 and impact value > 0). The sorted pathways were matched with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database to visualize the pathways.
3. Results and discussion
3.1. Physicochemical and microbial characteristics of chicken breast during storage
Meat color is an essential quality parameter that influences consumer choice (Sujiwo et al., 2018; Jung et al., 2022). The storage period significantly increased lightness (L* value) and decreased redness (a* value) and yellowness (b* value) (Fig. 1a and b, P < 0.05). Changes in meat color have been reported to occur because of the oxidation of myoglobin, which converts myoglobin to metmyoglobin, turning the meat brown (Sujiwo et al., 2018). Jung et al. (2022) reported that an increase in exudate during storage might affect meat brightness. Therefore, meat color is influenced by oxidation of myoglobin and increased drip loss.
Fig. 1.
Change in quality characteristics of the chicken breast during storage at 4 °C (N = 40, n = 5). The changes in appearance (a), color changes by CIE value (b), pH value (c), cumulative drip loss (d), total aerobic bacterial count (TAB) (e), and volatile basic nitrogen (VBN) (f) (mg/100 g) of chicken breast during storage. a-f Means with different letters indicate significant differences (P < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
pH is related to the freshness of the chicken, as it reflects the degree of protein degradation and accumulation of spoilage metabolites (Lee et al., 2022). However, as shown in Fig. 1c, the pH value of chicken breast meat ranged from 6.10 to 6.16 during storage, with no significant differences among storage days in the present study.
Refrigeration may affect the structure of proteins and the amount of drip exudation (Hong et al., 2015). The cumulative drip loss increased with storage over time (Fig. 1d). On days 9, 11, and 13, the cumulative drip loss percentage was significantly higher (P < 0.05) than that on the first day.
VBN is an indicator of meat quality. A higher VBN value indicates increased microbial spoilage because VBN compounds, such as ammonia and biogenic amines, are formed mainly by the action of microorganisms and proteolytic enzymes (Lee et al., 2022). Meat with a VBN value of 20 mg/100 g or higher is considered spoiled meat (Sujiwo et al., 2018). In this study, the VBN values of chicken breast meat were significantly increased by storage, and the VBN value exceeded 20 mg/100 g on day 7 (Fig. 1g, P < 0.05).
The initial TAB count (day 0) was 4.10 log CFU/g (Fig. 1e). The storage period significantly affected the TAB count (P < 0.05). After day nine, the TAB count exceeded 7.0 log CFU/g. Meat with a microbial count more than 7.0 log CFU/g is considered spoiled by the International Food and Microbiology Commission (Kim et al., 2018). Additionally, the change in microbial composition of chicken breast meat on days 0, 7, and 13 was confirmed by 16S rRNA analysis (Fig. S1; Table S1). As the storage period increased, the relative abundances of Carnobacterium, Lactococcus, Serratia, and Hafnia tended to increase, while those of Acinetobacter and Chryseobacterium species tended to decrease (Fig. S1). Carnobacterium is a type of lactic acid bacteria frequently appearing as the predominant species in meat, fish, and dairy products (Leisner et al., 2007). Lactococcus is a type of lactic acid bacteria that can contribute to protein hydrolysis, including protein degradation, peptide transport, peptide degradation, and amino acid catabolism (Vesanto et al., 1996). It produces organic acids via anaerobic metabolism (Wang et al., 2021). Serratia is an abundant microorganism in moist environments that can grow under refrigerated conditions (Bhadra et al., 2005). Hafnia species area thermophilic microorganisms found in seafood, meat, and dairy products (Li et al., 2019). Among them, Hafnia alvei has been reported to inhibit the growth of other microorganisms without altering the pH or lactic acid concentration (Delbès-Paus et al., 2013). In this study, the composition of lactic acid microorganisms such as Carnobacterium and Lactococcus increased; however, there was no significant difference in pH during storage (Fig. 1c). This could be explained by the volatile nitrogenous compounds produced by Carnobacterium, Lactococcus, Serratia, and Hafnia (Bhadra et al., 2005; Li et al., 2019; Vesanto et al., 1996) (Fig. 1g). In addition, the increased decarboxylase secreted by these microorganisms may be partially responsible for the increase in the VBN value.
Therefore, the quality of chicken breast meat decreased during storage for 13 days and, in particular, day 7 was considered the spoilage point because it exceeded the freshness standard for VBN value (>20 mg/100 g; MFDS, 2022). The samples were divided into the fresh group (0, 1, 3, and 5 day) and the spoiled group (7, 9, 11, and 13 day). Afterward, we examined the metabolites during the storage period and analyzed the correlation between each metabolite.
