Abstract
A substantial body of evidence has established the contributions of both mitochondrial dynamics and lipid metabolism to the pathogenesis of diabetic kidney disease (DKD). However, the precise interplay between these two key metabolic regulators of DKD is not fully understood. Here, we uncover a link between mitochondrial dynamics and lipid metabolism by investigating the role of carbohydrate-response element-binding protein (ChREBP), a glucose-responsive transcription factor and a master regulator of lipogenesis, in kidney podocytes. We find that inducible podocyte-specific knockdown of ChREBP in diabetic db/db mice improves key biochemical and histological features of DKD in addition to significantly reducing mitochondrial fragmentation. Because of the critical role of ChREBP in lipid metabolism, we interrogated whether and how mitochondrial lipidomes play a role in ChREBP-mediated mitochondrial fission. Our findings suggest a key role for a family of ether phospholipids in ChREBP-induced mitochondrial remodeling. We find that overexpression of glyceronephosphate O-acyltransferase, a critical enzyme in the biosynthesis of plasmalogens, reverses the protective phenotype of ChREBP deficiency on mitochondrial fragmentation. Finally, our data also points to Gnpat as a direct transcriptional target of ChREBP. Taken together, our results uncover a distinct mitochondrial lipid signature as the link between ChREBP-induced mitochondrial dynamics and progression of DKD.
Keywords: lipid metabolism, mitochondria, phospholipid, kidney metabolism, diabetic nephropathy
Lipids play a central role in cellular energy production, membrane structure, and signaling, and their dysregulated metabolism has been recognized as a significant contributor to the pathogenesis of several pathologies, including diabetic kidney disease (DKD) (1, 2, 3, 4). However, the molecular mechanisms underlying the link between lipid metabolism and the pathogenesis of DKD remain incompletely understood.
A growing body of evidence suggests that the interplay between mitochondrial dynamics and lipid metabolism is a crucial factor in the pathogenesis of metabolic diseases (5, 6, 7, 8, 9). Indeed, both impaired mitochondrial dynamics and aberrant lipid metabolism have been associated with DKD pathogenesis and disease progression (4, 10, 11, 12, 13, 14). Mitochondrial dynamics is a concept that describes the dynamic nature of mitochondrial morphology and continuous changes in mitochondrial structure that ultimately controls mitochondrial function and is tightly regulated by a tight balance among “mitochondrial shaping” proteins that are involved in mitochondrial fission (fragmentation) and fusion (15, 16, 17). Mitochondrial fission is essential for mitochondrial transport and distribution within the cell and elimination of damaged mitochondria, whereas mitochondrial fusion is crucial for mitochondrial mixing and enhanced oxidative phosphorylation (18, 19). But how mitochondrial dynamics is linked to lipid metabolism is largely unknown. Recent studies have suggested that mitochondrial dynamics is controlled not only by fusion-fission machinery proteins but also by the cellular lipid milieu (5, 20, 21, 22). However, little is known about the underlying molecular mechanisms by which aberrant lipid metabolism impacts key features of diabetes-induced mitochondrial dynamics that could lead to the progression of DKD.
Here, we examined the interaction between lipid metabolism and mitochondrial dynamics in progression of DKD through interrogating the role of carbohydrate-response element-binding protein (ChREBP), a whole-body glucose sensor and one of the major transcription factors that regulate both glycolysis and lipogenesis (23). Having previously showed that ChREBP expression is significantly increased in podocytes induced by high glucose (HG) milieu (24), we now examine the role of ChREBP onprogression of DKD in vivo and its role as a link between mitochondrial dynamics and lipid metabolism by generating an inducible podocyte-specific ChREBP KO mouse model on a diabetic db/db background. Our findings indicate that diabetic mice with conditional ChREBP knockdown (KD) in podocytes are protected against progression of DKD. Importantly, this protective effect was associated with decreased mitochondrial fission in podocytes. We also identified that altered peroxisomal-derived ether phospholipids linked ChREBP to mitochondrial dynamics in DKD. These findings also highlight a surprising role of ChREBP in coordinating the HG-mediated crosstalk among different organelles, including peroxisomes, endoplasmic reticulum (ER), and mitochondria.
Results
Generation and characterization of mice with inducible podocyte-specific ChREBP KD
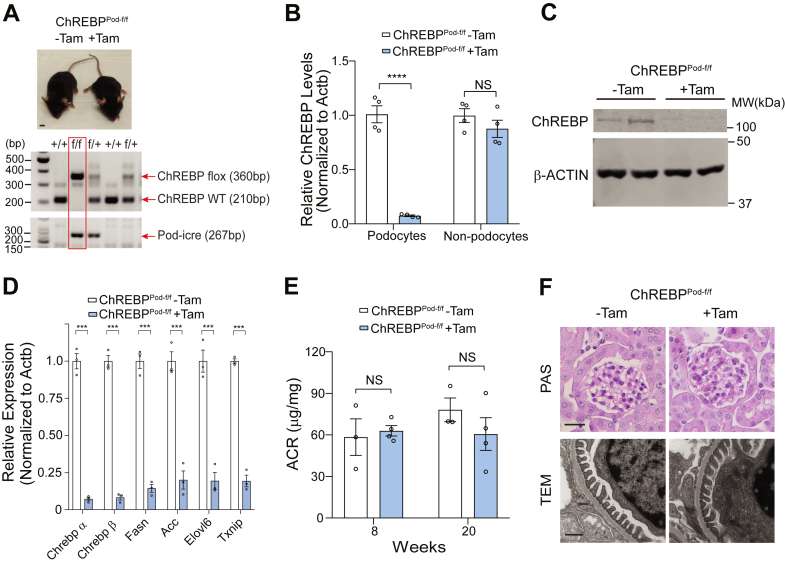
We have previously found that ChREBP expression is upregulated in podocytes under HG conditions (24). To validate these results in vivo, we isolated primary podocytes from diabetic db/db mice, an established experimental model of type 2 diabetes, at 24 weeks of age and confirmed that ChREBP expression was significantly increased in kidney podocytes from db/db mice compared to nondiabetic dbm littermate mice (Fig. S1, A–C). To gain further insights into the role of ChREBP on podocyte homeostasis in vivo, we generated an inducible podocyte-specific ChREBP KO mouse model by crossing floxed ChREBP mice (ChREBPflox/flox) with tamoxifen-inducible Cre transgenic mice which carry a Cre recombinase driven by human podocin gene promoter (podocin-iCreERT2) previously established in our laboratory (Fig. 1A) (25, 26). At 8 weeks of age, ChREBPflox/flox; podocin-iCreERT2 mice (referred herein as ChREBPPod-f/f) were induced with tamoxifen as previously described (25). Robust reduction of ChREBP in podocytes was confirmed by qPCR and Western blot analysis of primary podocytes isolated from ChREBPPod-f/f mice (Fig. 1, B and C). Both isoforms of ChREBP, ChREBPα and ChREBPβ, were deleted in podocytes from tamoxifen-induced ChREBPPod-f/f mice (Fig. 1D). We also found multiple previously established downstream targets of ChREBP, including lipogenic genes, such as Fasn (fatty acid synthase), Acc (acetyl-CoA carboxylase), Elovl6 (elongation of very long chain fatty acids protein 6), and redox regulator, Txnip (thioredoxin-interacting protein), were also significantly downregulated in podocytes from tamoxifen-induced ChREBPPod-f/f mice (Fig. 1D). ChREBPPod-f/f mice did not show overall gross abnormalities (Fig. 1A), and we did not observe significant changes in the urinary albumin to creatinine ratio, kidney histology or podocyte morphology assessed by Periodic-Acid Schiff’s staining, and transmission electron microscopy (TEM) after tamoxifen induction at 20 weeks of age (Fig. 1, E and F). These findings suggest that conditional ChREBP KD in podocytes does not elicit major biochemical or histologic consequences in the kidney in the absence of significant metabolic stress.
Figure 1.
