Abstract
Purpose
To present fully automated DL-based prostate cancer detection system for prostate MRI.
Methods
MRI scans from two institutions, were used for algorithm training, validation, testing. MRI-visible lesions were contoured by an experienced radiologist. All lesions were biopsied using MRI-TRUS-guidance. Lesions masks, histopatho-logical results were used as ground truth labels to train UNet, AH-Net architectures for prostate cancer lesion detection, segmentation. Algorithm was trained to detect any prostate cancer ≥ ISUP1. Detection sensitivity, positive predictive values, mean number of false positive lesions per patient were used as performance metrics.
Results
525 patients were included for training, validation, testing of the algorithm. Dataset was split into training (n = 368, 70%), validation (n = 79, 15%), test (n = 78, 15%) cohorts. Dice coefficients in training, validation sets were 0.403, 0.307, respectively, for AHNet model compared to 0.372, 0.287, respectively, for UNet model. In validation set, detection sensitivity was 70.9%, PPV was 35.5%, mean number of false positive lesions/patient was 1.41 (range 0–6) for UNet model compared to 74.4% detection sensitivity, 47.8% PPV, mean number of false positive lesions/patient was 0.87 (range 0–5) for AHNet model. In test set, detection sensitivity for UNet was 72.8% compared to 63.0% for AHNet, mean number of false positive lesions/patient was 1.90 (range 0–7), 1.40 (range 0–6) in UNet, AHNet models, respectively.
Conclusion
We developed a DL-based AI approach which predicts prostate cancer lesions at biparametric MRI with reason-able performance metrics. While false positive lesion calls remain as a challenge of AI-assisted detection algorithms, this system can be utilized as an adjunct tool by radiologists.
Keywords: Prostate cancer, Artificial intelligence, Magnetic resonance imaging, Deep learning
Introduction
Since the introduction of population-based screening in the early 1990’s, prostate cancer has been diagnosed by systematic biopsy prompted by an elevated serum prostate specific antigen (PSA) level or abnormal digital rectal exam [1]. Wide use of screening has led to a significant shift from metastatic to localized disease at the time of the initial diagnosis and a decrease in disease-specific mortality [2]. This has been attributed to early detection and treatment of localized prostate cancer. However, due to the screening-related increase in disease incidence, the morbidity and toxicity of prostate cancer related treatment has raised concerns of overdiagnosis and overtreatment. The serum PSA level is organ-specific but not necessarily cancer-specific and can be elevated for a variety of reasons such as benign prostatic hyperplasia or prostatitis [3, 4]. Furthermore, transrectal ultrasound (TRUS) was only used to guide the biopsy needle into predefined systematic templated locations within the prostate rather than to detect and sample prostate cancer suspicious lesions [5]. The combination of different anatomic and functional magnetic resonance imaging (MRI) sequences has led to the development of multiparametric MRI, which has been proven to be capable of detecting and visualizing prostate cancer lesions [6]. However, like all imaging techniques, its diagnostic performance is highly dependent on radiologist training and expertise. Thus, an international panel of experts proposed the Prostate Imaging-Reporting and Data System (PI-RADS) in 2011 to achieve more standardization in image evaluation and reporting. PI-RADS has been widely accepted as the standard in prostate multiparametric MRI [7]. Nevertheless, recent data still suggests significant disparities in detection and classification performance especially among readers with different experience [8]. Many factors have been discussed but a major contributor to this performance mismatch is human subjectivity. The PI-RADS guidelines are based on qualitative criteria which cannot be objectively verified.
Machine Learning (ML) solutions can assist human beings in solving everyday problems due to their ability to process large amount of data much faster than human beings are naturally capable of. Deep Learning (DL) is a subbranch of ML and Artificial Intelligence (AI) which uses Deep Neural Networks for many different applications. Due to recent advancements in algorithm theory, computer processor technology and the abundance of big data, DL has undergone significant progress over the last decade and has become a significant part in social media, image processing, self-driving cars, industrial robotics, security, medicine and many other fields of everyday life [9]. Recent literature evidence also suggests that prostate MRI can benefit from DL-based AI approaches for intraprostatic lesion detection and classification tasks [10–12]. However, majority of current literature is limited in terms of data size and diversity for their training and testing populations [13].
In this work, we aim to present a new fully automated DL-based prostate cancer detection system for prostate MRI using a large-scale, diverse expert-annotated training dataset.
Material and methods
Study population
This retrospective study includes 525 patients from two different institutions who underwent multiparametric prostate MRI and subsequent MRI-targeted prostate biopsy for clinical suspicion of prostate cancer. Imaging protocols and biopsy technique differed among the centers.
PROSTATEx open source dataset
This dataset is publicly available and was used for an international contest between November 2016 and January 2017 and consists of 347 exams of 344 patients (three patients had repeated exams). From this population 145 patients underwent MRI-targeted biopsy [14, 15]. These patients were included in this study for model training (n = 109), validation (n = 18) and testing (n = 18). All exams were performed at the Radboud University Medical Center in Nijmegen, The Netherlands where patients underwent T2-weighted, proton density-weighted, dynamic contrast enhanced and diffusion-weighted MR imaging using the 3 Tesla MAGNETOM Trio and Skyra scanner systems (Siemens Healthineers, Erlangen, Germany) without the use of an endorectal coil (Supplement Table 1). T2-weighted images were obtained using a turbo spin echo sequence and had a resolution of 0.5 mm inplane and a slice thickness of 3.6 mm. The DWI series were obtained with a single-shot echo planar imaging sequence with a resolution of 2 mm in-plane and 3.6 mm slice thickness and with diffusion-encoding gradients in three directions. Three b-values were acquired (50, 400, and 800 s/mm2), and ADC maps and b = 1400 DWI were calculated by the scanner software. MRI-guided biopsies were performed by medical experts with multiple years of experience in MRI-guided prostate biopsies. At the start of the biopsy procedure a T2-weighted volume and an ADC map were acquired. After lesions have been identified, a needle guide was inserted transrectally. Consecutive sagittal and transversal MRIs were made during repositioning of the needle guide to confirm the correct position. Once the correct position has been reached a biopsy needle was inserted and a biopsy core was obtained. To verify the biopsy location, sagittal and transversal images were made with the needle in situ. Subsequently, biopsies were histopathologically processed, inspected and graded by an experienced uropathologist with 17 years of experience in prostate pathology.