3.2. Metabolites identification and multivariate analysis of metabolites in the chicken breast during storage
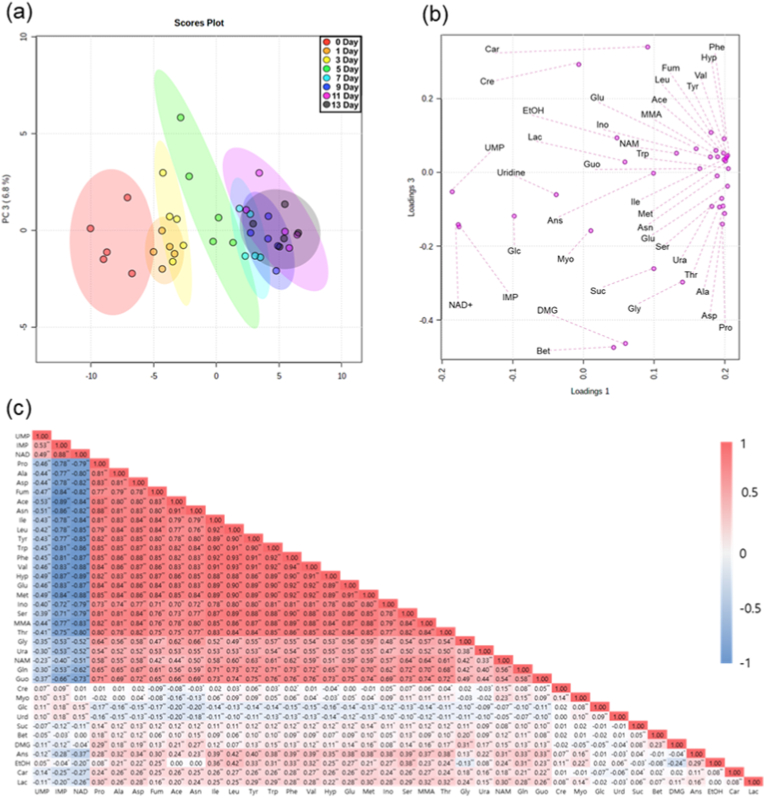
NMR spectroscopy is an analytical technique that leverages the atomic nuclear characteristics, specifically spin, along with chemical shifts influenced by a molecule's local electronic environment. While generally considered less sensitive than Mass Spectrometry (MS)-based methods, NMR spectroscopy can identify and quantify metabolites without the limitations of polarity, volatility, or chromophore content (Kim et al., 2022). NMR spectroscopy has several advantages compared to chromatography, such as easy sample preparation, short run times, and only one reference compound (Kim et al., 2020). Also, Kim et al. (2019) revealed that metabolite quantification using NMR spectroscopy has less than 5% of the relative standard deviations compared to HPLC quantification in chicken breast meat. In this study, a total of 38 metabolites were identified in the chicken breast meat during storage by NMR spectroscopy (Table S2). The identified metabolites were quantitatively analyzed using 1D 1H NMR spectroscopy (Table S3). PCA results showed that each storage day cluster (days 0, 1, 3, 5, 7, and 9) could be clearly distinguished by principal component (PC) 1 (60.8%) and PC 2 (6.8%) (Fig. 2a). On day 0, the sample had the lowest value for PC 1, which increased with the storage period. Loading plots showed that the contribution of the variables under investigation for the discrimination of storage days was evaluable by loading 1 (Fig. 2b). By comparing the loading plot, it was found that phenylalanine, hypoxanthine, fumarate, valine, leucine, tyrosine, acetate, methylmalonate, glutamine, inosine, niacinamide, tryptophan, guanosine, isoleucine, methionine, asparagine, glutamate, serine, uracil, threonine, glycine, alanine, aspartic acid, and proline substantially contributed to positive loading 1, However, uridine 5′-monophosphate (UMP), inosine 5′-monophosphate (IMP), and NAD + contributed prominently to the negative loading 1. These results were similar to those of our previous study (Kim et al., 2022).
Fig. 2.
Principal component analysis (PCA) (a), loading plot (b). and correlation analysis (c) of metabolites in chicken breast meat during storage at 4 °C (N = 40, n = 5). The PCA, loading plot, and correlation analysis were calculated for log10 transformed ratios of the auto-scaling. *P < 0.05. **P < 0.0001. UMP, uridine 5′-monophosphate; IMP, inosine 5′-monophosphate; NAD, nicotinamide adenine dinucleotide; Pro, proline; Ala, alanine; Asp, aspartate; Fum, fumarate; Ace, acetate; Asn, asparagine; Ile, isoleucine; Leu, leucine; Tyr, tyrosine; Trp, tryptophan; Phe, phenylalanine; Val, valine; Hyp, hypoxanthine; Glu, glutamate; Met, methionine; Ino, inosine; Ser, serine; MMA, methylmalonate; Thr, threonine; Gly, glycine; Ura, uracil; NAM, niacinamide; Gln, glutamine; Guo, guanosine; Cre, creatine; Myo, myo-inositol; Glc, glucose; Urd, uridine; Suc, succinate; Bet, betaine; DMG, N,N-dimethylglycine; Ans, anserine; EtOH, ethanol; Car, carnosine; Lac, lactate.
The values of the Pearson pairwise correlation coefficient among the selected metabolites were calculated and visualized in a heat map (Fig. 2c). A total of 703 correlations were analyzed, of which 658 were significant (P < 0.01). Of these 658 significant correlations, 502 were positive and 156 were negative with correlation values. Among them, 252 positive correlations and 49 negative correlations have a high correlation (r > 0.5 or −0.5 > r) according to Jung et al. (2019). The highest correlation was between valine and phenylalanine (r = 0.938), and the lowest value of correlation was between acetate and IMP (r = −0.889). To simplify the elucidation, metabolites were grouped by compound class (nucleotides: UMP, IMP, NAD, hypoxanthine, inosine, uracil, niacinamide, guanosine, and uridine; amino acids: proline, alanine, aspartate, asparagine, isoleucine, leucine, tyrosine, tryptophan, phenylalanine, valine, glutamate, methionine, serine, threonine, glycine, glutamine, betaine, N,N-dimethylglycine, anserine, and carnosine; organic acids: fumarate, acetate, methylmalonate, creatine succinate, ethanol, lactate, myoinositol, and glucose). Among the 252 positive correlations, 118 were highly correlated between two amino acids (0.516 < r < 0.938), 47 between amino acids and organic acids (0.558 < r < 0.906), 74 between amino acids and nucleotides (0.501 < r < 0.906), and 13 between nucleotides and organic acids (0.519 < r < 0.870). Among the 49 negative correlations, 33 were between nucleotides and amino acids (−0.880 < r < −0.509), seven were between nucleotides and organic acids (−0.893 < r < −0.526), and nine were between two nucleotides (−0.889 < r < −0.507).
