ABSTRACT
Understanding the evolutionary strategies of the SARS-CoV-2 omicron variant is crucial for comprehending the COVID-19 pandemic and preventing future coronavirus pandemics. In this study, we determined the crystal structures of the receptor-binding domains (RBDs) from currently circulating omicron subvariants XBB.1 and XBB.1.5 (also the emerging XBB.1.9.1), each complexed with human ACE2. We studied how individual RBD residues evolved structurally in omicron subvariants, specifically how they adapted to human ACE2. Our findings revealed that residues 493 and 496, which exhibited good human ACE2 adaptation in pre-omicron variants, evolved to poor adaptation in early omicron subvariants (but with good adaption to mouse ACE2) and then reverted to good adaptation in recent omicron subvariants. This result is consistent with the hypothesis that non-human animals facilitated the evolution of early omicron subvariants. Additionally, residue 486, which exhibited good human ACE2 adaptation in early omicron subvariants, evolved to poor adaptation in later omicron subvariants and then returned to good adaptation in recent omicron subvariants. This result is consistent with the hypothesis that immune evasion facilitated the evolution of later omicron subvariants. Thus, our study suggests that both non-human animals and immune evasion may have contributed to driving omicron evolution at different stages of the pandemic.
IMPORTANCE
The sudden emergence and continued evolution of the SARS-CoV-2 omicron variant have left many mysteries unanswered, such as the origin of early omicron subvariants and the factors driving omicron evolution. To address these questions, we studied the crystal structures of human ACE2-bound receptor-binding domains (RBDs) from omicron subvariants XBB.1 and XBB.1.5 (XBB.1.9.1). Our in-depth structural analysis sheds light on how specific RBD mutations adapt to either human or mouse ACE2 and suggests non-human animals and immune evasion may have influenced omicron evolution during different stages of the pandemic. These findings provide valuable insights into the mechanisms underlying omicron evolution, deepen our understanding of the COVID-19 pandemic, and have significant implications for preventing future coronavirus pandemics.
KEYWORDS: COVID-19, omicron subvariants, receptor-binding domain (RBD), receptor-binding motif (RBM), angiotensin-converting enzyme 2, surface plasmon resonance, X-ray crystallography
INTRODUCTION
The sudden emergence of the SARS-CoV-2 omicron variant in the fall of 2021 marked a significant turning point in the COVID-19 pandemic (1 - 4). Compared to pre-omicron strains like the prototypic strain, the early omicron subvariants BA.1 and BA.2 contained many new mutations and spread more rapidly, quickly replacing them as the dominant strain in human populations (1 - 4). Presently, XBB.1 and XBB.1.5 are the omicron subvariants circulating in humans (5 - 7), while XBB.1.9.1 is emerging (8, 9). Understanding the evolution of the omicron subvariants is crucial for comprehending the COVID-19 pandemic and preventing potential future coronavirus pandemics.
The binding interface between coronavirus receptor-binding domains (RBDs) and their host receptor has been established as an excellent structural platform for studying coronavirus evolution at the atomic level (10, 11). Both SARS-CoV-2 and the related SARS-CoV-1 recognize human ACE2 (hACE2) as their entry receptor (12 - 14). Previously, we determined the crystal structures of hACE2-bound RBDs from SARS-CoV-1, the prototypic SARS-CoV-2 strain, and the early omicron subvariants BA.1 and BA.2 (15 - 18). These structures revealed that both SARS-CoV-1 and SARS-CoV-2 RBDs contain a core structure and a receptor-binding motif (RBM), with the latter mediating receptor binding. Three virus-binding hotspots have been discovered at the RBM/ACE2 interface for both viruses, which center on Lys31 and Lys353 in hACE2 and a receptor-binding ridge in the RBD (named hotspot-31, hotspot-353, and hotspot-ridge, respectively) (10, 11, 16, 18). These hotspots are also the hotpots for viral mutations. The concept of the hotspots has been instrumental to understanding how RBM mutations affect ACE2 binding, and consequently, how SARS-CoV-1 and SARS-CoV-2 adapt to host infections.
