Abstract
New approach methodologies (NAMs) that make use of in vitro screening and in silico approaches to inform chemical evaluations rely on in vitro toxicokinetic (TK) data to translate in vitro bioactive concentrations to exposure metrics reflective of administered dose. With 1,364 per- and polyfluoroalkyl substances (PFAS) identified as of interest under Section 8 of the U.S. Toxic Substances Control Act (TSCA) and concern over the lack of knowledge regarding environmental persistence, human health and ecological effects, the utility of NAMs to understand potential toxicities and toxicokinetics across these data-poor compounds is being evaluated. To address the TK data deficiency, 71 PFAS selected to span a wide range of functional groups and physico-chemical properties were evaluated for in vitro human plasma protein binding (PPB) by ultracentrifugation with liquid chromatography-mass spectrometry analysis. For the 67 PFAS successfully evaluated by ultracentrifugation, fraction unbound in plasma (fup) ranged from less than 0.0001 (pentadecafluorooctanoyl chloride) to 0.7302 (tetrafluorosuccinic acid), with over half of the PFAS showing PPB exceeding 99.5% (fup <0.005). Category-based evaluations revealed that perfluoroalkanoyl chlorides and perfluorinated carboxylates (PFCAs) with 6–10 carbons were the highest bound, with similar median values for alkyl, ether, and polyether PFCAs. Interestingly, binding was lower for the PFCAs with a carbon chain length of ≥ 11. Lower binding also was noted for fluorotelomer carboxylic acids when compared to their carbon-equivalent perfluoroalkyl acids. Comparisons of fup value derived using two PPB methods, ultracentrifugation or rapid equilibrium dialysis (RED), revealed RED failure for a subset of PFAS of high mass and/or predicted octanol-water partition coefficients exceeding 4 due to failure to achieve equilibrium. Bayesian modeling was used to provide uncertainty bounds around fup point estimates for incorporation into TK modeling. This PFAS PPB evaluation and grouping exercise across 67 structures greatly expands our current knowledge and will aid in PFAS NAM development.
Keywords: PFAS, plasma protein binding, toxicokinetics, LC-MS
Introduction
Manufactured since the late 1940s, commercial production of per- and polyfluoroalkyl substances (PFAS) grew considerably in the 1980s as their functionality became increasingly recognized 1, 2. Comprised of high-energy carbon-fluorine bonds, many PFAS uniquely and efficiently repel both water and oils, possess surface tension lowering properties, and are thermally stable3 providing them great utility in household product and industrial applications ranging from stain repellants, food contact surface coating, and aqueous film-forming foams 4, 5. Unfortunately, the very properties that make PFAS so useful render them resistant to biodegradation, resulting in widespread accumulation in aquatic environments, biota, wildlife, and humans 6.
Whereas estimates of the number of PFAS in existence are subject to the specific definitions employed in conjunction with the scope of interest 7, a recent database released by the Organisation for Economic Co-operation and Development (OECD) has listed 4,729 that meet the common definition of containing at least one perfluoroalkyl moiety 8. In the United States (US), where commercial PFAS are regulated under the Toxic Substances Control Act (TSCA), a recently proposed TSCA section 8 rule would require manufacturers and importers to report the identity of any PFAS manufactured since January 1, 2011, as well as byproducts from the manufacturing process, categories of use, production volumes, disposal information, worker exposures, and any information pertaining to environmental and health effects 9. At the US Environmental Protection Agency (EPA), the Office of Pollution Prevention and Toxics applies the following working definition when identifying PFAS on the TSCA inventory: a structure that contains the unit R-CF2-CF(R’)(R”), where R, R’ and R” do not equal “H” and the carbon-carbon bond is saturated (note: branching, heteroatoms, and cyclic structures are included) (see https://www.epa.gov/pesticides/pfas-packaging). As of April 2021, the US EPA has identified at least 1,364 PFAS matching this definition that would potentially be subject to this rule. Although much has already been learned about “legacy” PFAS – perfluoroalkyl acids (PFAAs) comprised of carboxylic acid (e.g., perfluorooctanoic acid (PFOA)) or sulfonate (e.g., perfluorohexanesulfonate (PFHxS)) functional groups – most PFAS categories have limited or nonexistent monitoring or toxicologic data. Resolution of these data gaps is a critical need to enable the US EPA and other global regulatory bodies a better understanding of the impact of these thousands of PFAS on human health and the environment.
As it is not feasible to employ traditional in vivo testing strategies to rapidly evaluate the thousands of PFAS lacking hazard information, the US EPA PFAS Strategic Roadmap 10 along with the National PFAS Testing Strategy (https://www.epa.gov/system/files/documents/2021–10/pfas-natl-test-strategy.pdf) describe strategies that employ New Approach Methods (NAMs) in conjunction with grouping strategies to evaluate hazard across the broader PFAS landscape. NAMs comprised of in vitro toxicity testing and toxicokinetics (TK) as well as in silico modeling will be employed to efficiently provide in vitro points of departure that will be useful in prioritizing those PFAS most likely to yield adverse health outcomes at anticipated exposures. Selection of representative PFAS to populate groupings containing the varied functional groups and structures present across all PFAS has been described and used to populate a PFAS screening library employed during NAMs data generation 11. Category-based evaluations of PFAS bioactivity and/or TK will inform the utility of a grouping or read-across approach in understanding hazard across the broader space of untested PFAS.
Within NAM frameworks, in vitro TK evaluations provide pivotal absorption, distribution, metabolism, and excretion (ADME) information to translate in vitro bioactivity measures to an external dose prediction that would be required to achieve in vivo concentrations equivalent to the bioactivity concentrations. Plasma protein binding (PPB), along with hepatic clearance, are key TK determinants of a widely adopted in vitro-in vivo extrapolation (IVIVE) approach to estimate such administered equivalent doses 12. PPB is arguably one of the more critical ADME metrics, as plasma interactions and high binding can heavily influence steady state concentrations in IVIVE 13, 14 and bioaccumulative potential of PFAAs 15. Widely evaluated by the pharmaceutical industry since the 1980s, four PPB methods are in common use: rapid equilibrium dialysis (RED), ultracentrifugation (UC), ultrafiltration, and solid phase microextraction 16–20. Although evaluations of performance, reproducibility, and throughput have settled on high-throughput dialysis methods such as the RED assay as the most efficient for drugs and many commercial chemicals, cross-platform comparisons of the RED and UC assays have shown good agreement 21, 22. With any method, incorporation of relevant controls and reference compounds is required to ensure the assay is functioning as intended 17, 23.
Despite widespread efforts to understand PPB contributions to PFAS bioaccumulation and mechanism of action, empirical in vitro data for TK modeling are still very limited. In vivo studies reporting preferential accumulation of PFAAs in the plasma and liver 24, 25 preceded plasma binding studies that primarily evaluated binding kinetics to albumin and fatty acid binding proteins 26–28, with in vitro PPB studies limited to isolated TK studies on specific PFAAs (e.g., PFOA, PFNA, PFOS) 29, 30. Building up our knowledge of in vitro PPB across dozens of structures within the PFAS space will greatly expand our ability to evaluate PFAS TK in addition to persistence, bioaccumulation, and fate predictions. This report describes our PPB evaluations of 71 PFAS that span a wide mass range (164–726 g/mol), predicted lipophilicity, and functional groups. This structural diversity, in addition to the unique C-F backbone imparting unique hydrophobic/hydrophilic regions, prompted performance and amenability comparisons between two widely used PPB assays. A Bayesian approach to incorporate experimental uncertainty was also performed to better understand anticipated uncertainty associated with these groups of PFAS and for downstream application in high-throughput toxicokinetic (HTTK) modeling efforts. These evaluations of experimental rigor and uncertainty provide a framework that can be readily applied to emerging contaminants and other data-poor chemicals for which assay performance may require greater scrutiny.
Experimental Procedures
PFAS Selection, Stock Preparation, and Plasma Pools.
The 71 PFAS evaluated in this effort were procured through a US EPA contract with Evotec Inc. (Branford, CT). These are a subset of a larger group of over 140 PFAS (the remainder evaluated in 31) that were selected to enable evaluation across the wide range of functional groupings and structures that comprise PFAS identified as of commercial interest based on their inclusion on the TSCA inventory. Criteria employed to select these PFAS was described in 11; the PFAS analyzed by liquid chromatography-mass spectrometry in this work and their functional group and OECD category assignments are described in Table S1. Substances were procured from vendors and solubilized at a target concentration of 30 mM in DMSO. Solubility evaluations were conducted on stock solutions; if precipitate was observed, then concentrations of 20 or 10 mM were used. DMSO stocks of all selected PFAS in this study had passed an analytical quality control (QC) evaluation (described in 32).
Pooled, mixed sex human plasma from de-identified donors was obtained from a commercial vendor that operates a U.S. Food and Drug Administration-licensed and inspected donor center (BioIVT, Westbury, NT). Donors were comprised of ten males and ten females ranging in age from 20 to 50 years old. Plasma was collected using anti-coagulant K2EDTA and was sterile filtered (0.2 μM) and stored under −70ºC until use in the ultracentrifugation and rapid equilibrium dialysis assays described below.
Ultracentrifugation (UC) Assay.