Thus, it was confirmed that the metabolites detected in the chicken meat extracts could be distinguished by the storage period. Next, we established a metabolite-based spoilage point prediction formula.
3.3. Uni- and multi-variate analysis for selecting biomarkers
First, potential biomarkers were first identified through both univariate and multivariate analyses. Univariate analysis is important for its ability to compare the means of individual variables, revealing significant differences that are often overlooked in multivariate analysis (Gandhi et al., 2022; Vinaixa et al., 2012). In this study, the t-test identified 37 metabolites that showed significant differences between the fresh and spoiled meat groups (Table 1, P < 0.01). All of these metabolites had False Discovery Rate (FDR) values below 0.05. Specifically, metabolites such as proline, methylmalonate, methionine, asparagine, tryptophan, phenylalanine, glutamate, tyrosine, valine, aspartate, threonine, NAD, serine, hypoxanthine, acetate, isoleucine, alanine, leucine, IMP, inosine, guanosine, fumarate, glycine, glutamine, niacinamide, uracil, and UMP, exhibited low FDR values of 0.00132. Further validation using one-way ANOVA confirmed these findings; specifically, those metabolites exhibiting low FDR values of 0.00132 also yielded p-values below 0.001, thus confirming their differential expression across the fresh and spoiled meat groups.
Table 1.
Quantified metabolites in chicken breast meat (μg/mL) and their univariate analysis.
| Metabolite | Freshness | Spoilage |
t-test |
FDR | ANOVA |
||
|---|---|---|---|---|---|---|---|
| t-value | p-value | F-value | p-value | ||||
| Acetate | 248.689 ± 316.007 | 316.007 ± 37.645 | −102.789 | <0.0001 | 0.00132 | 53.8180 | <0.0001 |
| Alanine | 85.066 ± 26.095 | 176.971 ± 43.625 | −115.287 | <0.0001 | 0.00132 | 70.7558 | <0.0001 |
| Asparagine | 290.681 ± 42.159 | 592.076 ± 141.308 | −132.898 | <0.0001 | 0.00132 | 110.2997 | <0.0001 |
| Aspartate | 8.122 ± 1.254 | 8.297 ± 1.177 | −132.043 | <0.0001 | 0.00132 | 101.7319 | <0.0001 |
| Fumarate | 250.431 ± 56.167 | 368.283 ± 23.875 | −88.688 | <0.0001 | 0.00132 | 37.8765 | <0.0001 |
| Glutamate | 966.141 ± 298.709 | 1185.097 ± 281.126 | −141.789 | <0.0001 | 0.00132 | 110.9478 | <0.0001 |
| Glutamine | 50.699 ± 3.694 | 61.194 ± 2.725 | −77.318 | <0.0001 | 0.00132 | 32.2485 | <0.0001 |
| Glycine | 111.801 ± 23.830 | 190.157 ± 23.233 | −77.616 | <0.0001 | 0.00132 | 38.3771 | <0.0001 |
| Guanosine | 4461.769 ± 310.159 | 4670.743 ± 260.777 | −101.765 | <0.0001 | 0.00132 | 53.7403 | <0.0001 |
| Hypoxanthine | 298.166 ± 39.890 | 361.186 ± 31.822 | −116.001 | <0.0001 | 0.00132 | 67.1264 | <0.0001 |
| IMP | 424.626 ± 89.210 | 634.660 ± 57.662 | 109.276 | <0.0001 | 0.00132 | 56.8843 | <0.0001 |
| Inosine | 106.972 ± 29.063 | 183.383 ± 24.283 | −122.993 | <0.0001 | 0.00132 | 83.1489 | <0.0001 |
| Isoleucine | 9.827 ± 2.310 | 15.888 ± 3.007 | −126.504 | <0.0001 | 0.00132 | 83.1432 | <0.0001 |
| Leucine | 1183.548 ± 203.091 | 730.347 ± 161.744 | −118.974 | <0.0001 | 0.00132 | 66.2471 | <0.0001 |
| Methionine | 3206.861 ± 251.107 | 3382.191 ± 254.628 | −148.671 | <0.0001 | 0.00132 | 119.4979 | <0.0001 |
| Methylmalonate | 311.500 ± 52.771 | 464.027 ± 65.480 | −142.782 | <0.0001 | 0.00132 | 119.5845 | <0.0001 |
| NAD | 109.151 ± 22.847 | 91.414 ± 42.132 | 91.536 | <0.0001 | 0.00132 | 38.8420 | <0.0001 |
| Niacinamide | 85.463 ± 9.433 | 87.447 ± 12.711 | −73.413 | <0.0001 | 0.00132 | 29.2467 | <0.0001 |
| Phenylalanine | 77.613 ± 11.240 | 81.226 ± 12.306 | −147.919 | <0.0001 | 0.00132 | 121.3030 | <0.0001 |
| Proline | 32.611 ± 5.003 | 66.074 ± 20.611 | −144.096 | <0.0001 | 0.00132 | 150.8123 | <0.0001 |