Institutional prostate MRI and biopsy dataset
For this dataset, patients were scanned between January 2015 and September 2019. This consecutive cohort includes patients recruited as part of one or more IRB approved protocols, including radiologic profiling of prostate cancer (Clinical Trials.gov Identifier: NCT03354416), patients undergoing MRI-fusion-guided prostate biopsy (NCT00102544), and patients undergoing surgical treatment for intermediate or high-risk prostate cancer (NCT02594202). All these patients signed informed consent for these clinical trials, and they were a part of a larger patient population whose MRI-targeted biopsy outcomes were published in a major publication [6]. This dataset additionally included patients who did not have any visible lesions on MRI with subsequent cancer negative 12-core TRUS guided systematic biopsies to avoid selection bias towards prostate cancer and more aggressive disease. After application of these inclusion and exclusion criteria a total of 380 patients were included in this study for model training (n = 259), validation (n = 61) and testing (n = 60).
Multiparametric MRI exams at the National Cancer Institute (NCI), National Institutes of Health (NIH) were performed using a 3 Tesla magnet (Achieva 3.0-T-TX, Philips Healthcare, Best, the Netherlands) with a 16-channel surface coil (SENSE, Philips Healthcare, Best, the Netherlands) and an endorectal coil (BPX-30, MEDRAD, Pittsburgh, Pennsylvania) inflated with perfluorocarbon (3 mol/l Fluorinert) to a volume of 45 ml in 235 patients. T2-weighted turbo spin echo acquisition sequences were obtained in the axial, sagittal, and coronal planes. Diffusion-weighted imaging was obtained for production of apparent diffusion coefficient (ADC) maps using a monoexponential decay model and a separate high b-value sequence (2000s/mm2 for scans with endorectal coil, 1500 s/mm2 for scans without endorectal coil). In 145 patients where an endorectal coil was not used, a 32-channel cardiac coil (SENSE, InVivo, Gainesville, Florida) was used instead. MRI acquisition parameters are summarized in Supplemental Table 1. Patients with lesions identified on MRI underwent a targeted biopsy performed by a highly trained urologist (PAP) and an interventional radiologist (BJW) with > 15 years of experience in MRI-targeted prostate biopsy [16]. Patients also underwent a standard systematic biopsy in the same session. The standard biopsy was typically 12 cores collected in an extended sextant template of biopsies from the lateral and medial aspects of the base, mid, and apical prostate on both sides. Using the UroNav® MRI/ultrasound fusion device (InVivo, Gainesville, Florida) the targeted biopsies were performed with the previously identified MRI lesions superimposed using the T2-weighted sequence on the real-time TRUS images. Each lesion was sampled both in axial and sagittal planes by an end-fire TRUS probe (Philips, Healthcare, Best, the Netherlands).
MRI evaluation and lesion annotation
All MRI exams were evaluated by a single genitourinary radiologist (BT) with over 15 years of experience and reading over 1500 multiparametric MRI exams per year. Suspicious areas were manually outlined using an in-house segmentation software on T2-weighted series, with visual correlation to diffusion-weighted series for lesion volume determination [17]. Histopathological reports of MRI-targeted biopsies were reviewed, and the highest ISUP (International Society of Urological Pathology) grade group was assigned as the outcome label for every lesion [18]. All in-house biopsy specimens were read by a dedicated genitourinary pathologist (MJM) with > 15 years of experience. For scans from the Prostate-X cohort, targeted biopsy outcomes were available as a point location. Any point location within 5 mm of a radiologist-identified region was considered a correlate. All contours were then saved in MIPAV VOI (volume of interest) format. For all subsequent image processing, analysis, and training, all DICOM images were converted to NIfTI format, maintaining original spatial resolution. Diffusion-weighted series (ADC and high b-value) were then resampled to spatial resolution of T2W imaging. Contours were converted to NIfTI format as binary masks, where value of 0 includes both background or suspicious lesions with negative biopsy result and value of 1 represents any lesion with cancer-positive findings. The masks were saved in equivalent spatial resolution as T2-weighted imaging. DCE MRI pulse sequence was not used for lesion annotation and AI model development due to significant mismatches between image acquisition parameters of the open source and institutional datasets.
AI algorithm development
We adopted a 3D UNet model [19] and an AH-Net model [20] independently for experiment benchmark. Both models are popular neural network architectures for 3D medical image segmentation. In the experiment, the problem is formulated as volume-to-volume mapping (2-channel foreground/background segmentation output from 3-channel multiparametric MRI input). To further reduce the problem complexity, we resampled all input MRI and label volumes towards uniform spacing/resolution 0.5 mm × 0.5 mm × 3.0 mm. Resampling imaging volumes is via linear interpolation, and resampling label volumes is based on nearest neighbor interpolation. The MRI input is pre-processed using Z-score standard normalization independently.