Generation of inducible podocyte-specific ChREBP KO mice.A, top, a representative image of ChREBPPod-f/f mice with or without tamoxifen induction. Scale bar represents 1 cm. Bottom, genotyping PCR detecting ChREBPf/f homozygosity and Pod-iCreERT2 as indicated in the red box. B and C, qPCR (B) and Western blot (C) analysis of ChREBP expression in isolated podocytes from tamoxifen-induced ChREBPPod-f/f and noninduced control mice. D, qPCR analysis of indicated ChREBP target genes expression in isolated primary podocytes. E, urinary ACR analysis of tamoxifen-induced and noninduced mice at 8 and 20 weeks. F, upper panel: representative images of kidney tissue sections from 20-week-old mice stained with PAS. Scale bar represents 50 μm. Bottom panel: representative TEM micrographs from indicated groups. Scale bar represents 0.5 μm. Results are presented as mean ± SEMs. ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ACR, albumin to creatinine ratio; ChREBP, carbohydrate-response element-binding protein; NS, not significant; PAS, Periodic-Acid Schiff’s; TEM, transmission electron microscopy.
Podocyte-specific ChREBP deficiency attenuates progression of DKD
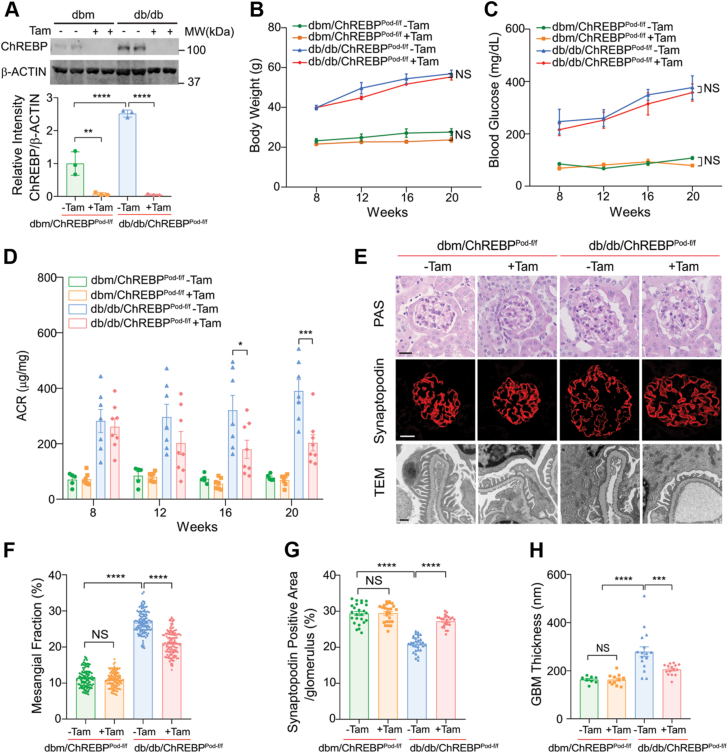
To investigate whether ChREBP deficiency in podocytes confers protection against progression of DKD, we crossed ChREBPPod-f/f mice with diabetic Leprdb/+ mice to generate diabetic Leprdb/db; ChREBPPod-f/f mice (hereafter referred to as db/db; ChREBPPod-f/f). While podocytes obtained from noninduced db/db; ChREBPPod-f/f mice exhibited markedly enhanced expression of ChREBP compared to dbm controls, a significant reduction of ChREBP protein level was observed in podocytes of tamoxifen-induced db/db; ChREBPPod-f/f mice (Fig. 2A). ChREBP deficiency in podocytes did not significantly affect body weight and blood glucose levels (Fig. 2, B and C). However, ChREBP KD in podocytes of tamoxifen-induced db/db; ChREBPPod-f/f mice markedly improved albuminuria (a decline of ∼50%, from 390.4 ± 40.8 μg/mg to 203.0 ± 29.0 μg/mg) as measured by albumin to creatinine ratio compared with noninduced diabetic control mice at 20 weeks of age (Fig. 2D).
Figure 2.
Conditional deletion of ChREBP in podocytes confers protection against DKD.A, Western blot analysis and quantification of ChREBP expression in podocytes isolated from tamoxifen-induced and noninduced dbm/ChREBPPod-f/f mice and db/db/ChREBPPod-f/f mice. B–D, body weight (B), blood glucose (C), and urinary ACR (D) of tamoxifen-induced dbm/ChREBPPod-f/f mice (n = 7) and noninduced dbm controls (n = 5) and tamoxifen-induced db/db/ChREBPPod-f/f mice (n = 8) and noninduced db/db controls (n = 7) at different ages. E, representative micrographs of PAS-stained kidney tissue sections, glomerular expression of synaptopodin, and TEM images showing podocyte morphology obtained from 20-week-old mice. Scale bars represent 50 μm in row 1 and 2; scale bar represents 0.5 μm in row 3. F, quantification of mesangial matrix expansion. n = 4 mice per group. G, quantification of synaptopodin-positive area per glomerulus. n=3 to 4 mice per group. H, quantification of GBM thickness. n = 3 or 4 mice per group. Results are presented as mean ± SEMs. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; NS, not significant. ACR, albumin to creatinine ratio; ChREBP, carbohydrate-response element-binding protein; DKD, diabetic kidney disease; GBM, glomerular basement membrane; PAS, Periodic-Acid Schiff’s; TEM, transmission electron microscopy.
Histologically, tamoxifen-induced db/db; ChREBPPod-f/f mice displayed a significant reduction of mesangial matrix expansion and restored expression of synaptopodin, a podocyte marker, compared to noninduced diabetic controls at 20 weeks of age (Fig. 2, E–G). Consistent with these findings, TEM revealed improved glomerular basement membrane thickening and podocyte foot process effacement in tamoxifen-induced db/db; ChREBPPod-f/f mice compared with their noninduced diabetic controls (Fig. 2, E and H). Taken together, these results suggest that podocyte-specific ChREBP deficiency improves key features of DKD. However, the molecular mechanisms by which ChREBP contributes to the pathogenesis of DKD remain unclear.
Plasmalogen phospholipids link ChREBP to mitochondrial remodeling
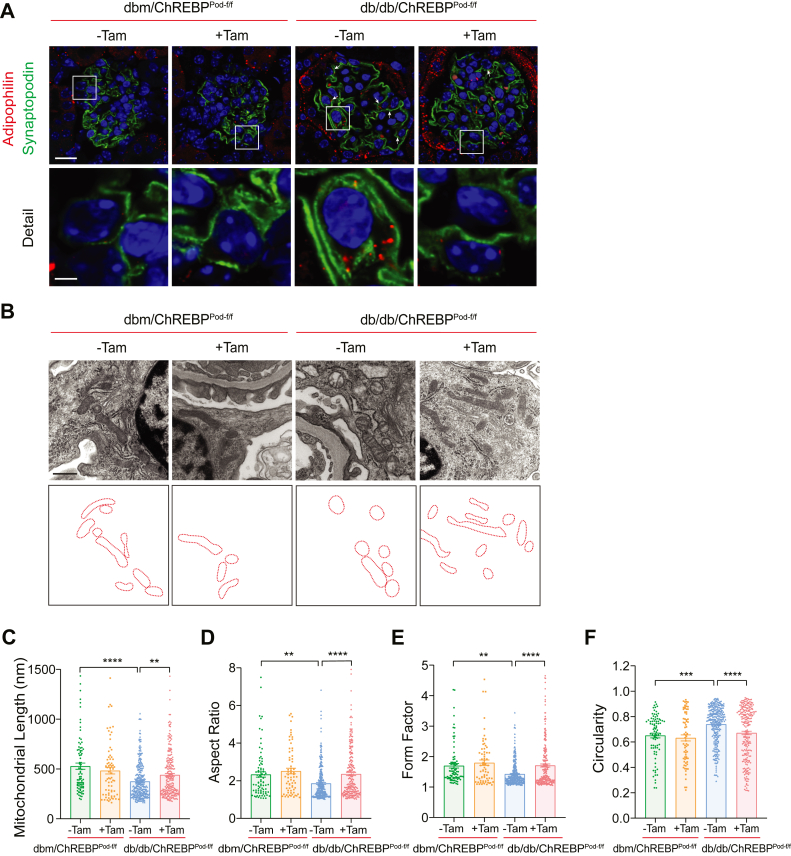
Both lipid dysregulation and mitochondrial remodeling have been shown to contribute to the development of DKD (11, 13, 27). However, the importance of the interplay between two key processes in DKD pathogenesis remains largely unknown. A possible link between lipogenesis and mitochondrial morphology in several biological activities of the cell has been recently proposed (5, 22, 28). Therefore, we examined whether ChREBP, a master regulator of lipogenesis, impacts the development of DKD by linking aberrant lipid metabolism with mitochondrial remodeling in DKD. To this aim, we first performed immunostaining of kidney tissue sections in our experimental diabetic mice and observed that compared to noninduced diabetic controls, tamoxifen-induced diabetic db/db; ChREBPPod-f/f mice had fewer lipid droplets in podocytes (Fig. 3A). Importantly, when we examined TEM micrographs using kidney tissues from diabetic mice, we found significant improvement in mitochondrial fission, a key feature of mitochondrial dynamics, in the podocytes of tamoxifen-induced diabetic db/db; ChREBPPod-f/f mice (Fig. 3, B–F) (11, 12, 26). Specifically, we noticed that podocytes from tamoxifen-induced db/db; ChREBPPod-f/f mice has more elongated mitochondria with increased aspect ratio and form factor and decreased circularity compared with mitochondria in podocytes from noninduced diabetic controls as assessed by quantitative analyses of TEM micrographs (Fig. 3, B–F). Taken together, these data suggest that ChREBP deficiency in podocytes from diabetic mice is associated with significant improvement in lipid accumulation and mitochondrial fission. However, the link between ChREBP-mediated lipogenesis and mitochondrial fission in podocytes remained unknown.