The UC assay to evaluate PFAS PPB was developed based on earlier publications with several modifications 18, 33 (Fig S1). Briefly, PFAS and reference compound, n-butylparaben, were grouped in sets of 3–8 chemicals based on analytical considerations and prepared as a 3 mM DMSO stock mixture. An aliquot of this stock was added to human plasma to achieve a final assay concentration of 10 μM (DMSO concentration at 0.3%), thoroughly mixed, and incubated at 37°C for 1 hr with shaking. After 1 hr pre-incubation, one aliquot was collected (T1hr), a second aliquot continued to incubate at 37°C for an additional 4 hr (T5hr), and the remainder (aqueous fraction, AF) was transferred to a polycarbonate tube and underwent ultracentrifugation at 850,000xg for 4 hr at 37°C in a Beckman OptimaMax ultracentrifuge (Beckman Coulter Inc., Brea, CA). After ultracentrifugation, AF and T5hr were each transferred to new tubes.
At the relevant time points, each volume of sample was matrix matched as needed: that is, AF was mixed with an equal volume of blank (i.e., no chemical added) plasma and the T1hr or T5hr samples were mixed with an equal volume of blank AF. Blank AF was prepared using Centriprep 30K centrifugal filter devices (Millipore, Burlington, MA) to separate plasma proteins from the plasma ultrafiltrate. One volume of each sample (T1hr, AF, and T5hr) was then mixed with 3 volumes of ice-cold acetonitrile containing mass-labeled PFAS standards (MPFAC-24ES; Wellington Laboratories, Guelph, Ontario, Canada) and Ring-13C6 labeled n-butyl paraben (Cambridge Isotope Laboratories, Tewksbury, MA) at a concentration of 16 ng/mL. The samples were mixed vigorously, stored for 10 min at −20°C, then centrifuged at 12,500xg for 10 min at 4°C. The supernatant was collected and stored below −70°C until mass spectrometry analysis. All assay samples were run with 3 experimental replicates.
Fraction unbound in plasma (fup) is calculated from these samples by dividing the AF concentrations by the T5hr concentration (Equation 1). Employing a cassette approach for UC assays is viable provided the number of chemicals evaluated and the concentrations employed do not saturate the PPB partners for the compounds of interest. Albumin, the PPB partner of PFAS 26, 34, is present in plasma at approximately 600 μM; a validation study demonstrated that compounds could be pooled as long as the total drug concentration was less than 1/5th the total albumin concentration 35. In addition to evaluations in this study confirming that equivalent fup values were reported, an independently published effort demonstrated that up to 20 analytes could be successfully combined with no saturation of binding 18. N-butylparaben was included in each assay as an assay reference compound to evaluate assay performance and to ensure no protein contamination of the AF occurred. Also, chemical stability in plasma over the assay time course was evaluated using the T1hr and T5hr samples (Equation 2). Any chemicals with less than 80% stability were removed from the analysis.
Rapid Equilibrium Dialysis (RED) Assay.
The RED assay was performed as previously described 36 but with some modifications (Fig S2). Similar to the UC assay, human plasma samples spiked with each 10 μM PFAS (evaluated individually) were incubated at 37°C for 1 hr with shaking prior to addition to the RED assay plate. Using a portion of each pre-incubated spiked sample, PFAS plasma stability was evaluated, resulting in T1hr and T5hr samples. Following the 1 hr pre-incubation, an aliquot of each PFAS-spiked plasma sample was added to respective wells on the RED assay plate and incubated at 37ºC for 4 hr at 150 rpm. Equilibrium check samples, comprised of analyte-spiked PBS in the red-ringed and blank PBS in the clear-ringed dialysis chamber, were included to assess the ability of the analytes to cross the membrane and achieve equilibrium during the 4 hr dialysis period. At the relevant time points, the samples were matrix matched and mixed with 3 volumes of ice-cold acetonitrile containing mass-labeled standards as described above for the UC assay. All assay samples were run with 3 experimental replicates, and n-butylparaben was included as a reference compound on each plate. For the RED assay, fup was determined as a ratio of the concentration of each analyte in the PBS (clear-ringed) well to the plasma containing (red-ringed) well across each dialysis membrane (Equation 3).
PFAS Quantitation by Ultra-High-Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC-MS/MS).
Samples from the PPB assays were analyzed using a Waters ACQUITY I-Class Ultra-High Performance Liquid Chromatography System (UPLC) coupled to a Xevo TQ-S micro triple quadrupole mass spectrometer (Waters Corporation, Milford, MA). The UPLC system was plumbed with the Waters’ PFAS Solution Installation Kit with solvent lines containing polyether ether ketone, stainless-steel filter, and an isolator column (ACQUITY BEH C18 Column 2.1×50 mm). For most PFAS analytes assessed, the chromatographic separation was carried out using a Waters CORTECS T3 reversed-phase column (3 mm x 100 mm, 2.7μm), a flow rate of 0.6 mL/min, and a binary mobile phase gradient with mobile phases A (95:5, 2.5 mM ammonium acetate: acetonitrile) and B (95:5, acetonitrile: 2.5 mM ammonium acetate). The gradient program was 6.5 min total and programmed as follows: 20% B (0.45 min), 20–50% B (0.15 min), 50–58% B (0.9 min), 58–66% B (0.75 min), 66–75% B (0.15 min), 75–80% B (1.2 min), 80–100% B (0.3 min), 100% B (1.74 min), 100–20% B (0.06 min), 20% B (0.8 min). The PFAS that eluted too early using the CORTECS T3 column were subsequently assessed via hydrophobic interaction liquid chromatography (HILIC) with an ACQUITY BEH Amide column (2.1 mm x 100 mm, 1.7 μm) using mobile phases A (95:5, 2.5 mM ammonium acetate: acetonitrile) and B (95:5, acetonitrile: 2.5 mM ammonium acetate) pH-adjusted to ~9.0. The gradient program was modified from a Waters application note for organic acids (WA60096, June 2009; https://www.waters.com/waters/library.htm?cid=511436&lid=10116208), being 7.5 min total. The gradient and associated flow rate varied over the method program: 99% B (0.3 mL/min, 1.5 min), 99% B (0.6 mL/min, 0.5 min), 99–60% B (0.6 mL/min, 1.5 min), 60–30% B (0.6 mL/min, 0.5 min), 30–99% B (0.6mL/min, 2.0 min), 99% B (0.3 mL/min, 1.5 min). 10 μL of each sample was injected regardless of chromatography employed.
The Waters Xevo TQ-S micro triple quadrupole mass spectrometer (MS) was operated in both positive (ESI+) and negative (ESI-) electrospray ionization modes. The source temperature was 150oC with desolvation temperature, desolvation gas flow, and cone gas flow at 500oC, 1000L/hr, 150 L/hr, respectively. Previously optimized, multiple reaction monitoring (MRM) transitions were used for each unique PFAS (Table S2) and often included a quantitation and confirmation transition. Internal standards were also optimized (Table S3). Blank matrix samples as well as instrument blanks were included to monitor for any potential PFAS contamination or low-level responses associated with the matrix and/or instrumentation. Samples were thawed to room temperature, vortexed briefly, and resuspended in 80:20 mobile phase A:mobile phase B in polypropylene autosampler vials. A 17-point calibration curve was prepared in an identical matrix to the assay sample calibration curve, ranging from at instrument concentrations of 0.17 nM to 250 nM. A quadratic regression fit was applied to the calibration curve with 1/x weighting to assign higher priority to lower end concentrations that are of interest for fup determinations.
For each assay set, solvent and matrix blanks were included to evaluate possible contamination from laboratory operation and method performance. Matrix blanks were prepared as either equal parts of AF and human plasma or PBS and human plasma for UC and RED assays, respectively. Blanks were assessed every sixth sample of the sample analysis worklist. All blanks were determined to have a concentration to be less than half the estimated method detection limit (eMDL; see below). Additionally, the recoveries of labeled standards were within 75–125%.
Estimated Method Detection Limits (eMDLs) of PFAS in Plasma.
The eMDL for each analyte in each plasma matrix was determined by UPLC-MS/MS, where this value is the minimum measured concentration of the chemical reported with a 99% confidence that it is distinguishable from a method blank modified from US EPA Method 821-R-16–00637. To determine this value, a set of the lower-end calibration curve points were injected seven times. The eMDL is calculated by multiplying the standard deviation of their reported concentration by the one-tailed t-distribution test (for seven samples with six degrees of freedom, t-value of 3.14 was used at 99% confidence level). Determined eMDLs guided blank contamination or instrumental issues if presented. Additionally, using these calibration curve points, the estimated limit of quantitation (eLOQ) was determined as the concentration where all seven measures were within 30% of the theoretical value.
Bayesian Modeling to Incorporate Experimental Uncertainty with Experimental Point Estimates.
To estimate measurement uncertainty, a Bayesian analysis was performed using Markov Chain Monte Carlo on both the UC and RED assay data. For each assay type, the data were organized into a single file for analysis (Tables S5-S6). The relationship between the parameters assumed to be involved in the measurement process was described as a graphic model in the JAGS language 38 interfaced through R 39. The basis of this model was the PPB Bayesian model 40, modified to reflect either UC or RED assay type and to include calibration curves as well as two statistical models being employed. The first statistical model describes the chemical-specific MS response factor (that is, the conversion factor between analyte peak ratio (to standard) and chemical concentration). It employs information provided from blanks, calibration curve points and variability of observations to estimate the relationship between chemical concentration and mass spectrometry peak area. The second model describes the relationship between the assay samples with the relevant assay method (UC or RED). All observations used contribute to an estimate of a distribution of plausible parameter values for both models. Both models are analyzed jointly by JAGS using 5 Markov chains. R package runjags 41 was used to repeatedly extend the Markov chains until the multivariate shrink factor calculated with all five chains was less than 1.05. Each time the chains were extended using a 50,000 iteration burn-in was followed by 50,000 iterations thinned to 2,000 samples. If measurements were made on multiple days, separate response calibrations were made but a single fup was estimated per chemical. The median and 95 percent credible interval (upper and lower bounds) were calculated from the final (converged) 10,000 samples from the five Markov chains. The analysis was performed using EPA-developed R package invitroTKstats, which is available upon request.