During the conventional model training, the input of the network are 3-channel patches (T2, ADC map, High-b map) with size 160 × 160 × 32, randomly cropped from imaging volumes. The optimization objective is the combination of soft dice loss [21] and cross-entropy loss. The model is trained for total 5,000 epochs using Adam optimizer [22], and the learning rate of optimizer is 0.0005. The overall batch size is 32 (8 per GPU). Necessary data augmentation techniques (e.g., random intensity shift, random flipping, random zooming, random Gaussian noise, random Gaussian smoothing) are used for training. The validation accuracy is measured with the Dice’s score after sliding-window inference with window size is 160 × 160 × 32. Then the trained model with the best validation accuracy is selected for final validation and testing. Our proposed approach is implemented with NVIDIA Clara Train SDK [23] and experimented on four NVIDIA V100 GPUs with 16 GB memory.
Statistical analysis
The detection performance was measured at the lesion level and patient level. The lesion level, calculation of true positive (TP), false positive (FP), and false negative (FN) was compared to targeted biopsy outcomes to evaluate sensitivity and positive predictive value (PPV). A TP is defined as any overlap with radiologist-defined ROI that was associated with positive biopsy outcome. A FP is defined as any prediction outside of radiologists defined ROIs or a positive prediction within a radiologist-defined ROI that was associated with a negative (benign) biopsy outcome. Given the lack of surgical correlation, true negatives cannot be evaluated at the lesion-level. At the patient level, the segmentation accuracy was measured with the Dice similarity coefficient score. Detection performance metrics were calculated based on the patient’s overall cancer status from biopsy. A True Negative is a patient with an overall negative (benign) biopsy result and no predicted AI areas. The number of false positives per patient were tabulated for reporting.
Results were reported separately for validation and testing datasets. Performance metrics were calculated from the entire set, where 95% confidence intervals (CI) were estimated from 2000 bootstrap samples by random sampling on the patient-level to account for intra- and inter-lesion correlations.
Results
Study population and data split
A total of 525 patients were included for training, validation and testing of the model. The dataset was split into a training cohort (n = 368, 70%), a validation cohort (n = 79, 15%) and a test cohort (n = 78, 15%). The patient demographics after data split for patients from our institution are shown in Table 1. Histopathology on the patient-level based on ISUP grade groups is summarized in Table 2. A total of 844 contoured lesions with histopathological confirmation were included for training, validation and testing of the models. Lesion histopathology based on ISUP grades after data split for the complete dataset are summarized in Table 3.
Table 1.
Patient demographics before and after data split for the National Cancer Institute MRI and biopsy dataset
| Patient characteristics Variable | Overall, n = 380 | Training, n = 259 | Validation, n = 61 | Test, n = 60 |
|---|---|---|---|---|
|
| ||||
| Age (years) | 65 (60–70) | 66 (60–70) | 66 (58–70) | 64 (61–69) |
| PSA (ng/ml) | 7.1 (5.1–11.0) | 6.9 (5.1–10.4) | 7.0 (5.2–12.2) | 9.0 (5.1–12.2) |
| Prostate volume (ml) | 51 (36–69) | 51 (36–70) | 51 (33–64) | 52 (39–68) |
| Ethnicity | ||||
| African American | 53 (14%) | 36 (14%) | 9 (15%) | 8 (13%) |
| Asian | 12 (3%) | 9 (3%) | 0 | 3 (5%) |
| White | 219 (58%) | 155 (60%) | 32 (52%) | 32 (53%) |
| Multiple race | 3 (< 1%) | 2(1%) | 0 | 1 (< 1%) |
| Unknown | 93 (24%) | 57 (22%) | 20 (33%) | 16 (27%) |
Table 2.
Histopathological outcomes on the patient-level after data split for the complete dataset
| Split | Patient ISUP Grade Groups |
Total | |||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
|
| |||||||
| Training | 74 (20%) | 42 (11%) | 110 (30%) | 59 (16%) | 53 (14%) | 30 (8%) | 368 |
| Validation | 15 (19%) | 11 (14%) | 21 (27%) | 13 (16%) | 15 (19%) | 4 (5%) | 79 |
| Test | 18 (23%) | 5 (6%) | 26 (33%) | 9 (12%) | 15 (19%) | 5 (6%) | 78 |
Italic values represent total number of lesions in training/validation/test)
ISUP International Society of Urological Pathology
Table 3.
Histopathological outcomes on the lesion-level after data split for the complete dataset
| Split | Radiologist-identified lesion TBx ISUP grade groups |
Total | Total cancer | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | |||
|
| ||||||||
| Training | 222 (39%) | 87 (15%) | 140 (24%) | 48 (8%) | 54 (9%) | 25 (4%) | 576 | 354 |
| Validation | 52 (38%) | 22 (16%) | 32 (23%) | 12 (9%) | 14 (10%) | 6 (4%) | 138 | 86 |
| Test | 49 (38%) | 20 (15%) | 30 (23%) | 12 (9%) | 13 (10%) | 6 (5%) | 130 | 81 |
Italic values represent total number of lesions in training/validation/test)
ISUP International Society of Urological Pathology
Patient‑level cancer detection performance
The detection performance of both trained models in the validation and testing sets based on ISUP grade groups is summarized in Table 4. Overall patient level cancer detection sensitivity was similar between the two models, with 92.2% and 95.3% Sensitivity for UNet and AHNet models, respectively. However, AHNet demonstrated a superior specificity of 26.7% compared to 6.7% in UNet. A similar result was observed in the testing set, with 93.3% and 91.7% sensitivity and 22.2% and 5.6% specificity values for AHNet and UNet, respectively.