Figure 3.
ChREBP deficiency inhibits lipogenesis and alleviates mitochondrial fission in podocytes in diabetic mice.A, representative micrographs showing glomerular expression of synaptopodin and adipophilin from 20-week-old mice. Scale bar represents 50 μm in row 1; scale bar represents 10 μm in row 2. B, representative TEM images showing mitochondrial morphology in podocytes from dbm/ChREBPPod-f/f and db/db/ChREBPPod-f/f mice obtained from 20-week-old mice. Scale bars represent 0.5 μm. The lowest panel shows tracings of mitochondria in the middle panel. C–F, mitochondrial morphology was quantitated from TEM micrographs as the (C) mitochondrial length, (D) mitochondrial aspect ratio, (E) mitochondrial form factor, (F) mitochondrial circularity. Results are presented as mean ± SEMs. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ChREBP, carbohydrate-response element-binding protein; NS, not significant; TEM, transmission electron microscopy.
Mounting evidence suggests that lipid composition of mitochondrial inner and outer membranes play important roles in coordinating mitochondrial fission and fusion (20, 21, 29). Phospholipids, an important class of lipids, are mainly responsible for the integrity of mitochondrial membranes (29). The major phospholipids found in mitochondrial membranes include phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylinositol, phosphatidylserine, phosphatidic acid, phosphatidylglycerol, and cardiolipin (29). PC and PE are the most abundant phospholipids in mitochondria, comprising 40 and 30% of total mitochondrial phospholipids, respectively (30). Importantly, among different classes of phospholipids, cardiolipin, phosphatidic acid, and plasmalogens, a special subclass of PC and PE lipids, have recently been implicated in the regulation of mitochondrial morphology (20, 21, 31).
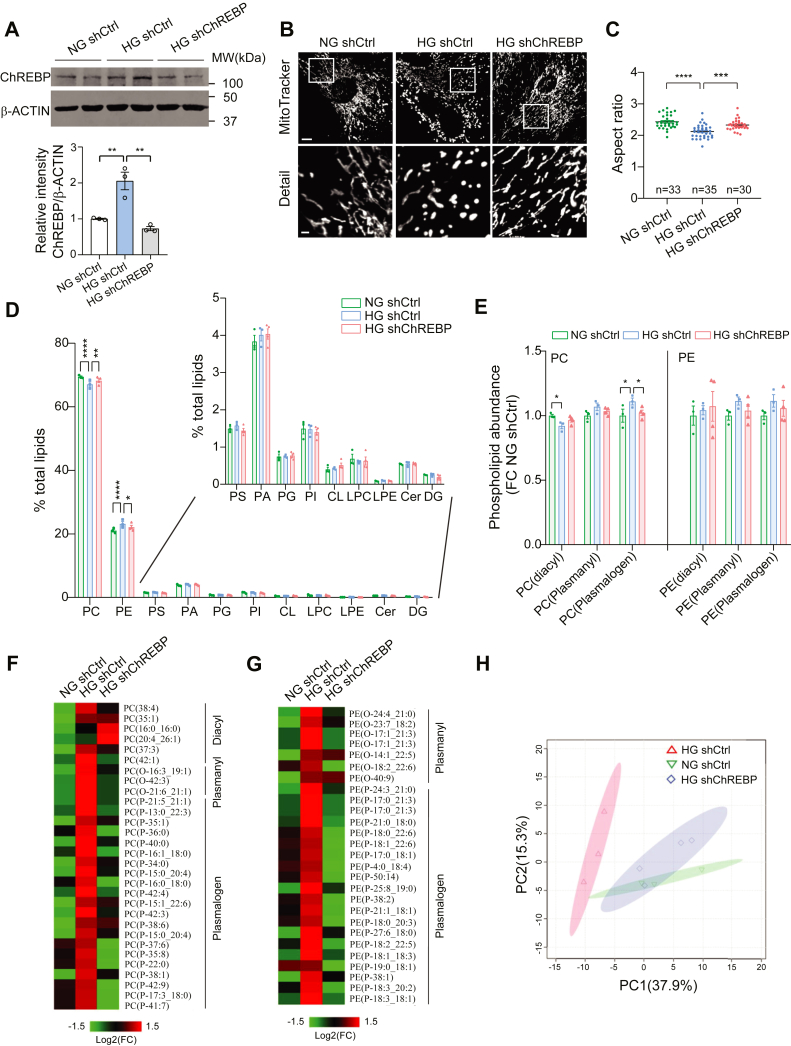
We therefore reasoned that ChREBP-induced changes in mitochondrial lipidomes could play a role in mitochondrial remodeling in podocytes. To test this hypothesis, we first examined the role of ChREBP on mitochondrial lipidomes using an unbiased approach and then examined whether a selective lipid species, downstream of ChREBP, could serve as a link between aberrant lipogenesis and mitochondrial remodeling in podocytes of diabetic mice. To develop an in vitro model to test our hypothesis, we generated stable ChREBP-knockdown (ChREBP-KD) in cultured podocytes transfected with shRNA against ChREBP (shChREBP). We found that shChREBP led to ∼65% reduction in ChREBP expression as compared to scramble shRNA (shCtrl) by Western blotting (Fig. 4A). Notably, we observed that while mitochondrial network was fragmented in shCtrl-transfected cultured podocytes under HG conditions (25 mM for 48 h), downregulation of ChREBP in cultured podocytes restored the tubular and elongated morphology of mitochondria, as evidenced by an increase in the mitochondrial aspect ratio (Fig. 4, B and C). We also found that ChREBP-KD improved the effect of HG on ATP and mitochondrial reactive oxygen species (ROS) production in cultured podocytes (Fig. S2, A and B). These results suggest that our ChREBP-KD cells recapitulate some of the key metabolic features of diabetic mouse models and could be used as an in vitro model to further investigate the role of ChREBP on mitochondrial lipidome.
Figure 4.
The abundance of plasmalogen phospholipids is increased in HG-induced podocytes and decreases with ChREBP-KD.A, Western blot analysis and quantification of ChREBP expression levels in control cells treated with NG or HG and ChREBP-KD cells treated with HG. B, representative immunofluorescence micrographs of podocytes stained with MitoTracker Red. Scale bar represents 10 μm in row 1; scale bar represents 2 μm in row 2. C, quantification of mitochondrial AR in each experimental condition. D, mitochondrial lipidome of control cells treated with NG (n = 3) or HG (n = 3) and ChREBP-KD cells treated with HG (n = 4). Inset shows expanded y axis for clarification. E, lipidomic analysis showing relative abundance of PC and PE lipids. F and G, heat map of mitochondrial PC (F) and PE (G) changes among three groups. p < 0.1. H, PCA of top differentially expressed PC and PE ether lipids (n = 124). Results are presented as mean ± SEMs. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. AR, aspect ratio; Cer, ceramide; ChREBP, carbohydrate-response element-binding protein; CL, cardiolipin; DG, diacylglycerol; HG, high glucose; KD, knockdown; LPC, lysoPC; LPE, lysoPE; NG, normal glucose; PCA, principal component analysis; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PA, phosphatidic acid; PG, phosphatidylglycerol; PI, phosphatidylinositol.