Results
Analytical Method Optimization and Performance.
Two barriers to measuring PPB are developing chemical-specific methods to detect the compound and further, for highly bound chemicals, finding a method sensitive enough to detect the unbound chemical. It was necessary to develop a universal UPLC-MS/MS method for determining fraction unbound values that included as many LC-amenable PFAS as possible within an acceptably short run time that also provided low detection limits for quantitating the unbound fraction of each chemical in plasma. Not only did this allow a more rapid turnaround time for high-throughput means, but also showed minimal impacts from matrix effects by incorporating a range of labeled PFAS internal standards that spanned the structural and physicochemical property diversity of the nearly 70 unique assessed analytes. The LC gradient and MS parameters utilized reflected the same conditions employed in our initial work that investigated the quality of these DMSO-solubilized PFAS solutions and provided consistency in our methodologies 32. Each PFAS standard in DMSO was optimized by selecting two multiple reaction monitoring (MRM) transitions when possible, where the most intense transition was designated for quantification (Table S2).
During initial optimization, several diprotic PFAS required additional LC gradient optimization as early elution was observed. These analytes were hexafluoroglutaryl chloride (DTXSID0060985), perfluoro-3,6-dioxaoctane-1,8-dioic acid (DTXSID20375106), perfluorohexanedioic acid (DTXSID4059833), hexafluoroglutaric acid (DTXSID8059926), and tetrafluorosuccinic acid (DTXSID8059928). HILIC chemistry improved retention and thereby allowed more accurate quantitation. The largest driver of this type of gradient was the basic pH, assisting with the ionization potential and analyte retention. In addition, this adapted chromatographic gradient permitted the mass spectrometer to perform more consistently without the concern of undesired material reaching the detector.
Calibration curves in associated media for UC and RED assays were assessed alongside each experiment. A three-order concentration range was incorporated as the aqueous fractions were expected to be very low nM quantities for many PFAS analytes. With quadratic fitting, the correlation coefficient for most analytes was greater than 0.98. The lower-end calibration curve points were utilized to determine the eMDL and eLOQ of under 0.5 nM (i.e., 0.036 pg/μL for PFBA, 0.070 pg/μL for PFOA, and 0.355 pg/μL for PFOS (Table S2), showcasing our confidence in accurately quantitating the fup values. Similar results were also found by others when utilizing LC-MS/MS for determining various PFAS in plasma34, 42, 43. It is also important to note that only protein precipitation with centrifugation was employed for sample clean-up and did not present any impacting interferences or significant instrument downtime.
PPB Evaluations by Ultracentrifugation Assay.
Of the original 71 PFAS evaluated, experimental fup values were successfully measured for 67. Initial PFAS selection was designed to span multiple structural and functional groupings to address data gaps in PFAS TK 11, 44, 45, making use of the PFAS-Map framework to populate the PFAS across five high-level categories originally described by OECD 8, 45. To provide a more comprehensive evaluation of LCMS-amenable PFAS studied here, more specific groupings as defined using conventions described in Buck et al., 2011 and by the OECD 2, 46 are also presented, where the PFAS evaluated spanned 18 groups. In general, the Buck/OECD categories are anchored to specific structural characteristics and functional group presence; whereas the PFAS-MAP groupings are more overarching with broader coverage combining PFAAs with varied composition (e.g., carboxylic acid, ether and polyether linkages combined) and based on synthesis processes (e.g., fluorotelomer precursors of PFAAs). Moreover, the “others” PFAS-Map category combines perfluorinated and polyfluorinated structures in one group. This analysis consists of reasonable coverage for 9 of the Buck/OECD functional groupings and all PFAS-Map categories, defined as 3 or more analytes in each. More information is provided in Table 1 and Table S1.
Table 1.
Structural Category Coverage for PFAS Analyzed in this Study
| Buck/OECD2, 46 | #a | Example | PFAS-Map8, 45 | # | Examples |
|---|---|---|---|---|---|
| Perfluoroalkyl carboxylates | 17 | PFOAb | Perfluoroalkyl acids (PFAAs) | 31 | PFOA, PFPE-7 |
|
| |||||
| Perfluoroalkane sulfonates | 9 | PFBS | PFAA precursors (per and poly) | 17 | 9H-PFNA |
|
| |||||
| Polyfluoroalkyl carboxylates | 5 | 4H-PFBA | Fluorotelomer PFAA precursors (poly) | 11 | 8:2 FTS |
|
| |||||
| Fluorotelomer carboxylates | 4 | 5:3 FTCA | FASA-based PFAA precursors (poly) | 4 | NMeFOSA |
|
| |||||
| Perfluoroalkyl ether carboxylates | 4 | PFPE-7 | Others (per and poly) | 8 | Perfluorononanoyl chloride |
|
| |||||
| Perfluoroalkyl polyether carboxylates | 4 | PFPE-3 | OECD Subcategory | # | Detail |
|
| |||||
| Perfluoroalkanoyl chlorides | 5 | Hexafluoroglutaryl chloride | lt8 | 30 | < 8 carbons |
|
| |||||
| Perfluoroalkane sulfonamides (FASAs) | 4 | PFHxSA | gte8 | 36 | ≥ 8 carbons |
|
| |||||
| Fluorotelomer sulfonates | 3 | 6:2 FTS | volatile | 5 | volatile |
|
| |||||
| Three groups | 2 | miscellaneous | |||
|
| |||||
| Six groups | 1 | miscellaneous | |||
#= Number of PFAS in each category.
Abbreviations: PFOA, perfluorooctanoic acid; PFBS, perfluorobutane sulfonate; 5:3 FTCA, 5:3 fluorotelomer carboxylic acid; PFPE-7, perfluoro(4-methoxybutanoic) acid; PFPE-3, Perfluoro-3,6-dioxaheptanoic acid; PFHxSA, perfluorohexanesulfonate; 6:2 FTS, fluorotelomer sulfonic acid; 9H-PFNA, 8:2 FTS, 8:2 fluorotelomer sulfonic acid; 9H-perfluorononanoic acid; NMeFOSA, N-Methylperfluorooctanesulfonamide.
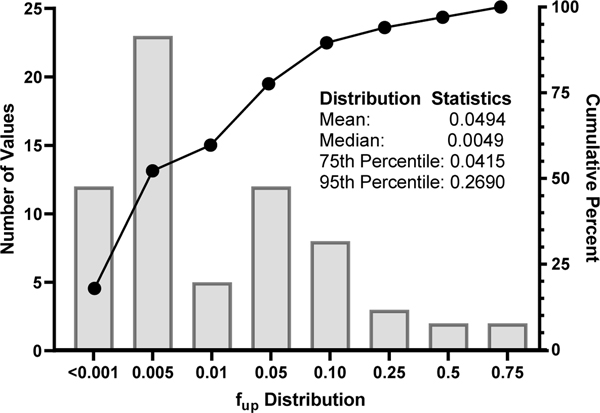
Across the 67 PFAS that were successfully analyzed, experimental fup values ranged from 0.0001 (or 99.99% bound) for pentadecafluorooctanoyl chloride (DTXSID40187142) to 0.7302 for tetrafluorosuccinic acid, a dioic acid (DTXSID8059928). Median fup of 0.0049 was 10-fold lower than the average value of 0.0494, demonstrating the strong trend of most PFAS being very highly bound. High binding rates (fup ≤0.0415) were observed for 75 percent of the analytes (Figure 1). Of the four that were withdrawn from the analysis, one, 3,3-Bis(trifluoromethyl)-2-propenoic acid (DTXSID30170109) was completely degraded before the end of the assay. Three PFAS containing an additional halide functional group - PFBS-F (DTXSID20861913), PFOS-F (DTXSID5027140) and 9Cl-PFNA (DTXSID30382104) - were highly unstable during analysis, preventing the generation of reliable data. Raw data are provided in Table S4.
Fig. 1. Distribution of PPB across 67 PFAS analyzed.
Uncertainty Analysis.
Table S6 provides the median and 95% credible intervals (upper and lower bounds) from the Bayesian analyses. Interval ranges were evaluated, with any exceeding three orders of magnitude flagged. For this experimental set, the measured fup for three of the 67 chemicals were found to be uncertain. The mean CV was 0.16 when the three uncertain chemicals were excluded. indicating an improvement over the mean CV of 0.4 for RED PPB assay in general 40. The three with uncertain values were ((perfluorooctyl)ethyl)phosphonic acid (DTXSID30627108); pentadecafluorooctanoyl chloride (DTXSID40187142); and PFOA (DTXSID8031865). The reason for the uncertainty was varied and could not be attributed to high binding alone, as many with higher binding exhibited low uncertainty, and the experimental fup values of these three were 0.007, 0.0001 and 0.001, respectively. For ((perfluorooctyl)ethyl)phosphonic acid, high experimental variability was also noted with highly variable replicate values noted (experimental fup = 0.007±0.0113 (mean ± SD); Table S4). For PFOA, PFOA signal was present in some of the instrument solvent blanks, albeit well below levels that would impact quantitation (analytical quality control procedures were followed during experimental data analysis to evaluate data acceptance). The Bayesian approach is designed to be more conservative and did not consider dilution factors in a similar manner as the analytical review, explaining this outcome.
