ABSTRACT
Arthropod-borne viruses (arboviruses) are an emerging and evolving global public health threat, with limited antiviral treatments or vaccines available. La Crosse virus (LACV) from the Bunyavirales order is responsible for pediatric encephalitis cases in the United States, yet little is known about the infectivity of LACV. Given the structural similarities between class II fusion glycoproteins of LACV and chikungunya virus (CHIKV), an alphavirus from the Togaviridae family, we hypothesized that LACV would share similar entry mechanisms with CHIKV. To test this hypothesis, we performed cholesterol-depletion and repletion assays and used cholesterol-modulating compounds to study LACV entry and replication. We found that LACV entry was cholesterol dependent, while replication was less affected by cholesterol manipulation. In addition, we generated single-point mutants in the LACV Gc ij loop that corresponded to known CHIKV residues important for virus entry. We found that a conserved histidine and alanine residue in the Gc ij loop impaired virus infectivity and attenuated LACV replication in vitro and in vivo. Finally, we took an evolution-based approach to explore how the LACV glycoprotein evolves in mosquitoes and mice. We found multiple variants that cluster in the Gc glycoprotein head domain, providing evidence for the Gc glycoprotein as a contributor to LACV adaptation. Together, these results begin to characterize the mechanisms of LACV infectivity and how the LACV glycoprotein contributes to replication and pathogenesis.
IMPORTANCE
Vector-borne viruses are significant health threats that lead to devastating disease worldwide. The emergence of arboviruses, in addition to the lack of effective antivirals or vaccines, highlights the need to study how arboviruses replicate at the molecular level. One potential antiviral target is the class II fusion glycoprotein. Alphaviruses, flaviviruses, and bunyaviruses encode a class II fusion glycoprotein that contains strong structural similarities at the tip of domain II. Here, we show that the bunyavirus La Crosse virus uses a cholesterol-dependent entry pathway similar to the alphavirus chikungunya virus, and residues in the ij loop are important for virus infectivity in vitro and replication in mice. These studies show that genetically diverse viruses may use similar pathways through conserved structure domains, suggesting that these viruses may be targets for broad-spectrum antivirals in multiple arboviral families.
KEYWORDS: bunyavirus, La Crosse, glycoprotein, adaptation, entry
INTRODUCTION
Arthropod-borne viruses (arboviruses) are significant human pathogens with epidemic potential (1 - 3). Arboviruses infect and replicate within humans and insects, yet the molecular mechanisms these viruses use for infection and replication are not completely understood. Arboviruses include a long list of human pathogens that span multiple virus families. These families include the positive-strand RNA alphaviruses and flaviviruses, as well as the segmented, negative-strand RNA bunyaviruses. While our knowledge of how alphaviruses and flaviviruses infect and replicate in mammals and insects has expanded in response to large outbreaks, little is known about how bunyaviruses infect these very disparate hosts.
The Bunyavirales order includes a number of significant human pathogens such as Rift Valley fever virus, La Crosse virus (LACV), Oropouche virus, and Crimean Congo hemorrhagic fever virus (4, 5). La Crosse virus is a member of the orthobunyavirus genus and is an enveloped, segmented, and negative-strand RNA virus that contains a tripartite genome consisting of S, M, and L segments. The S segment encodes for the nucleoprotein (N) and NSs protein (4, 6). The M segment encodes for the viral glycoproteins Gn and Gc (7, 8) and nonstructural protein NSm. The L segment encodes for the viral RNA-dependent RNA polymerase (4). LACV is transmitted by the Aedes species of mosquito (9, 10) and can cause severe encephalitis, as it is the most common cause of pediatric neuroinvasive arboviral disease in children in the United States (11 - 14).