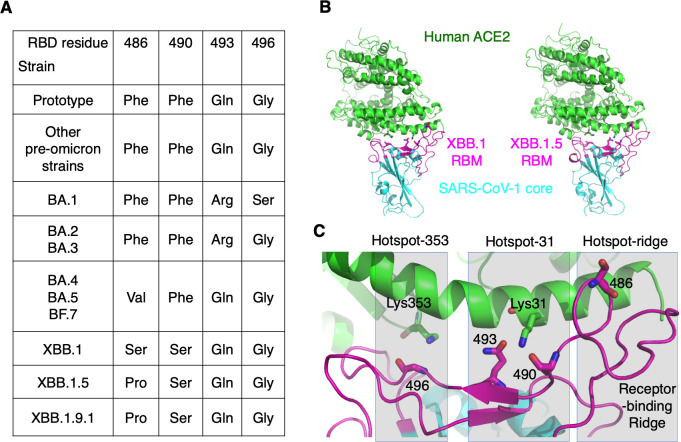
There are two theories that explain how the omicron variant evolved (19). The first theory proposes that the RBD mutations in early omicron subvariants occurred in immunocompromised people, where the virus was unable to be cleared and instead developed mutations to evade the immune system. The second theory suggests that the mutations took place in non-human animals, where the virus developed mutations to adapt to its host and then transmitted to humans. Recently, we examined the crystal structure of BA.2 RBD bound to mouse ACE2 (mACE2), which showed that several mutations in BA.2 RBM were viral adaptations to mACE2 but were incompatible with hACE2. This finding suggested that rodents might have played a role in the emergence of early omicron subvariants. This hypothesis was supported by the discovery that 16.5% of rats in New York City had been infected with SARS-CoV-2 (20). The pre-omicron variants and early omicron subvariants have had their receptor recognition well documented (15 - 22 - 21 - 22), but little information is available for the structural evolution of recent omicron subvariants. To date, omicron subvariants have evolved at four RBM residues: 486, 490, 493, and 496 (Fig. 1A) (5, 6, 23). Understanding the structural roles of each of these four residues and their mutations in receptor binding is crucial for comprehending the evolutionary history and future of the omicron variant.
Fig 1.
Overview of omicron evolution in the receptor-binding motif. (A) Four residues in the RBM have undergone evolution from early omicron subvariants to recent ones and are listed here. (B) Overall structures of the chimeric XBB.1 RBD complexed with human ACE2 (left) and of the chimeric XBB.1.5 RBD complexed with human ACE2 (right). Human ACE2 is in green. The core structures in the two RBDs (in cyan) are from SARS-CoV-1 and the RBMs (in magenta) are from XBB.1 and XBB.1.5, respectively. (C) Structure of the interface between XBB.1 RBM and human ACE2. RBM residues that have undergone mutations from the early omicron subvariants to the recent one are shown as sticks. Three mutational hotspots are highlighted: hotspot-353 centers on Lys353 in human ACE2, hotspot-31 centers on Lys31 in human ACE2, and hotspot-ridge centers on the receptor-binding ridge in the RBD.
In this study, we analyzed the crystal structures of the RBDs from two recent omicron subvariants, XBB.1 and XBB.1.5, each bound to hACE2. It should be noted that emerging XBB.1.9.1 shares the same RBD as XBB.1.5. Our goal was to understand how these subvariants have evolved to bind hACE2 and mACE2. Specifically, we aimed to achieve the following three objectives: shed light on the evolutionary tactics employed by omicron, gain insight into the origins of early omicron subvariants, and identify the factors driving the evolution of later and more recent omicron subvariants.
RESULTS
To investigate the interactions between omicron RBM and hACE2, we created two chimeric RBDs (Fig. 1B). These RBDs contained the core structure from SARS-CoV-1 and the RBM from XBB.1 and XBB.1.5 (XBB.1.9.1), respectively. This chimeric RBD design has been previously described in detail and repeatedly proven successful for studying how SARS-CoV-2, SARS-CoV-1, and related viruses recognize receptors (15 - 18 - 24). In essence, to design these chimeric RBDs, we took advantage of the easy formation of high-quality crystals of the SARS-CoV-1 RBD/hACE2 complex. We preserved the crystal contact regions involving the SARS-CoV-1 RBD core structure and modified the non-crystal contact regions to include the omicron RBM. As expected, both chimeric RBDs bound to hACE2 and formed high-quality crystals under the same conditions as the hACE2-bound SARS-CoV-1 RBD. We determined the structures of both chimeric RBDs using molecular replacement and refined them to a resolution of 2.7 Å and 3.1 Å for XBB.1 and XBB.1.5, respectively (Fig. 1B; Table 1; Fig. S1 and S2). These structures serve as the foundation for comprehending omicron mutations.
TABLE 1.