Category-Based Evaluations of PFAS for Plasma Protein Binding.
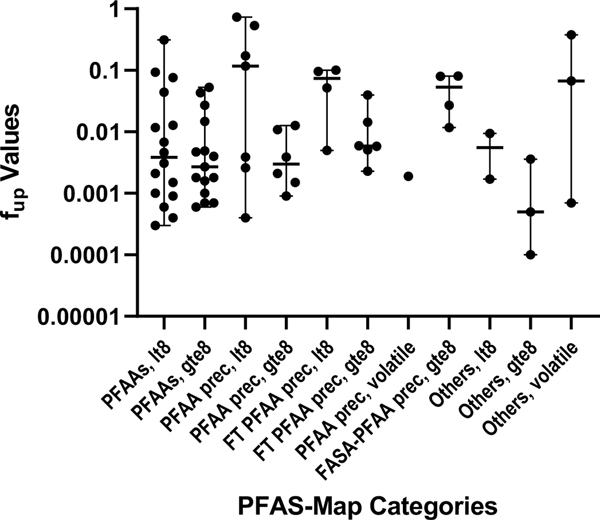
Using the structural features and physico-chemical properties represented across the analyzed PFAS, PPB binding was evaluated for category-based trends. Using the PFAS-Map categories, which divide the 5 general categories into 10 grouping based on carbon number (i.e., <8 or ≥8), evaluation of fup across the categories did not indicate any group that had distinct binding difference from the others (Figure 2). Groups with 8 or more carbons exhibited higher binding; but as the PFAS-Map classifications are still broad, no strong patterns emerged. Initial evaluations using the structural and functional grouping categories showed better discrimination by virtue of having greater specificity in the groupings, but gains were limited by no qualification of carbon number.
Fig. 2. PFAS-Map Category-Based Evaluations of PPB.
Abbreviations defined in Table 1 and in the text.
Closer review of values with regard to carbon (C) number and status as either a perfluorinated or polyfluorinated compound led to interesting observations (Table 2.) Although the perfluoro carboxylates are widely expected to exhibit increased PPB with increasing C number, the highest binding was observed for Perfluoroheptanoic acid (PFHpA, DTXSID1037303, 7 carbons (C7); fup = 0.0004; or 99.96%). Although still quite high for PFOA (C8) fup levels were 2.5X higher (fup=0.0010). The trend continued out to perfluorotridecanoic acid (PFTrDA, DTXSID90868151, C13; fup=0.0433), where values were 100X higher than PFHpA. Although still exhibiting 96% binding, these differences could provide useful information in understanding bioaccumulative potential. Binding comparisons between perfluoroalkyl substances, in which all fluorine atoms are attached to every possible binding site along the carbon chain (except for the carbon site to which the functional group is attached), to the non-fully fluorinated polyfluoroalkyl substances revealed differences when structure carbon number was considered (Table 2). Four comparisons were possible between PFCAs and fluorotelomer carboxylates (FTCA); five were possible between PFCAs and polyfluorinated carboxylates. Fup values for 3:3 FTCA and 5:3 FTCA values were 10 or 6-fold higher, respectively, than perfluorohexanoic acid (PFHxA, DTXSID3031862) and PFOA. Fup values for 4H-PFBA, 7H-PFHpA and 8H-PFOA were also higher than the respective carbon-equivalent PFCAs, but the trend was not maintained for longer carbon chain lengths. For perfluoroalkyl sulfonates (PFSAs), this pattern was observed for the C6 and C8 comparisons (5 or 3-fold higher, respectively), but not the C10.
Table 2.
Influence of Structural Groupings on PPB for PFAS Carboxylates and Sulfonates
| Carboxylates | Sulfonates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Perfluorinated (Per) | Polyfluorinated (Poly) | Per | Poly | |||||||
| # C |
Per | fup | FT | fup | Poly | fup | Per | fup | FT | fup |
| 3 | PFPA | 0.3108 | -- | -- | -- | -- | -- | -- | ||
| 4 | PFBA | 0.0930 | -- | -- | 4H-PFBA | 0.1714 | PFBS | 0.0046 | -- | -- |
| 5 | PFPeA | 0.0440 | -- | -- | -- | -- | -- | -- | ||
| 6 | PFHxA | 0.0068 | 3:3FTCA | 0.0518 | PFHxS | 0.0009 | 4:2FTS | 0.0050 | ||
| 7 | PFHpA | 0.0004 | -- | -- | 7H-PFHpA | 0.0026 | PFHpS | 0.0006 | -- | -- |
| 8 | PFOA | 0.0010 | 5:3FTCA | 0.0059 | 8H-PFOA | 0.0021 | PFOSa | 0.0045 | 6:2FTS | 0.0143 |
| 9 | PFNA | 0.0016 | 6:3FTCA | 0.0023 | 9H-PFNA | 0.0009 | -- | -- | -- | -- |
| 10 | PFDA | 0.0027 | 7:3FTCA | 0.0051 | PFDS | 0.0528 | 8:2FTS | 0.0396 | ||
| 11 | PFUnDA | 0.0148 | -- | -- | 11H-PFUnDA | 0.0015 | -- | -- | -- | -- |
| 13 | PFTrDA | 0.0433 | -- | -- | -- | -- | -- | -- | ||
| 14 | PFTeDA | 0.0271 | -- | -- | -- | -- | -- | -- | ||
C= carbons; FT=fluorotelomer. Full names for PFAS listed in Table S1. fup = fraction unbound in plasma;
a: PFOS shown as an average of 3 independent values derived for K+ PFOS, PFOS, and PFOS anion. Per and FT comparisons are in bold. PFCAs with at least 11 Cs and that exhibit reduced binding are italicized.
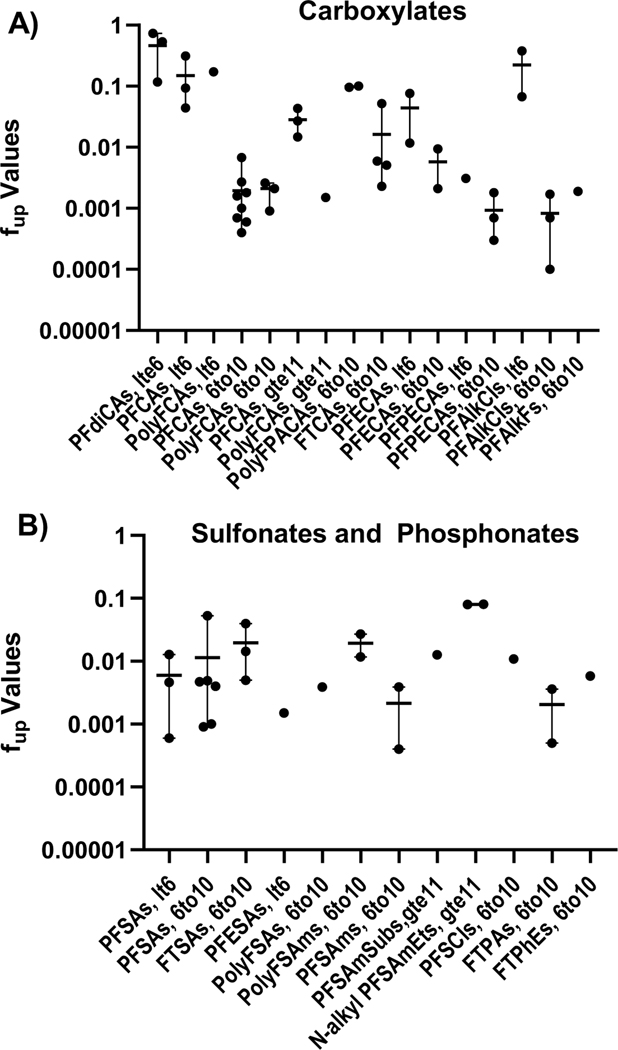
Leveraging the observation that PFAS PPB diverged based on C number, clustering in three bins, category-based evaluations were next employed using the structural and functional use groupings split into those with less than 6 (lt6); less than or equal to 6 (lte6; for dioic acids only), 6 to 10 (6to10); or greater than or equal to 11 (gte11) carbons, resulting in 13 groupings for the PFCAs and 10 for the PFSAs and 2 for the phosphonate containing PFAS (Figures 3A-B). Coverage was better for the carboxylates, where 7 of the groups had a minimum of 3 structures. Only 3 of the 10 PFSA categories had at least three structures; and none for the phosphonate-PFAS groups.
Fig. 3. Category-Based Evaluations of PFAS PPB.
Fup point estimates, mean and range (horizontal and vertical lines, respectively) displayed. Grouped by functional group presence and carbon number. A) PFAS with carboxylates (CAs) PF=perfluoro; PFdiCAs, PF dioic acids; PolyFCAs, polyfluoro CAs; PolyFPACAs, polyfluro polyalkyl CAs; FTCAs, fluorotelomer CAs; PFECAs, PF alkyl ether CAs; PFPECAs, PF alkyl polyether CAs; PFAlkCls, PF alkanoyl chlorides; PFAlkFs, PF alkanoyl fluorides. B) PFAS with sulfonate and phosphonate functional groups.. SAs = sulfonates; PFESAs, PF ethyl sulfonamides; PFSAms, PF alkane sulfonamides; PFSAmSubs, PF alkane sulfonamido substances; N-alkyl PFSAmEts, N-alkyl PF alkane sulfonamidoethanols; PFSCls; PF alkane sulfonyl chlorides; FTPAs, FT phosphonates; FTPhEs FT phosphate esters.