While our knowledge of bunyavirus entry is limited, we can use what is known from alphaviruses and flaviviruses to infer how bunyaviruses establish infections. Alphaviruses, flaviviruses, and bunyaviruses encode class II fusion glycoproteins required for assembly, entry, and virion stability (15 - 17). While the amino acid sequences between viral families show minimal sequence homology, the class II glycoproteins of these three genetically distant viral families share striking structural homology in domain II, which includes the fusion loop required for membrane fusion (17) (Fig. 1A). In addition, each class II fusion protein contains an ij and bc loop flanking the fusion loops. Extensive work with alphaviruses and flaviviruses has shown that the ij loop is critical for virus infectivity (18), cholesterol-dependent entry (19, 20 - 23), and transmission (24, 25). In addition, the ij loop and β-strand c have been shown to be key sites of adaptive evolution by leading to increased transmission in mosquitoes, while β-strand c has been associated with enhanced virulence (26, 27). Given the structural similarities between these viral proteins, we hypothesize that the bunyavirus ij loop will also play key roles in virus infectivity, entry, and replication.
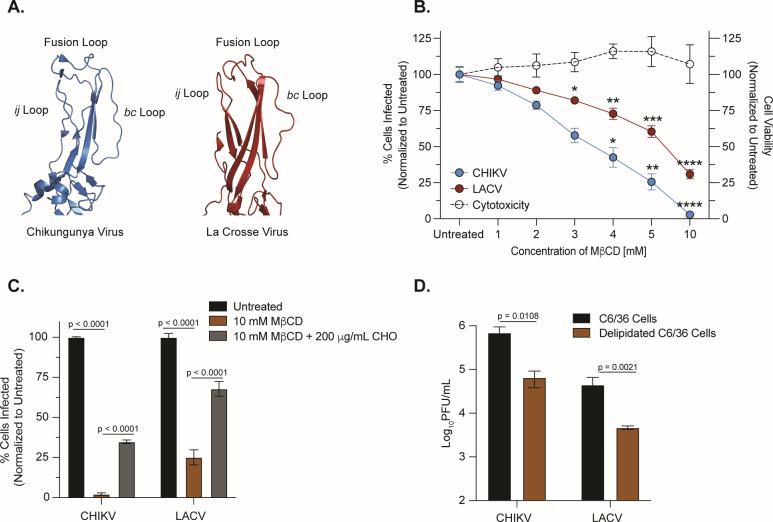
FIG 1.
LACV entry is cholesterol dependent. (A) Crystal structures of domain II of the CHIKV E1 (PDB: 3N42) and LACV Gc (PDB: 7A57) fusion glycoproteins with the ij loop, fusion loop, and bc loop labeled. (B) BHK-21 cells were pretreated with MβCD for 1 h, and cholesterol-depleted cells were infected with CHIKV-ZsGreen or LACV (Human/78) at a multiplicity of infection (MOI) of 1 before adding 20-mM NH4Cl. Cells were fixed at 24 h post-infection. LACV infections were stained with primary antisera and a goat anti-rabbit IgG-Alexa488 secondary antibody, and both infections were stained with 4′,6-diamidino-2-phenylindole. The number of infected cells was quantified using a CX7 high-content microscope. Data are normalized to an untreated control and represent three independent trials with internal duplicates. A one-way analysis of variance (ANOVA) test was performed with P values representing *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) BHK-21 cells were pretreated with 10-mM MβCD and were incubated with either Dulbecco’s Modified Eagle Medium or 200-µg/mL water-soluble cholesterol. Cholesterol-depleted, complemented, or untreated cells were infected with CHIKV-ZsGreen or LACV (Human/78) at an MOI of 1 for 1 h before adding 20-mM NH4Cl. Data represent two independent experiments with internal triplicates. A two-way ANOVA test was performed and P values are shown. (D) Delipidated C6/36 insect cells were infected with CHIKV (MOI of 0.01) or LACV (MOI of 0.1). After a phosphate-buffered saline wash and adding back complete media, viral supernatants were harvested 24 h post-infection and used to quantify infectious particles by plaque assay. Data represent two independent experiments with internal triplicates. Mann-Whitney tests were performed and P values are shown. The average and standard error of the mean are shown for all data.
