ABSTRACT
SARS-CoV-2 can enter cells after its spike protein is cleaved by either type II transmembrane serine proteases (TTSPs), like TMPRSS2, or cathepsins. It is now widely accepted that the Omicron variant uses TMPRSS2 less efficiently and instead enters cells via cathepsins, but these findings have yet to be verified in more relevant cell models. Although we could confirm efficient cathepsin-mediated entry for Omicron in a monkey kidney cell line, experiments with protease inhibitors showed that Omicron (BA.1 and XBB1.5) did not use cathepsins for entry into human airway organoids and instead utilized TTSPs. Likewise, CRISPR-edited intestinal organoids showed that entry of Omicron BA.1 relied on the expression of the serine protease TMPRSS2 but not cathepsin L or B. Together, these data force us to rethink the concept that Omicron has adapted to cathepsin-mediated entry and indicate that TTSP inhibitors should not be dismissed as prophylactic or therapeutic antiviral strategy against SARS-CoV-2.
IMPORTANCE
Coronavirus entry relies on host proteases that activate the viral fusion protein, spike. These proteases determine the viral entry route, tropism, host range, and can be attractive drug targets. Whereas earlier studies using cell lines suggested that the Omicron variant of SARS-CoV-2 has changed its protease usage, from cell surface type II transmembrane serine proteases (TTSPs) to endosomal cathepsins, we report that this is not the case in human airway and intestinal organoid models, suggesting that host TTSP inhibition is still a viable prophylactic or therapeutic antiviral strategy against current SARS-CoV-2 variants and highlighting the importance of relevant human in vitro cell models.
KEYWORDS: SARS-CoV-2, Omicron, serine protease, TMPRSS2, cathepsin, airway organoids, intestinal organoids, CRISPR/Cas9, coronavirus
INTRODUCTION
In late 2021, the SARS-CoV-2 Omicron variant emerged and rapidly replaced the Delta variant globally. Omicron contained a highly mutated spike glycoprotein, which shows extensive antigenic drift. In the past 2 years, Omicron has continued to evolve into various subvariants that antigenically have drifted even further away from the initial SARS-CoV-2 variants (e.g., 614D/G, Alpha, Beta, and Delta), highlighting antibody neutralization as a major evolutionary pressure (1 - 4). Besides changing antigenically, Omicron variants may also have changed in other ways. Several studies showed that Omicron infects hamsters less efficiently and is less pathogenic in these animals (5 - 7). In contrast, in humans, Omicron appears to be more transmissible, with viral loads comparable to previous variants (8 - 10).
SARS-CoV-2 enters cells after proteolytic cleavage of spike to activate it for membrane fusion. This step in the replication cycle is crucial for virus infectivity and, therefore, is an attractive drug target. After spike contacts the target cell by binding to the receptor, ACE2, a motif directly adjacent to the fusion peptide in spike can be cleaved by either the type II transmembrane serine protease (TTSP) TMPRSS2 at the cell surface, or cathepsin B/L in the endosome (11, 12). Although many TTSPs can potentially be used when they are overexpressed (11, 13), SARS-CoV-2, as well as SARS-CoV and MERS-CoV, rely specifically on TMPRSS2 for entry into human intestinal organoids (12). In contrast, several studies have now shown that the SARS-CoV-2 Omicron (BA.1) variant that emerged at the end of 2021 has altered its preference for TMPRSS2 and instead uses endosomal cathepsins more efficiently (14 - 18). Multiple studies attribute the poor TMPRSS2 utilization of Omicron to its reduced cleavage of the S1/S2 site (also known as furin cleavage site), which is cleaved by the virus-producing cell. Although these findings have now become generally accepted (19, 20), they have yet to be verified in relevant cell models. This is important because drugs targeting TTSPs are promising antiviral therapies with a broad activity against coronaviruses and other virus families (13, 21 - 34) when administered early or prophylactically (35 - 39), and their dismissal could potentially harm further (pre-) clinical development. Therefore, we investigated which entry route SARS-CoV-2 Omicron takes in relevant cell models of the human airway and intestine.
RESULTS
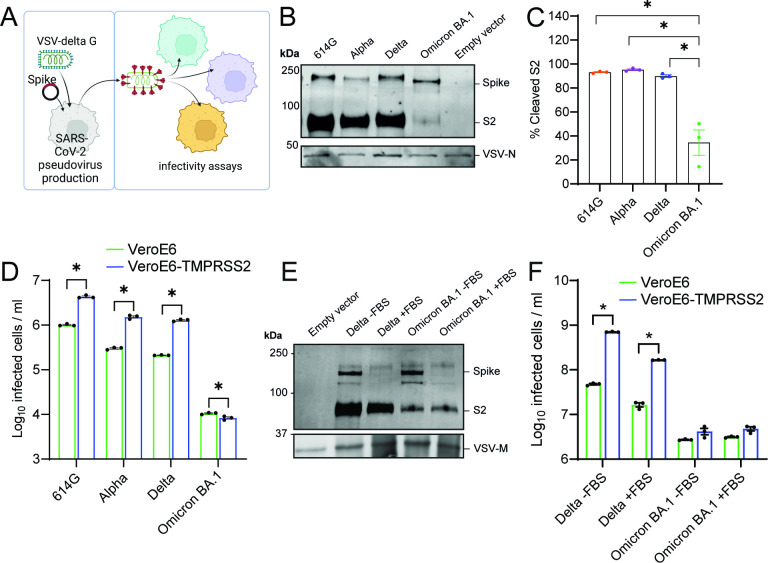
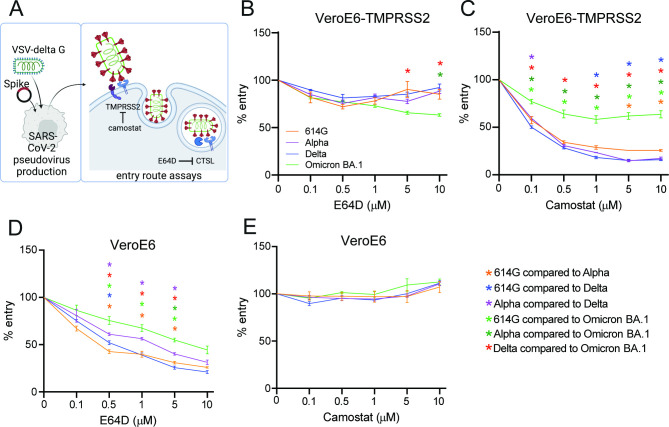
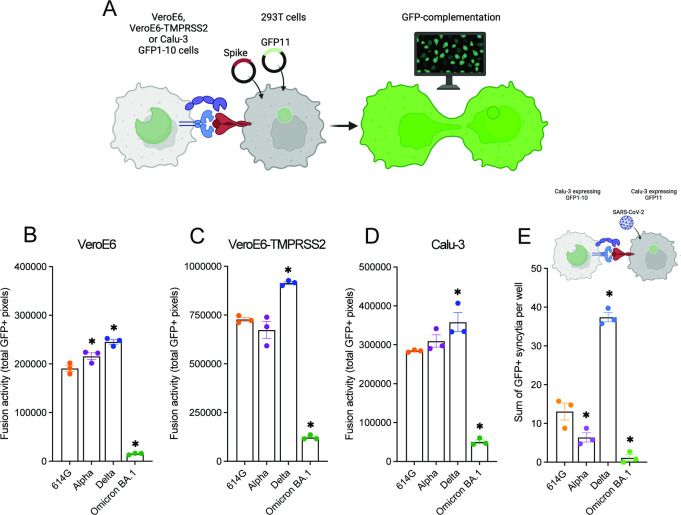
First, we investigated whether we could confirm that Omicron has switched entry pathway from TMPRSS2 to cathepsins. For this purpose, we used the monkey kidney cell line VeroE6, which possesses an active cathepsin-mediated entry pathway (25). Human TMPRSS2 can be expressed in these cells to establish an active TTSP-mediated entry pathway. We used these cells in combination with vesicular stomatitis virus (VSV)-based spike pseudoviruses to determine viral infectivity in the presence and absence of TMPRSS2 (Fig. 1A). We confirmed that spike is efficiently expressed in our pseudoviruses, although to a lesser extent for Omicron BA.1 and found that Omicron BA.1 spike is poorly cleaved (Fig. 1B and C). Ectopic TMPRSS2 expression in target cells effectively increased the infectivity of 614G, Alpha, and Delta pseudoviruses but not Omicron BA.1 pseudoviruses (Fig. 1D). Next, we determined whether decreased TMPRSS2 usage was due to inefficient S1/S2 cleavage. We found that during pseudovirus production, the presence of 10% fetal bovine serum (FBS) resulted in a cleaved Omicron BA.1 spike; however, despite this, cleaved Omicron BA.1 pseudoviruses did not infect VeroE6-TMPRSS2 cells more efficiently than VeroE6 cells (Fig. 1E and F). We confirmed Omicron’s low utilization of TMPRSS2 by investigating the entry route taken by different spike pseudoviruses (Fig. 2A). In VeroE6-TMPRSS2 cells, TMPRSS2-mediated entry was inhibited using the TTSP inhibitor camostat, whereas cathepsin B- and L-dependent entry was inhibited using E64D (Fig. 2B and C). Whereas 614G, Alpha, and Delta pseudoviruses entered these cells exclusively through TMPRSS2, Omicron BA.1 pseudoviruses used this entry route inefficiently, leading to partial cathepsin-mediated entry. In VeroE6 lacking TMPRSS2, all pseudotyped variants entered through the cathepsin-mediated pathway (Fig. 2D and E). These results confirm that Omicron BA.1 does not efficiently use TMPRSS2. We also confirmed earlier findings (14 - 16) that Omicron BA.1 was less fusogenic compared with earlier SARS-CoV-2 variants in a spike overexpression system (Fig. 3A through D) as well as a live virus fusion system using green fluorescent protein (GFP) complementation (Fig. 3E).