| Serine | 503.302 ± 110.330 | 841.432 ± 101.661 | −122.867 | <0.0001 | 0.00132 | 78.2253 | <0.0001 |
| Threonine | 2.391 ± 0.794 | 4.533 ± 1.331 | −129.937 | <0.0001 | 0.00132 | 100.7048 | <0.0001 |
| Tryptophan | 170.660 ± 31.154 | 274.534 ± 39.141 | −146.734 | <0.0001 | 0.00132 | 124.3470 | <0.0001 |
| Tyrosine | 2517.473 ± 331.987 | 2546.035 ± 250.215 | −145.133 | <0.0001 | 0.00132 | 113.7579 | <0.0001 |
| UMP | 56.820 ± 3.484 | 62.673 ± 3.638 | 133.768 | <0.0001 | 0.00132 | 88.0218 | <0.0001 |
| Uracil | 78.899 ± 30.171 | 32.763 ± 6.321 | −91.173 | <0.0001 | 0.00132 | 43.4488 | <0.0001 |
| Valine | 37.741 ± 5.775 | 40.234 ± 4.821 | −148.490 | <0.0001 | 0.00132 | 119.4142 | <0.0001 |
| Succinate | 19.433 ± 6.051 | 4.865 ± 2.940 | −33.894 | <0.0001 | 0.03684 | 5.8198 | 0.0208 |
| Carnosine | 303.408 ± 42.096 | 407.210 ± 31.875 | −33.606 | <0.0001 | 0.03816 | 4.6149 | 0.0381 |
| Lactate | 89.246 ± 19.953 | 147.614 ± 14.434 | −32.342 | <0.0001 | 0.03947 | 6.3831 | 0.0158 |
| Anserine | 181.286 ± 20.146 | 266.084 ± 31.864 | −30.947 | <0.0001 | 0.04079 | 3.9236 | 0.0549 |
| Glucose | 24.919 ± 6.076 | 45.295 ± 6.330 | 23.807 | <0.0001 | 0.04211 | 2.7032 | 0.1084 |
| DMG | 27.688 ± 8.018 | 34.191 ± 9.141 | −20.777 | <0.0001 | 0.04342 | 4.5918 | 0.0386 |
| Betaine | 237.558 ± 38.386 | 347.011 ± 36.629 | −13.720 | <0.0001 | 0.04474 | 1.5737 | 0.2173 |
| myo-Inositol | 13.223 ± 3.560 | 13.440 ± 7.204 | −7.990 | <0.0001 | 0.04605 | 0.3116 | 0.5800 |
| Ethanol | 5.806 ± 2.228 | 11.313 ± 3.147 | −6.421 | <0.0001 | 0.04737 | 0.0166 | 0.8981 |
| Creatine | 127.445 ± 34.941 | 233.194 ± 27.677 | −4.293 | <0.0001 | 0.04868 | 0.0567 | 0.8130 |
| Uridine | 136.764 ± 29.144 | 221.416 ± 21.881 | −1.747 | 0.0808 | 0.05 | 0.0524 | 0.8201 |
NAD, nicotinamide adenine dinucleotide; IMP, inosine 5′-monophosphate; UMP, uridine 5′-monophosphate; DMG, N,N-dimethylglycine.
Multivariate analysis complements univariate methods by capturing inter-related patterns among variables, providing explanations for otherwise ambiguous results seen in univariate analysis (Vinaixa et al., 2012). PLS-DA results showed that the fresh and spoiled groups could be clearly distinguished based on component 1, which explained 60.8% of the variance, and component 2, accounting for 6.8% (Fig. 3a). Generally, the data set in PLS-DA is highly statistically significant when R2 and Q2 are more than 0.67 and 0, respectively (Henseler et al., 2009). In this study, PLS-DA cross-validation of the two components showed high significance (R2 = 0.86; Q2 = 0.76; Fig. 3c). Following the evaluation of model fit with high R2 and Q2 values, the significance of individual metabolites to the separation of fresh and spoiled clusters was assessed using Variable Importance in the Projection (VIP) scores. A VIP score above 1.00 indicates that it contributed significantly to cluster separation (Galindo-Prieto et al., 2014). Twenty-six metabolites (proline, asparagine, methylmalonate, methionine, tryptophan, aspartate, glutamate, phenylalanine, tyrosine, threonine, valine, serine, alanine, inosine, acetate, isoleucine, UMP, hypoxanthine, NAD, leucine, guanosine, uracil, IMP, fumarate, glycine, and glutamine) contributed to the significant separation of fresh and spoiled clusters using the VIP score (Fig. 3b).
Fig. 3.
Multivariate analysis of metabolites in chicken breast meat for selection of the spoilage point biomarkers. Partial least squares-discriminant analysis (PLS-DA) cluster between fresh and spoiled (a), variable influence on projection (VIP) score of PLS-DA (b), Random forest classification (RF) between fresh and spoiled (c), and mean decrease accuracy of RF (d).