Table 4.
Patient-level detection performance
| Split | model | Sensitivity | Specificity | PPV | detection rate by patient-level ISUP Grade Groups |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||||||
|
| |||||||||||
| Validation | UNet | 0.922 | 0.067 | 0.808 | 14/15 | 11/11 | 19/21 | 12/13 | 14/15 | 3/4 | |
| AHNet | 0.953 | 0.267 | 0.847 | 11/15 | 10/11 | 20/21 | 13/13 | 14/15 | 4/4 | ||
| test | UNet | 0.917 | 0.056 | 0.764 | 17/18 | 4/5 | 25/26 | 8/9 | 13/15 | 5/5 | |
| AHNet | 0.933 | 0.222 | 0.800 | 14/18 | 5/5 | 24/25 | 7/9 | 15/15 | 5/5 | ||
ISUP International Society of Urological Pathology
Lesion‑level cancer detection performance
The Dice Similarity Coefficient (DSC) in the training and validation sets were 0.403 and 0.307, respectively, for the AHNet model compared to 0.372 and 0.287, respectively, for the UNet model. Cases with highest DSC and representative case from those with lowest Dice scores in the testing set are shown in Figs. 1 and 2, respectively. Overall lesion-based detection performance is summarized in Table 5. In the validation set, overall detection sensitivity was 70.9%, PPV was 35.5% and the mean number of false positive lesions per patient was 1.41 (range 0 – 6) for the UNet model compared to 74.4% detection sensitivity, 47.8% PPV, and mean number of false positive lesions per patient was 0.87 (0–5) for the AHNet model. Performance analysis in the test set demonstrated an increased false positive rate in both models, increasing to mean 1.90 (range 0–7) and mean 1.40 (range 0–6) false positive lesions per patient in UNet and AHNet models, respectively (Table 5). An example prediction demonstrating false positive is shown in Fig. 3. Detection sensitivity in the testing set remained similar to validation set for UNet (72.8%) compared to a decrease for the AHNet model (63.0%).
Fig. 1.
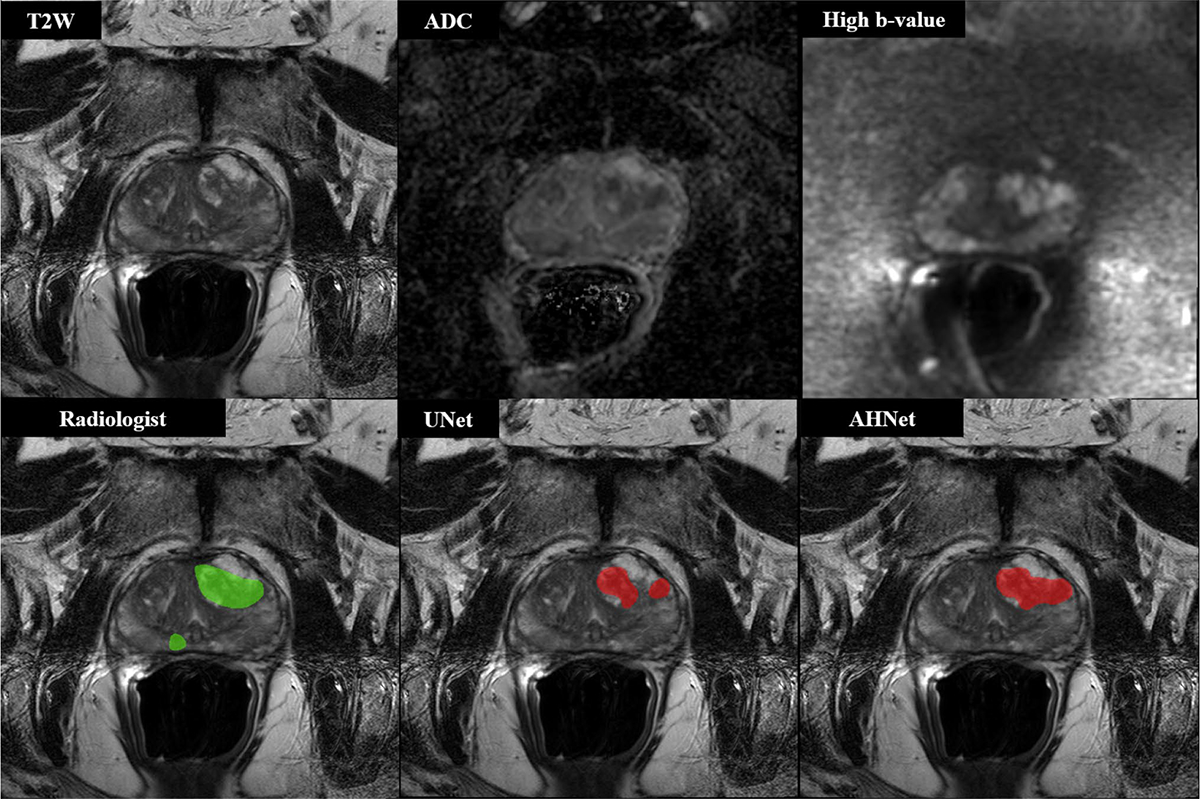
68-year-old male with serum PSA 10.57 ng/ml who underwent targeted biopsy of MR findings, revealing Gleason 3 + 3 in right midline peripheral zone lesion and Gleason 4 + 4 in left anterior transition zone lesion. Both UNet and AHNet produced true positive segmentation in left anterior transition zone lesion and false negative result in region of right midline peripheral zone lesion. In addition to biopsy positive lesions, AHNet produced 1 false positive and overall Dice score of 0.80922. UNet overall Dice score was 0.72229
Fig. 2.