Crystallography data collection and refinement statistics a
| XBB.1 | XBB.1.5 | |
|---|---|---|
| Data collection | ||
| Space group | P1211 | P1211 |
| Unit cell dimensions | ||
| a, b, c (Å) | 79.74, 116.37, 111.66 | 78.37, 116.21, 108.77 |
| α, β, γ (°) | 90, 92.81, 90 | 90, 96.60, 90 |
| Resolution (Å) | 80.5–2.7 (2.8–2.7) | 77.9–3.1 (3.2–3.1) |
| R sym or R merge | 0.152 (0.930) | 0.121 (0.881) |
| I/σI | 5.3 (1.7) | 6.8 (1.6) |
| Completeness (%) | 85.5 (67.2) | 86.8 (52.6) |
| Redundancy | 4.1 (3.8) | 3.9 (3.8) |
| CC1/2 | 0.993 (0.549) | 0.995 (0.653) |
| CC* b | 0.998 (0.910) | 0.999 (0.914) |
| Refinement | ||
| Resolution (Å) | 80.5–2.7 (2.8–2.7)* | 77.9–3.1 (3.2–3.1) |
| No. of reflections | 24,960 (178) | 24,811(100) |
| R work/R free | 0.219/0.283 | 0.190/0.241 |
| No. of atoms | 13,162 | 13,107 |
| Protein | 12,772 | 12,765 |
| Ligand/ion | 361 | 339 |
| Water | 29 | 3 |
| B-factor | 71.67 | 78.35 |
| Protein | 71.24 | 77.60 |
| Ligand/ion | 88.72 | 106.87 |
| Water | 49.54 | 38.48 |
| Ramachandran plot | ||
| Favored (%) | 94.45 | 94.82 |
| Allowed (%) | 5.42 | 4.80 |
| Outliers (%) | 0.13 | 0.38 |
| RMS c deviations | ||
| Bond lengths (Å) | 0.004 | 0.002 |
| Bond angles (°) | 0.61 | 0.49 |
Statistics for the highest-resolution shell are shown in parentheses.
CC*, correlation coefficient calculated between final merged dataset and unknown true intensities
RMS, root mean square
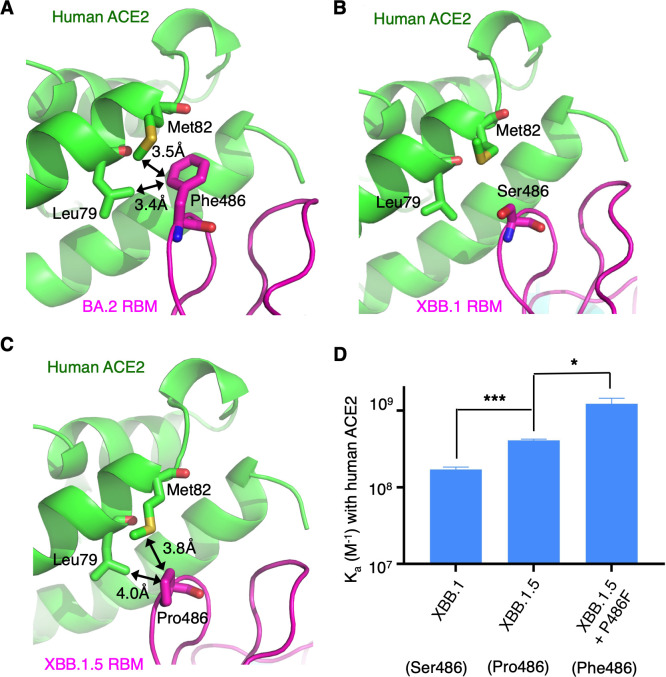
RBM residue 486 is situated in the hotspot-ridge area (Fig. 1C). In both pre-omicron variants and early omicron subvariants, it was a phenylalanine, which evolved into a valine in later omicron subvariants, then a serine in XBB.1, and finally a proline in XBB.1.5 (Fig. 1A). When RBM residue 486 is a phenylalanine, it strongly interacts with Leu79 and Met82 from hACE2 through hydrophobic interactions (Fig. 2A). However, when it is a serine, there are no interactions with Leu79 and Met82 (Fig. 2B). The proline also interacts hydrophobically with Leu79 and Met82 from hACE2 (Fig. 2C), but these interactions are not as strong as those formed by a phenylalanine. This is because a phenylalanine makes more extensive hydrophobic contacts than a proline does. To verify the above structural analysis, we conducted surface plasmon resonance (SPR) experiments to determine the binding affinity between hACE2 and RBDs containing different residues at position 486 (Fig. 2D; Table S1; Fig. S3). The binding affinity increased from XBB.1 RBD, which had Ser486, to XBB.1.5 RBD, which had Pro486, and then to XBB.1.5 RBD, which had Phe486. Therefore, both structural and biochemical data reveal that a phenylalanine is best adapted to hACE2 at position 486, followed by a proline and then a serine.
Fig 2.