Using an unbiased ultrahigh liquid chromatography coupled with mass spectrometry, we examined the impact of ChREBP on mitochondrial lipidome using enriched mitochondrial fractions from shCtrl- and shChREBP-transfected podocytes treated with normal glucose (NG, 5 mM for 48 h) or HG (25 mM for 48 h). The enrichment of mitochondrial fraction and the purity of isolated mitochondria were first confirmed (Fig. S3A). The untargeted mitochondrial lipidomic profiling recovered major lipid classes in mitochondrial membranes, but only the abundance of PC and PE, the two most abundant phospholipids in mitochondrial membranes, were significantly changed with the abundance of PE significantly increased, while PC abundance was significantly decreased in mitochondrial lipidomes in HG-treated podocytes relative to NG (Fig. 4D). Notably, these HG-induced changes in mitochondrial lipidomes were reversed with ChREBP KD. (Fig. 4D).
PC and PE are made up of diacyl, alkyl (plasmanyl), and alkenyl (plasmenyl, also known as plasmalogens) subclasses (Fig. S3B) (32, 33). To examine exactly which subclass was responsible for the overall changes observed in ChREBP and HG-induced PC and PE levels, we measured the abundance of diacyl, plasmanyl, and plasmalogen subclasses in our samples. We found that plasmalogen-PC abundance was significantly increased by HG and reversed with ChREBP KD (Fig. 4E). A similar trend was observed in plasmalogen-PE levels among the three groups, albeit it did not reach a statistical significance. Detailed analysis revealed that HG increased plasmalogen-PC and PE with varied acyl side chain length in a ChREBP-dependent manner, suggesting a possible modulatory role of ChREBP on plasmalogen-PC and plasmalogen-PE synthesis pathways (Fig. 4, F and G). Collectively, these results suggest that ChREBP plays an important role in HG-mediated mitochondrial lipidomic remodeling, probably through regulating plasmalogen synthesis. Importantly, we found that these results were consistent among samples in each group as evidenced by principal component analysis of the lipidomes with multivariate clustering of top differentially expressed PC- and PE-ether lipids revealing two distinct lipidomic profiles with NG- and HG-treated podocytes, whereas HG-treated ChREBP-KD podocytes exhibited a lipidomic shift back towards NG conditions (Fig. 4H).
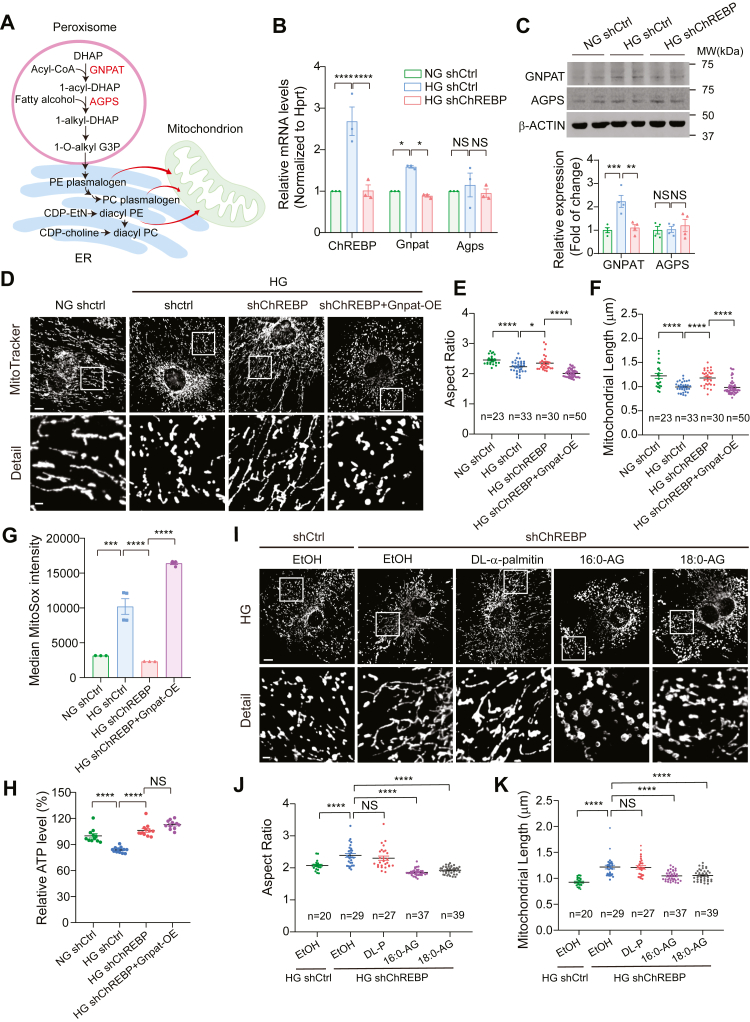
Plasmalogens are basic components of mitochondrial bilayer, whose biosynthesis initiates in peroxisomes but culminates in the ER (Fig. 5A) (33). Plasmalogens biosynthesis begins with two key enzymes, glyceronephosphate O-acyltransferase (GNPAT) and alkyl-glyceronephosphate synthase (AGPS). Plasmalogens are then translocated from peroxisomes to mitochondria where they are incorporated into mitochondrial membrane where they play important roles in regulating mitochondrial dynamics through their ability to modulate physical properties of lipid bilayers (31, 34). Since plasmalogen metabolism involves interaction among several subcellular compartments, we reasoned that it could serve as a link between ChREBP-mediated lipogenesis and mitochondrial fission. Therefore, we next focused on key enzymes involved in plasmalogen synthesis that could be directly regulated by ChREBP. To this end, we examined the expression of two key enzymes in the biosynthesis of plasmalogens in the peroxisome, GNPAT, and AGPS (35) and found a significant increase in the protein and mRNA expression of GNPAT in podocytes treated with HG (Fig. 5, B and C). Interestingly, this increase was reversed upon targeted KD of ChREBP. In contrast, the protein and mRNA expression of AGPS enzyme remained unchanged (Fig. 5, B and C). We then validated whether Gnpat is a HG- and ChREBP-responsive gene in vivo. We found that Gnpat expression is significantly higher in podocytes obtained from db/db mice than in nondiabetic dbm mice (Fig. S4A). However, Gnpat expression was significantly reduced in primary podocytes isolated from tamoxifen-induced ChREBPpod-f/f mice compared to podocytes from noninduced ChREBPpod-f/f control mice (Fig. S4B). These findings suggest that Gnpat could be a HG- and ChREBP-responsive gene.
Figure 5.
GNPAT and plasmalogen phospholipids link ChREBP to mitochondrial dynamics.A, biosynthetic pathway of plasmalogens. GNPAT catalyzes the initial peroxisomal step by utilizing acyl-CoA to acylate dihydroxyacetone phosphate (DHAP) at the sn-1 position to form 1-acyl-DHAP. AG is the ether lipid precursor and generated in the final peroxisomal step. B and C, qPCR (B) and Western blot (C) analysis of Gnpat and Agps expression level in control cells treated with NG or HG and ChREBP-KD cells treated with HG. D, representative confocal micrographs showing MitoTracker red staining of control podocytes treated with NG or HG and HG-induced ChREBP-KD podocytes with or without Gnpat overexpression. Scale bar represents 10 μm in row 1; scale bar represents 2 μm in row 2. E and F, quantification of mitochondrial aspect ratio (E) and mitochondrial length (F) in cells described in (D). G and H, mitochondrial function as assessed by MitoSox fluorescence intensity (G) and ATP production (H) in indicated cell lines. I, MitoTracker red staining of control podocytes and ChREBP-KD podocytes treated with 0.05% ethanol (EtOH), DL-α-palmitin (DL-P), 16:0-AG, 18:0-AG in the presence of high glucose. Scale bar represents 10 μm in row 1; scale bar represents 2 μm in row 2. J and K, mitochondrial morphology was quantitated from confocal micrographs as mitochondrial aspect ratio (J) and mitochondrial length (K) in cells described in (I). Scale bar represents 10 μm. Results are presented as mean ± SEMs. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; 1-O-alkyl-G3P (AG), 1-O-alkyl-2-hydroxylsn-glycerol-3-phosphate; AG, alkylglycerols; ChREBP, carbohydrate-response element-binding protein; GNPAT, glyceronephosphate O-acyltransferase; HG, high glucose; KD, knockdown; NG, normal glucose; NS, not significant.