In this study, we use LACV as a model orthobunyavirus to investigate entry and the role of the ij loop in the bunyavirus life cycle. We show that while LACV entry is cholesterol dependent, similar to the alphavirus chikungunya virus (CHIKV), CHIKV replication relies on cholesterol more than LACV replication. Using an infectious clone, we generated point mutants in the LACV ij loop and found that specific residues contributed to overall infectivity as well as cholesterol-dependent entry. Importantly, the ij loop variants were attenuated in vitro in BHK-21 cells, and the variant H1193A was attenuated in a newborn mouse model of infection. Finally, given that the alphavirus glycoprotein has been found as fundamental for adaptive evolution, we addressed the evolution of the LACV glycoproteins. In mice and mosquitoes, we found a number of mutations in the Gc head domain, suggesting its importance for host adaptation. Together, our data show that LACV enters cells through a cholesterol-dependent pathway and may use similar domains and amino acids for virus entry as CHIKV. Moreover, we have identified the LACV Gc head domain as a hotspot for virus adaptation in vivo. These studies and future work dissecting the bunyavirus glycoprotein are critical to our global understanding of class II glycoprotein function and viral pathogenesis.
RESULTS
Host cholesterol contributes to La Crosse virus entry
Alphaviruses, flaviviruses, and bunyaviruses all encode a class II fusion glycoprotein that plays a central role in mediating viral attachment and membrane fusion (7, 15, 16). However, while our understanding of alphavirus and flavivirus glycoprotein biology has grown significantly, our understanding of vector-borne bunyavirus glycoprotein function is less understood. To begin to understand how the bunyavirus glycoprotein functions, we first explored the structural similarities between the class II fusion glycoprotein of the orthobunyavirus LACV and the alphavirus chikungunya virus (Fig. 1A). Given the structural similarities in the tip of domain II between these two glycoproteins, particularly the fusion loop, ij loop, and bc loop, we hypothesized that LACV would use a cholesterol-dependent mechanism of entry similar to what has been shown for CHIKV. To investigate the cholesterol-dependent entry of LACV, we depleted mammalian BHK-21 cells of cholesterol using increasing concentrations of methyl-β-cyclodextrin (MβCD) for 1 h, which we found had minimal cytotoxicity between 1 and 10 mM (Fig. 1B). We then infected cells with either wild-type (WT) CHIKV-ZsGreen (06-049 strain) or LACV (Human/78 strain) (Fig. 1B), washed the cells with phosphate-buffered saline (PBS), and added back complete media containing 20 mM ammonium chloride, which was used to neutralize the endosomal and lysosomal pH to further block viral fusion and spread (27). We found that both CHIKV and LACV were sensitive to cholesterol depletion compared to their untreated controls, as the number of infected cells significantly decreased with higher concentrations of MβCD. Specifically, the number of CHIKV-infected cells decreased ~25-fold, and the number of LACV-infected cells decreased ~4-fold at 10-mM MβCD. Notably, we were able to restore CHIKV or LACV infection when we added back cholesterol to BHK-21 cells initially depleted with 10-mM MβCD, yet not to the same extent as our untreated controls (Fig. 1C). Finally, since both CHIKV and LACV infect Aedes (Ae.) mosquitoes, we wanted to test their cholesterol-dependent entry in C6/36 Ae. albopictus cells. We grew C6/36 cells in the presence or absence of delipidated serum and tested whether delipidation affected viral entry (Fig. 1D). We found both viruses sensitive to delipidation, indicating that LACV infection is cholesterol dependent in both mammalian and mosquito cells. These data show that both CHIKV and LACV utilize cholesterol for entry, as depleting levels of cholesterol and lipids decrease the infectivity of each virus.
Cholesterol-modulating compounds differentially impact CHIKV and LACV replication in Vero cells
As an alternative approach to understanding the role of cholesterol in the LACV life cycle, we used the cholesterol-modulating compounds atorvastatin, lovastatin, and imipramine. Atorvastatin and lovastatin are Food and Drug Administration (FDA)-approved hydroxymethylglutaryl coenzyme A (HMG-CoA) reductase inhibitors that decrease cholesterol levels in the liver while also stimulating low-density lipoprotein receptors expression on hepatic cells (28, 29). Imipramine is an FDA-approved antidepressant that disrupts cholesterol trafficking within the cell. As some of these compounds have been shown to inhibit the entry and replication of CHIKV and other arboviruses (30 - 33), we hypothesized they may also alter LACV infection as well. We first tested the cytotoxicity of each compound using a cell viability and proliferation MTT assay and observed limited cytotoxicity in Vero cells until 100 µM (Fig. 2A). However, because the EC-50 of imipramine was estimated to be around 100 µM based on our MTT assay, we chose to only continue with concentrations up to 50 µM to limit the role cytotoxicity could play in our experiments. To test how these compounds affect our viruses, we pretreated Vero cells with each drug at increasing concentrations for 1 h and then infected the cells with CHIKV Zs-Green or LACV diluted in Dulbecco’s Modified Eagle Medium (DMEM) and each compound. After the incubation, we added media containing each compound for 24 h.