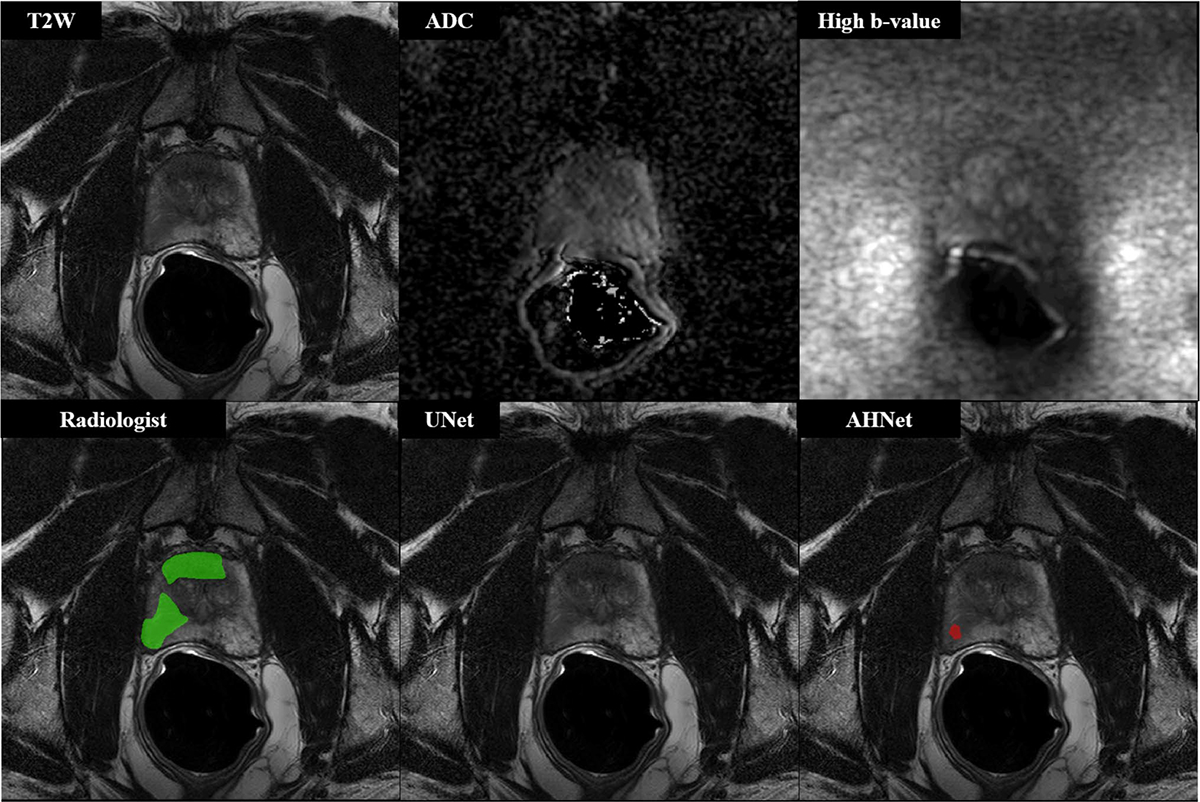
57-year-old male with serum PSA 6.68 ng/ml who underwent targeted biopsy of MR findings, revealing Gleason 3 + 3 in all targeted cores (n = 3 lesions, 2 shown in this figure). UNet results demonstrated false negative on the patient-level (Dice = 0, n = 3 False Negative lesions), while AHNet underestimated the volume of disease (Dice = 0.0385, n = 1 True Positive lesion, n = 2 False Negative lesions)
Table 5.
Lesion-level detection performance
| Split | Model | Sensitivity (95% CI) | PPV (95% CI) | Mean FPs/exam (range) | Detection rate by lesion-level ISUP Grade Groups |
||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||||
|
| |||||||||
| Validation | UNet | 0.709 (0.60,0.81) | 0.355 (0.27,0.45) | 1.41 (0–6) | 14/22 | 27/32 | 8/12 | 9/14 | 3/6 |
| AHNet | 0.744 (0.66,0.83) | 0.478 (0.38,0.58) | 0.87 (0–5) | 15/22 | 25/32 | 9/12 | 11/14 | 4/6 | |
| Test | UNet | 0.728 (0.62,0.83) | 0.285 (0.21,0.36) | 1.90 (0–7) | 11/20 | 22/30 | 10/12 | 10/13 | 6/6 |
| AHNet | 0.630 (0.53–0.73) | 0.319 (0.24,0.41) | 1.40 (0–6) | 12/20 | 18/30 | 7/12 | 9/13 | 5/6 | |
95% Confidence Intervals produced from bootstrap analysis on the patient-level
ISUP International Society of Urological Pathology
Fig. 3.