Fig 1.
Omicron BA.1 uses TMPRSS2 less efficiently regardless of pseudovirus spike cleavage status. (A) Graphical depiction of VSV pseudovirus production and subsequent infectivity assays (made with Biorender). (B) Immunoblotting of concentrated 614G, Alpha, Delta, and Omicron BA.1 pseudovirus stocks. (C) Quantification of blots in (B). (D) Infectivity of pseudoviruses in VeroE6 and VeroE6-TMPRSS2 cells. (E) Immunoblotting the S2 of Delta and Omicron BA.1 pseudovirus grown in the presence or absence of 10% FBS and concentrated by high-speed centrifugation. (F) Infectivity of pseudoviruses produced in (E) in VeroE6 and VeroE6-TMPRSS2 cells. Groups were compared by two-way ANOVA. *P < 0.05. Error bars depict SEM. Panels D and F indicate three technical replicates. Panel C indicates quantification from three independent pseudovirus productions.
Fig 2.
Omicron BA.1 partially utilizes the cathepsin-mediated entry pathway in VeroE6-TMPRSS2 cells, and all variants enter VeroE6 cells using the cathepsin-mediated pathway. (A) Graphical depiction of VSV pseudovirus production and subsequent entry route assays (made with Biorender). (B and C) Percentage entry of pseudoviruses in VeroE6-TMPRSS2 cells pretreated with a concentration range of either E64D (B) or camostat (C). (D and E) Percentage entry of pseudoviruses in VeroE6 cells pretreated with a concentration range of either E64D (D) or camostat (E). Groups were compared by two-way ANOVA. *P < 0.05. Error bars depict SEM. Panels B–E indicate three technical replicates.
Fig 3.
Omicron BA.1 is less fusogenic compared with earlier SARS-CoV-2 variants. (A) Graphical depiction of the GFP-complementation fusion assay (made with Biorender). (B to D) Quantification of 614G, Alpha, Delta, and Omicron BA.1 S-mediated cell-cell fusion by measuring the sum of all GFP + pixels per well on VeroE6, VeroE6-TMPRSS2, and Calu-3 cells 18 h post transfer. (E) Quantification of 614G, Alpha, Delta, and Omicron BA.1 authentic virus GFP-complementation fusion assay on Calu-3 cells by counting the sum of all GFP + syncytia per well at 24 h p.i. Groups were compared by one-way ANOVA comparing variants to 614G. *P < 0.05. Error bars depict SEM. Panels B−E indicate three technical replicates. A representative from at least two independent experiments is shown.
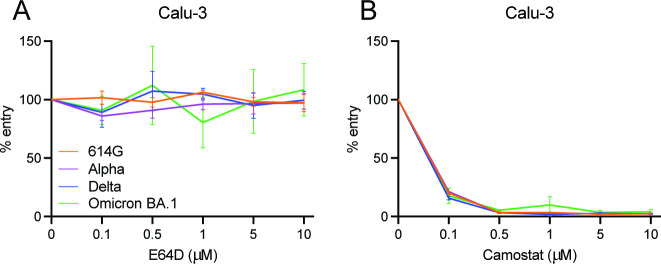
Next, we determined the infectivity of Omicron BA.1 pseudoviruses on Calu-3 cells, a human lung epithelial cell line in which the ancestral 614G SARS-CoV-2 is known to enter using TMPRSS2 (40) (Fig. S1A). In these cells, Omicron BA.1 pseudovirus entry was fully TTSP mediated (Fig. 4A and B; Fig. S1B and C). The data indicate that the preferred SARS-CoV-2 entry route depends on the type of cell line being used.
Fig 4.
Omicron BA.1 uses the TTSP-mediated entry route in Calu-3 cells. (A and B) Percentage entry of pseudoviruses in Calu-3 cells pretreated with a concentration range of either E64D (A) or camostat (B). Groups were compared by two-way ANOVA. Error bars depict SEM. Each panel indicates an experiment performed in triplicate.
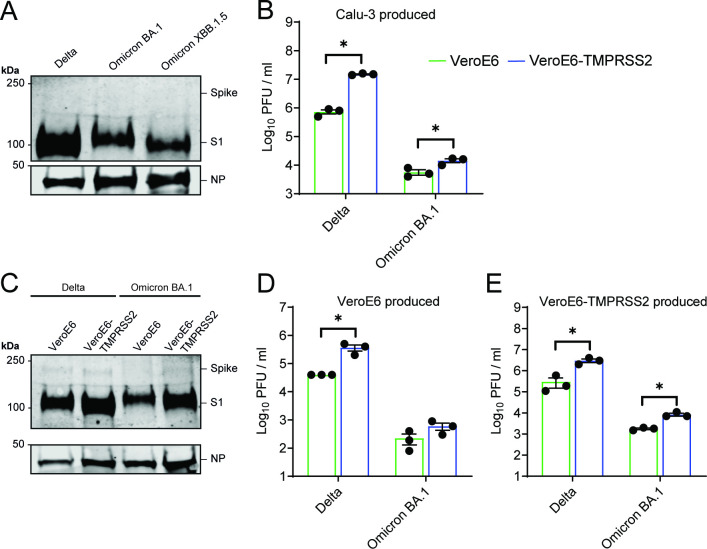
To confirm our findings with authentic SARS-CoV-2, we generated virus stocks on Calu-3 cells. We confirmed equal spike expression and observed efficient cleavage of all variants, including the currently dominant Omicron XBB1.5 variant (Fig. 5A; Fig. S2A and B; Fig. S3). Omicron BA.1 was only slightly more infectious on VeroE6-TMPRSS2 cells compared to VeroE6 cells, benefitting much less from TMPRSS2 than Delta (Fig. 5B). To confirm that efficient S1/S2 cleavage was not a result of the readily available TMPRSS2 on Calu-3 cells, we passaged Delta and Omicron BA.1 once on VeroE6 or VeroE6-TMPRSS2 cells and harvested the viruses. Regardless of the cell line used to propagate virus, spike cleavage of both variants remained unchanged, suggesting that TMPRSS2 is not essential for S1/S2 cleavage (Fig. 5C). Additionally, VeroE6 and VeroE6-TMPRSS2 Omicron BA.1 stocks were only slightly more infectious on VeroE6-TMPRSS2 cells compared to VeroE6 cells (Fig. 5D and E).
Fig 5.
Omicron BA.1 grown on Calu-3, VeroE6, or VeroE6-TMPRSS2 cells is equally cleaved and benefits less from TMPRSS2. (A) Immunoblotting the S1 of Delta, Omicron BA.1, and Omicron XBB.1.5 viruses grown on Calu-3 cells. (B) Viral titers on VeroE6 and VeroE6-TMPRSS2 cells of stocks produced on Calu-3 cells. (C) Immunoblotting of Delta and Omicron BA.1 stocks passaged once on VeroE6 or VeroE6-TMPRSS2 cells. (D and E) Viral titers on VeroE6 and VeroE6-TMPRSS2 cells of stocks produced on either VeroE6 (D) or VeroE6-TMPRSS2 (E) cells. Groups were compared using a two-way ANOVA. *P < 0.05. Error bars depict SEM. NP = nucleocapsid protein. Panels B, D–E indicate three technical replicates. Panel B is a representative from two independent experiments.