Structural evolution of RBM residue 486. (A) Structural interface between the RBM of omicron subvariant BA.2 and human ACE2 (PDB 7UFK). Black double arrows indicate favorable hydrophobic interactions. (B) Structural interface between the RBM of omicron subvariant XBB.1 and human ACE2. (C) Structural interface between the RBM of omicron subvariant XBB.1.5 and human ACE2. (D) Surface plasmon resonance assay for the binding of RBDs to human ACE2. The data are presented as mean ± SEM (n = 3). A Student’s two-tailed t-test was performed to analyze the statistical difference between the XBB.1 RBD and XBB.1.5 RBD (which differ from each other only at position 486) and between XBB.1.5 RBD and XBB.1.5 RBD containing a P486F mutation. ***P < 0.001; *P < 0.05.
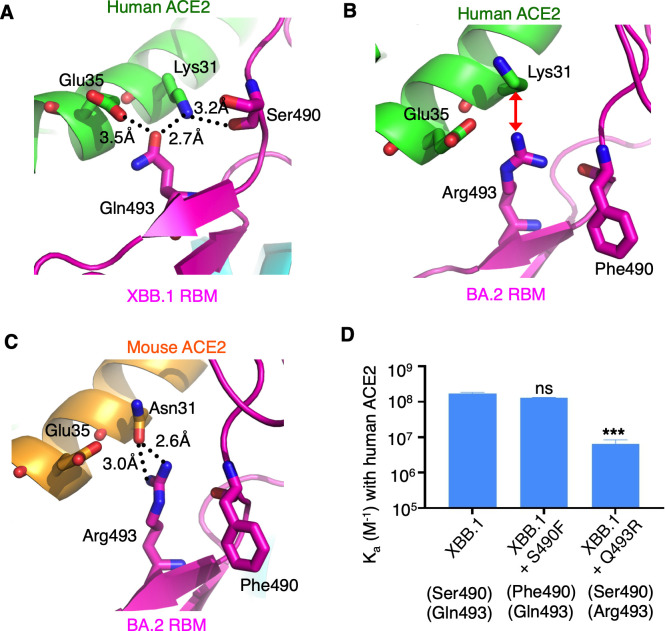
RBM residue 493 is located in the hotspot-31 area (Fig. 1C). It was a glutamine in pre-omicron variants, including the prototypic strain. Then, it changed to an arginine in early omicron subvariants but reverted to a glutamine in later and recent omicron subvariants (Fig. 1A). In pre-omicron variants and recent omicron subvariants, Gln493 forms favorable hydrogen bonds with Lys31 and Glu35 from hACE2 (Fig. 3A). However, in early omicron subvariants, Arg493 creates unfavorable charge repulsion with Lys31 from hACE2 (Fig. 3B) while forming favorable bifurcated hydrogen bonds with Asn31 from mACE2 (Fig. 3C). Previously, we used SPR to demonstrate that the Q493R mutation decreased the binding affinity of prototypic RBD for hACE2 but increased its binding affinity for mACE2 (18). Here, also using SPR, we further demonstrated that the Q493R mutation decreased the binding affinity of XBB.1 RBD for hACE2 (Fig. 3D; Table S1; Fig. S4). Therefore, both structural and biochemical data reveal that Gln493 is better adapted to hACE2 than Arg493 is, while Arg493 is better adapted to mACE2 than Gln493 is.
Fig 3.
Structural evolution of RBM residues 493 and 490. (A) Structural interface between the RBM of omicron subvariant XBB.1 and human ACE2. Black dotted lines indicate favorable hydrogen bonds. (B) Structural interface between the RBM of omicron subvariant BA.2 and human ACE2 (PDB 7UFK). Red double arrow indicates unfavorable charge repulsion. (C) Structural interface between the RBM of omicron subvariant BA.2 and mouse ACE2 (PDB 7UFL). (D) Surface plasmon resonance assay for the binding of RBDs to human ACE2. The data are presented as mean ± SEM (n = 3). A Student’s two-tailed t-test was performed to analyze the statistical difference between the XBB.1 RBD and XBB.1 RBD containing an S490F mutation and between XBB.1 RBD and XBB.1 RBD containing a Q493R mutation. ***P < 0.001; ns, not significant.
RBM residue 490 is also situated in the hotspot-31 area (Fig. 1C). It has gone through evolution from a phenylalanine in pre-omicron and early/later omicron subvariants to a serine in recent omicron subvariants (Fig. 1A). Structural data revealed that the main chain of residue 490, but not its side chain, directly interacts with hACE2 (Fig. 3A), suggesting that its side chain does not play a crucial role in hACE2 binding. The SPR data supported that the S490F mutation did not cause a significant effect on the binding affinity of XBB.1 RBD for hACE2 (Fig. 3D; Table S1; Fig. S4). Therefore, Phe490 and Ser490 are comparably adapted to hACE2.