RF analysis was performed to identify the differentiating metabolites between each fresh and spoiled group (Fig. 3c–d). In the RF analysis, 5000 trees were grown, and six features were randomly selected at each node (Gandhi et al., 2022). The highest RF misclassification occurred at the bag (OOB) error of 0.025. Based on the mean decrease in accuracy (MDA), 15 metabolites (leucine, proline, tryptophan, methylmalonate, phenylalanine, valine, asparagine, methionine, tyrosine, glutamate, acetate, inosine, aspartate, alanine, and isoleucine) were selected for variable importance (MDA>0.01; Table S4)31. Thus, 27 metabolites (proline, asparagine, methylmalonate, methionine, tryptophan, aspartate, glutamate, phenylalanine, tyrosine, threonine, valine, serine, alanine, inosine, acetate, isoleucine, UMP, hypoxanthine, NAD, leucine, guanosine, uracil, IMP, fumarate, glycine, glutamine, and valine) were selected using PLS-DA and RF.
Therefore, twenty-seven selected metabolites through univariate and multivariate analyses were used as variables in the regression equation to establish a spoilage point prediction equation.
3.4. Estimation of potential biomarkers for spoilage point of chicken breast meat during storage using binary logistic regression
The training data for establishing the prediction formula was prepared by augmenting the data using the Monte Carlo simulation technique (Jang et al., 2020). Monte Carlo simulation is a practical strategy for creating a prediction model to avoid the risk of overfitting when multiple biomarkers are used (Xu & Liang, 2001). A total of 8000 training data points were augmented by generating 1000 samples for each storage day (Mundform et al., 2011). After data augmentation, 27 metabolites were selected using univariate and multivariate analyses to establish a spoilage point prediction model. Using the BLR algorithm, we identified proline, methionine, glutamate, threonine, acetate, UMP, hypoxanthine, glycine, and glutamine as the best predictors of spoilage points in the BLR model (Table S5). Finally, the estimated BLR model is as follows:
This model was statistically significant, as determined by the likelihood ratio test (χ2 = 6.443, P < 0.05). The Cox & Snell R-Square and Nagelkerke R-Square values were 0.697 and 0.931, respectively. Odds ratios greater than one are reported to have significantly higher probabilities (Tsoukalas et al., 2019). The odds ratios indicated that proline (1.0097), methionine (1.0203), glutamate (1.0027), threonine (1.0057), acetate (1.1228), UMP (0.7799), hypoxanthine (1.0279), glycine (1.0081), and glutamine (1.0080) were associated with spoilage points (Table S5). The accuracy of the prediction formula is 95.3% (Table S6).
The prediction formula was validated using the ROC curve. The results showed an AUC of 0.992 (95% CI:0.991–0.994; Fig. 4). Using a cut-off value of 0.391, the sensitivity and specificity were 0.948 and 0.948, respectively.
Fig. 4.
Receiver operating characteristic (ROC) curve analysis for the predictive power of selected biomarker model for distinguishing the spoilage point. The final binary logistic model included nine metabolites: proline, methionine, glutamate, threonine, acetate, uridine 5′-monophosphate, hypoxanthine, glycine, and glutamine.
Furthermore, pathway analysis was performed to determine which metabolic pathway affected the nine metabolites (proline, methionine, glutamate, threonine, acetate, UMP, hypoxanthine, glycine, and glutamine) that contributed to the logistic regression analysis for predicting spoilage point of chicken breast meat.
3.5. Identification of pathways to contribute to the biomarkers
For pathway analysis, the results of metabolites on days 0 and 7 (initial pathway) and days 7 and 13 (late pathway) were used (Fig. S2). As a result, the seven metabolisms identified in the initial pathway were confirmed as follows: alanine, aspartate, and glutamate metabolism; glycine, serine, and threonine metabolism; pyrimidine metabolism; arginine and proline metabolism; purine metabolism; pyruvate metabolism; citrate cycle (TCA cycle) (Fig. S2a). These metabolisms were also confirmed in the late pathway, except for glycine, serine, and threonine metabolism, which were not significantly different in the late pathway. The impact values of the initial and late pathways were similar (0.3–0.5), however, the -log P value was 3.39–6.57 for the initial pathway which was higher than the late pathway at 1.73–4.16. It can be interpreted that the metabolites in the initial pathway changed more dramatically than those in the later pathway. It has been reported that meat undergoes extreme metabolic changes immediately after slaughter (Hambrecht et al., 2005; Terlouw et al., 2021). The difference in - log P values appears to be influenced by the meat, which exhibits extreme metabolic changes after slaughter.
The illustrated pathway results showed that metabolites affected the initial and late pathways (Fig. 5). The nine metabolites (proline, methionine, glutamate, threonine, acetate, UMP, hypoxanthine, glycine, and glutamine) selected by the BLR model were confirmed to be related to the number of 1 energy metabolism (pyruvate metabolism), 4 amino acid metabolism (arginine and proline metabolism, cysteine and methionine metabolism, alanine, aspartate and glutamate metabolism, glycine, serine, and threonine metabolism), and 2 nucleotide metabolisms (pyrimidine metabolism and purine metabolism).
Fig. 5.