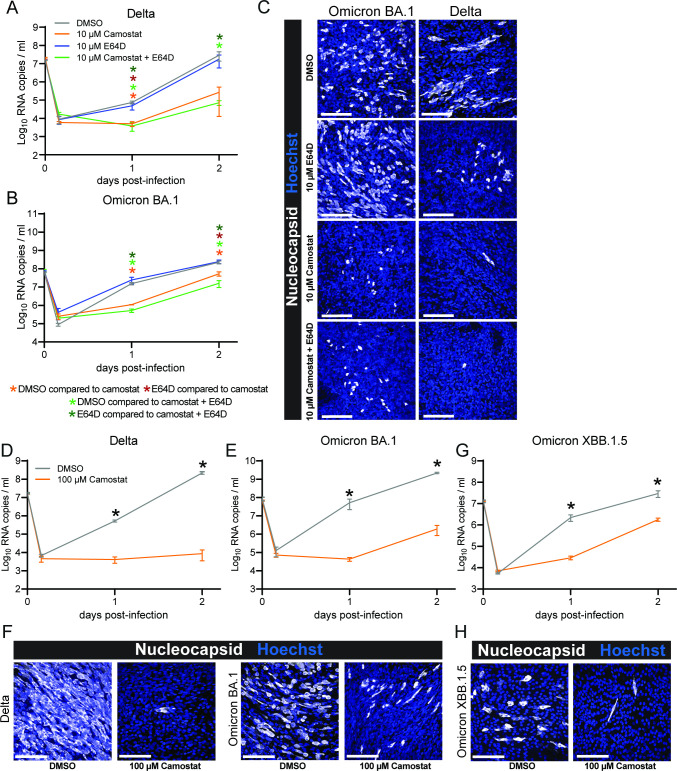
As different cell line models led to different results, we tested which entry route is used by Omicron and Delta authentic viruses in a more relevant cell model, human airway organoids. Both viruses were effectively inhibited by camostat (10 µM) but not E64D (10 µM). The combination of both resulted in similar kinetics as camostat alone (Fig. 6A through C), indicating that both viruses use the TTSP but not the cathepsin-mediated entry pathway in these cells. These findings were reproduced in a second experiment using a higher camostat concentration (100 µM) (Fig. 6D through F). Next, we investigated the currently dominant Omicron variant XBB1.5 (at the time of writing) and found that XBB1.5 infection was also inhibited by camostat in human airway organoids (100 µM) (Fig S3; Fig. 6G and H). These findings show that in human airway cells, Omicron variants still rely on a TTSP for entry. However, due to the broad activity of camostat against serine proteases (22) and the wide range of TTSPs expressed by airway cells (12), we could not determine whether Omicron specifically requires TMPRSS2 or other TTSPs.
Fig 6.
Omicron uses the TTSP-mediated entry route in air-liquid interface human airway organoids. (A and B) Replication of Delta (A) and Omicron BA.1 (B) in the presence of dimethyl sulfoxide (DMSO, vehicle), camostat (10 µM), E64D (10 µM), or the combination of camostat and E64D. (C) Representative images of 2 days post infection air-liquid interface human airway organoids from panels A and B stained for nuclei with Hoechst (blue) and stained for virus nucleocapsid (white). (D and E) Replication of Delta (D) and Omicron BA.1 (E) in the presence of DMSO (vehicle) or camostat (100 µM). (F) Representative images of 3 days post infection air-liquid interface human airway organoids from panels D and E stained for nuclei with Hoechst (blue) and stained for virus nucleocapsid (white). (G) Replication of Omicron XBB.1.5 in the presence of DMSO (vehicle) or camostat (100 µM). (H) Representative images of 2 days post infection air-liquid interface human airway organoids from panel G stained for nuclei with Hoechst (blue) and stained for virus nucleocapsid (white). Groups were compared by two-way ANOVA. *P < 0.05. Error bars depict SEM. Panels A–B, D–E, and G indicate three technical replicates. A representative from two independent experiments is shown.
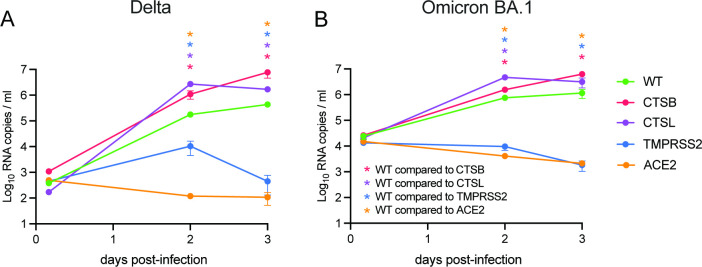
To confirm our findings in another relevant organoid system and to simultaneously address whether Omicron still specifically uses TMPRSS2 for entry, we made use of a human intestinal organoid CRISPR/Cas9 knockout biobank that we established previously (12). As a positive control, replication of both Delta and Omicron was completely inhibited when ACE2 was deleted (Fig. 7A and B). The deletion of TMPRSS2 had a similar effect to the deletion of ACE2, whereas the deletion of cathepsin L and B had no negative effect on replication. These findings indicate that in human intestinal cells Omicron still uses TMPRSS2.
Fig 7.
Omicron relies on TMPRSS2, and not cathepsin L or B, for entry into human intestinal organoids. (A and B) Replication of Delta (A) and Omicron (B) in CRISPR/Cas9-edited knockout clones deleted for the indicated genes. Groups were compared by two-way ANOVA. *P < 0.05. Error bars depict SEM. Each dot shows data from three individual organoid wells, whereby each time point represents three independent organoid wells.
DISCUSSION
For SARS-CoV-2 to enter a cell, its fusogenic spike glycoprotein needs to be proteolytically activated by host proteases. Early SARS-CoV-2 variants were shown to use the TTSP-mediated entry pathway for entry into human airway and intestinal cells, whereas cathepsin-mediated entry was only observed in some cell lines lacking TTSPs (12, 25, 41). In contrast, it is now widely accepted that the Omicron variant uses TTSPs less efficiently and instead enters cells via cathepsins (14 - 16), but this concept solely relies on data obtained in cell lines. Therefore, we aimed to verify this in more relevant cell models.
Here, we could replicate that Omicron indeed relied more on cathepsins for entry into VeroE6 cells ectopically expressing the serine protease TMPRSS2. In addition, the expression of TMPRSS2 did not increase the infectivity for Omicron, whereas it did for earlier SARS-CoV-2 variants. This confirmed earlier findings (14 - 16) that Omicron uses TMPRSS2 less efficiently, but in Calu-3 cells, Omicron still used the TTSP-mediated entry pathway and is not inhibited by E64D. In addition to blocking cathepsins, E64D inhibits cysteine proteinases and has been shown to inhibit Mouse hepatitis virus gene 1 polyprotein processing, resulting in reduced viral replication (42). However, E64D does not prevent SARS-CoV-2 viral replication in Calu-3 cells or airway organoids, suggesting that its role in blocking SARS-CoV-2 replication is limited at the concentrations used. Differences in entry routes in different cell lines raise the question: Which cell line model represents the human situation in vivo?
To address this, we used organoid models of the human airway and intestine. This unambiguously showed that the Delta and Omicron BA.1 and XBB1.5 variants still rely on TTSPs for airway cell entry and that cathepsins B and L were not involved.