RBM residue 496 is located in the hotspot-353 area (Fig. 1C) and has been a glycine in all pre-omicron variants and omicron subvariants except BA.1, where it was a serine (Fig. 1A). In pre-omicron variants and XBB.1, the main chain of Gly496 and the side chain of Lys353 from hACE2 both form a favorable hydrogen bond with Tyr501 in the RBM (Fig. 4A). In BA.1, Tyr501 forms two hydrogen bonds with both the main chain and side chain of Ser496 and can no longer form a hydrogen bond with Lys353 from hACE2, forcing Lys353 to be oriented toward a different direction (Fig. 4B). In mACE2, however, residue 353 becomes a histidine, which is compatible with Ser496 and Tyr501 from BA.1 RBD (Fig. 4C and D). Using SPR data, we further demonstrated that the S496G mutation increased the binding affinity of BA.1 RBD for hACE2 but had no significant impact on its binding affinity for mACE2 (Fig. 4D; Table S1; Fig. S5). Hence, Gly496 is better adapted to hACE2 than Ser496 is, while both Gly496 and Ser496 are comparably adapted to mACE2.
Fig 4.
Structural evolution of RBM residue 496. (A) Structural interface between the RBM of omicron subvariant XBB.1 and human ACE2. Black dotted lines indicate favorable hydrogen bonds. (B) Structural interface between the RBM of omicron subvariant BA.1 and human ACE2 (PDB 7U0N). (C) Structural interface between the RBM of omicron subvariant BA.2 and mouse ACE2 (PDB 7UFL). (D) Surface plasmon resonance assay for the binding of RBDs to mouse ACE2 (top) or to human ACE2 (bottom). The data are presented as mean ± SEM (n = 3). A Student’s two-tailed t-test was performed to analyze the statistical difference between the RBDs in each graph. *P < 0.05; ns, not significant.
In addition to the detailed characterization of how individual omicron mutations affected receptor recognition, we conducted analyses of the RBM/ACE2 interfaces for omicron subvariants. First, we conducted an electrostatic analysis of these interfaces. We observed that, compared to the prototypic strain, the RBMs of omicron subvariants (BA.1, BA.2, XBB.1, and XBB.1.5) exhibit a higher positive charge around hotspot-31 and hotspot-353 but a lower negative charge around hotspot-ridge (Fig. S6). On the virus-binding surface of hACE2, hotspot-31 and hotspot-353 display a negative charge, whereas hotspot-ridge shows a slightly positive charge (Fig. S6). Consequently, it is possible that the omicron subvariants have evolved to recognize hACE2 through stronger electrostatic interactions, which are long range and potentially enable virus particles to more rapidly attach to the receptor (25). Second, we calculated the buried surface areas and contact residues for the RBM/ACE2 interfaces. Comparing the omicron subvariants to the prototypic strain, we observed slightly smaller buried surface areas and slightly fewer ACE2-contacting residues in the RBMs (Tables S2 and S3). However, BA.1 and BA.2 RBDs both exhibited a slightly stronger binding affinity to hACE2 than the prototypic RBD did (17, 18). These findings suggest that the omicron subvariants might have undergone changes to reduce their receptor-binding surface area and the number of receptor-contacting residues but managed to maintain, or even slightly increase, their affinity for hACE2, potentially by enhancing the strengths of these receptor-binding interactions. It is important to note that the above observations were based on structural models and would require further investigation through in-depth computational analysis.
DISCUSSION
Since it first emerged in the fall of 2021, the omicron variant has continued to evolve within human populations. In order to understand how omicron has been evolving, we conducted an investigation into the structural changes of the omicron subvariants in the RBM region. Specifically, we analyzed the crystal structures of the XBB.1 RBD and XBB.1.5 RBD, each complexed with human ACE2. It should be noted that emerging XBB.1.9.1 shares the same RBD as XBB.1.5. The omicron subvariants have evolved at four RBM residues (486, 490, 493 and 496), which are located at three virus-binding hotspots within the RBM/ACE2 interface. Two of these hotspots, known as hotspot-31 and hotspot-353, involve stabilizing two lysines from human ACE2 (Lys31 and Lys353). The third hotspot, known as hotspot-ridge, involves interactions between the N-terminal helix from human ACE2 and a receptor-binding ridge from the RBM. At hotspot-ridge, Phe486 is the most compatible with Leu79 and Met82 from human ACE2, followed by Pro486, with Ser486 being the least compatible with human ACE2. At hotspot-31, Gln493 is more compatible with Lys31 from human ACE2 than Arg493 is, whereas Phe490 and Ser490 are equally compatible with human ACE2. At hotspot-353, Gly496 is more compatible with Lys353 from human ACE2 than Ser496 is. Therefore, of the four RBM residues that have undergone evolution, residues 493 and 496 have become better adapted to human ACE2 over time, residue 486 first became less compatible with human ACE2 before evolving to become more compatible, and the evolution of residue 490 has no significant impact on its compatibility with human ACE2.