Perfluoroalkyl structures possessing 6–10 C, regardless of functional group, were the most highly bound of all tested groups. Perfluoroalkanoyl chlorides (PFAlkCls) are the most highly bound group, with mean fup of 0.0008 for three compounds, followed by perfluoroalkyl polyether carboxylates (PFPECAs, mean fup = 0.0009; n=3). PFCAs, the largest group tested, had a mean of 0.0019. Five of the 6 PFSAs with 6–10 C were also highly bound with a mean of 0.0031 but a mean of 0.0114 when including Na+ PFDS, (fup = 0.0528).
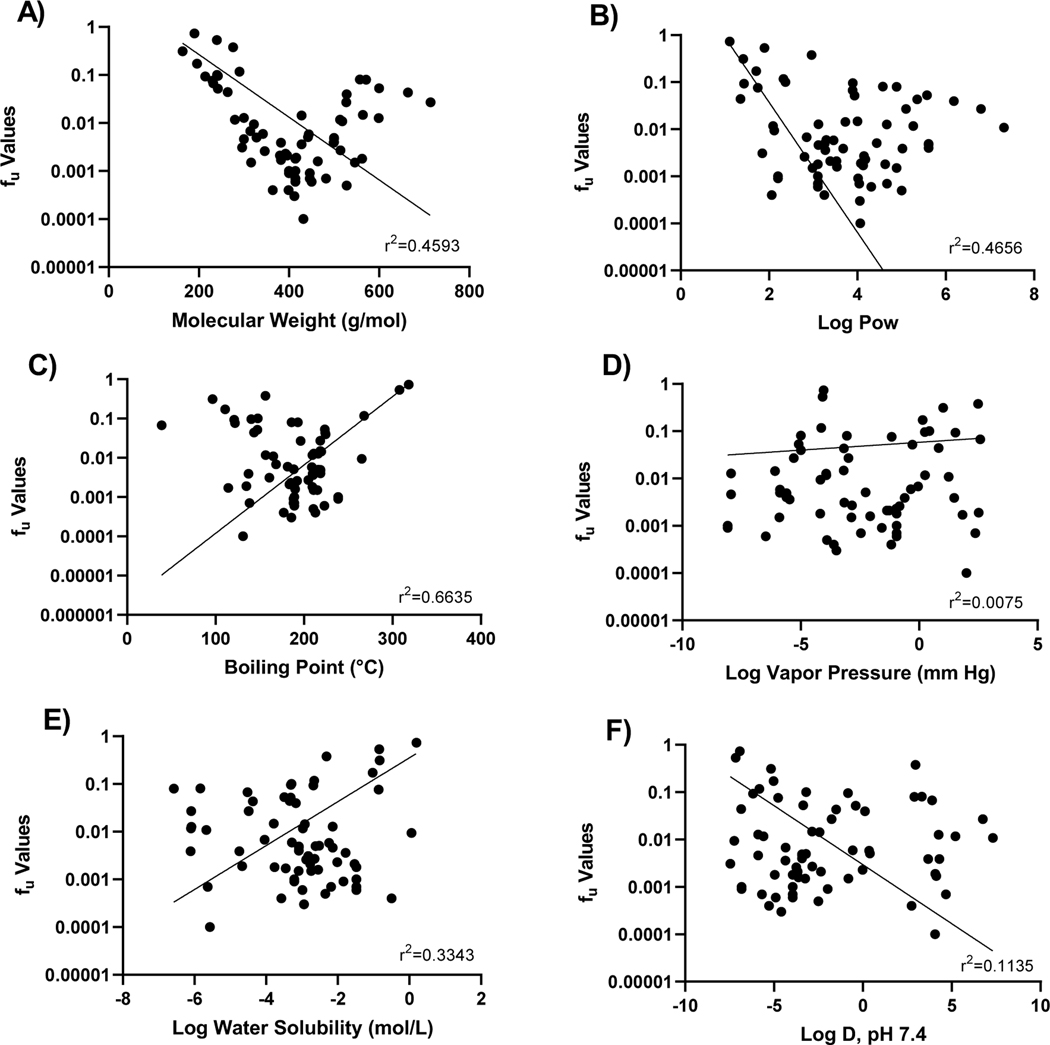
Trends analysis of PPB across several physico-chemical properties was also performed (Figure 4). Apart from molecular mass, OPERA predicted values were used 47, 48. The U-shaped trend noted between molecular weight and PPB (Table 2) was confirmed in this analysis across all evaluated PFAS, with the highest binding observed at a mass of 400 g/mol prior to a decrease at higher masses (Figure 4A). Similar patterns were observed for the water-octanol partition coefficient (LogPow); fup comparison (i.e., a decrease in fup values with increasing LogPow up to ~4.5; then a rebound in fup), was consistent with the relationship between increasing weight and increasing lipophilicity (Figure 4B). No strong trends were noted for boiling point, Log vapor pressure, Log water solubility, or Log D at pH 7.4 (Figure 4C–4F).
Fig. 4. Trend evaluation of fu and physico-chemical properties.
Experimental fu values for 65 PFAS were plotted against A) mass and OPERA predicted B) Log Pow; C) boiling point; D) vapor pressure; E) water solubility; and F) Log D at pH 7.4.
Measurement and Performance Comparisons for PFAS PPB: UC vs. RED.
Of the 71 PFAS successfully analyzed by UC assay, 22 were selected for evaluation using the RED assay to compare the two assay types for performance, sensitivity, and reproducibility. The selected PFAS spanned a range of physicochemical properties (molecular weights from 214–714 g/mol and LogPows from 1.35 – 6.80) (Table 3). To review performance in the UC assay, a reference compound is included in all incubations to ensure the aqueous fraction is free of contaminating protein, which if present will lead to an overestimation of fup. Quantifiable measures were obtained for all UC assay sample types, including the AF where levels can be as low as 0.164 pg/μL. Review of the RED assay data indicated that quantifiable measures in the PBS dialysis chamber (analogous to the UC AF sample) were not attainable for 4 of the 22 PFAS. Raw data are provided in Table S4.
Table 3.
Comparisons of RED and UC Assay fup Results
| Common Name | UC-fup | RED-fup | Avg Mass | Log Pow | Equilib-rium in RED? | RED/UC |
|---|---|---|---|---|---|---|
| 8:2 Fluorotelomer sulfonic acid (8:2 FTS) | 0.0396 | 0.0012 | 528.18 | 6.18 | Yes | 0.0311 |
| 3:3 Fluorotelomer carboxylic acid (3:3 FTCA) | 0.0518 | 0.0761 | 242.09 | 3.94 | Yes | 1.4706 |
| Perfluoroheptanoic acid (PFHpA) | 0.0004 | 0.0013 | 364.06 | 2.06 | Yes | 3.6214 |
| Perfluorodecanoic acid (PFDA) | 0.0027 | 0.0001 | 514.09 | 4.15 | Yes | 0.0349 |
| Perfluorohexanoic acid (PFHxA) | 0.0068 | 0.0106 | 314.05 | 2.85 | Yes | 1.5605 |
| Perfluorooctanesulfonic acid (PFOS) | 0.0049 | 0.0003 | 500.13 | 5.61 | Yes | 0.0600 |
| Perfluorooctanesulfonamide (PFOSA) | 0.0039 | 0.0008 | 499.14 | 5.02 | Yes | 0.2102 |
| 4:2 Fluorotelomer sulfonic acid (4:2 FTS) | 0.0050 | 0.0079 | 328.15 | 3.26 | Yes | 1.5846 |
| Perfluorobutanoic acid (PFBA) | 0.0930 | 0.1453 | 214.04 | 1.43 | Yes | 1.5624 |
| Perfluorobutanesulfonic acid (PFBS) | 0.0128 | 0.0121 | 300.09 | 3.12 | Yes | 0.9450 |
| Perfluoro-3,6,9-trioxatridecanoic acid (PFPE-6) a | 0.0018 | 0.00003 | 562.08 | 4.63 | No | 0.0167 |
| Perfluoropentanoic acid (PFPeA) | 0.0440 | 0.0455 | 264.05 | 1.35 | Yes | 1.0341 |
| Sodium Perfluorodecanesulfonate (Na + PFDS) | 0.0528 | 0.00 * | 622.12 | 5.58 | No | NA * |
| N-Methyl-N-(2-hydroxyethyl) perfluorooctanesulfonamide (NMeFOSE) | 0.0803 | 0.00 * | 557.22 | 4.58 | No | NA * |
| Perfluorohexanesulfonic acid (PFHxS) | 0.0009 | 0.0004 | 400.11 | 2.20 | No | 0.3973 |
| Perfluorononanoic acid (PFNA) | 0.0016 | 0.0004 | 464.08 | 3.54 | Yes | 0.2754 |
| Perfluorooctanoic acid (PFOA) | 0.0010 | 0.0011 | 414.07 | 3.11 | Yes | 1.0935 |
| Perfluoro-3,6,9-trioxadecanoic acid (PFPE-4) | 0.0003 | 0.0011 | 412.06 | 4.06 | Yes | 4.1570 |
| Perfluoroundecanoic acid (PFUnDA) | 0.0148 | 0.0001 | 564.09 | 4.00 | No | 0.0064 |
| Perfluoroheptanesulfonic acid (PFHpS) | 0.0006 | 0.00* | 450.12 | 4.31 | Yes | NA* |
| Perfluorotetradecanoic acid (PFTeDA) | 0.0271 | 0.0016 | 714.12 | 5.10 | No | 0.0574 |
| Perfluorotridecanoic acid (PFTrDA) | 0.0433 | 0.000048 | 664.11 | 5.35 | No | 0.0011 |
Values could not be accurately determined by UPLC-MS/MS and noted as 0.00.