A limitation of the airway model is that it is not amenable to genetic engineering using CRISPR/Cas9. Therefore, we were not able to test whether these variants specifically rely on TMPRSS2 in these cells. To solve this, we made use of a human intestinal organoid CRISPR/Cas9 knockout biobank that we established previously (12). Knocking out TMPRSS2 reduced viral replication of Delta and Omicron to levels similar to knocking out the entry receptor, ACE2, whereas deleting cathepsin L or B had no negative effect on replication. Our data are in line with recent findings showing that Omicron still relies on TMPRSS2 for replication in mice, but these findings may be murine specific since Omicron is human adapted and replicates poorly in mouse models (43, 44). One publication observed limited Omicron replication in human intestinal organoids compared to previously circulating variants; however, we observed no dramatic differences in replication of Omicron BA.1 and Delta (45). In fact, Omicron has been shown to cause gastrointestinal symptoms in some patients (46, 47). Nonetheless, Omicron variants predominantly replicate in the respiratory tract; therefore, the role of TMPRSS2 during infection of the airway epithelium should be elucidated in future studies. In particular, considering that a recent Genome-wide association study () has identified an association with the TMPRSS2 gene and COVID-19 disease severity (48). We conclude that there is no solid evidence for Omicron using the cathepsin-mediated entry pathway in relevant human cell models, despite the fact that we could replicate that Omicron uses cathepsin-mediated entry in VeroE6-TMPRSS2 cells. Instead, we provide ample evidence that Omicron variants still rely on the TTSP-mediated entry route for entry into relevant human cells. This includes the currently circulating XBB.1.5 variant (at the time of writing). Multiple studies have characterized the replication kinetics, binding, and entry into primary airway cultures of recent Omicron variants, such as BA.5 and XBB; however, few studies have assessed the entry route utilized by these variants (49 - 52). These data force us to rethink the current concept that Omicron has adapted to cathepsin-mediated entry and indicate that TTSP inhibitors should not be dismissed as prophylactic or therapeutic antiviral strategy against SARS-CoV-2.
MATERIALS AND METHODS
Cell lines
VeroE6 and Calu-3 cells were purchased from ATCC. VeroE6-TMPRSS2 cells were generated as described previously (25). Briefly, TMPRSS2 cDNA (Genscript, NM_005656; OHu13675D) was fused to a C-terminal HA tag and cloned into pQXCIH (Clontech) to obtain the vector used for transducing VeroE6 cells. Transduced cells were selected and maintained in medium containing hygromycin (Invitrogen). VeroE6 and VeroE6-TMPRSS2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), HEPES (20 mM, Lonza), sodium pyruvate (1 mM, Gibco), penicillin (100 IU/mL), and streptomycin (100 IU/mL) at 37°C in a humidified CO2 incubator. Calu-3 cells (ATCC HTB-55) were maintained in Opti-MEM I (1×) + GlutaMAX (Gibco) supplemented with 10% FBS, penicillin (100 IU/mL), and streptomycin (100 IU/mL) at 37°C in a humidified CO2 incubator. HEK-293T cells were cultured in DMEM supplemented with 10% FBS, sodium pyruvate (1 mM, Gibco), non-essential amino acids (1×, Lonza), penicillin (100 IU/mL), and streptomycin (100 IU/mL) at 37°C in a humidified CO2 incubator. TMPRSS2, GFP11, and GFP1-10 stable cells were maintained in a medium containing hygromycin (Invitrogen), puromycin (Invivogen), and geneticin (Invitrogen), respectively. Cell lines were tested negative for mycoplasma.
Organoids
Human adult airway stem cells were isolated from lung parenchyma as described previously using a protocol adapted from Sachs and colleagues (53, 54). Cells were differentiated using commercially available Pneumacult-ALI medium (complete base medium with 1× maintenance supplement; Stemcell) as described before (54). Cells were differentiated at the air-liquid interface for 3–4 weeks. Medium was replaced every 4–5 days. Adult human lung tissue was obtained from non-tumor lung tissue of patients undergoing lung resection surgery for lung cancer. The Medical Ethical Committee of the Erasmus MC Rotterdam granted permission for this study (METC 2012-512).
Human ileum intestinal organoids were grown and CRISPR/Cas9 edited as described before (12, 55).
Plasmids
Codon-optimized SARS-CoV Wuhan-D614G, Alpha, and Delta spike expression plasmids (pLV) were ordered from Invivogen. The pCAGGS-Omicron expression plasmid was kindly provided by Dr. Berend Jan Bosch. All spike expressing plasmids contained a deletion of the last 19 amino acids containing the Golgi retention signal of the SARS-CoV spike protein. The plasmids were used for spike pseudovirus production and the GFP-complementation fusion assay. The pCAGGS-β-Actin-P2A-7xGFP11-BFP plasmid was cloned into pQCXIP and used for retrovirus production and the subsequent generation of the GFP11 stable cells as previously described by Mykytyn et al. (25).
Pseudovirus assays
Pseudovirus production, infectivity, and entry assays were performed as described before (25). Briefly, pseudoviruses were generated using spike pseudotyping vectors from Invivogen. Pseudoviruses were either produced in the presence of 10% FBS or absence of serum. To compare the titers between these, pseudoviruses were concentrated as described below, and medium was replaced for Opti-MEM I (1×) + GlutaMAX without serum. Pseudoviruses expressing 614G, Alpha, Delta, and Omicron BA.1 spike were titrated in triplicate by preparing 10-fold serial dilutions in Opti-MEM I (1×) + GlutaMAX (Gibco). Thirty microliters of each dilution were transferred to 2 × 104 VeroE6 and VeroE6-TMPRSS2 cell monolayers or 8 × 104 Calu-3 cell monolayers in the same medium in a 96-well plate. The cells were incubated at 37°C overnight and then scanned on the Amersham Typhoon Biomolecular Imager (channel Cy2; resolution 10 µm; GE Healthcare). Pseudovirus entry routes were determined by pretreating VeroE6, VeroE6-TMPRSS2, and Calu-3 cell monolayers with a concentration range of camostat mesylate (Sigma) or E64D (MedChemExpress) diluted in Opti-MEM I (1×) + GlutaMAX (Gibco) for 2 h prior to infection with approximately 1 × 103 pseudoviruses. The cells were incubated at 37°C overnight and scanned on the Amersham Typhoon Biomolecular Imager (channel Cy2; resolution 10 mm; GE Healthcare). All pseudovirus experiments were quantified using ImageQuant TL 8.2 image analysis software (GE Healthcare).
Pseudovirus concentration
Pseudovirus stocks were concentrated on a 10% sucrose cushion (10% sucrose, 15 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA) at 20,000 × g for 1.5 h at 4°C. Supernatant was decanted, and particles were resuspended in Opti-MEM I (1×) + GlutaMAX to achieve a 100-fold concentration. Particles were used either for titrations, immunoblotting, or silver staining.
Western blots
The expression and cleavage of spike were assessed using immunoblotting. Pseudovirus samples were boiled in 1× Laemmli buffer containing 5% beta mercaptoethanol for 30 min, whereas virus stocks were boiled in 2× Laemmli buffer containing 10% beta mercaptoethanol for 30 min. All samples were used for dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis using 10% wt/vol TGX gels (Bio-Rad). Gels were run in Tris-glycine-SDS buffer at 80 V for 30 min, followed by 200 V for 45 min. Transfer was performed at 300 mA for 1 h onto 0.45 µm Immobilon-FL PVDF membranes in Tris-glycine buffer containing 20% methanol. Membranes were blocked in 5% milk in PBS containing 1% Tween. Spike was stained using either rabbit anti-SARS-CoV S1 (1:1,000, Sino Biological) or mouse anti-SARS-CoV-2 S2 (1:1,000, GeneTex). VSV-N was stained with mouse anti-VSV-N (1:1,000, Absolute Antibody), and SARS-CoV-2 nucleocapsid protein was stained with rabbit anti-SARS-CoV NP (1:1,000, Sino Biological). All blots were stained with infrared-labeled secondary antibodies goat anti-mouse or goat anti-rabbit (1:20,000, Licor). Western blots were scanned on an Odyssey CLx and analyzed using Image Studio Lite Ver 5.2 software.
Silver staining
Quality and yield of pseudovirus productions between batches were assessed using either immunoblotting against VSV-N or silver stains. Boiled samples used for immunoblotting were run on 10% wt/vol TGX gels as described above. Gels were transferred into ultrapure water, and silver stains were performed according to manufacturer’s instructions (Silver Stain for Mass Spectrometry Kit, Pierce). Images were taken on the Amersham AI600 (GE Healthcare).
Viruses
SARS-CoV-2 isolates, 614G/Bavpat-1 (OM304632), Alpha (MW947280), Delta (OM287123), BA.1 (OM287553), and XBB1.5 (GISAID: EPI_ISL_16640568), were grown to passage 3 on Calu-3 (ATCC HTB-55) cells in Advanced DMEM/F12 (Gibco), supplemented with HEPES, Glutamax, penicillin (100 IU/mL) (AdDF+++), and streptomycin (100 IU/mL) at 37°C in a humidified CO2 incubator. Where indicated an additional passage was done on VeroE6 or VeroE6-TMPRSS2 cells in the same medium. Virus isolates at passage 3 were deep sequenced and were confirmed to be genetically identical to the original material. Infections were performed at a multiplicity of infection (moi) of 0.01, and virus was harvested after 72 h. The culture supernatant was cleared by centrifugation at 1,000 × g for 5 min and stored at −80°C in aliquots.