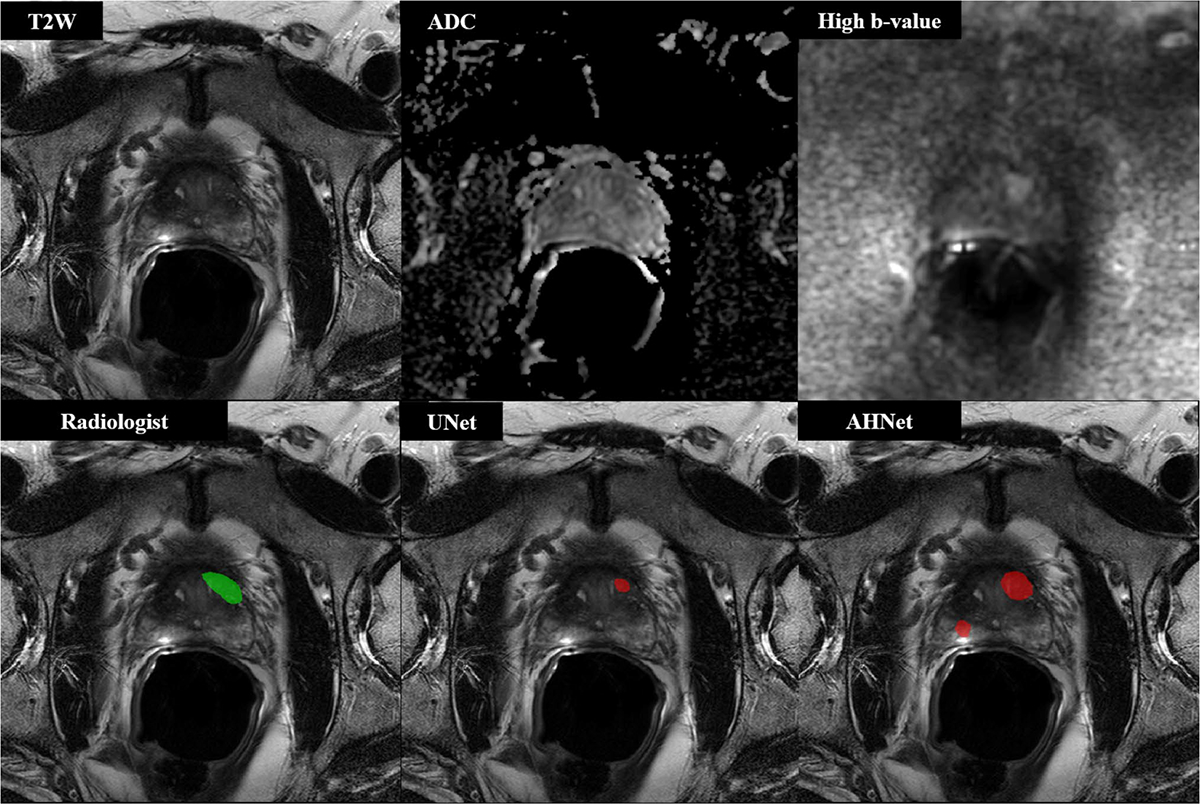
55-year-old male with serum PSA 10.97 ng/ml who underwent targeted biopsy of MR findings, revealing Gleason 3 + 4 in one lesion located in left anterior transition zone, which was targeted at biopsy. UNet results demonstrated true positive detection with 1 false positive penalty (DSC = 0.2057). AHNet produced 1 true positive detection with 3 false positive penalty lesions (DSC = 0.4776)
Analysis of false positive lesions
Several false positive lesions in each model cohort were found to correspond to radiologist-detected lesions with a benign result (n = 52 in validation set, n = 49 in testing set; Table 3). For the UNet model, 16/111 false positives in the validation set and 21/148 false positives in the validation set corresponded to radiologist-detected cancer suspicious but tumor negative areas. For the AHNet model, 12/70 false positives in the validation set and 19/109 false positives in the testing set corresponded to radiologist-detected cancer suspicious but tumor negative regions. The prospectively assigned PIRADSv2 score during clinical assessment (i.e., prior to targeted biopsy) for these benign results are shown in Fig. 4. Relatively, a higher proportion of PIRADSv2 3–5 lesions were detected by AI compared to PIRADSv2 1–2 lesions. Remaining false positive lesions in all cohorts did not correspond to areas suspicious for cancer by PIRADSv2 criteria.
Fig. 4.
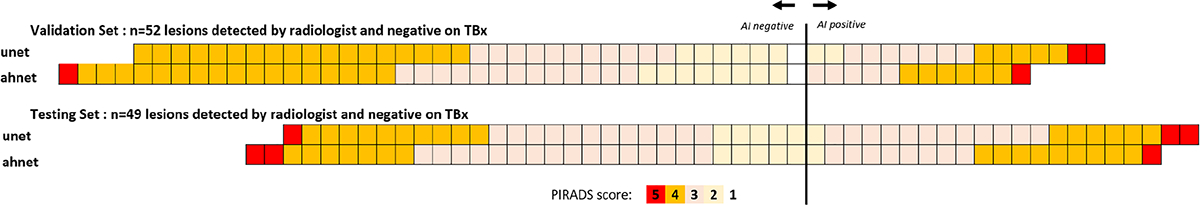
Outcome of AI predictions in targeted biopsies with benign result
Discussion
In this work, we present the outcomes of a DL-based cascaded fully automated detection model for prostate cancer on biparametric MRI using two different architectures. The model was trained by imaging and histopathology information from two different centers, one in the USA and one in The Netherlands. The systems can automatically detect prostate cancer lesions at biparametric MRI and demonstrated a detection sensitivity ranging 63.0–72.8% in an independent test set from both centers. This came at the cost of an average of 1.4–1.9 false positive lesions per patient. False positive lesion calls still remain a major challenge for fully automated detection algorithms, on average approximately 15% of false positives correspond to prospectively identified regions by a radiologist but were negative on targeted biopsy. Nevertheless, although not ripe for use as a fully automated system, we believe the model will be helpful for prostate multiparametric MRI readers as an assistive tool during the reading process. Studies have shown that ML-based models tend to improve the detection performance in prostate MRI reading for prostate cancer. This has been shown to be most pronounced in less experienced readers while highly experienced readers seem to benefit less from Machine Learning-based assistance [24].