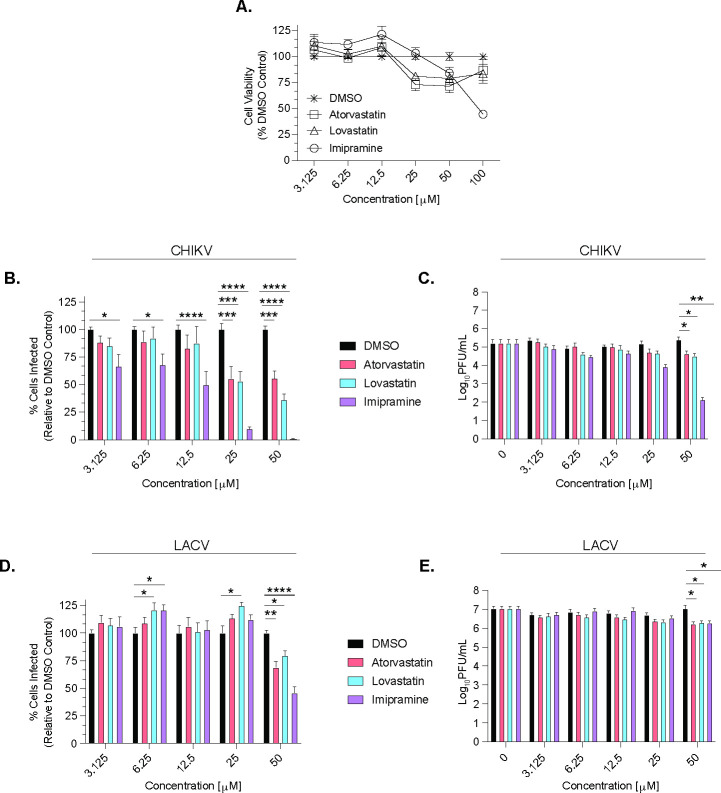
FIG 2.
CHIKV and LACV are sensitive to cholesterol-modulating compounds. (A) An MTT assay was used to determine cytotoxicity from dimethylsulfoxide (DMSO), atorvastatin, lovastatin, and imipramine in Vero cells at increasing concentrations at 24 h post-treatment, and is shown as a dotted line on the graphs for each respective compound. Vero cells were pre-treated with atorvastatin, lovastatin, or imipramine at increasing concentrations and then infected with CHIKV-ZsGreen (B and C) or LACV (Human/78) (D and E) at an MOI of 0.1 for 1 h. Cells were then washed with PBS and incubated in Vero media with the same drug concentrations for an additional 24 h. After incubation, cells were fixed and stained for analysis using a CX7 high-content microscope (B and D), and supernatants were collected for a plaque assay to determine infectious particle titers (C and E). Data are normalized to a DMSO control and represent at least three independent experiments with internal duplicates. A two-way ANOVA test was performed with P values representing *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. The average and SEM are shown for all data.
We then measured virus replication by (i) intracellular staining to look at the potential impacts on virus spread and (ii) plaque assay to address impacts on infectious particle production. We found that by intracellular staining in Vero cells, CHIKV was sensitive to all three compounds, although at varying degrees (Fig. 2B and C). However, this reduction in infection was only reflected in infectious particle production at 50 µM of each compound. In contrast, LACV replication, as measured by intracellular staining, showed slight increases in infection at low concentrations of specific compounds and was only reduced at 50 µM of each compound assayed by intracellular staining and infectious particles (Fig. 2D and E). Taken together, these results suggest that although both LACV and CHIKV are dependent on cholesterol for entry (Fig. 1), they use cholesterol differently for replication.