For plaque assay titrations, virus stocks or experimental samples were thawed and diluted in 10-fold serial dilutions in 200 µL Opti-MEM I (1×) + GlutaMAX. Hundred microliters of each dilution were added to monolayers of 1 × 106 Calu-3 cells in the same medium in a 12-well plate. Cells were incubated with inoculums at 37°C for 4 h, and then medium was replaced for 1.2% Avicel (FMC biopolymers) in Opti-MEM I (1×) + GlutaMAX for 2 days. Next, cells were fixed in 4% formalin for 20 min, permeabilized in 70% ice-cold ethanol, and washed in PBS. Cells were blocked in 0.6% BSA (bovine serum albumin; Sigma) in PBS and stained with rabbit anti-nucleocapsid (Sino biological; 1:2,000) in PBS containing 0.6% BSA, washed thrice in PBS, and stained with goat anti-rabbit Alexa Fluor 488 (Invitrogen; 1:4,000) in PBS containing 0.6% BSA. Cells were washed twice in PBS, and plates were scanned on the Amersham Typhoon Biomolecular Imager (channel Cy2; resolution 10 µm; GE Healthcare). All staining steps were performed at room temperature for 1 h. Plaque assay analysis was performed using ImageQuant TL 8.2 software (GE Healthcare). All work with infectious SARS-CoV-2 was performed in a Class II Biosafety Cabinet under BSL-3 conditions at Erasmus Medical Center.
SARS-CoV-2 infections
Prior to SARS-CoV-2 infection of 2D airway organoid cultures, cells were washed three times with AdDF+++ medium. To assess the effect of camostat mesylate (Sigma) and E64D (MedChemExpress) on viral replication, cultures were pretreated in the basal and apical compartment for 1 h with either compound or the combination of both, or dimethyl sulfoxide (DMSO) in the negative control at the indicated concentrations. Cells were incubated with inoculums for 2–4 h at 37°C in a humidified CO2 incubator at an moi of 0.1. Washing was performed three times at 4 h post infection in AdDF+++ after which a fourth wash was performed and collected as the 4 h post infection baseline.
For 2D airway organoid cultures, samples were taken at the indicated time points as follows: 500 µL for 12 mm inserts or 200 µL for 6.5 mm inserts of AdDF+++ was added to the apical side of the cells, and cells were incubated for 10 min at 37°C in a humidified CO2 incubator after which supernatants were pipetted up and down on the cells twice and transferred to a microvial. All supernatant samples were stored at −80°C until further processing for viral titer determinations. After collecting, for experiments with camostat mesylate or E64D, the basal compartment medium was replaced daily with medium containing fresh compound.
Intestinal organoids infection experiments were performed as described before (12).
Immunofluorescent staining
Transwell inserts with 2D airway organoids were fixed in 4% formalin, permeabilized in 70% ethanol, and blocked for 60 min in blocking buffer (10% filtered normal goat serum; MP Biomedicals) in PBS. Cells were incubated with rabbit anti-nucleocapsid protein (1:500; Sino biological) overnight at 4°C in blocking buffer, washed twice with PBS, incubated with Alexa 488-conjugated anti-rabbit IgG (1:500; Invitrogen) in blocking buffer for 2 h at room temperature, washed two times with PBS, incubated with Hoechst (ThermoFisher) for 30 min, washed twice with PBS, and mounted in Prolong Antifade (Invitrogen) mounting medium. Images were taken on an LSM700 confocal microscope using ZEN software.
Determination of virus titers using qRT-PCR
SARS-CoV-2 RNA was extracted as described previously (55), and RNA genome copies (E-gene) were determined by qRT-PCR. Briefly, supernatant samples were thawed and centrifuged at 2,000 × g (supernatant) for 3 min. A sample of 60 µL was lysed in 90 µL MagnaPure LC Lysis buffer (Roche) at RT for 10 min. RNA was extracted by incubating samples with 50 µL Agencourt AMPure XP beads (Beckman Coulter) for 15 min at room temperature, washing beads twice with 70% ethanol on a DynaMag-96 magnet (Invitrogen) and eluting in 30 µL DEPC-treated water. RNA copies per mL were determined by qRT-PCR using primers targeting the E gene and comparing the Ct values to a counted standard curve derived from a 614G stock.
GFP complementation cell-cell fusion assays
The GFP complementation cell-cell fusion assay with transient spike protein expression was performed as previously described. Briefly, 1.5 µg pCAGGS-spike DNA and pCAGGS-β-Actin-P2A-7xGFP11-BFP DNA or empty vector DNA were transfected into HEK-293T cells with PEI (Polysciences) in a ratio of 1:3 (DNA: PEI). Transfected HEK-293T cells were transferred in three technical replicates to GFP1-10 expressing VeroE6, VeroE6-TMPRSS2, and Calu-3 cell monolayers in a ratio of 1:80 (HEK-293T cells:GFP1-10 expressing cells). Cell-cell fusion events were quantified at 18 h post transfer using Amersham Typhoon Biomolecular Imager (channel Cy2; resolution 10 µm; GE Healthcare). Data were analyzed using the ImageQuant TL 8.2 image analysis software (GE Healthcare) by calculating the sum of all GFP + pixels per well.
For the live/authentic virus GFP complementation cell-cell fusion assay, Calu-3 cells expressing GFP1-10 and Calu-3 cells expressing GFP11 were seeded in a 1:1 ratio in 96-well Cell Carrier Ultra plates (Perkin Elmer). The cells were then incubated at 37°C in a humidified CO2 incubator until confluent. Virus was diluted in twofold serial dilutions starting with 20,000 viral particles based on 8 h infectious viral titers in 200 µL Opti-MEM I (1×) + GlutaMAX. Hundred microliters of each dilution were added to the monolayers of 1 × 105 Calu-3 cells. Cells were incubated with inoculums at 37°C for 24 h in a humidified CO2 incubator, and cells were fixed in 4% paraformaldehyde for 20 min. The cells were washed three times in PBS and stained for Hoechst 33343 for 30 min and washed again three times in PBS. Next, cells were imaged using the Opera phenix spinning disk confocal HCS system (Perkin Elmer) equipped with a 10× air objective (NA 0.3) and 405 and 488 nm solid state lasers. Hoechst and GFP were detected using 435–480 nm and 500–550 nm emission filters, respectively. Nine fields per well were imaged covering approximately 50% of the individual wells. The sum of GFP positive syncytia per well were quantified using the Harmony software (version 4.9, Perkin Elmer). Cellular origin of the GFP positive areas was confirmed by eye, using the Hoechst labeled nuclei.
Statistical analysis
Statistical analysis was performed with the GraphPad Prism 9 software. Groups were compared by one- or two-way ANOVA, followed by a Benjamini and Hochberg (original FDR method; q = 0.05, whereby P is adjusted P-value.) multiple-comparison test, respectively.
ACKNOWLEDGMENTS
We thank Robbert Rottier for providing human adult lung material.
This work was supported by Netherlands Organization for Health Research and Development (10150062010008; B.L.H.), PPP allowance (LSHM19136; B.L.H.). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 874735 (VEO). This manuscript was part of the research program of the Netherlands Centre for One Health (NCOH).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conceptualization, M.M.L., B.L.H.; methodology, A.Z.M, T.I.B., M.M.L., M.E.R., J.B., M.G., H.C., P.D., D.S., R.A., J.Z.; formal analysis, A.Z.M, T.I.B., M.M.L.; investigation, A.Z.M, T.I.B., M.M.L., B.L.H.; resources, B.L.H.; writing – original draft, M.M.L.; writing – review and editing, A.Z.M, T.I.B., M.M.L., B.L.H., J.B., M.G., H.C., M.E.R., P.D., D.S., R.A., J.Z.
Contributor Information
Bart L. Haagmans, Email: b.haagmans@erasmusmc.nl.
Mart M. Lamers, Email: mart@duke-nus.edu.sg.
Stacey Schultz-Cherry, St Jude Children's Research Hospital, Memphis, Tennessee, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00851-23.