This DL-based model demonstrates a higher detection sensitivity compared to classical Machine Learning feature-based algorithms. However, direct comparison of performance metrics are not plausible and relevant. Ideally, model performance should be compared to the gold standard in the same patient population. The gold standard for automated systems is human performance. Therefore, the next step in the evaluation of this model will be a prospective comparative study of human readers of different experience backgrounds comparing the detection performance of radiologists with and without model assistance.
There has been a rise of DL technology in many domains including medicine over the last past years and is replacing traditional Machine Learning techniques. In medicine, DL-based applications have demonstrated super-human performance for several clinical applications [25, 26]. Therefore, DL-based prediction models have become a matter of intensive research recently. Saha et al. from the Radboud Medical university in Nijmegen, The Netherlands recently published an article presenting the outcomes of a DL-based detection model using a Deep attention mechanism combined with a false positive lesion filter [27]. They used a large dataset of 1950 patient scans from their institution for training the model which was tested on 486 patient scans from their own institution and 296 patient scans from a separate center in the same country using the same MRI vendor and model. Lesions were biopsied using MRI-guided biopsy technique and prostate cancer histopathology > Gleason 3 + 3 were defined as clinically significant. Based on these criteria, 1092 and 97 prostate cancer lesions were included in the study. They reported sensitivity of 92.29% with a mean number of 1.69 false positive lesions per patient in the institutional test set and a sensitivity of 84.6% with a mean number 2.22 false positive lesions per patient in the external test set. Furthermore, model performance was compared to a consensus of radiologists from different backgrounds. Radiologists achieved 90.72% detection sensitivity with a mean number of 0.3 false positive lesions per patient demonstrating the main challenge for Machine Learning-based algorithms remain as false positive lesion calls. The main strength of this work is the large dataset, the application of a false positive lesion filter and direct comparison to human performance. However, this comparison was done retrospectively and as mentioned earlier prospective comparative studies are needed to evaluate the additional value of a prediction model as an assistant tool. Our model demonstrated a slightly lower detection sensitivity per lesion level with similar false positive rates. As we highlighted earlier direct comparisons are inherently flawed, but we believe the lower detection sensitivity might be related to lesion selection. Our study used a more conservative approach utilizing all cancer-positive lesions as ground truth annotations including ISUP grade group 1 lesions. It is well-known that low-grade prostate cancer is less visible on multiparametric MRI for human readers and machines alike. Furthermore, the smaller dataset may have additionally contributed to lower detection sensitivity.
Mehta et al. developed a model for patient-level prediction of clinically significant prostate cancer using publicly available imaging datasets [28]. Overall, 215 patients from the PICTURE dataset and 282 patients from the PROSTATEx dataset were used for model training and testing. The model was designed as a patient-level classification algorithm for clinically significant prostate cancer defined as ISUP grade group > 1. Seven 3D ResNets were trained, and forward feature selection was used to train a support vector machine (SVM). Furthermore, clinical features were also used for training another SVM. Both SVMs were then combined to produce the final patient classification probability. The model was trained on 170 and tested in 40 patient scans. Patient-level sensitivity ranged from 75 to 95% and specificity from 35 to 55% depending on the threshold. Direct comparison to our model is limited by significant differences in methodology. Our model was trained as a detection system for histopathologically confirmed prostate cancer lesions rather than patient-based prostate cancer classification. However, our patient level sensitivity result for cancer detection (80%) is similar to reported results by Mehta et al. The main strengths of our study are the diversity of patients and imaging data which is rooted on the multi-center nature of the study and the application of DL technology which has proven to be very powerful especially with the availability of large representative imaging data.
This study has five major limitations. First, as mentioned is the “in silico” rather than “in vivo” nature of the study. There was only one highly experienced genitourinary radiologist involved in image reevaluation and annotation which is due to the institutional structure of our center and although this may seem like a limitation, a prostate cancer detection AI model trained on a highly focused radiologist can be advantageous for consistent radiology evaluations. Second, although two different centers with different radiologists, pathologists and MRI scanners were involved, institution-based overfitting can only be ruled out by including more data and model testing on data from different centers and MRI scanners around the world. Third, our histopathology verification is based on targeted biopsies but not surgical specimens, which did not enable us to formally study specificity of our AI model. However, we are aware of that fact that including surgical cases only would have artificially boosted our performance metrics since surgical patients have higher risk prostate cancers, where AI may not be needed as much compared to pre-biopsy use. Fourth, our AI model was developed using biparametric MRI but not multiparametric MRI. DCE MRI was not incorporated into the model due to inconsistent image acquisition in our study population. Considering the increasing role of biparametric MRI in prostate imaging, the approach we took with AI can be more advantageous for biparametric MRI users. Fifth, our AI model was not tested in an interaction with a radiologist in a real life read out scenario, which we plan to conduct prospectively as a near future research goal.
In conclusion, we developed a DL-based AI approach which predicts prostate cancer lesions at biparametric MRI with reasonable performance metrics. This system can be utilized as an adjunct tool for radiologists during the reading process.
Supplementary Material
Funding
This research is funded by intramural research program of NIH.
Footnotes
Conflict of interest Author BJW is supported by the Intramural Research Program of the NIH and the NIH Center for Interventional Oncology and NIH Grant # Z1A CL040015–08. NIH and Philips/In-Vivo Inc have a cooperative Research and Development Agreement. NIH and Philips/InVivo Inc have a patent license agreement and NIH and BJW, BT, PAP, PLC may receive royalties. DY, DZ, HR, ZX are NVIDIA Cooperation employees. The remaining authors have no disclosures.