Chemicals not achieving equilibrium during the RED assay are in bold font.
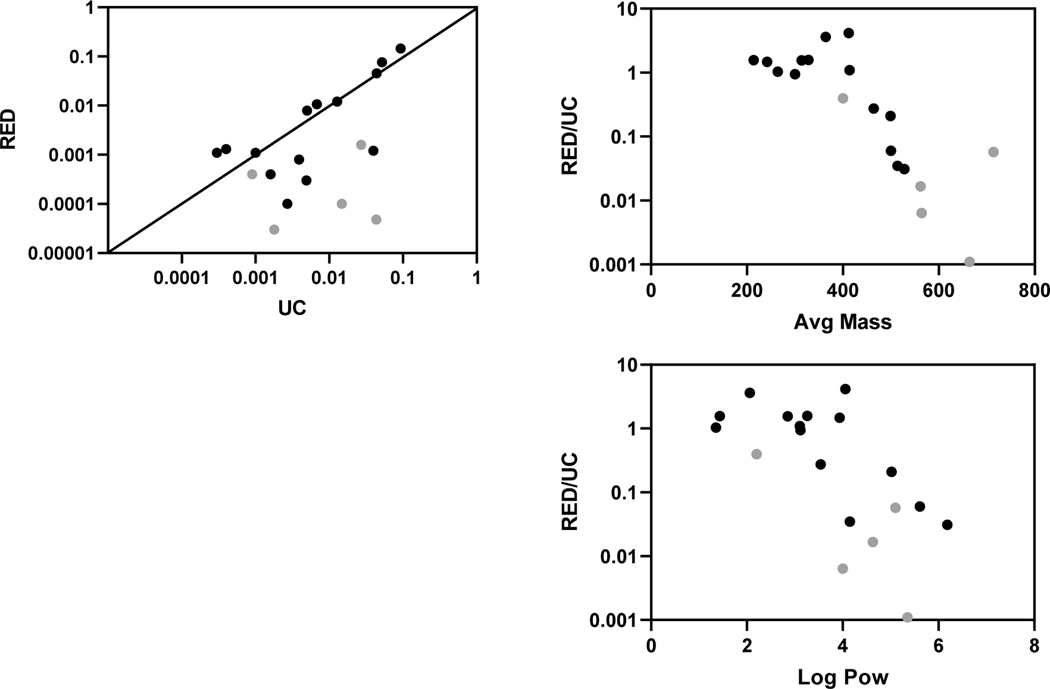
Evaluation of the RED assay performance involved review of the equilibrium control samples, where PBS was added to both sides of the membrane. If equilibrium has been achieved during the assay timeframe, the analyte on both sides of the membrane should be equivalent. This evaluation indicated that 7 did not achieve equilibrium. Although likelihood that a chemical would not achieve equilibrium appears related to molecular weights exceeding 500 g/mol and LogPow over 4, there were exceptions as noted in Figure 5 and Table 3. Perfluorohexane sulfonic acid, for instance, (M.W. 400.11 g/mol; Log Pow 2.2) did not achieve equilibrium; conversely, 8:2 fluorotelomer sulfonic acid (M.W. 528.18 g/mol; Log Pow 6.183) and perfluorooctane sulfonate (PFOS; 500.13 g/mol; LogPow 5.609) did. It should be noted that when equilibrium is not achieved, the RED assay will return artifactually low fup values (as less compound has crossed the membrane to the “PBS” (or protein-free) chamber before reaching equilibrium).
Fig.5. RED vs. UC assay comparisons.
Data for 19 PFAS with quantitative measures to derive fu in the two ssay types were evaluated. A) RED vs. UC-derived fu; RED/UC agreement based on mass; and RED/UC agreement by Log Pow. Gray circles indicate those that did not achieve
Omitting the 8 PFAS where fup was unmeasurable and/or equilibrium was not achieved in the RED assay, comparison of the 14 RED and UC fup values, showed that seven were within 3-fold of each other. Nine of the 14 had higher fup values in the RED assay when compared to the UC assay results whereas 5 had lower fup values. However, the fold-difference was greater when RED-derived fup values were lower: ranging up to 29-fold lower for PFDA compared to 4.2-fold higher for PFPE-4 (Table 3). Evaluations of PFAS molecular weight and Log Pow revealed that although both of these properties led to increased differences in fup from the two platforms, the trend associated with higher molecular weight was stronger (Figure 5).
Discussion
PPB is an important metric that can inform bioaccumulative potential and xenobiotic dosimetry evaluations. Highly bound chemicals are retained in the plasma, sequestered from metabolic enzymes that facilitate their clearance from an organism. Legacy PFAS such as PFOA, PFNA, and PFOS, already characterized for their extremely high binding and low to no hepatic clearance, are recognized as highly persistent chemicals, being widely detected in humans and ecological species 6. To date, PPB evaluations have only been completed for the more commonly studied carboxylates and PFOS 29, 30. With over 4,700 PFAS structures recognized by the OECD, this in vitro evaluation across 67 PFAS provides useful information as scientists strive to evaluate the larger data-poor space. In addition to providing PPB measures and grouping-based binding evaluations across several functional groups, this effort also provides useful methodologic considerations in developing sensitive targeted analytical methods, in generating reliable and reproducible PPB data, and in understanding the range of uncertainty around PPB point estimate values as we look toward NAM application.
The 67 PFAS analyzed here has provided data across 25 distinct groups, with 10 and 6 groups possessing data for at least three and two structures, respectively (Figure 3). Six groups with the lowest mean fup, were the perfluoroalkanoyl chlorides (PFAlkCls) (mean= 0.0008), perfluoropolyether carboxylates (PFPECAs) (mean=0.0009), PFCAs (mean=0.0019), fluorotelomer phosphonates (mean = 0.0021), and perfluoro ether carboxylates (mean = 0.0058). Although there are no reports on usage and/or fate for ether and polyether containing PFAS (PFECAs and PFPECAs) have gained commercial usage as a fluorinated alternative to long-chain PFAAs. The presence of repeating units of 2–3 perfluorinated C atoms per O atom purportedly ensures that degradation will not lead to long-chain PFCA formation thus reducing their persistence, although degradation studies have thus far been insufficient in confirming such decreased persistence 49. Marketed as surface treatments for a range of items including textiles, metal, leather, paper and paperboard treatment for food-contact applications 2, PFECAs and/or chlorinated PFPECAs have recently been discovered in nontargeted analyses in both environmental and biomonitoring samples including in the waters of southwestern NJ and in human follicular fluid samples 50, 51.
Almost complete coverage of PFAS carboxylates possessing 3 to 14 Cs revealed an intriguing U-shaped response in PPB not previously reported (Table 2). Short-chain PFCAs with up to 6 Cs possessed a mean fup =0.31. The mean fup value for PFCAs containing 6–10 Cs decreased by over 100-fold to 0.0019, with the highest binding observed for C7. For PFCAs with 11 or more Cs, the fup decreased 10-fold to a mean fup = 0.0217. Although data were more limited for PFSAs, a similar trend was noted with Na+ PFDS, the largest PFSA tested, returning fup = 0.0528, 16-fold lower than for the 6–9 group (fup =0.0031). Long-chain definitions proposed by OECD 46 define PFSAs with 6 or more Cs compared to PFCAs with 7 or more carbons to provide a more C-F equivalent comparison, since one C is part of the carboxylate functional group for the PFCAs. This decrease in binding is consistent with a meta-analysis of albumin binding affinity studies that reported a plateau or decrease in the binding affinity between 6 and 9 perfluorinated Cs 15. Our findings in conjunction with these earlier studies suggest that size differences and/or conformational changes of the longer chain PFCAs are hindering access to the albumin binding sites.
Comparative evaluations of the RED and UC PPB assay platforms underscored the importance of including reference compounds and assay checks as methods are applied to emerging contaminants that are beyond the scope of the chemicals commonly screened in such assays. The amphiphilic nature of PFAS and concern for unique binding interactions that may confound successful use of a membrane-based system led to the selection of UC as the method of choice. Reliant solely on centrifugal force to separate plasma proteins, lipoproteins, and fatty acid complexes from the aqueous fraction, UC assay showed successful quantitation of PFAS levels in all samples. Concentrations in the T1hr samples were typically within 90% of the target (10 μM) indicating minimal loss to non-specific binding in the system. Use of reference compound evaluations ensured AF samples were cleanly isolated, free from contaminating protein that may artifactually increase fup values 18. Similarly, use of equilibrium controls in the RED assay are vital to ensuring equilibrium is indeed achieved, particularly for high mass and highly lipophilic compounds. For high molecular weight and highly lipophilic chemicals, RED assay -derived PPB estimates were prone to an overestimation of binding, due to the inability of chemical to traverse to and achieve equilibrium within the dialysis chamber.