LACV Gc ij loop is important for LACV replication in vitro and in vivo
Domain II of the class II fusion glycoproteins of alphaviruses, flaviviruses, and bunyaviruses contains the fusion loop (cd loop), the ij loop, and the bc loop. While the fusion loop of La Crosse has been studied extensively (7, 34 - 36), how the other loops function in virus entry has not been explored. In particular, the ij loop of alphaviruses and flaviviruses has been shown to be critical for virus assembly, entry, and transmission (18, 23, 25). In addition, we have shown that a conserved alanine residue in the Zika virus and yellow fever virus ij loop is critical for virion assembly and envelope protein accumulation (37). Therefore, we hypothesized that the orthobunyavirus ij loop would also play important roles in assembly and infectivity. To address this hypothesis, we first looked at the conservation of the ij loop between orthobunyaviruses (Fig. 3A). Interestingly, LACV and several other orthobunyaviruses have an alanine at the tip of the ij loop, similar to alphaviruses and flaviviruses. In addition, there is a conserved histidine and arginine flanking the ij loop tip, suggesting these may be important for glycoprotein function. Given this conservation between viruses and viral families, we generated point mutations in the ij loop of the Gc glycoprotein of LACV (LACV/78/NC-cl strain), which we hypothesized would affect infection in vitro and in vivo (Fig. 3A and B). To investigate the role of these residues in the LACV life cycle, we first performed growth curves in BHK-21 cells and found that all three ij loop point mutants were attenuated at 16 and 24 h post- infection, indicating that the ij loop is important in the LACV life cycle (Fig. 3C). We then asked whether each mutant is attenuated in vivo by infecting 7- to 8-day-old C57BL/6 mice with LACV WT or each ij loop mutant and quantifying viral titers in the brain 3 days post-infection (Fig. 3D). We found that within each trial, the H1193A mutant was consistently and significantly attenuated in mice compared to the other mutants. These results indicate that the ij loop is important for LACV infection in vitro and in vivo, similar to what has been found for LACV fusion loop variants (35, 36).
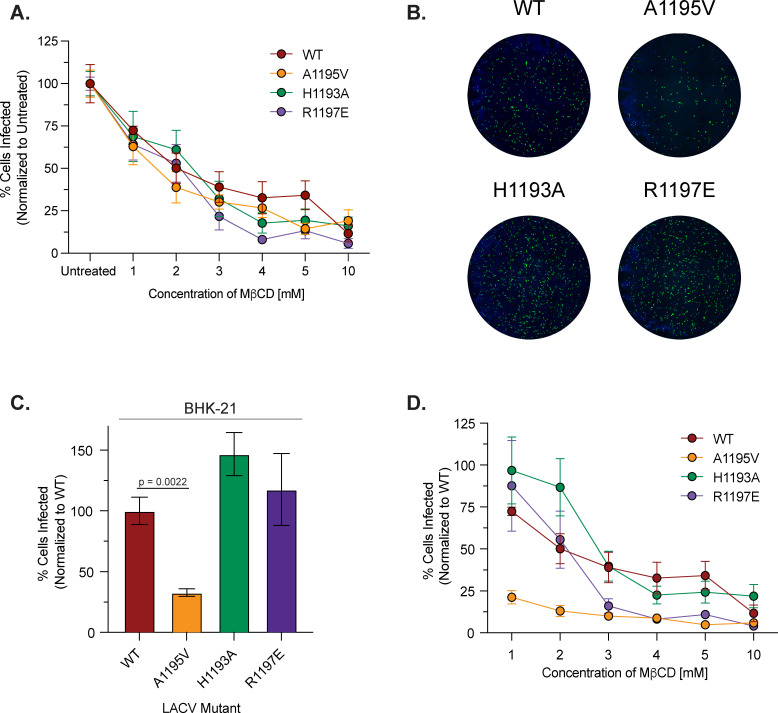
FIG 3.