We next examined whether GNPAT couples ChREBP with mitochondrial morphology. To this end, we generated a Gnpat-overexpressing construct to rescue GNPAT expression in ChREBP-KD–cultured podocytes (Fig. S5, A–C). As expected and consistent with our previous observations (11), we observed increased mitochondrial fission in HG-treated podocytes transfected with a nontargeted shRNA (shCtrl) (Fig. 5D). However, ChREBP-KD podocytes treated with HG exhibited significantly less mitochondrial fission, whereas concurrent overexpression (OE) of GNPAT in ChREBP-KD podocytes (ChREBP-KD/Gnpat OE) resulted in enhanced mitochondrial fission, with a morphology shift towards a more fragmented network consistent with a higher proportion of shorter and rounder mitochondria (Fig. 5, D–F). Importantly, mitochondrial ROS as assessed by MitoSox fluorescence was also significantly higher in ChREBP-KD/Gnpat OE–transfected podocytes than in targeted ChREBP-KD podocytes when both exposed to HG (Fig. 5G). Surprisingly, GNPAT OE did not significantly affect the protective effect of ChREBP-KD on ATP production (Fig. 5H). Furthermore, while we observed that ChREBP-KD effectively restored both the basal and maximal oxygen consumption rate levels which were reduced by HG, cells with GNPAT-OE exhibited no significant additional impact beyond the levels obtained with ChREBP-KD (Fig. S6, A–C).
To further examine the effect of plasmalogens on mitochondrial remodeling, we next tested whether restoration of plasmalogen levels by direct supplementation of alkylglycerols (AG) could reverse the protective effect of ChREBP-KD on mitochondrial phenotype in podocytes since AGs have previously been shown to enter the synthetic pathway of plasmalogens, enhancing plasmalogen levels (36, 37). We treated ChREBP-KD podocytes with two compounds containing either 16- or 18-carbon AGs, 1-O-Hexadecyl-rac-glycerol (16:0-AG) or 1-O-Octadecyl-rac-glycerol (18:0-AG) (31). We observed that supplementation with either 16:0-AG or 18:0-AG led to significantly enhanced mitochondrial fragmentation in ChREBP-KD podocytes cultured in HG. In contrast, treatment with DL-α-palmitin, a control substance that contains an acyl group instead of the ether group, had no effect on mitochondrial morphology in ChREBP-KD podocytes (Fig. 5, I–K).
ChREBP binds to Gnpat promoter and activates its transcription
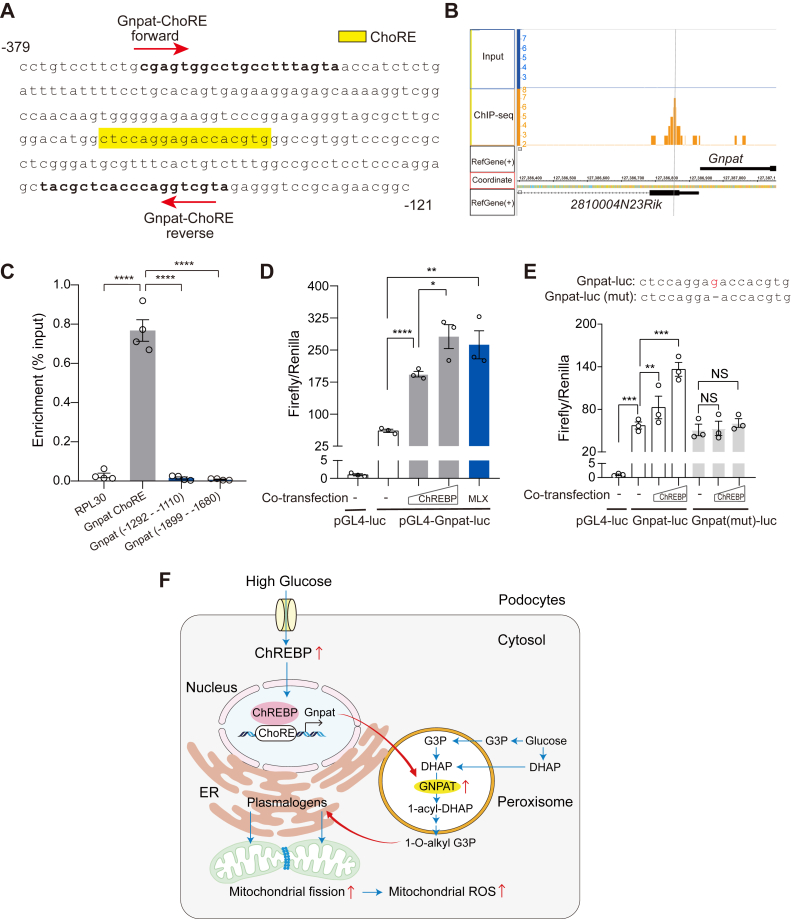
Finally, we examined whether GNPAT is a direct transcriptional target of ChREBP. Consistent with our hypothesis, we found one putative carbohydrate responsive element (ChoRE) in the promoter of mouse Gnpat gene (Fig. 6A). We then re-analyzed our ChIP-Seq data previously generated from adipose tissue (38) identified a peak of ChREBP binding upstream of Gnpat promoter, which was absent in the input control (Fig. 6B). To validate the ChIP-seq results in podocytes, we performed chromatin immunoprecipitation and qPCR using primers targeting various regions of the Gnpat promoter. We found that ChREBP specifically interacts with a proximal region of Gnpat promoter that covers the putative ChoRE region (Fig. 6C). To explore whether this interaction could affect Gnpat promoter activity, we generated a luciferase reporter construct driven by the mouse Gnpat proximal promoter and assessed the promoter activity with transient expression of ChREBP. We found that OE of ChREBP increased the luciferase activity of Gnpat promoter in a dose-dependent manner. Similarly, OE of Max-like protein (MLX), a functional obligatory partner of ChREBP, also enhanced the promoter activity of Gnpat (Fig. 6D). Furthermore, we generated a ChoRE mutant construct to test whether it can abolish ChREBP binding. Indeed, the mutant ChoRE failed to show activation of Gnpat promoter activity (Fig. 6E). These results strongly indicate that ChREBP directly regulates transcription of Gnpat gene. Furthermore, our findings demonstrate that ChREBP–Gnpat–plasmalogen axis is a novel regulator of mitochondrial dynamics in podocytes.
Figure 6.
ChREBP binds to Gnpat promoter and activates its transcription.A, mouse Gnpat promoter sequence from −121 bp to −379 bp relative to transcription start site. The putative ChoRE for ChREBP binding is highlighted in yellow. Primers used for ChIP assay are indicated within the sequence. B, ChIP-Seq analysis of mouse white adipose tissues using anti-ChREBP antibody identified a specific peak in the Gnpat proximal promoter. C, ChIP-qPCR validation of ChREBP binding to Gnpat promoter in cultured mouse podocyte, using primers to amplify various regions of the Gnpat promoter. D and E, firefly-to-renilla luciferase activities for the Gnpat gene promoter (D) or its ChoRE mutant (E) in HEK293T cells after transiently transfected with increasing amounts of ChREBP or MLX plasmids. F, proposed model depicting the link between mitochondrial lipidomes and mitochondrial morphology by ChREBP in podocytes under high glucose conditions. High glucose induces ChREBP expression and promotes its nuclear translocation and binding to the ChoRE element present on the promoter of Gnpat gene. Elevated expression of GNPAT results in accumulation of peroxisomal-derived plasmalogens, which are transported to mitochondria and incorporated into mitochondrial membranes. The subsequent changes in the membrane phospholipid composition will lead to mitochondrial fission and enhanced mitochondrial ROS production. Results are presented as mean ± SEMs. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. ChREBP, carbohydrate-response element-binding protein; ChIP-qPCR, chromatin immunoprecipitation and qPCR; ChoRE, carbohydrate responsive element; GNPAT, glyceronephosphate O-acyltransferase; ROS, reactive oxygen species.
Discussion
This study identifies a novel role for the transcription factor ChREBP in linking mitochondrial lipidomes with mitochondrial dynamics and the progression of DKD. Using podocyte-specific ChREBP-deficient mice, we provide in vivo evidence that the deletion of ChREBP in podocytes mitigates mitochondrial fission and improves key features of DKD. Furthermore, we define plasmalogen phospholipids as the mechanistic link between ChREBP-mediated lipid metabolism and mitochondrial remodeling. We also identify GNPAT, a critical enzyme in plasmalogen biosynthesis, as a direct transcriptional target of ChREBP. Our findings underscore the crucial role of ChREBP in mediating inter-organelle interactions across multiple cellular compartments, revealing ChREBP as a key regulator of cellular homeostasis. Specifically, our study suggests that ChREBP functions as a link between the HG environment and a coordinated response between peroxisomes and the ER, ultimately regulating and controlling mitochondrial morphology (Fig. 6F).