Figures S1 to S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Mykytyn AZ, Rissmann M, Kok A, Rosu ME, Schipper D, Breugem TI, van den Doel PB, Chandler F, Bestebroer T, de Wit M, van Royen ME, Molenkamp R, Oude Munnink BB, de Vries RD, GeurtsvanKessel C, Smith DJ, Koopmans MPG, Rockx B, Lamers MM, Fouchier RAM, Haagmans BL. 2022. Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Sci Immunol 7:eabq4450. doi: 10.1126/sciimmunol.abq4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mykytyn AZ, Rosu ME, Kok A, Rissmann M, van Amerongen G, Geurtsvankessel C, de Vries RD, Munnink BBO, Smith DJ, Koopmans MPG, Lamers MM, Fouchier RAM, Haagmans BL. 2023. Antigenic mapping of emerging SARS-CoV-2 Omicron variants BM.1.1.1, BQ.1.1, and XBB.1. Lancet Microbe 4:e294–e295. doi: 10.1016/S2666-5247(22)00384-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dejnirattisai W, Shaw RH, Supasa P, Liu C, Stuart AS, Pollard AJ, Liu X, Lambe T, Crook D, Stuart DI, Mongkolsapaya J, Nguyen-Van-Tam JS, Snape MD, Screaton GR, Com-COV2 study group . 2022. Reduced neutralisation of SARS-CoV-2 Omicron B.1.1.529 variant by post-immunisation serum. Lancet 399:234–236. doi: 10.1016/S0140-6736(21)02844-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. GeurtsvanKessel CH, Geers D, Schmitz KS, Mykytyn AZ, Lamers MM, Bogers S, Scherbeijn S, Gommers L, Sablerolles RSG, Nieuwkoop NN, Rijsbergen LC, van Dijk LLA, de Wilde J, Alblas K, Breugem TI, Rijnders BJA, de Jager H, Weiskopf D, van der Kuy PHM, Sette A, Koopmans MPG, Grifoni A, Haagmans BL, de Vries RD. 2022. Divergent SARS-CoV-2 Omicron–reactive T and B cell responses in COVID-19 vaccine recipients. Sci Immunol 7:eabo2202. doi: 10.1126/sciimmunol.abo2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, Darling TL, Joshi A, Loeber S, Singh G, Foster SL, Ying B, Case JB, Chong Z, Whitener B, Moliva J, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Warang P, Gagne M, Li R, Sakai-Tagawa Y, Liu Y, Larson D, Osorio JE, Hernandez-Ortiz JP, Henry AR, Ciuoderis K, Florek KR, Patel M, Odle A, Wong L-YR, Bateman AC, Wang Z, Edara V-V, Chong Z, Franks J, Jeevan T, Fabrizio T, DeBeauchamp J, Kercher L, Seiler P, Gonzalez-Reiche AS, Sordillo EM, Chang LA, van Bakel H, Simon V, Douek DC, Sullivan NJ, Thackray LB, Ueki H, Yamayoshi S, Imai M, Perlman S, Webby RJ, Seder RA, Suthar MS, García-Sastre A, Schotsaert M, Suzuki T, Boon ACM, Diamond MS, Kawaoka Y, Consortium Mount Sinai Pathogen Surveillance (PSP) study group . 2022. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603:687–692. doi: 10.1038/s41586-022-04441-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuan S, Ye Z-W, Liang R, Tang K, Zhang AJ, Lu G, Ong CP, Man Poon VK, Chan C-S, Mok B-Y, Qin Z, Xie Y, Chu A-H, Chan W-M, Ip JD, Sun H, Tsang J-L, Yuen T-T, Chik K-H, Chan C-Y, Cai J-P, Luo C, Lu L, Yip C-Y, Chu H, To K-W, Chen H, Jin D-Y, Yuen K-Y, Chan J-W. 2022. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science 377:428–433. doi: 10.1126/science.abn8939 [DOI] [PubMed] [Google Scholar]
- 7. Boon ACM, Darling TL, Halfmann PJ, Franks J, Webby RJ, Barouch DH, Port JR, Munster VJ, Diamond MS, Kawaoka Y, Dvorin JD. 2022. Reduced airborne transmission of SARS-CoV-2 BA.1 Omicron virus in Syrian hamsters. PLoS Pathog 18:e1010970. doi: 10.1371/journal.ppat.1010970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puhach O, Adea K, Hulo N, Sattonnet P, Genecand C, Iten A, Jacquérioz F, Kaiser L, Vetter P, Eckerle I, Meyer B. 2022. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS-CoV-2. Nat Med 28:1491–1500. doi: 10.1038/s41591-022-01816-0 [DOI] [PubMed] [Google Scholar]
- 9. Boucau J, Marino C, Regan J, Uddin R, Choudhary MC, Flynn JP, Chen G, Stuckwisch AM, Mathews J, Liew MY, Singh A, Lipiner T, Kittilson A, Melberg M, Li Y, Gilbert RF, Reynolds Z, Iyer SL, Chamberlin GC, Vyas TD, Goldberg MB, Vyas JM, Li JZ, Lemieux JE, Siedner MJ, Barczak AK. 2022. Duration of shedding of culturable virus in SARS-CoV-2 Omicron (BA.1) infection. N Engl J Med 387:275–277. doi: 10.1056/NEJMc2202092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuasa S, Nakajima J, Takatsuki Y, Takahashi Y, Tani-Sassa C, Iwasaki Y, Nagano K, Sonobe K, Yoshimoto T, Nukui Y, Takeuchi H, Tanimoto K, Tanaka Y, Kimura A, Ichimura N, Tohda S. 2022. Viral load of SARS-CoV-2 Omicron is not high despite its high infectivity. J Med Virol 94:5543–5546. doi: 10.1002/jmv.27974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beumer J, Geurts MH, Lamers MM, Puschhof J, Zhang J, van der Vaart J, Mykytyn AZ, Breugem TI, Riesebosch S, Schipper D, van den Doel PB, de Lau W, Pleguezuelos-Manzano C, Busslinger G, Haagmans BL, Clevers H. 2021. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat Commun 12:5498. doi: 10.1038/s41467-021-25729-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Arora P, Sørensen LK, Søgaard OS, Hasselstrøm JB, Winkler M, Hempel T, Raich L, Olsson S, Danov O, Jonigk D, Yamazoe T, Yamatsuta K, Mizuno H, Ludwig S, Noé F, Kjolby M, Braun A, Sheltzer JM, Pöhlmann S. 2021. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 65:103255. doi: 10.1016/j.ebiom.2021.103255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meng B, Abdullahi A, Ferreira I, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, Zepeda SK, Papa G, Kemp SA, Ikeda T, Toyoda M, Tan TS, Kuramochi J, Mitsunaga S, Ueno T, Shirakawa K, Takaori-Kondo A, Brevini T, Mallery DL, Charles OJ, CITIID-NIHR BioResource COVID-19 Collaboration, Genotype to Phenotype Japan (G2P-Japan) Consortium, Ecuador-COVID19 Consortium, Bowen JE, Joshi A, Walls AC, Jackson L, Martin D, Smith KGC, Bradley J, Briggs JAG, Choi J, Madissoon E, Meyer KB, Mlcochova P, Ceron-Gutierrez L, Doffinger R, Teichmann SA, Fisher AJ, Pizzuto MS, de Marco A, Corti D, Hosmillo M, Lee JH, James LC, Thukral L, Veesler D, Sigal A, Sampaziotis F, Goodfellow IG, Matheson NJ, Sato K, Gupta RK. 2022. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603:706–714. doi: 10.1038/s41586-022-04474-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D, Scott S, Logan N, Ashraf S, Manali M, Szemiel A, Cowton V, Vink E, Harvey WT, Davis C, Asamaphan P, Smollett K, Tong L, Orton R, Hughes J, Holland P, Silva V, Pascall DJ, Puxty K, da Silva Filipe A, Yebra G, Shaaban S, Holden MTG, Pinto RM, Gunson R, Templeton K, Murcia PR, Patel AH, Klenerman P, Dunachie S, PITCH Consortium, COVID-19 Genomics UK (COG-UK) Consortium, Haughney J, Robertson DL, Palmarini M, Ray S, Thomson EC. 2022. Publisher correction: SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol 7:1709. doi: 10.1038/s41564-022-01241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du X, Tang H, Gao L, Wu Z, Meng F, Yan R, Qiao S, An J, Wang C, Qin F-F. 2022. Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct Target Ther 7:45. doi: 10.1038/s41392-022-00903-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shuai H, Chan J-W, Hu B, Chai Y, Yuen T-T, Yin F, Huang X, Yoon C, Hu J-C, Liu H, Shi J, Liu Y, Zhu T, Zhang J, Hou Y, Wang Y, Lu L, Cai J-P, Zhang AJ, Zhou J, Yuan S, Brindley MA, Zhang B-Z, Huang J-D, To K-W, Yuen K-Y, Chu H. 2022. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603:693–699. doi: 10.1038/s41586-022-04442-5 [DOI] [PubMed] [Google Scholar]