As exemplified by the RED vs. UC assay methodologic comparisons discussed above, considerations of robust experimental design and execution are critical to reduce experimental uncertainty. Quantification of uncertainty around experimental point estimates using Bayesian modeling provides an unbiased and transparent way to capture and display the uncertainty for use in downstream applications. In this study, it might be speculated that the high number of highly bound analytes would lead to a greater degree of uncertainty in this test set, as quantitative limits of the analytical method in conjunction with the outsize effect that may result with differences in between low fup replicate values. Review of the outputs did not support this speculation: only two of the 67 PFAS were classed as highly uncertain (defined as possessing a 95% credible interval spanning three orders of magnitude), and experimental CVs typically aligned with the modeled CVs. An important consideration addressed in the Bayesian model is the presence of signal in blank samples, an indication of the presence of contaminants in either the instrumentation or the experimental system. This led to the flagging of PFOA. Experimentally, it bears noting that neither PFOA nor any other PFAS analyte triggered a contaminant signal in the blanks that was sufficiently high to fail an independent analytical quality assurance criterion (blank signal ≤ ½ eMDL). As such, the flagging in the uncertainty analysis is deemed to be conservative; consideration will be given to modifying the manner in which dilution factors are considered as more data with similar issues become available. Overall, these results confirm that, across the larger set of PFAS, our analytical methods were sufficiently sensitive for successful quantitation across all stable PFAS in the UC assay.
With PPB being a critical determinant in steady state chemical pharmacokinetics 13, the 7,300-fold difference between the lowest and highest bound PFAS will have a similarly wide influence on internal dose estimation when IVIVE approaches are employed. To communicate the impact that an expected range of fup and hepatic clearance rates observed for PFAS would have on steady state predictions, twelve TK input scenarios selected based on experimental PFAS TK data, using four fup values (minimum, median, 25th and 75th percentiles of data presented here (Figure 1; Table 4)). For estimates of hepatic in vitro clearance (Clin vitro), bins were created that capture low (0), moderate (3) and high (10) Clin vitro (μL/min*106 cells) after review of data for a comparable set of PFAS 12, 52. Simply put, when hepatic clearance is identical, a 400-fold difference in fup yields a 400-fold difference in Css. PFAS with no clearance compared to moderate or high clearance will exhibit Css levels that are 6- or 17-fold higher, respectively. Consequently, the ability to accurately measure fup is extremely important to estimations of internal systemic concentrations are as accurate as possible. Moreover, the vast difference in binding noted is very informative in relating PPB to bioaccumulative potential if biomonitoring efforts are expanded to evaluate more PFAS.
Table 4.
Impact of PPB and Hepatic Clearance on PFAS Css Estimation
| PFAS Binding | fup | fub | Clin vitro (μl/min*106 cells) | Clhepatic (L/hr) | Css (μM) | fup Fold-Difference | Css Fold-Difference |
|---|---|---|---|---|---|---|---|
| Minimum fup | 0.0001 | 0.00018 | 0 | 0.0000 | 5710.51 | 1 | 1 |
| 3 | 0.0057 | 1000.23 | 1 | 6 | |||
| 10 | 0.0191 | 342.05 | 1 | 17 | |||
|
| |||||||
| 25th percentile fup | 0.0018 | 0.00327 | 0 | 0.0000 | 317.25 | 18 | 18 |
| 3 | 0.1033 | 55.62 | 18 | 103 | |||
| 10 | 0.3434 | 19.07 | 18 | 299 | |||
|
| |||||||
| Median fup | 0.005 | 0.00909 | 0 | 0.0000 | 114.21 | 50 | 50 |
| 3 | 0.2864 | 20.06 | 50 | 285 | |||
| 10 | 0.9475 | 6.91 | 50 | 827 | |||
|
| |||||||
| 75th percentile fup | 0.04 | 0.07273 | 0 | 0.0000 | 14.28 | 400 | 400 |
| 3 | 2.2410 | 2.55 | 400 | 2237 | |||
| 10 | 7.0599 | 0.92 | 400 | 6187 | |||
Css values were predicted using a generic TK model described previously (Wetmore et al., 2015), using 1mg/kg/day dose rate and TK inputs above. PFAS fup distributions calculated for test set. Blood:plasma ratio used to calculate fu in blood (fub); glomerular filtration rate (GFR) of 6.71 L/hr used to estimate renal clearance (GFR*fub).
This report provides in vitro PPB data across 67 PFAS to inform TK modeling, NAMs development, read-across grouping strategies, and ongoing evaluations of PFAS persistence and bioaccumulation. Leveraging new knowledge gained for these data-poor PFAS presents valuable insight as scientists seek to identify and prioritize PFAS groupings for robust exposure characterization, environmental fate and degradation studies, biomonitoring studies and/or toxicity testing.
Supplementary Material
Acknowledgments and Disclaimers
The authors would like to thank Katherine Coutros (USEPA) for managing the Evotec contract; Larry McMillan, Evgenia Korol-Bexell, and Matt Phillips for their technical assistance, and Charles Lowe, Elaina Kenyon, and Hisham El-Masri for their technical review of this manuscript. The views expressed in this manuscript are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the United States Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement for use.
Footnotes
Supporting Information
Analyte list, LC-MS method details, plasma protein binding assay data, and uncertainty estimations (XLSX)
Assay illustrations (DOC)
References
- 1.Prevedouros K; Cousins IT; Buck RC; Korzeniowski SH, Sources, fate and transport of perfluorocarboxylates. Environ Sci Technol 2006, 40 (1), 32–44. [DOI] [PubMed] [Google Scholar]
- 2.Buck RC; Franklin J; Berger U; Conder JM; Cousins IT; de Voogt P; Jensen AA; Kannan K; Mabury SA; van Leeuwen SP, Perfluoroalkyl and polyfluoroalkyl substances in the environment: terminology, classification, and origins. Integr Environ Assess Manag 2011, 7 (4), 513–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmler HJ, Synthesis of environmentally relevant fluorinated surfactants--a review. Chemosphere 2005, 58 (11), 1471–96. [DOI] [PubMed] [Google Scholar]
- 4.Gluge J; Scheringer M; Cousins IT; DeWitt JC; Goldenman G; Herzke D; Lohmann R; Ng CA; Trier X; Wang Z, An overview of the uses of per- and polyfluoroalkyl substances (PFAS). Environ Sci Process Impacts 2020, 22 (12), 2345–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z; DeWitt JC; Higgins CP; Cousins IT, A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)? Environ Sci Technol 2017, 51 (5), 2508–2518. [DOI] [PubMed] [Google Scholar]
- 6.Evich MG; Davis MJB; McCord JP; Acrey B; Awkerman JA; Knappe DRU; Lindstrom AB; Speth TF; Tebes-Stevens C; Strynar MJ; Wang Z; Weber EJ; Henderson WM; Washington JW, Per- and polyfluoroalkyl substances in the environment. Science 2022, 375 (6580), eabg9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams AJ; Gaines LGT; Grulke CM; Lowe CN; Sinclair GFB; Samano V; Thillainadarajah I; Meyer B; Patlewicz G; Richard AM, Assembly and Curation of Lists of Per- and Polyfluoroalkyl Substances (PFAS) to Support Environmental Science Research. Front Environ Sci 2022, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.OECD, Toward a New Comprehensive Global Database of Per and Polyfluoroalkyl Substances (PFASs): Summary Report on Updating the OECD 2007 List of Per and Polyfluoroalkyl Substances (PFASs). OECD Environment, Health and Safety Publication Series on Risk Management 2018, 39. [Google Scholar]
- 9.USEPA, Toxic Substances Control Act Reporting and Recordkeeping Requirements for Perfluoroalkyl and Polyfluoroalkyl Substances. Federal Register: 2021; Vol. 40 CFR Part 705, pp 33926–33966. [Google Scholar]
- 10.USEPA, PFAS Strategic Roadmap: EPA’s Commitments to Action 2021–2024. 2021. [Google Scholar]
- 11.Patlewicz G; Richard AM; Williams AJ; Grulke CM; Sams R; Lambert J; Noyes PD; DeVito MJ; Hines RN; Strynar M; Guiseppi-Elie A; Thomas RS, A Chemical Category-Based Prioritization Approach for Selecting 75 Per- and Polyfluoroalkyl Substances (PFAS) for Tiered Toxicity and Toxicokinetic Testing. Environ Health Perspect 2019, 127 (1), 14501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetmore BA; Wambaugh JF; Allen B; Ferguson SS; Sochaski MA; Setzer RW; Houck KA; Strope CL; Cantwell K; Judson RS; LeCluyse E; Clewell HJ; Thomas RS; Andersen ME, Incorporating High-Throughput Exposure Predictions With Dosimetry-Adjusted In Vitro Bioactivity to Inform Chemical Toxicity Testing. Toxicol Sci 2015, 148 (1), 121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wambaugh JF; Wetmore BA; Pearce R; Strope C; Goldsmith R; Sluka JP; Sedykh A; Tropsha A; Bosgra S; Shah I; Judson R; Thomas RS; Setzer RW, Toxicokinetic Triage for Environmental Chemicals. Toxicol Sci 2015, 147 (1), 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis LJ; Houston JB; Hallifax D, Impact of Plasma Protein Binding in Drug Clearance Prediction: A Data Base Analysis of Published Studies and Implications for In Vitro-In Vivo Extrapolation. Drug Metab Dispos 2021, 49 (3), 188–201. [DOI] [PubMed] [Google Scholar]
- 15.Ng CA; Hungerbuhler K, Bioaccumulation of perfluorinated alkyl acids: observations and models. Environ Sci Technol 2014, 48 (9), 4637–48. [DOI] [PubMed] [Google Scholar]