As RBM residues 493 and 496 evolved, an evolutionary gap emerged between pre-omicron variants and early omicron subvariants (Fig. 5). At these two positions, the residues well adapted to human ACE2 in pre-omicron variants abruptly became less adapted in early omicron subvariants. Two other RBM residues, 498 and 505, also underwent this transition. However, residues 493 and 496 have since reverted to the same residues as in pre-omicron variants that are better adapted to human ACE2. Residues 498 and 505, in contrast, have remained unchanged across all omicron subvariants. Our previous structural study showed that Arg493, Arg498, and His505 in early omicron subvariants are well adapted to mouse ACE2 but are incompatible with human ACE2; the structural data were confirmed using both RBD/ACE2-binding assay and SARS-CoV-2 pseudovirus entry assay (18). These data together led to the hypothesis that rodents played a role in the evolution of early omicron subvariants. The current study demonstrated that Ser496 is also better adapted to mouse ACE2 than to human ACE2, further supporting the hypothesis. The hypothesis is also supported by a surveillance study, which revealed that 16.5% of rats in New York City were infected by SARS-CoV-2 and indicated that the virus has been evolving in rodents (20). The hypothesis is further bolstered by the reversion mutations observed at residues 493 and 496, indicating that the virus, which had previously adapted for rodent infectivity, is now undergoing changes to enhance its ability to infect humans once again. Additionally, another report demonstrated that some of these mutated residues are involved in stabilizing interactions within and between RBDs in the spike protein (26); this suggests that the aforementioned mutations may impact the function of the spike protein in more ways than one, extending beyond their impact on receptor recognition alone.
Fig 5.
Proposed mechanisms for structural evolution of omicron RBM and potential driving forces for the evolution. The values in this graph are not drawn to scales.
As RBM residues 486 and 490 evolved, an evolutionary gap appeared not between the pre-omicron variants and early omicron subvariants but rather between early omicron subvariants and more recent ones (Figure 5). At these positions, the residues that had been well adapted to human ACE2 in pre-omicron variants and early omicron subvariants became less adapted in later and recent omicron subvariants. There could be two possible mechanisms for this evolution: either non-human animals were involved in the evolution of recent omicron subvariants or there is another driving force for viral evolution besides host ACE2 adaptation. Unlike early omicron subvariants, where numerous mouse ACE2-adapted mutations appeared abruptly and simultaneously, evolution of later and recent omicron subvariants appeared to be gradual and sporadic, making the second scenario more likely. The second scenario is supported by reports indicating that the F486S and F490S mutations enabled the omicron variant to evade pre-existing neutralizing antibody responses (Table S4) (5, 27). While these mutations did not improve the affinity of the RBD for human ACE2, they still benefited the virus through immune evasion. On the contrary, the R493Q mutation in the XBB.1 subvariant increased its vulnerability to pre-existing neutralizing antibodies, aligning with our previous argument that the R493Q mutation emerged through the receptor recognition mechanism rather than immune evasion (Table S4) (5). Notably, in recent omicron subvariants, the S486P mutation partially restored the binding affinity of the RBD to human ACE2; this implies that the virus is adapting to strike a balance between maintaining a strong receptor-binding affinity and evading immune detection.
In conclusion, the structural evolution of the omicron RBM has disclosed multiple evolutionary strategies employed by the virus. These include reinforcing the three virus-binding hotspots at the RBM/receptor interface to improve adaptation to human ACE2 and striking a balance between high receptor-binding affinity and immune evasion. These strategies work together to drive the evolution of the RBM. It will be intriguing to observe how the omicron variant will continue to evolve in the future.
MATERIALS AND METHODS
Plasmids
The genes encoding the spike protein of SARS-CoV-2 prototypic strain (GenBank accession number: QHD43416), human ACE2 (GenBank accession number: NM_021804), and mouse ACE2 (GenBank accession number: NM_027286) were synthesized (GenScript Biotech). The genes encoding the spike protein of SARS-CoV-2 omicron subvariants (XBB.1—GISAID: EPI_ISL_14917652; XBB.1.5—GISAID: EPI_ISL_15921922; XBB.1.9.1—GISAID: EPI_ISL_16093023) were constructed by site-directed mutagenesis of the gene encoding the spike protein of SARS-CoV-2 prototypic strain.
For biochemical assays, the genes encoding the RBDs of omicron subvariants (residues 319–535) and their mutants, human ACE2 (residues 1–615) and mouse ACE2 (residues 1–615), were constructed from the above full-length genes, respectively. The RBDs of omicron subvariants and their mutants were subcloned into pLenti-transfer vector (Addgene) with an N-terminal tissue plasminogen activator signal peptide and a C-terminal His tag. Human ACE2 and mouse ACE2 were subcloned into the same vector except that a C-terminal human IgG4 Fc region replaced the His tag.