- 18. Zhao H, Lu L, Peng Z, Chen L-L, Meng X, Zhang C, Ip JD, Chan W-M, Chu A-H, Chan K-H, Jin D-Y, Chen H, Yuen K-Y, To K-W. 2022. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 11:277–283. doi: 10.1080/22221751.2021.2023329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pia L, Rowland-Jones S. 2022. Omicron entry route. Nat Rev Immunol 22:144. doi: 10.1038/s41577-022-00681-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomson EC, Willett BJ. 2022. Omicron: a shift in the biology of SARS-CoV-2. Nat Microbiol 7:1114–1115. doi: 10.1038/s41564-022-01149-1 [DOI] [PubMed] [Google Scholar]
- 21. Chupp G, Spichler-Moffarah A, Søgaard OS, Esserman D, Dziura J, Danzig L, Chaurasia R, Patra KP, Salovey A, Nunez A, May J, Astorino L, Patel A, Halene S, Wang J, Hui P, Patel. P, Lu J, Li F, Gan G, Parziale S, Katsovich L, Desir GV, Vinetz JM. 2022. A phase 2 randomized, double-blind, placebo-controlled trial of oral camostat mesylate for early treatment of COVID-19 outpatients showed shorter illness course and attenuation of loss of smell and taste. medRxiv. doi: 10.1101/2022.01.28.22270035 [DOI]
- 22. Breining P, Frølund AL, Højen JF, Gunst JD, Staerke NB, Saedder E, Cases-Thomas M, Little P, Nielsen LP, Søgaard OS, Kjolby M. 2021. Camostat mesylate against SARS-CoV-2 and COVID-19-rationale, dosing and safety. Basic Clin Pharmacol Toxicol 128:204–212. doi: 10.1111/bcpt.13533 [DOI] [PubMed] [Google Scholar]
- 23. Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R Jr, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G. 2015. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 116:76–84. doi: 10.1016/j.antiviral.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karolyi M, Pawelka E, Omid S, Koenig F, Kauer V, Rumpf B, Hoepler W, Kuran A, Laferl H, Seitz T, Traugott M, Rathkolb V, Mueller M, Abrahamowicz A, Schoergenhofer C, Hecking M, Assinger A, Wenisch C, Zeitlinger M, Jilma B, Zoufaly A. 2022. Camostat mesylate versus lopinavir/ritonavir in hospitalized patients with COVID-19-results from a randomized, controlled, open label, platform trial (ACOVACT). Front Pharmacol 13:870493. doi: 10.3389/fphar.2022.870493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mykytyn AZ, Breugem TI, Riesebosch S, Schipper D, van den Doel PB, Rottier RJ, Lamers MM, Haagmans BL. 2021. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. Elife 10. doi: 10.7554/eLife.64508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shirato K, Kawase M, Matsuyama S. 2013. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87:12552–12561. doi: 10.1128/JVI.01890-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Böttcher E, Matrosovich T, Beyerle M, Klenk H-D, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80:9896–9898. doi: 10.1128/JVI.01118-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84:12658–12664. doi: 10.1128/JVI.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abe M, Tahara M, Sakai K, Yamaguchi H, Kanou K, Shirato K, Kawase M, Noda M, Kimura H, Matsuyama S, Fukuhara H, Mizuta K, Maenaka K, Ami Y, Esumi M, Kato A, Takeda M. 2013. TMPRSS2 is an activating protease for respiratory parainfluenza viruses. J Virol 87:11930–11935. doi: 10.1128/JVI.01490-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bertram S, Dijkman R, Habjan M, Heurich A, Gierer S, Glowacka I, Welsch K, Winkler M, Schneider H, Hofmann-Winkler H, Thiel V, Pöhlmann S. 2013. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol 87:6150–6160. doi: 10.1128/JVI.03372-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, Nakajima N, Sekizuka T, Shirato K, Suzaki Y, Ainai A, Nakatsu Y, Kanou K, Nakamura K, Suzuki T, Komase K, Nobusawa E, Maenaka K, Kuroda M, Hasegawa H, Kawaoka Y, Tashiro M, Takeda M. 2014. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 88:5608–5616. doi: 10.1128/JVI.03677-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. 2019. TMPRSS2 contributes to virus spread and Immunopathology in the airways of murine models after coronavirus infection. J Virol 93:e01815-18. doi: 10.1128/JVI.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neary M, Box H, Sharp J, Tatham L, Curley P, Herriott J, Kijak E, Arshad U, Hobson JJ, Rajoli R, Pertinez H, Valentijn A, Dhaliwal K, McCaughan F, Rannard SP, Kipar A, Stewart JP, Owen A. 2021. Evaluation of intranasal nafamostat or camostat for SARS-CoV-2 chemoprophylaxis in Syrian golden hamsters. bioRxiv:2021.07.08.451654. doi: 10.1101/2021.07.08.451654 [DOI] [Google Scholar]
- 34. Lee MG, Kim KH, Park KY, Kim JS. 1996. Evaluation of anti-influenza effects of camostat in mice infected with non-adapted human influenza viruses. Arch Virol 141:1979–1989. doi: 10.1007/BF01718208 [DOI] [PubMed] [Google Scholar]
- 35. Quinn TM, Gaughan EE, Bruce A, Antonelli J, O’Connor R, Li F, McNamara S, Koch O, MacKintosh C, Dockrell D, Walsh T, Blyth KG, Church C, Schwarze J, Boz C, Valanciute A, Burgess M, Emanuel P, Mills B, Rinaldi G, Hardisty G, Mills R, Findlay EG, Jabbal S, Duncan A, Plant S, Marshall ADL, Young I, Russell K, Scholefield E, Nimmo AF, Nazarov IB, Churchill GC, McCullagh JSO, Ebrahimi KH, Ferrett C, Templeton K, Rannard S, Owen A, Moore A, Finlayson K, Shankar-Hari M, Norrie J, Parker RA, Akram AR, Anthony DC, Dear JW, Hirani N, Dhaliwal K. 2022. Randomised controlled trial of intravenous nafamostat mesylate in COVID pneumonitis: phase 1B/2A experimental study to investigate safety, pharmacokinetics and pharmacodynamics. EBioMedicine 76:103856. doi: 10.1016/j.ebiom.2022.103856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gunst JD, Staerke NB, Pahus MH, Kristensen LH, Bodilsen J, Lohse N, Dalgaard LS, Brønnum D, Fröbert O, Hønge B, Johansen IS, Monrad I, Erikstrup C, Rosendal R, Vilstrup E, Mariager T, Bove DG, Offersen R, Shakar S, Cajander S, Jørgensen NP, Sritharan SS, Breining P, Jespersen S, Mortensen KL, Jensen ML, Kolte L, Frattari GS, Larsen CS, Storgaard M, Nielsen LP, Tolstrup M, Sædder EA, Østergaard LJ, Ngo HTT, Jensen MH, Højen JF, Kjolby M, Søgaard OS. 2021. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with COVID-19-a double-blind randomized controlled trial. EClinicalMedicine 35:100849. doi: 10.1016/j.eclinm.2021.100849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kinoshita T, Shinoda M, Nishizaki Y, Shiraki K, Hirai Y, Kichikawa Y, Tsushima K, Shinkai M, Komura N, Yoshida K, Kido Y, Kakeya H, Uemura N, Kadota J. 2022. A multicenter, double-blind, randomized, parallel-group, placebo-controlled study to evaluate the efficacy and safety of mamostat mesilate in patients with COVID-19 (CANDLE study). BMC Med 20:342. doi: 10.1186/s12916-022-02518-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim YS, Jeon SH, Kim J, Koh JH, Ra SW, Kim JW, Kim Y, Kim CK, Shin YC, Kang BD, Kang SJ, Park CH, Lee B, Lee JY, Lee CH, Choi JP, Kim JY, Yu SN, Peck KR, Kim SH, Heo JY, Kim HA, Park HJ, Choi J, Han J, Kim J, Kim HJ, Han SH, Yoon A, Park M, Park S, Kim Y, Jung M, Oh MD. 2023. A double-blind, randomized, placebo-controlled, phase II clinical study to evaluate the efficacy and safety of camostat mesylate (DWJ1248) in adult patients with mild to moderate COVID-19. Antimicrob Agents Chemother 67:e0045222. doi: 10.1128/aac.00452-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shapira T, Monreal IA, Dion SP, Buchholz DW, Imbiakha B, Olmstead AD, Jager M, Désilets A, Gao G, Martins M, Vandal T, Thompson CAH, Chin A, Rees WD, Steiner T, Nabi IR, Marsault E, Sahler J, Diel DG, Van de Walle GR, August A, Whittaker GR, Boudreault P-L, Leduc R, Aguilar HC, Jean F. 2022. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 605:340–348. doi: 10.1038/s41586-022-04661-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang S-M, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen H, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB. 2021. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184:76–91. doi: 10.1016/j.cell.2020.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lamers MM, Mykytyn AZ, Breugem TI, Wang Y, Wu DC, Riesebosch S, van den Doel PB, Schipper D, Bestebroer T, Wu NC, Haagmans BL. 2021. Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. Elife 10:e66815. doi: 10.7554/eLife.66815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim JC, Spence RA, Currier PF, Lu X, Denison MR. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64D. Virology 208:1–8. doi: 10.1006/viro.1995.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Metzdorf K, Jacobsen H, Greweling-Pils MC, Hoffmann M, Lüddecke T, Miller F, Melcher L, Kempf AM, Nehlmeier I, Bruder D, Widera M, Ciesek S, Pöhlmann S, Čičin-Šain L. 2023. TMPRSS2 is essential for SARS-CoV-2 Beta and Omicron infection. Viruses 15:271. doi: 10.3390/v15020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Iwata-Yoshikawa N, Kakizaki M, Shiwa-Sudo N, Okura T, Tahara M, Fukushi S, Maeda K, Kawase M, Asanuma H, Tomita Y, Takayama I, Matsuyama S, Shirato K, Suzuki T, Nagata N, Takeda M. 2022. Essential role of TMPRSS2 in SARS-CoV-2 infection in murine airways. Nat Commun 13:6100. doi: 10.1038/s41467-022-33911-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyakawa K, Machida M, Kawasaki T, Nishi M, Akutsu H, Ryo A. 2022. Reduced replication efficacy of severe acute respiratory syndrome coronavirus 2 Omicron variant in "mini-gut" organoids. Gastroenterology 163:514–516. doi: 10.1053/j.gastro.2022.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maruyama S, Wada D, Oishi T, Saito F, Yoshiya K, Nakamori Y, Kuwagata Y. 2023. A descriptive study of abdominal complications in patients with mild COVID-19 presenting to the emergency department: a single-center experience in Japan during the Omicron variant phase. BMC Gastroenterol 23:43. doi: 10.1186/s12876-023-02681-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Menni C, Valdes AM, Polidori L, Antonelli M, Penamakuri S, Nogal A, Louca P, May A, Figueiredo JC, Hu C, Molteni E, Canas L, Österdahl MF, Modat M, Sudre CH, Fox B, Hammers A, Wolf J, Capdevila J, Chan AT, David SP, Steves CJ, Ourselin S, Spector TD. 2022. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of Omicron and Delta variant dominance: a prospective observational study from the ZOE COVID study. Lancet 399:1618–1624. doi: 10.1016/S0140-6736(22)00327-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pairo-Castineira E, Rawlik K, Bretherick AD, Qi T, Wu Y, Nassiri I, McConkey GA, Zechner M, Klaric L, Griffiths F, Oosthuyzen W, Kousathanas A, Richmond A, Millar J, Russell CD, Malinauskas T, Thwaites R, Morrice K, Keating S, Maslove D, Nichol A, Semple MG, Knight J, Shankar-Hari M, Summers C, Hinds C, Horby P, Ling L, McAuley D, Montgomery H, Openshaw PJM, Begg C, Walsh T, Tenesa A, Flores C, Riancho JA, Rojas-Martinez A, Lapunzina P, Yang J, Ponting CP, Wilson JF, Vitart V, Abedalthagafi M, Luchessi AD, Parra EJ, Cruz R, Carracedo A, Fawkes A, Murphy L, Rowan K, Pereira AC, Law A, Fairfax B, Hendry SC, Baillie JK, GenOMICC Investigators, SCOURGE Consortium, ISARICC Investigators, 23andMe COVID-19 Team . 2023. GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature 619:764–768. doi: 10.1038/s41586-023-06383-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kimura I, Yamasoba D, Tamura T, Nao N, Suzuki T, Oda Y, Mitoma S, Ito J, Nasser H, Zahradnik J, Uriu K, Fujita S, Kosugi Y, Wang L, Tsuda M, Kishimoto M, Ito H, Suzuki R, Shimizu R, Begum MM, Yoshimatsu K, Kimura KT, Sasaki J, Sasaki-Tabata K, Yamamoto Y, Nagamoto T, Kanamune J, Kobiyama K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Shirakawa K, Takaori-Kondo A, Kuramochi J, Schreiber G, Ishii KJ, Hashiguchi T, Ikeda T, Saito A, Fukuhara T, Tanaka S, Matsuno K, Sato K. 2022. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 185:3992–4007. doi: 10.1016/j.cell.2022.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamura T, Ito J, Uriu K, Zahradnik J, Kida I, Anraku Y, Nasser H, Shofa M, Oda Y, Lytras S, Nao N, Itakura Y, Deguchi S, Suzuki R, Wang L, Begum MM, Kita S, Yajima H, Sasaki J, Sasaki-Tabata K, Shimizu R, Tsuda M, Kosugi Y, Fujita S, Pan L, Sauter D, Yoshimatsu K, Suzuki S, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Yamamoto Y, Nagamoto T, Schreiber G, Maenaka K, Phenotype Japan C, Hashiguchi T, Ikeda T, Fukuhara T, Saito A, Tanaka S, Matsuno K, Takayama K, Sato K. 2023. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat Commun 14:2800. doi: 10.1038/s41467-023-38435-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li C, Huang J, Yu Y, Wan Z, Chiu MC, Liu X, Zhang S, Cai J-P, Chu H, Li G, Chan JF-W, To KK-W, Yang Z, Jiang S, Yuen K-Y, Clevers H, Zhou J. 2023. Human airway and nasal organoids reveal escalating replicative fitness of SARS-CoV-2 emerging variants. Proc Natl Acad Sci U S A 120:e2300376120. doi: 10.1073/pnas.2300376120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chiu MC, Zhang S, Li C, Liu X, Yu Y, Huang J, Wan Z, Zhu X, Zhou J. 2023. Apical-out human airway organoids modeling SARS-CoV-2 infection. Viruses 15:1166. doi: 10.3390/v15051166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Böttinger L, Klay D, Weeber F, Huelsz-Prince G, Iakobachvili N, Amatngalim GD, de Ligt J, van Hoeck A, Proost N, Viveen MC, Lyubimova A, Teeven L, Derakhshan S, Korving J, Begthel H, Dekkers JF, Kumawat K, Ramos E, van Oosterhout MF, Offerhaus GJ, Wiener DJ, Olimpio EP, Dijkstra KK, Smit EF, van der Linden M, Jaksani S, van de Ven M, Jonkers J, Rios AC, Voest EE, van Moorsel CH, van der Ent CK, Cuppen E, van Oudenaarden A, Coenjaerts FE, Meyaard L, Bont LJ, Peters PJ, Tans SJ, van Zon JS, Boj SF, Vries RG, Beekman JM, Clevers H. 2019. Long-term expanding human airway organoids for disease modeling. EMBO J 38:e100300. doi: 10.15252/embj.2018100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lamers MM, van der Vaart J, Knoops K, Riesebosch S, Breugem TI, Mykytyn AZ, Beumer J, Schipper D, Bezstarosti K, Koopman CD, Groen N, Ravelli RBG, Duimel HQ, Demmers JAA, Verjans GMGM, Koopmans MPG, Muraro MJ, Peters PJ, Clevers H, Haagmans BL. 2021. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J 40:e105912. doi: 10.15252/embj.2020105912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. 2020. SARS-CoV-2 productively infects human gut enterocytes. Science 369:50–54. doi: 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S3.