Code availability The code is available upon request.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate Informed consent was obtained from all individual participants included in the study.
Consent for publication All coauthors are aware of submission of this work and they approved the submission.
Supplementary Information The online version of this article contains supplementary material available https://doi.org/10.1007/s00261-022-03419-2.
Availability of data and material
ProstateX data are publicly available ((https://www.aapm.org/GrandChallenge/PROSTATEx-2/default.asp); NCI dataset is not publicly available.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018. [DOI] [PubMed] [Google Scholar]
- 2.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Zappa M, Nelen V, et al. Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet. 2014;384:2027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han C, Zhu L, Liu X, Ma S, Liu Y, Wang X. Differential diagnosis of uncommon prostate diseases: combining mpMRI and clinical information . Insights into imaging. 2021;12(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker SM, Turkbey B. Role of mpMRI in Benign Prostatic Hyperplasia Assessment and Treatment. Current urology reports. 2020;21(12):55. [DOI] [PubMed] [Google Scholar]
- 5.Brown AM, Elbuluk O, Mertan F, Sankineni S, Margolis DJ, Wood BJ, et al. Recent advances in image-guided targeted prostate biopsy. Abdominal imaging. 2015;40(6):1788–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahdoot M, Wilbur AR, Reese SE, Lebastchi AH, Mehralivand S, Gomella PT, et al. MRI-Targeted, Systematic, and Combined Biopsy for Prostate Cancer Diagnosis. N Engl J Med. 2020;382(10):917–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. European Urology. 2019;0232:1–12. [DOI] [PubMed] [Google Scholar]
- 8.Girometti R, Giannarini G, Greco F, Isola M, Cereser L, Como G, et al. Interreader agreement of PI-RADS v. 2 in assessing prostate cancer with multiparametric MRI: A study using whole-mount histology as the standard of reference. Journal of Magnetic Resonance Imaging. 2019;49:546–55. [DOI] [PubMed] [Google Scholar]
- 9.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 10.Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology. 2019;293(3):607–17. [DOI] [PubMed] [Google Scholar]
- 11.Winkel DJ, Tong A, Lou B, Kamen A, Comaniciu D, Disselhorst JA, et al. A Novel Deep Learning Based Computer-Aided Diagnosis System Improves the Accuracy and Efficiency of Radiologists in Reading Biparametric Magnetic Resonance Images of the Prostate: Results of a Multireader, Multicase Study. Investigative radiology. 2021;56(10):605–13. [DOI] [PubMed] [Google Scholar]
- 12.Hosseinzadeh M, Saha A, Brand P, Slootweg I, de Rooij M, Huisman H. Deep learning-assisted prostate cancer detection on biparametric MRI: minimum training data size requirements and effect of prior knowledge. Eur Radiol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syer T, Mehta P, Antonelli M, Mallett S, Atkinson D, Ourselin S, et al. Artificial Intelligence Compared to Radiologists for the Initial Diagnosis of Prostate Cancer on Magnetic Resonance Imaging: A Systematic Review and Recommendations for Future Studies. Cancers. 2021;13(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-Aided Detection of Prostate Cancer in MRI. IEEE Transactions on Medical Imaging. 2014;33(5):1083–92. [DOI] [PubMed] [Google Scholar]
- 15.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging. 2013;26(6):1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui MM, Rais-Bahrami S, Turkbey B, George AK, Rothwax J, Shakir N, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313:390–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turkbey B, Fotin SV, Huang RJ, Yin Y, Daar D, Aras O, et al. Fully automated prostate segmentation on MRI: comparison with manual segmentation methods and specimen volumes. AJR Am J Roentgenol. 2013;201:W720–9. [DOI] [PubMed] [Google Scholar]
- 18.Epstein JI, Zelefsky MJ, Sjoberg DD, Nelson JB, Egevad L, Magi-Galluzzi C, et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol. 2016;69:428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, editors. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation2016; Cham: Springer International Publishing. [Google Scholar]
- 20.Liu S, Xu D, Zhou SK, Pauly O, Grbic S, Mertelmeier T, et al. , editors. 3D Anisotropic Hybrid Network: Transferring Convolutional Features from 2D Images to 3D Anisotropic Volumes2018; Cham: Springer International Publishing. [Google Scholar]
- 21.Milletari F, Navab N, Ahmadi S-AJFICoDV. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016:565–71. [Google Scholar]
- 22.Diederik P, Kingma JB Adam: A Method for Stochastic Optimization. ICLR (Poster). 2015. [Google Scholar]
- 23.Documentation CTS. Clara Train SDK Documentation. [Google Scholar]
- 24.Greer MD, Lay N, Shih JH, Barrett T, Bittencourt LK, Borofsky S, et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: an international multi-reader study. Eur Radiol. 2018;28(10):4407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94. [DOI] [PubMed] [Google Scholar]
- 27.Saha A, Hosseinzadeh M, Huisman H. End-to-end prostate cancer detection in bpMRI via 3D CNNs: Effects of attention mechanisms, clinical priori and decoupled false positive reduction. Medical image analysis. 2021;73:102155. [DOI] [PubMed] [Google Scholar]
- 28.Mehta P, Antonelli M, Ahmed HU, Emberton M, Punwani S, Ourselin S. Computer-aided diagnosis of prostate cancer using multiparametric MRI and clinical features: A patient-level classification framework. Med Image Anal. 2021;73:102153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ProstateX data are publicly available ((https://www.aapm.org/GrandChallenge/PROSTATEx-2/default.asp); NCI dataset is not publicly available.