Major metabolic pathways of metabolites in chicken breast meat between fresh and spoilage. The biomarker for distinguishing fresh/spoiled was selected through correlation analysis, multivariate analysis and binary regression model (N = 40, n = 5). The relative abundance of metabolites is indicated by a colored scale from blue (low) to red (high) in the box. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
After slaughter, the oxygen supply is stopped owing to the end of the blood circulation (Terlouw et al., 2021). When oxygen is depleted due to the stoppage of oxygen supply, oxidative metabolism by the citrate cycle and electron transport chain is stopped, and starts to anaerobic metabolism (Terlouw et al., 2021). When glycolysis starts due to anaerobic metabolism, threonine and glycine are increased by glycine, serine, and threonine metabolism, and are used for pyruvate metabolism. This increases NADH production and proton motive force by the respiratory chain to accumulate organic acids, such as acetate (Hambrecht et al., 2005; Ye et al., 2018). When energy metabolism is stopped due to glycogen depletion, ATP is sequentially catabolized into adenosine diphosphate (ADP), adenosine monophosphate (AMP), adenosine, inosine, and hypoxanthine using purine metabolism (Yin et al., 2018). This may support an increase in the hypoxanthine levels. In addition, glutamine is converted to glutamate when it produces IMP and UMP (Yoo et al., 2020).
Microbial contamination can also affect the production of organic acids (Hambrecht et al., 2005; Terlouw et al., 2021; Wang et al., 2021). Carnobacterium and Lactococcus are lactic acid bacteria that produce acetate through anaerobic metabolism (Wang et al., 2021). When the pH of meat decreases, proteolytic enzymes in the lysosome are released, and the protein is degraded (Hambrecht et al., 2005). Carnobacterium, Lactococcus and Serratia could contribute to protein hydrolysis, which affects protein degradation, peptide transport, and amino acid catabolism (Marquis et al., 1987; Mahlen, 2011; Vesanto et al., 1996; Wang et al., 2021).
In this study, amino acid metabolism may have increased due to protein degradation by proteolytic enzymes. Anaerobic metabolism that occurs after slaughter generates free radicals, such as reactive oxygen species, in meat (Terlouw et al., 2021). Proline is involved in the regulation of scavenging oxidants (Wu et al., 2011), and this antioxidant property may explain the increased concentration of proline caused by arginine and proline metabolism in response to oxidative stress in meat.
Overall, the nine metabolites selected through the BLR model might be affected by one energy metabolism, four amino acid metabolisms, two nucleotide metabolisms, and the metabolism of contaminating microorganisms.
4. Conclusion
During the storage period, the overall quality attributes of the chicken breast decreased. In particular, it was judged to be spoiled after 7 days of storage based on the VBN value. Establishing a prediction model for the spoilage point of chicken breast meat included selecting proper biomarkers and deducing formula via data augmentation. Twenty-four candidate metabolites were selected as biomarkers using univariate and multivariate analyses. A predictive regression equation was established using a binary logistic regression model. This equation contained proline, methionine, glutamate, threonine, acetate, uridine 5′-monophosphate, hypoxanthine, glycine, and glutamine, with a high AUC value of 0.992. Importantly, the predictive equation and selected metabolites have the potential for application in commercial slaughtering to assess meat freshness and spoilage. However, it is important to note that the size and range of samples in this study are limited. Therefore, the further studies are required before implementing these findings in commercial slaughtering and distribution processes with a larger sample size and extended sample area is needed.
CRediT authorship contribution statement
Hyun-Jun Kim: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Hye-Jin Kim: Validation, Data curation, Writing – original draft, Writing – review & editing. Hyun Cheol Kim: Conceptualization, Methodology, Formal analysis, Investigation. Dongheon Lee: Formal analysis, Investigation. Hyun Young Jung: Formal analysis, Investigation. Taemin Kang: Formal analysis, Investigation. Cheorun Jo: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this study.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2022R1A6A3A01085938). This study was also partially supported by the “Cooperative Research Program for Agriculture Science and Technology Development" (Project No. PJ016201), Rural Development Administration, Republic of Korea.