ChREBP has long been recognized to play a pivotal role in cell response to carbohydrate and glucose cues from the environment (23). While the impact of ChREBP global knockout in the kidneys of diabetic streptozotocin mice has been recently reported (39), the role of ChREBP KD in the kidney cells and the underlying molecular mechanism of its modulatory effect were unknown. Here, we demonstrate that conditional deletion of ChREBP in podocytes significantly attenuates albuminuria, mesangial matrix expansion, glomerular basement membrane thickening, and podocyte effacement in db/db diabetic mice. We found that these improvements are, at least in part, associated with reduced lipogenesis and improved mitochondrial fission in podocytes with ChREBP KD.
Recent advances in lipidomic technologies have empowered a better understanding of the interplay between lipid metabolism and mitochondrial dysfunction. For instance, several reports have recently suggested that the lipid composition in mitochondrial membranes may play a critical role in regulating and maintaining mitochondrial shape and function (29, 34, 40). However, the link between lipids and mitochondrial function in podocytes and its role in the progression of DKD remain poorly understood. In the present study, we used ultra-high-performance liquid chromatography coupled with high resolution mass spectrometry to demonstrate that mitochondrial plasmalogen phospholipids are differentially regulated by ChREBP. Notably, we found that elevated levels of plasmalogens in podocytes, achieved either through OE of GNPAT or by direct supplementation of plasmalogen precursors, could reverse the protective effect of ChREBP KD on mitochondrial fission in the diabetic milieu.
A key finding of this study is the identification of ChREBP as a novel transcriptional regulator of GNPAT expression. We provide evidence supporting the potential binding of ChREBP to Gnpat promoter and activating its transcription. Our findings indicate that ChREBP transcriptionally activates Gnpat expression in response to HG, resulting in increased plasmalogens content in mitochondrial lipidome.
Taken together, our results indicate that the transcription factor ChREBP links lipid metabolism with mitochondrial dynamics and the progression of DKD. We also identify the key role of plasmalogens, a vinyl-ether subclass of glycerophospholipids, in mediating ChREBP-induced mitochondrial remodeling in the diabetic environment. These findings support previously published work suggesting that plasmalogens, as major lipid components of biological membranes, play important roles in the lipid composition of mitochondrial membranes (31, 33, 34), and changes in their abundance are associated with mitochondrial membrane remodeling (31, 34, 41). Based on our findings, we propose that HG induces ChREBP expression and promotes its nuclear translocation and binding to the ChoRE element present on the promoter of Gnpat gene (Fig. 6F). Elevated expression of GNPAT results in the accumulation of peroxisomal-derived plasmalogens, which are transported to mitochondria and incorporated into mitochondrial membranes. The subsequent changes in the plasmalogen content could trigger mitochondrial membrane remodeling by several mechanisms as previously suggested, importantly by altering the biophysical properties of mitochondrial membrane, leading to increased mitochondrial fission (31, 41). Furthermore, our data suggests that targeting plasmalogen content by pharmacological approaches may present a novel therapeutic approach in DKD.
Experimental procedures
Tissue culture
Conditionally immortalized mouse podocytes were cultured as previously reported (11, 42). Podocytes were treated with 20 μM of DL-a-palmitin (Sigma-Aldrich), 1-O-Hexadecyl-rac-glycerol (16:0-AG, Santa Cruz), 1-O-Octadecyl-rac-glycerol (18:0-AG, Sigma-Aldrich), or 0.05% ethanol (vehicle) for 48 h. Human embryonic kidney fibroblast 293T cells (CRL-3216) were obtained from American Type Culture Collection and cultured according to the instructions.
Animal work
All animal experiments were conducted in accordance with the “Principles of Laboratory Animal Care” (NIH publication No. 85023, revised 1985) and the guidelines of the IACUC of The University of Texas MD Anderson Cancer Center. Diabetic db/db mice and their control littermates dbm (Strain #000642, BKS.Cg-Dock7m+/+Leprdb/J) were obtained from the Jackson Laboratory. Conditionally, ChREBP-targeted mice were described previously (43). The ChREBP-floxed mice were backcrossed ten generations on C57BLKS background. Podocin-iCreERT2 transgenic mice were previously generated in our laboratory (44). Mice were genotyped by PCR analysis of tail DNA with primers listed in Table S1. Mice with appropriate genotypes were administered 4 mg/d tamoxifen (Sigma-Aldrich, T5648) dissolved in sesame oil intraperitoneally for 10 days. Urinary albumin was measured using a mouse albumin ELISA kit (Excocell). Urinary creatinine concentration was measured using QuantiChrom Creatinine Assay Kit (BioAssay Systems).
Primary podocytes isolation
Primary podocytes were isolated from mouse kidneys as previously described (45). In brief, podocytes were ex vivo selected by biotin-labeled anti-Kirrel3 and podocalyxin antibodies (R&D Systems BAF4910 and R&D Systems BAF1556, respectively), then isolated using magnetic, streptavidin-labeled Dynabeads (Thermo Fisher Scientific).
Kidney histopathology
Formalin-fixed, paraffin-embedded tissues were sectioned at 5 μm. Periodic-Acid Schiff’s staining was performed according to manufacturer’s instructions (Sigma Aldrich). At least 30 glomeruli were examined per mouse.
Transmission electron microscopy
Kidneys removed from anesthetized mice were immediately trimmed, fixed, and processed as previously described (12). Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques). Glomerular basement membrane thickness was analyzed using ImageJ Software (version 1.53 g; National Institutes of Health; https://imagej.nih.gov/ij/download.html).
Mitochondrial morphology assessment
Mitochondrial shape in TEM micrographs were analyzed by ImageJ as previously described (11, 12). The morphology of at least 80 mitochondria was determined for each condition.
Immunofluorescence
Podocytes were plated at 5 x 103 seeding density on BioCoa Collagen I 22 mm coverslips in 6-well plates. After treatment, cells were incubated with 100 nM MitoTracker Red CMXRos (Life technologies) for 15 min. Cells were washed with DPBS, then fixed with ice-cold 1:1 methanol/acetone (v/v) and mounted. Images were acquired on a FV1200 confocal microscope (Olympus), with a x60 immersion objective.
Immunohistochemistry
Immunostaining of paraffin-embedded kidney sections was performed as previously described (12). Primary and secondary antibodies applied in this study were listed in Table S2. Images were acquired on a FV1200 confocal microscope (Olympus) with the same settings for each channel. Synaptopodin expression levels were determined using ImageJ software.
Mitochondrial ROS, ATP, and oxygen consumption rate
Podocytes were incubated with HBSS buffer containing 3 μM MitoSox (Life Technologies) for 20 min at 37 °C in a non-CO2 incubator. MitoSox fluorescence was analyzed by flow cytometry with help from UT-MDACC FCCICF Core, as previously described (25). Cellular ATP levels were measured using the CellTitre-Glo Luminescent Cell Viability Assay Kit (Promega). ATP levels were normalized by DNA concentration in each well. DNA concentration was measured using CyQUANT Cell Proliferation Assay Kit (Molecular Probes). Oxygen consumption rate was measured using Seahorse XFe96 Analyzer (Agilent) as previously described (25) and subsequently normalized using the CyQUANT Cell Proliferation Assay Kit for DNA concentration.
Western blot analysis
Western blotting and quantitative analyses were performed as previously described (25). The primary antibodies and secondary antibodies used for Western blotting were listed in Table S2.
RNA extraction and RT-qPCR
RNA extraction and RT-qPCR were performed as previously described (25). We calculate fold changes in gene expression normalized to Hprt or Actb using 2-ΔΔCT method. The specific primers for target genes in this study are listed in Table S1.