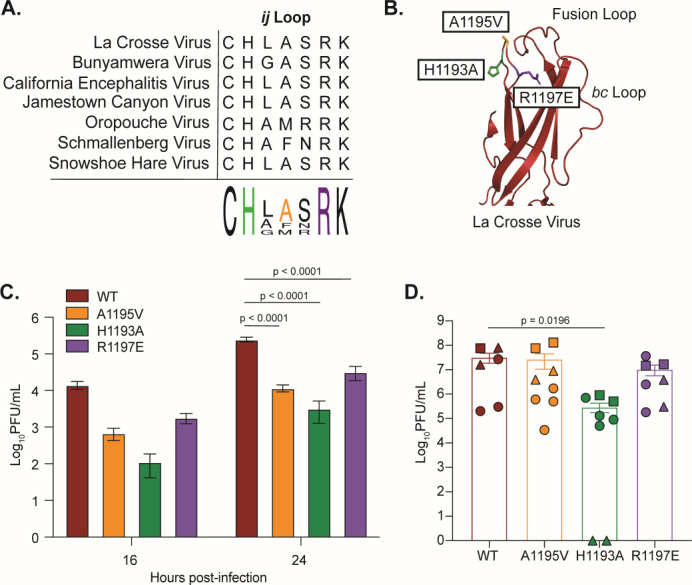
The orthobunyavirus Gc ij loop is highly conserved and contributes to LACV infection. (A) Protein alignment of the ij loop of different orthobunyaviruses. (B) Crystal structure of the LACV Gc domain II showing individual ij loop mutations (PDB: 7A57). (C) BHK-21 cells were infected with WT LACV (78/NC-cl) or each variant at an MOI of 0.1 for 1 h. Culture supernatants were collected at the indicated time points and viral titers were quantified by plaque assay. Data represent at least two independent experiments with internal duplicates. A two-way ANOVA test was performed and P values are shown. (D) Seven- to eight-day-old C57BL/6 J mice were infected subcutaneously with 50 PFU of either LACV WT or each ij loop mutant. Mice were euthanized 3 days post inoculation; the brain was harvested and used for a plaque assay to quantify infectious particles. Shapes correspond to a different cage of pups that were infected and represent three independent experiments with at least n = 6 mice for each condition. A Mann-Whitney test was performed and data with statistical significance were labeled with their corresponding P value. The average and SEM are shown for all data.
The LACV Gc ij loop contributes to infectivity in vitro
The alphavirus ij loop has been shown to contribute to cholesterol-dependent entry, infectivity, and transmission. Notably, the emergence of an A226V variant in the ij loop of the E1 glycoprotein of CHIKV contributed to the 2005 La Reunion epidemic and has been shown to contribute to cholesterol-dependent entry (22, 25, 38). Given these findings, we asked whether the LACV ij loop behaved similarly. To begin, we first addressed cholesterol-dependent entry by depleting BHK-21 cells of cholesterol using MβCD and infecting cells with either LACV WT or each ij loop mutant at a multiplicity of infection (MOI) of 1. When normalizing our data to untreated controls of each virus, we found differences between various concentrations of MβCD, where the mutants are more sensitive to cholesterol levels for entry (Fig. 4A). However, when looking at our untreated infected controls, images from the CX7 high-content microscope (Fig. 4B) showed reduced infectivity of the A1195V variant, while the H1193A and R1197E variants increased infectivity compared to WT, which we also see when quantifying the number of infected cells in these wells (Fig. 4C). Given this reduction in initial infectivity, we then normalized the cholesterol depletion assay to WT LACV and found that while the infectivity of the mutants varied, variants H1193A and R1197E showed a decline in infection after treatment similar to a wild-type virus, while the A1195V did not reduce infection any further (Fig. 4D). Our findings show that changes to the ij loop lead to variation in infectivity in BHK-21 cells in vitro compared to WT, while cholesterol-dependence was not significantly impacted by the variants tested. These results reiterate the importance of the ij loop for LACV infection and suggests that while the ij loop of LACV Gc is important for infectivity, it may play less of a role in cholesterol-dependent entry.
FIG 4.