Handling Editor: Dr. Maria Corradini
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2023.100590.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Bae Y.S., Lee J.C., Jung S., Kim H.J., Jeon S.Y., Park D.H., Lee S.K., Jo C. Differentiation of deboned fresh chicken thigh meat from the frozen-thawed one processed with different deboning conditions. Korean J. Food Sci. Animal Res. 2014;34(1):73–79. doi: 10.5851/kosfa.2014.34.1.73. 10.5851/kosfa.2014.34.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhit A.E.D.A., Holman B.W., Giteru S.G., Hopkins D.L. Total volatile basic nitrogen (TVB-N) and its role in meat spoilage: a review. Trends Food Sci. Technol. 2021;109:280–302. doi: 10.1016/j.tifs.2021.01.006. [DOI] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bhadra B., Roy P., Chakraborty R. Serratia ureilytica sp. nov., a novel urea-utilizing species. Int. J. Syst. Evol. Microbiol. 2005;55(5):2155–2158. doi: 10.1099/ijs.0.63674-0. [DOI] [PubMed] [Google Scholar]
- Delbès-Paus C., Miszczycha S., Ganet S., Hélinck S., Veisseire P., Pochet S., Thevenot D., Montel M.C. Behavior of Escherichia coli O26: H11 in the presence of Hafnia alvei in a model cheese ecosystem. Int. J. Food Microbiol. 2013;160(3):212–218. doi: 10.1016/j.ijfoodmicro.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Deng Y., Lu L., Aponte L., Angelidi A.M., Novak V., Karniadakis G.E., Mantzoros C.S. Deep transfer learning and data augmentation improve glucose levels prediction in type 2 diabetes patients. NPJ Digital Med. 2021;4(1):1–13. doi: 10.1038/s41746-021-00480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-Prieto B., Eriksson L., Trygg J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS) J. Chemometr. 2014;28:623–632. doi: 10.1002/cem.2627. [DOI] [Google Scholar]
- Gandhi S., Chinnadurai V., Bhadra K., Gupta I., Kanwar R.S. Urinary metabolic modulation in human participants residing in Siachen: a 1H NMR metabolomics approach. Sci. Rep. 2022;12(1):1–16. doi: 10.1038/s41598-022-13031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambrecht E., Eissen J.J., Newman D.J., Smits C.H.M., Verstegen M.W.A., Den Hartog L.A. Preslaughter handling effects on pork quality and glycolytic potential in two muscles differing in fiber type composition. J. Anim. Sci. 2005;83(4):900–907. doi: 10.2527/2005.834900x. [DOI] [PubMed] [Google Scholar]
- Henseler J., Ringle C.M., Sinkovics R.R. The use of partial least squares path modeling in international marketing. New Challeng. Int. Market. 2009;20:277–319. doi: 10.1108/S1474-7979(2009)0000020014. [DOI] [Google Scholar]
- Hong G.E., Kim J.H., Ahn S.J., Lee C.H. Changes in meat quality characteristics of the sous-vide cooked chicken breast during refrigerated storage. Korean J. Food Sci. Anim. Resour. 2015;35(6):757. doi: 10.5851/kosfa.2015.35.6.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang A., Kim H.J., Kim M. Deep learning-based analysis of meat freshness measurement. J. Broadcast Eng. 2020;25(3):418–427. doi: 10.5909/JBE.2020.25.3.418. [DOI] [Google Scholar]
- Jung S.E., Lim S.M., Hong S.R., Lee E.H., Shin K.J., Lee H.Y. DNA methylation of the ELOVL2, FHL2, KLF14, C1orf132/MIR29B2C, and TRIM59 genes for age prediction from blood, saliva, and buccal swab samples. Forensic Sci. Int.: Genetics. 2019;38:1–8. doi: 10.1016/j.fsigen.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Jung D.Y., Lee D., Lee H.J., Kim H.J., Jung J.H., Jang A., Jo C. Comparison of chicken breast quality characteristics and metabolites due to different rearing environments and refrigerated storage. Poultry Sci. 2022;101(7) doi: 10.1016/j.psj.2022.101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim D., Kim H.J., Song S.O., Song Y.H., Jang A. Evaluation of the microbiological status of raw beef in Korea: considering the suitability of aerobic plate count guidelines. Food Sci. Animal Res. 2018;38(1):43–51. doi: 10.5851/kosfa.2018.38.1.043. 10.5851/kosfa.2018.38.1.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.C., Ko Y.J., Kim M., Choe J., Yong H.I., Jo C. Optimization of 1D 1H quantitative NMR (nuclear magnetic resonance) conditions for polar metabolites in meat. Food Sci. Animal Resour. 2019;39(1):1–12. doi: 10.5851/kosfa.2018.e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.C., Baek K.H., Ko Y.J., Lee H.J., Yim D.G., Jo C. Characteristic metabolic changes of the crust from dry-aged beef using 2D NMR spectroscopy. Molecules. 2020;25(13):3087. doi: 10.3390/molecules25133087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.C., Ko Y.J., Jo C. Potential of 2D qNMR spectroscopy for distinguishing chicken breeds based on the metabolic differences. Food Chem. 2021;342(128316):1–9. doi: 10.1016/j.foodchem.2020.128316. [DOI] [PubMed] [Google Scholar]
- Kim H.C., Baek K.H., Lee Y.E., Kang T., Kim H.J., Lee D., Jo C. Using 2D qNMR analysis to distinguish between frozen and frozen/thawed chicken meat and evaluate freshness. npj Sci. Food. 2022 doi: 10.1038/s41538-022-00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Choe J., Yoon J.W., Kim S., Oh H., Yoon Y., Jo C. Determination of salable shelf-life for wrap-packaged dry-aged beef during cold storage. Korean J. Food Sci. Animal Resour. 2018;38(2):251–258. doi: 10.5851/kosfa.2018.38.2.251. 10.5851/kosfa.2018.38.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Lee H.J., Jung D.Y., Kim H.J., Jang A., Jo C. Effect of an animal-friendly raising environment on the quality, storage stability, and metabolomic profiles of chicken thigh meat. Food Res. Int. 2022;155 doi: 10.1016/j.foodres.2022.111046. [DOI] [PubMed] [Google Scholar]