For crystallography studies, the genes encoding the RBDs of chimeric omicron subvariants (residues 319–535) were constructed by site-directed mutagenesis of the genes encoding the chimeric prototypic RBD (16, 17). They were then subcloned into pFastBac I vector (Life Technologies) with an N-terminal honeybee melittin signal peptide and a C-terminal His tag. Human ACE2 (residues 1–615) was subcloned in the same way as the chimeric RBDs.
Protein expression and purification
For biochemical assays, the RBDs of omicron subvariants and their mutants, human ACE2 and mouse ACE2, were prepared from 293F mammalian cells (28). Briefly, lentiviral particles were packaged for the construction of stable cell lines expressing one of the above proteins. These stable cell lines were selected in the presence of Puromycin (Gibco). Each of the proteins was collected from cell culture medium, purified on Ni-NTA column for His-tagged proteins or on Protein A column for Fc-tagged proteins, and purified further on Superdex 200 gel filtration column (Cytiva).
For crystallography studies, the RBDs of chimeric omicron subvariants and human ACE2 were prepared from sf9 insect cells using the Bac-to-Bac system (Life Technologies) (16). Subsequently, the His-tagged proteins were harvested from cell culture medium, purified on Ni-NTA column, and purified further on Superdex 200 gel filtration column (Cytiva).
Surface plasmon resonance assay
Binding interactions between RBDs and ACE2 molecules were measured by surface plasmon resonance using a Biacore S200 system (Cytiva) (17). Briefly, ACE2 was immobilized to a CM5 sensor chip through chemical cross-linking (Cytiva). Serial dilutions of purified recombinant RBD were injected at different concentrations. Binding kinetics were calculated using Biacore Evaluation Software (Cytiva).
Crystallization and structure determination
The complexes of the chimeric omicron XBB.1 RBD and human ACE2 and of the chimeric XBB.1.5 RBD and human ACE2 were each purified on gel filtration chromatography. Crystals of each of the complexes were grown at room temperature over wells containing 100 mM Tris (pH 8.5), 18%–24% PEG 6000, and 100 mM NaCl. X-ray diffraction data were collected at NECAT (24-IDC and 24-IDE) beamlines. HKL2000 was used for data processing (29). Both of the structures were determined by molecular replacement using the structure of prototypic chimeric RBD complexed with human ACE2 as the search template (PDB 6VW1). PHENIX and CCP4 were used for molecular replacement and model refinement (30, 31). COOT was used for model building (32). PYMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) was used for making structural figures. Structure data and refinement statistics are shown in Table 1.
ACKNOWLEDGMENTS
This study was supported by NIH grants R01AI089728 and R01AI110700 (to F.L.) and R35GM118047 (to H.A.). The Biacore S200 system was supported by NIH ORIP grant 1S10OD021539.
X-ray diffraction data were collected at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, which are funded by NIH (NIGMS P30 GM124165).
Contributor Information
Hideki Aihara, Email: aihar001@umn.edu.
Fang Li, Email: lifang@umn.edu.
Tom Gallagher, Loyola University Chicago, Maywood, Illinois, USA .
DATA AVAILABILITY
Coordinates and structure factors have been deposited in the Protein Data Bank: accession number 8SPH for chimeric XBB.1 RBD complexed with human ACE2 and accession number 8SPI for chimeric XBB.1.5 RBD complexed with human ACE2.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00822-23.