LACV Gc ij loop variants contribute to LACV infectivity. BHK-21 cells were pretreated with MβCD, and cholesterol-depleted cells were infected with LACV WT or each ij loop mutant at an MOI of 1. Cells were then treated with 20 mM NH4Cl for 24 h and were then fixed, stained, and quantified as before. Data are normalized to an untreated WT or untreated control within each mutant and represent three independent experiments with internal duplicates. Data are normalized to each virus’ untreated control (A) or WT LACV (D) and represent three independent trials with internal duplicates. (B) Representative images from a CX7 high-content microscope with the nuclei of BHK-21 cells stained blue with 4′,6-diamidino-2-phenylindole and LACV infected cells stained green from the antisera primary antibody and a goat anti-rabbit IgG-Alexa488 secondary antibody. (C) BHK-21 cells were infected with either WT LACV (78/NC-cl) or each ij loop mutant at an MOI of 1 for 1 h. After incubation, 20-mM NH4Cl was added and the cells incubated for 24 h before being fixed and stained for analysis. A Mann-Whitney test was performed and data with statistical significance are labeled with their corresponding P value. The average and SEM are shown for all data.
In vivo evolution of LACV highlights the Gc head domain as potential hotspot for viral adaptation
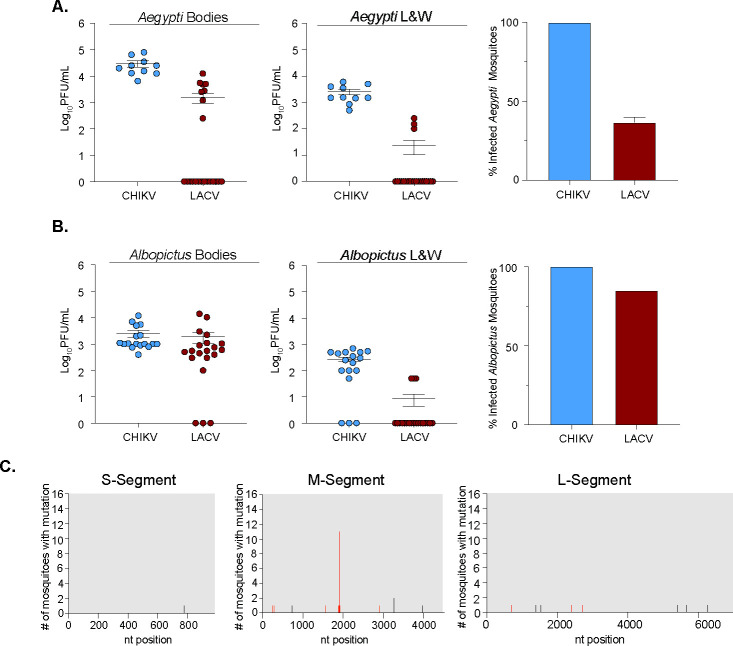
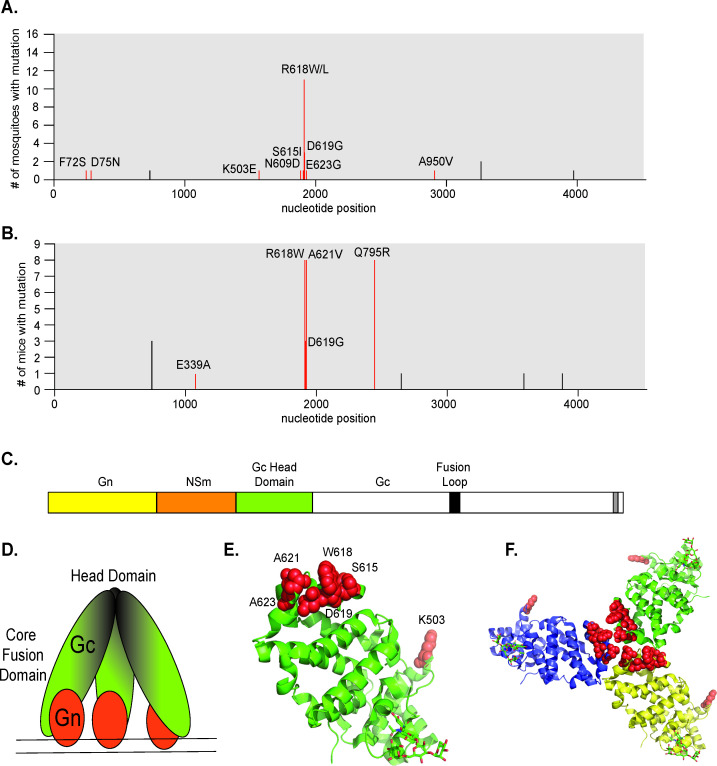
The arbovirus glycoproteins are key determinants of virus adaptation (24, 39). Viral glycoproteins have been shown to adapt in vivo both in nature and in research experiments (26, 27, 37, 38, 40), providing key snapshots into glycoprotein function. Therefore, we hypothesized that in vivo LACV infections may shed light on how LACV may evolve. Specifically, mosquitoes play a fundamental role in the evolution of emerging arboviruses, as the interaction with the host vector can cause various mutations to emerge in nature and change the biology of these viruses. Through the surveillance of different areas in the United States, LACV has been found to infect Aedes (Ae.) species of mosquitoes, including Ae. albopictus (41). To address the in vivo evolution of LACV (Human/78 strain), we infected Ae. aegypti (Mexico) and Ae. albopictus (New York) via an artificial blood meal. As a control for infection, we included CHIKV, as its primary vectors are Ae. aegypti and Ae. albopictus. At 7 days post-infection, we harvested individual mosquitoes and separated bodies (infection) from legs and wings (dissemination), then determined infectious titers via plaque assay (Fig. 5A and B). While CHIKV infected and disseminated in both species, LACV preferentially infected Ae. albopictus yet poorly disseminated in both species. We then extracted the RNA from the bodies of the LACV infected Ae. mosquitoes and Sanger sequenced the full LACV genome (Fig. 5C). Interestingly, we found a few synonymous and nonsynonymous mutations in the S