Chromatin immunoprecipitation and qPCR
ChIP assay was conducted using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology). Briefly, mouse podocytes were crosslinked with 1% formaldehyde for 10 min and quenched by 5 min incubation with 200 mM glycine. Nuclei were harvested and chromatin was digested by micrococcal nuclease to release nucleosome. Chromatin size and concentration were determined before incubation with ChIP-grade anti-ChREBP antibody (Novus, NB400-135) at 4 °C overnight. The next day ChIP-grade protein G magnetic beads were added to each reaction and incubated for 4 h with rotation. After a series of low salt and high salt wash, chromatin was eluted, and crosslink was reversed. DNA was then purified by spin column and quantitated by qPCR using specific primers spanning the putative ChREBP-binding elements in Gnpat promoter. Primers sequences used for ChIP-PCR are listed in Table S1.
Luciferase reporter assay
The 0.5kb proximal promoter of murine Gnpat gene (−400 nt to +101 nt relative to TSS) was amplified from genomic DNA of C57BL/6J mouse by PCR (Table S2). The 501 bp PCR product was subcloned between KpnI and XhoI sites of a promoter-less luciferase reporter pGL4.10 [luc2] (Promega). Site-directed mutagenesis was carried out with QuickChange II Site-Directed Mutagenesis Kit (Agilent). The putative carbohydrate responsive element (ChoRE) in Gnpat promoter (CTCCAGGAGACCACGTG) was mutated into CTCCAGGAACCACGTG, where the 5-nt spacer (underlined) between the two E-boxes was shortened to 4-nt as previously described (24). HEK293T cells were seeded in 12-well plates and transfected with firefly luciferase report, pGL4-luc vector or pGL4-Gnpat-luc promoter construct, and Renilla luciferase internal control pRL-TK (Promega), together with pcDNA3.1-ChREBP (24), or pcDNA3.1-MLX (24), using Lipofectamine 2000 (Thermo Fisher Scientific). Firefly-to-Renilla luciferase activities were assessed using a Dual-Luciferase Assay System (BPS Bioscience).
Reverse phase liquid chromatography
Mitochondria were isolated from mouse podocytes using Mitochondria Isolation Kit (Thermo Fisher Scientific). Mitochondria were pelleted by centrifugation at 3000g for 15 min at 4 °C, flash-frozen, and stored at −80 °C until use. Two hundred microliters of ethanol containing 1% 10 mM butylated hydroxytoluene in methanol and 2% Avanti SPLASH LIPIDOMIX Mass Spec Standards, pre-cooled to −80 °C, was added to each mitochondria sample. The tubes were vortexed for 10 min, placed on ice for 10 min, then centrifuged at 4 °C for 10 min at 17,000g. The supernatants were then collected for LC-MS analysis. Mobile phase A was 40:60 acetonitrile: 0.1% formic acid in 10 mM ammonium formate. Mobile phase B (MPB) was 90:9:1 isopropanol: acetonitrile: 0.1% formic acid in 10 mM ammonium formate. The chromatographic method included a Thermo Fisher Scientific Accucore C30 column (2.6 μm, 150 × 2.1 mm) maintained at 40 °C, a mobile phase flowrate of 0.200 ml/min, and a gradient elution program as follows: 0 to 3 min, 30% MPB; 3 to 13 min, 30 to 43% MPB; 13.1 to 33 min, 50 to 70% MPB; 33 to 48 min, 70 to 99% MPB; 48 to 55 min, 99% MPB; 55.1 to 60 min, 30% MPB.
Mass spectrometry
A Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer with heated electrospray ionization source was operated in data-dependent acquisition mode, in both positive and negative ionization modes, with scan ranges of 150 to 827 and 825 to 1500 m/z. An Orbitrap resolution of 120,000 (FWHM) was used for MS1 acquisition, and spray voltages of 3.6 kV and −2.9 kV were used for positive and negative ionization modes, respectively. For MS2 and MS3 fragmentation, a hybridized HCD/CID approach was used. Each sample was analyzed using 4 × 10 μl injections making use of the two scan ranges, in both ionization modes. Data were analyzed using Thermo Scientific LipidSearch software (version 5.0.63; https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/lipid-search-software.html) and R scripts written in house. The intensity of each peak was normalized to total lipid signal and to the internal standard.
shRNA and OE experiments
The coding region of Gnpat gene was amplified from a mouse Gnpat cDNA clone (Origene MC205395, NM_010322) by PCR (Table S1). The PCR product was inserted into CMV enhancer/promoter-driven vector Zeo-pT-MCS-GFP-T2A-Puro, a modified PiggyBac transposon system. An engineered miR-30–based ChREBP KD construct was assembled by PCR using previously validated ChREBP shRNA clone (24), followed by an optimized miR-E backbone (46), into Zeo-pT-MCS-GFP-IRES-BSD, another PiggyBac transposon system. Stable podocyte cell lines with ChREBP KD, or with ChREBP KD/Gnpat OE, were generated by cotransfection of above-mentioned constructs, together with PiggyBac transposase. After selection with 0.5 μg/ml of blasticidin alone or in combination with 1 μg/ml puromycin, GFP-positive cells were sorted by FACS, and the top 30% population were collected.
Statistical analysis
Group data are expressed as mean ± SEM. Comparisons of multiple groups were performed using one-way ANOVA followed by post hoc Tukey’s test. Comparisons between two groups were performed using two-tailed student’s t test. A p value of less than 0.05 was considered statistically significant. Statistical analyses were performed with GraphPad Prism version 8.0 (GraphPad; https://www.graphpad.com/features).
Data availability
The lipidomic data generated in this study have been deposited to Metabolomics Workbench data repository with project identifier PR001359.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank all members of the Danesh laboratory for their helpful discussions. We acknowledge the High Resolution Electron Microscopy Facility at MD Anderson Cancer Center (CCSG NIH P30CA016672) for helping with TEM experiments; the Metabolic Core Facility at MD Anderson Cancer Center (Cancer Prevention and Research Institute of Texas grant RP130397, NIH grants S10OD012304-01 and P30CA016672) for performing lipidomic studies; the Flow Cytometry and Cellular Imaging Core Facility at MD Anderson Cancer Center(NCI grant P30CA16672) for advice and help in flow cytometry experiments.
Author contributions
L. L., J. L., P. K. S., and B. H. C. methodology; L. L., J. L., K. M., N. P., L. T., P. K. S., and F. R. D. investigation; L. L., J. L., and F. R. D. writing–original draft; J. L., Y. S. K., and B. H. C. conceptualization; N. P. and P. K. S. resources; L. L., J. L., K. M., N. P., P. L. L., I. M., and L. T. formal analysis; P. L. L. data curation; I. M. visualization; B. H. C. project administration; F. R. D. supervision; B. H. C. and F. R. D. writing–review and editing; F. R. D. funding acquisition.