- Leisner J.J., Laursen B.G., Prévost H., Drider D., Dalgaard P. Carnobacterium: positive and negative effects in the environment and in foods. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2007;31(5):592–613. doi: 10.1111/j.1574-6976.2007.00080.x. 10.1111/j.1574-6976.2007.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Mei Y., He B., Sun X., Li J. Reducing quorum sensing-mediated virulence factor expression and biofilm formation in hafnia alvei by using the potential quorum sensing inhibitor L-carvone. Front. Microbiol. 2019;9:3324. doi: 10.3389/fmicb.2018.03324. 10.3389/fmicb.2018.03324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlen S.D. Serratia infections: from military experiments to current practice. Clin. Microbiol. Rev. 2011;24(4):755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis R.E., Bender G.R., Murray D.R., Wong A. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 1987;53(1):198–200. doi: 10.1128/aem.53.1.198-200.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MFDS Standards and specification for general foods. Ministry of food and drug safety. 2022. https://various.foodsafetykorea.go.kr/fsd/#/ext/Document/FC Available from:
- Moreno-Barea F.J., Franco L., Elizondo D., Grootveld M. Application of data augmentation techniques towards metabolomics. Comput. Biol. Med. 2022;148 doi: 10.1016/j.compbiomed.2022.105916. [DOI] [PubMed] [Google Scholar]
- Mundform D.J., Schaffer J., Kim M.J., Shaw D., Thongteeraparp A., Supawan P. Number of replications required in Monte Carlo simulation studies: a synthesis of four studies. J. Mod. Appl. Stat. Methods. 2011;10(1):4. 10.22237/jmasm/1304222580. [Google Scholar]
- Mussa N.J., Kibonde S.F., Boonkum W., Chankitisakul V. The comparison between Tanzanian indigenous (ufipa breed) and commercial broiler (ross chicken) meat on the physicochemical characteristics, collagen and nucleic acid contents. Food Sci. Animal Resour. 2022;42(5):833–848. doi: 10.5851/kosfa.2022.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival B.C., Latour Y.L., Tifft C.J., Grootveld M. Rapid identification of new biomarkers for the classification of GM1 type 2 gangliosidosis using an unbiased 1H NMR-linked metabolomics strategy. Cells. 2021;10(3):572. doi: 10.3390/cells10030572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukchon C., Nopwinyuwong A., Trevanich S., Jinkarn T., Suppakul P. Development of a food spoilage indicator for monitoring freshness of skinless chicken breast. Talanta. 2014;130:547–554. doi: 10.1016/j.talanta.2014.07.048. [DOI] [PubMed] [Google Scholar]
- Seleshe S., Kang S.N. Effect of Different Pediococcus pentosaceus and Lactobacillus plantarum strains on quality characteristics of dry fermented sausage after completion of ripening period. Food Sci. Animal Resour. 2021;41(4):636–649. doi: 10.5851/kosfa.2021.e21. 10.5851/kosfa.2021.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujiwo J., Kim D., Jang A. Relation among quality traits of chicken breast meat during cold storage: correlations between freshness traits and torrymeter values. Poultry Sci. 2018;97(8):2887–2894. doi: 10.3382/ps/pey138. [DOI] [PubMed] [Google Scholar]
- Sujiwo J., Kim H.J., Song S.O., Jang A. Relationship between quality and freshness traits and torrymeter value of beef loin during cold storage. Meat Sci. 2019;149:120–125. doi: 10.1016/j.meatsci.2018.11.017. [DOI] [PubMed] [Google Scholar]
- Terlouw E.C., Picard B., Deiss V., Berri C., Hocquette J.F., Lebret B., Lefèvre F., Hamill R., Gagaoua M. Understanding the determination of meat quality using biochemical characteristics of the muscle: stress at slaughter and other missing keys. Foods. 2021;10(1):84. doi: 10.3390/foods10010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoukalas D., Fragoulakis V., Sarandi E., Docea A.O., Papakonstaninou E., Tsilimidos G., Anamaterou C., Fragkiadaki P., Aschner M., Tsatsakis A., Drakoulis N., Calina D. Targeted metabolomic analysis of serum fatty acids for the prediction of autoimmune diseases. Front. Mol. Biosci. 2019;6:120. doi: 10.3389/fmolb.2019.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesanto E., Peltoniemi K., Purtsi T., Steele J.L., Palva A. Molecular characterization, over-expression and purification of a novel dipeptidase from Lactobacillus helveticus. Appl. Microbiol. Biotechnol. 1996;45(5):638–645. doi: 10.1007/s002530050741. [DOI] [PubMed] [Google Scholar]
- Vinaixa M., Samino S., Saez I., Duran J., Guinovart J.J., Yanes O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites. 2012;2(4):775–795. doi: 10.3390/metabo2040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu J., Lv M., Shao Z., Hungwe M., Wang J., Bai X., Xie J., Wang Y., Geng W. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021;378:1–19. doi: 10.3389/fbioe.2021.612285. 10.3389/fbioe.2021.612285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Bazer F.W., Burghardt R.C., Johnson G.A., Kim S.W., Knabe D.A., Li P., Li X., McKnight J.R., Satterfield M.C., Spencer T.E. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids. 2011;40(4):1053–1063. doi: 10.1007/s00726-010-0715-z. 10.1007/s00726-010-0715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q.S., Liang Y.Z. Monte Carlo cross validation. Chemometr. Intell. Lab. Syst. 2001;56(1):1–11. doi: 10.1016/S0169-7439(00)00122-2. [DOI] [Google Scholar]
- Ye J.Z., Lin X.M., Cheng Z.X., Su Y.B., Li W.X., Ali F.M., Jun Z., Peng B. Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida. J. Proteonomics. 2018;183:34–44. doi: 10.1016/j.jprot.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Yin J., Ren W., Huang X., Deng J., Li T., Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 2018;9:1697. doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.C., Yu Y.C., Sung Y., Han J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020;52(9):1496–1516. doi: 10.1038/s12276-020-00504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.