- 16.Nakai D; Kumamoto K; Sakikawa C; Kosaka T; Tokui T, Evaluation of the protein binding ratio of drugs by a micro-scale ultracentrifugation method. J Pharm Sci 2004, 93 (4), 847–54. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson KC; Luo YS; Rusyn I; Chiu WA, Comparative analysis of Rapid Equilibrium Dialysis (RED) and solid phase micro-extraction (SPME) methods for In Vitro-In Vivo extrapolation of environmental chemicals. Toxicol In Vitro 2019, 60, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieltyka K; McAuliffe B; Cianci C; Drexler DM; Shou W; Zhang J, Application of Cassette Ultracentrifugation Using Non-labeled Compounds and Liquid Chromatography-Tandem Mass Spectrometry Analysis for High-Throughput Protein Binding Determination. J Pharm Sci 2016, 105 (3), 1036–42. [DOI] [PubMed] [Google Scholar]
- 19.Wang C; Williams NS, A mass balance approach for calculation of recovery and binding enables the use of ultrafiltration as a rapid method for measurement of plasma protein binding for even highly lipophilic compounds. J Pharm Biomed Anal 2013, 75, 112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waters NJ; Jones R; Williams G; Sohal B, Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J Pharm Sci 2008, 97 (10), 4586–95. [DOI] [PubMed] [Google Scholar]
- 21.Ryu S; Riccardi K; Patel R; Zueva L; Burchett W; Di L, Applying Two Orthogonal Methods to Assess Accuracy of Plasma Protein Binding Measurements for Highly Bound Compounds. J Pharm Sci 2019, 108 (11), 3745–3749. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J; Pikul G; Horch J; Konovalov D; Li S; Cvijic ME; Shou W; Weller H, Enabling direct and definitive free fraction determination for highly-bound compounds in protein binding assay. J Pharm Biomed Anal 2021, 194, 113765. [DOI] [PubMed] [Google Scholar]
- 23.Wang H; Zrada M; Anderson K; Katwaru R; Harradine P; Choi B; Tong V; Pajkovic N; Mazenko R; Cox K; Cohen LH, Understanding and reducing the experimental variability of in vitro plasma protein binding measurements. J Pharm Sci 2014, 103 (10), 3302–9. [DOI] [PubMed] [Google Scholar]
- 24.Vanden Heuvel JP; Kuslikis BI; Peterson RE, Covalent binding of perfluorinated fatty acids to proteins in the plasma, liver and testes of rats. Chem Biol Interact 1992, 82 (3), 317–28. [DOI] [PubMed] [Google Scholar]
- 25.Rand AA; Mabury SA, Protein binding associated with exposure to fluorotelomer alcohols (FTOHs) and polyfluoroalkyl phosphate esters (PAPs) in rats. Environ Sci Technol 2014, 48 (4), 2421–9. [DOI] [PubMed] [Google Scholar]
- 26.Han X; Snow TA; Kemper RA; Jepson GW, Binding of perfluorooctanoic acid to rat and human plasma proteins. Chem Res Toxicol 2003, 16 (6), 775–81. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L; Ren XM; Guo LH, Structure-based investigation on the interaction of perfluorinated compounds with human liver fatty acid binding protein. Environ Sci Technol 2013, 47 (19), 11293–301. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y; Zhang H; Kang Y; Cao J, Effects of perfluorooctane sulfonate on the conformation and activity of bovine serum albumin. J Photochem Photobiol B 2016, 159, 66–73. [DOI] [PubMed] [Google Scholar]
- 29.Loccisano AE; Campbell JL Jr.; Butenhoff JL; Andersen ME; Clewell HJ 3rd, Comparison and evaluation of pharmacokinetics of PFOA and PFOS in the adult rat using a physiologically based pharmacokinetic model. Reprod Toxicol 2012, 33 (4), 452–467. [DOI] [PubMed] [Google Scholar]
- 30.Ohmori K; Kudo N; Katayama K; Kawashima Y, Comparison of the toxicokinetics between perfluorocarboxylic acids with different carbon chain length. Toxicology 2003, 184 (2–3), 135–40. [DOI] [PubMed] [Google Scholar]
- 31.Kreutz A, Clifton MS, Henderson MW, Phillips M, Korol-Bexell E, McMillan L, Wetmore BA, , Category-Based Toxicokinetic Evaluations of Data-Poor Per and Polyfluoroalkyl Substances (PFAS) by Gas Chromatography Coupled with Mass Spectrometry. in preparation 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smeltz MG, Clifton MS, Henderson WM, McMillan L, Wetmore BA, Targeted Per- and Polyfluoroalkyl Substances (PFAS) Assessments for High Throughput Screening: Analytical and Testing Considerations to Inform a PFAS Stock Quality Evaluation Framework. Toxicology and Applied Pharmacology 2022, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brockman AH; Oller HR; Moreau B; Kriksciukaite K; Bilodeau MT, Simple method provides resolution of albumin, lipoprotein, free fraction, and chylomicron to enhance the utility of protein binding assays. J Med Chem 2015, 58 (3), 1420–5. [DOI] [PubMed] [Google Scholar]
- 34.Forsthuber M; Kaiser AM; Granitzer S; Hassl I; Hengstschlager M; Stangl H; Gundacker C, Albumin is the major carrier protein for PFOS, PFOA, PFHxS, PFNA and PFDA in human plasma. Environ Int 2020, 137, 105324. [DOI] [PubMed] [Google Scholar]
- 35.Wan H; Rehngren M, High-throughput screening of protein binding by equilibrium dialysis combined with liquid chromatography and mass spectrometry. J Chromatogr A 2006, 1102 (1–2), 125–34. [DOI] [PubMed] [Google Scholar]
- 36.Wetmore BA; Wambaugh JF; Ferguson SS; Sochaski MA; Rotroff DM; Freeman K; Clewell HJ 3rd; Dix DJ; Andersen ME; Houck KA; Allen B; Judson RS; Singh R; Kavlock RJ; Richard AM; Thomas RS, Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol Sci 2012, 125 (1), 157–74. [DOI] [PubMed] [Google Scholar]
- 37.USEPA, Definition and Procedure for the Determination of the Method Detection Limit, Revision 2. Water O. o., Ed. 2016; pp 1–8. [Google Scholar]
- 38.Core TR A language and environment for statistical computing; Vienna Austria, 2022. [Google Scholar]
- 39.Plummer M, JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. Proceedings of the Third International Workshop on Distributed Statistical Computing (DSC2003) 2003, Vienna, Austria. [Google Scholar]
- 40.Wambaugh JF; Wetmore BA; Ring CL; Nicolas CI; Pearce RG; Honda GS; Dinallo R; Angus D; Gilbert J; Sierra T; Badrinarayanan A; Snodgrass B; Brockman A; Strock C; Setzer RW; Thomas RS, Assessing Toxicokinetic Uncertainty and Variability in Risk Prioritization. Toxicol Sci 2019, 172 (2), 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denwood MJ, runjags: An R Package Providing Interface Utilities, Model Templates, Parallel Computing Methods and Additional Distributions for MCMC Models in JAGS. J Stat Softw 2016, 71 (9), 1–25. [Google Scholar]
- 42.Frigerio G; Cafagna S; Polledri E; Mercadante R; Fustinoni S, Development and validation of an LC-MS/MS method for the quantitation of 30 legacy and emerging per- and polyfluoroalkyl substances (PFASs) in human plasma, including HFPO-DA, DONA, and cC6O4. Anal Bioanal Chem 2022, 414 (3), 1259–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrington LM, Analysis of perfluoroalkyl and polyfluoroalkyl substances in serum and plasma by solvent precipitation-isotope dilution-direct injection-LC/MS/MS. Analytical Methods 2017, 9 (3), 473–481. [Google Scholar]
- 44.Patlewicz G, Richard AM, Williams AJ, Judson RS, Thomas RS, Towards Reproducible Structure-Based Chemical Categories of PFAS to Inform and Evaluate Toxicity and Toxicokinetic Testing. Comput Toxicol 2022, 24, 100250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su A; Rajan K, A database framework for rapid screening of structure-function relationships in PFAS chemistry. Sci Data 2021, 8 (1), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.OECD Reconciling Terminology of the Universe of Per- and Polyfluoroalkyl Substances: Recommendations and Practical Guidance; Environment Directorate: Paris, France, 2021. [Google Scholar]
- 47.Mansouri K; Cariello NF; Korotcov A; Tkachenko V; Grulke CM; Sprankle CS; Allen D; Casey WM; Kleinstreuer NC; Williams AJ, Open-source QSAR models for pKa prediction using multiple machine learning approaches. J Cheminform 2019, 11 (1), 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mansouri K; Grulke CM; Judson RS; Williams AJ , OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminform 2018, 10 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z; Cousins IT; Scheringer M; Hungerbuehler K, Hazard assessment of fluorinated alternatives to long-chain perfluoroalkyl acids (PFAAs) and their precursors: status quo, ongoing challenges and possible solutions. Environ Int 2015, 75, 172–9. [DOI] [PubMed] [Google Scholar]
- 50.Kang Q; Gao F; Zhang X; Wang L; Liu J; Fu M; Zhang S; Wan Y; Shen H; Hu J, Nontargeted identification of per- and polyfluoroalkyl substances in human follicular fluid and their blood-follicle transfer. Environ Int 2020, 139, 105686. [DOI] [PubMed] [Google Scholar]
- 51.McCord JP; Strynar MJ; Washington JW; Bergman EL; Goodrow SM, Emerging Chlorinated Polyfluorinated Polyether Compounds Impacting the Waters of Southwestern New Jersey Identified by Use of Nontargeted Analysis. Environ Sci Technol Lett 2020, 7 (12), 903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crizer DM, Zhou JR, Lavrich KS, Wetmore BA, Ferguson SS, DeVito MJ, Merrick BA In In Vitro Hepatic Clearance of Per- and Polyfluoroalkyl Substances (PFAS), Society of Toxicology, San Diego, CA, San Diego, CA, 2022; p Abstract No. 3089. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.