Funding and additional information
This work was supported by National Institutes of Health grants R01DK078900 and R01DK091310 (to F. R. D.). L. L. is supported by a scholarship from the China Scholarship Council (202006370196). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Supporting information
References
- 1.Harayama T., Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 2.Yoon H., Shaw J.L., Haigis M.C., Greka A. Lipid metabolism in sickness and in health: emerging regulators of lipotoxicity. Mol. Cell. 2021;81:3708–3730. doi: 10.1016/j.molcel.2021.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Jiang T., Li J., Proctor G., McManaman J.L., Lucia S., et al. Regulation of renal lipid metabolism, lipid accumulation, and glomerulosclerosis in FVBdb/db mice with type 2 diabetes. Diabetes. 2005;54:2328–2335. doi: 10.2337/diabetes.54.8.2328. [DOI] [PubMed] [Google Scholar]
- 4.Ducasa G.M., Mitrofanova A., Mallela S.K., Liu X., Molina J., Sloan A., et al. ATP-binding cassette A1 deficiency causes cardiolipin-driven mitochondrial dysfunction in podocytes. J. Clin. Invest. 2019;129:3387–3400. doi: 10.1172/JCI125316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammerschmidt P., Ostkotte D., Nolte H., Gerl M.J., Jais A., Brunner H.L., et al. CerS6-Derived sphingolipids interact with Mff and promote mitochondrial fragmentation in obesity. Cell. 2019;177:1536–1552.e1523. doi: 10.1016/j.cell.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Zhou L., Yu M., Arshad M., Wang W., Lu Y., Gong J., et al. Coordination among lipid droplets, peroxisomes, and mitochondria regulates energy expenditure through the CIDE-ATGL-PPARα pathway in adipocytes. Diabetes. 2018;67:1935–1948. doi: 10.2337/db17-1452. [DOI] [PubMed] [Google Scholar]
- 7.Field C.S., Baixauli F., Kyle R.L., Puleston D.J., Cameron A.M., Sanin D.E., et al. Mitochondrial integrity regulated by lipid metabolism is a cell-intrinsic checkpoint for Treg suppressive function. Cell Metab. 2020;31:422–437.e425. doi: 10.1016/j.cmet.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viader A., Sasaki Y., Kim S., Strickland A., Workman C.S., Yang K., et al. Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron. 2013;77:886–898. doi: 10.1016/j.neuron.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Q., Zhang H., Gutiérrez Cortés N., Wu D., Wang P., Zhang J., et al. Increased drp1 acetylation by lipid overload induces cardiomyocyte death and heart dysfunction. Circ. Res. 2020;126:456–470. doi: 10.1161/CIRCRESAHA.119.315252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galvan D.L., Green N.H., Danesh F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92:1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayanga B.A., Badal S.S., Wang Y., Galvan D.L., Chang B.H., Schumacker P.T., et al. Dynamin-related protein 1 deficiency improves mitochondrial fitness and protects against progression of diabetic nephropathy. J. Am. Soc. Nephrol. 2016;27:2733–2747. doi: 10.1681/ASN.2015101096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvan D.L., Long J., Green N., Chang B.H., Lin J.S., Schumacker P., et al. Drp1S600 phosphorylation regulates mitochondrial fission and progression of nephropathy in diabetic mice. J. Clin. Invest. 2019;129:2807–2823. doi: 10.1172/JCI127277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y., Sun Y., Wang M., Hou Y., Huang W., Zhou D., et al. Elevation of JAML promotes diabetic kidney disease by modulating podocyte lipid metabolism. Cell Metab. 2020;32:1052–1062.e1058. doi: 10.1016/j.cmet.2020.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Falkevall A., Mehlem A., Palombo I., Heller Sahlgren B., Ebarasi L., He L., et al. Reducing VEGF-B signaling ameliorates renal lipotoxicity and protects against diabetic kidney disease. Cell Metab. 2017;25:713–726. doi: 10.1016/j.cmet.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Mishra P., Chan D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan M., Brooks C., Liu F., Sun L., Dong Z. Mitochondrial dynamics: regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 2013;83:568–581. doi: 10.1038/ki.2012.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao C.H., Wang R., Wang Y., Kung C.P., Weber J.D., Patti G.J. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. Elife. 2019;8:e41351. doi: 10.7554/eLife.41351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban T., Ishihara T., Kohno H., Saita S., Ichimura A., Maenaka K., et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 2017;19:856–863. doi: 10.1038/ncb3560. [DOI] [PubMed] [Google Scholar]
- 21.Adachi Y., Itoh K., Yamada T., Cerveny K.L., Suzuki T.L., Macdonald P., et al. Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell. 2016;63:1034–1043. doi: 10.1016/j.molcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labbé K., Mookerjee S., Le Vasseur M., Gibbs E., Lerner C., Nunnari J. The modified mitochondrial outer membrane carrier MTCH2 links mitochondrial fusion to lipogenesis. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdul-Wahed A., Guilmeau S., Postic C. Sweet sixteenth for ChREBP: established roles and future goals. Cell Metab. 2017;26:324–341. doi: 10.1016/j.cmet.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Long J., Galvan D.L., Mise K., Kanwar Y.S., Li L., Poungavrin N., et al. Role for carbohydrate response element-binding protein (ChREBP) in high glucose-mediated repression of long noncoding RNA Tug1. J. Biol. Chem. 2020;295:15840–15852. doi: 10.1074/jbc.RA120.013228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Long J., Mise K., Galvan D.L., Overbeek P.A., Tan L., et al. PGC1α is required for the renoprotective effect of lncRNA Tug1 in vivo and links Tug1 with urea cycle metabolites. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Wang Y., Long J., Wang J., Haudek S.B., Overbeek P., et al. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 2012;15:186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afshinnia F., Nair V., Lin J., Rajendiran T.M., Soni T., Byun J., et al. Increased lipogenesis and impaired β-oxidation predict type 2 diabetic kidney disease progression in American Indians. JCI Insight. 2019;4 doi: 10.1172/jci.insight.130317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., Zhang T., Wang L., Cai Y., Zhong X., He X., et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017;36:1330–1347. doi: 10.15252/embj.201695417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kameoka S., Adachi Y., Okamoto K., Iijima M., Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018;28:67–76. doi: 10.1016/j.tcb.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schenkel L.C., Bakovic M. Formation and regulation of mitochondrial membranes. Int. J. Cell Biol. 2014;2014 doi: 10.1155/2014/709828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H., He A., Tan M., Johnson J.M., Dean J.M., Pietka T.A., et al. Peroxisome-derived lipids regulate adipose thermogenesis by mediating cold-induced mitochondrial fission. J. Clin. Invest. 2019;129:694–711. doi: 10.1172/JCI120606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez-Rojo N., Riezman H. On the road to unraveling the molecular functions of ether lipids. FEBS Lett. 2019;593:2378–2389. doi: 10.1002/1873-3468.13465. [DOI] [PubMed] [Google Scholar]
- 33.Dean J.M., Lodhi I.J. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett C.F., O'Malley K.E., Perry E.A., Balsa E., Latorre-Muro P., Riley C.L., et al. Peroxisomal-derived ether phospholipids link nucleotides to respirasome assembly. Nat. Chem. Biol. 2021;17:703–710. doi: 10.1038/s41589-021-00772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagan N., Zoeller R.A. Plasmalogens: biosynthesis and functions. Prog. Lipid Res. 2001;40:199–229. doi: 10.1016/s0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 36.Phuyal S., Skotland T., Hessvik N.P., Simolin H., Øverbye A., Brech A., et al. The ether lipid precursor hexadecylglycerol stimulates the release and changes the composition of exosomes derived from PC-3 cells. J. Biol. Chem. 2015;290:4225–4237. doi: 10.1074/jbc.M114.593962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergan J., Skotland T., Sylvänne T., Simolin H., Ekroos K., Sandvig K. The ether lipid precursor hexadecylglycerol causes major changes in the lipidome of HEp-2 cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poungvarin N., Chang B., Imamura M., Chen J., Moolsuwan K., Sae-Lee C., et al. Genome-wide analysis of ChREBP binding sites on male mouse liver and white adipose chromatin. Endocrinology. 2015;156:1982–1994. doi: 10.1210/en.2014-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen N., Mu L., Yang Z., Du C., Wu M., Song S., et al. Carbohydrate response element-binding protein regulates lipid metabolism via mTOR complex1 in diabetic nephropathy. J. Cell Physiol. 2021;236:625–640. doi: 10.1002/jcp.29890. [DOI] [PubMed] [Google Scholar]
- 40.MacVicar T., Ohba Y., Nolte H., Mayer F.C., Tatsuta T., Sprenger H.G., et al. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature. 2019;575:361–365. doi: 10.1038/s41586-019-1738-6. [DOI] [PubMed] [Google Scholar]
- 41.Koivuniemi A. The biophysical properties of plasmalogens originating from their unique molecular architecture. FEBS Lett. 2017;591:2700–2713. doi: 10.1002/1873-3468.12754. [DOI] [PubMed] [Google Scholar]
- 42.Mundel P., Reiser J., Zúñiga Mejía Borja A., Pavenstädt H., Davidson G.R., Kriz W., et al. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp. Cell Res. 1997;236:248–258. doi: 10.1006/excr.1997.3739. [DOI] [PubMed] [Google Scholar]
- 43.Jois T., Chen W., Howard V., Harvey R., Youngs K., Thalmann C., et al. Deletion of hepatic carbohydrate response element binding protein (ChREBP) impairs glucose homeostasis and hepatic insulin sensitivity in mice. Mol. Metab. 2017;6:1381–1394. doi: 10.1016/j.molmet.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Wang Y., Long J., Chang B.H., Wilson M.H., Overbeek P., et al. Tamoxifen-inducible podocyte-specific iCre recombinase transgenic mouse provides a simple approach for modulation of podocytes in vivo. Genesis. 2010;48:446–451. doi: 10.1002/dvg.20635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long J., Badal S.S., Ye Z., Wang Y., Ayanga B.A., Galvan D.L., et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 2016;126:4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lipidomic data generated in this study have been deposited to Metabolomics Workbench data repository with project identifier PR001359.