Fig. S1 to S5; Tables S1 to S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Karim SSA, Karim QA. 2021. Omicron SARS-Cov-2 variant: a new chapter in the COVID-19 pandemic. Lancet 398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. 2022. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 omicron wave-reply. JAMA 327:2148. doi: 10.1001/jama.2022.5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saxena SK, Kumar S, Ansari S, Paweska JT, Maurya VK, Tripathi AK, Abdel-Moneim AS. 2022. Characterization of the novel SARS-CoV-2 omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol 94:1738–1744. doi: 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 4. Gu H, Krishnan P, Ng DYM, Chang LDJ, Liu GYZ, Cheng SSM, Hui MMY, Fan MCY, Wan JHL, Lau LHK, Cowling BJ, Peiris M, Poon LLM. 2022. Probable transmission of SARS-CoV-2 omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg Infect Dis 28:460–462. doi: 10.3201/eid2802.212422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, Valdez R, Lauring AS, Sheng Z, Wang HH, Gordon A, Liu L, Ho DD. 2023. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186:279–286. doi: 10.1016/j.cell.2022.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yue C, Song W, Wang L, Jian F, Chen X, Gao F, Shen Z, Wang Y, Wang X, Cao Y. 2023. ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect Dis 23:278–280. doi: 10.1016/S1473-3099(23)00010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zou J, Kurhade C, Patel S, Kitchin N, Tompkins K, Cutler M, Cooper D, Yang Q, Cai H, Muik A, Zhang Y, Lee DY, Şahin U, Anderson AS, Gruber WC, Xie X, Swanson KA, Shi PY. 2023. Neutralization of BA.4-BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with bivalent vaccine. N Engl J Med 388:854–857. doi: 10.1056/NEJMc2214916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. CIDRAP . Available from: https://www.cidrap.umn.edu/covid-19/XBB.116-XBB.191-gain-more-ground-united-states)
- 9. WHO . Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants)
- 10. Li F. 2015. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89:1954–1964. doi: 10.1128/JVI.02615-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. doi: 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. Addendum: a pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588:E6. doi: 10.1038/s41586-020-2951-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:e00127-20. doi: 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li F, Li WH, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. doi: 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 16. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. doi: 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geng Q, Shi K, Ye G, Zhang W, Aihara H, Li F. 2022. Structural basis for human receptor recognition by SARS-CoV-2 omicron variant BA.1. J Virol 96:e0024922. doi: 10.1128/jvi.00249-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang W, Shi K, Geng Q, Ye G, Aihara H, Li F. 2022. Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant. Proc Natl Acad Sci U S A 119:e2206509119. doi: 10.1073/pnas.2206509119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mallapaty S. 2022. Where did omicron come from? Three key theories. Nature 602:26–28. doi: 10.1038/d41586-022-00215-2 [DOI] [PubMed] [Google Scholar]
- 20. Wang Y, Lenoch J, Kohler D, DeLiberto TJ, Tang CY, Li T, Tao YJ, Guan M, Compton S, Zeiss C, Hang J, Wan X-F, Lednicky JA. 2023. SARS-Cov-2 exposure in Norway rats (Rattus Norvegicus) from New York City. mBio 14:e0362122. doi: 10.1128/mbio.03621-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. doi: 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 22. Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, Han P, Li X, Peng Q, Su C, Huang B, Li D, Zhang R, Tian M, Fu L, Gao Y, Zhao X, Liu K, Qi J, Gao GF, Wang P. 2022. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 185:630–640. doi: 10.1016/j.cell.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qu P, Faraone JN, Evans JP, Zheng YM, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Panchal AR, Saif LJ, Oltz EM, Zhang B, Zhou T, Xu K, Gumina RJ, Liu SL. 2023. Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants. Cell Rep 42:112443. doi: 10.1016/j.celrep.2023.112443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu KL, Peng GQ, Wilken M, Geraghty RJ, Li F. 2012. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem 287:8904–8911. doi: 10.1074/jbc.M111.325803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vascon F, Gasparotto M, Giacomello M, Cendron L, Bergantino E, Filippini F, Righetto I. 2020. Protein electrostatics: from computational and structural analysis to discovery of functional fingerprints and biotechnological design. Comput Struct Biotechnol J 18:1774–1789. doi: 10.1016/j.csbj.2020.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stalls V, Lindenberger J, Gobeil SM-C, Henderson R, Parks R, Barr M, Deyton M, Martin M, Janowska K, Huang X, May A, Speakman M, Beaudoin E, Kraft B, Lu X, Edwards RJ, Eaton A, Montefiori DC, Williams WB, Saunders KO, Wiehe K, Haynes BF, Acharya P. 2022. Cryo-EM structures of SARS-CoV-2 omicron BA.2 spike. Cell Rep 39:111009. doi: 10.1016/j.celrep.2022.111009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao Y, Jian F, Wang J, Yu Y, Song W, Yisimayi A, Wang J, An R, Chen X, Zhang N, Wang Y, Wang P, Zhao L, Sun H, Yu L, Yang S, Niu X, Xiao T, Gu Q, Shao F, Hao X, Xu Y, Jin R, Shen Z, Wang Y, Xie XS. 2023. Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution. Nature 614:521–529. doi: 10.1038/s41586-022-05644-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geng Q, Tai W, Baxter VK, Shi J, Wan Y, Zhang X, Montgomery SA, Taft-Benz SA, Anderson EJ, Knight AC, Dinnon KH, Leist SR, Baric RS, Shang J, Hong S-W, Drelich A, Tseng C-T, Jenkins M, Heise M, Du L, Li F, Subbarao K. 2021. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog 17:e1009897. doi: 10.1371/journal.ppat.1009897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 30. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75:861–877. doi: 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5; Tables S1 to S4.
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank: accession number 8SPH for chimeric XBB.1 RBD complexed with human ACE2 and accession number 8SPI for chimeric XBB.1.5 RBD complexed with human ACE2.