and L segments of individual mosquitoes (Fig. 5C). However, in the M segment, where the glycoprotein is encoded, we found a number of mutations including one (R618W/L) that was found in multiple mosquitoes (Fig. 5C and 6A). Finally, in addition to mosquitoes, we infected 5- to 10-day-old mice with LACV (Human/78 strain) to understand LACV evolution in mammals. In mice, we found no nonsynonymous or synonymous mutations in the S or L segment. However, in the M segment, we found a number of mutations including several in the Gc head domain (R618W, D619G, A621V, and Q795R) present in nearly all infected mice (Fig. 6B). When we mapped these variants onto the crystal structure of the Gc head domain, we found these mutants to cluster at the interface of the head domain, which is important for contact between Gc monomers of the trimer (42) (Fig. 6C through F). Together, these results indicate that the M segment, specifically the Gc head domain, may be a potential hotspot for viral evolution, as the frequency of these mutations suggests the importance of these residues for infection and transmission in nature.
FIG 5.
LACV mosquito infections and evolution. Aedes (Ae.) aegypti (A) or Ae. albopictus (B) mosquitoes were infected with 106 PFU/mL of CHIKV or LACV (Human/78) via an artificial blood meal. Mosquitoes were harvested and dissected 7 days post-infection, and infectious titers were determined by plaque assay in the bodies or legs and wings. Percentage of infected mosquitoes are shown on the right. Data represent at least one independent experiment with at least n = 10 mosquitoes for each condition. The average and SEM are shown for all data. (C) The full LACV genome of individual mosquitoes was Sanger sequenced, and graphs represent the frequency of mutations found within each LACV genome segment. Black tick marks indicate a synonymous mutation, and red tick marks indicate a nonsynonymous mutation with its position labeled.
FIG 6.
The LACV Gc head domain is a potential hotspot for evolution. Frequency of mutations found in the LACV M segment by Sanger sequencing from mosquitoes (A) and mice (B). Black tick marks indicate a synonymous mutation, and red tick marks indicate a nonsynonymous mutation with its position on the glycoprotein labeled. (C) A schematic showing the LACV M segment, as well as a depiction of the Gc trimer head domain and Gn spike (D). Crystal structure of the Gc head domain with red spheres indicating the mutations found from the in vivo Sanger sequencing results as a monomer (E) or trimer (F) (PDB: 6H3W).
DISCUSSION
Arboviruses are genetically diverse pathogens that can cause devastating disease worldwide. There are limited antivirals and vaccines that target these viral threats, highlighting the necessity to study arbovirus biology at the molecular level. The majority of human pathogenic arboviruses fall into two prominent viral families, Togaviridae (alphavirus genus) and Flaviviridae (flavivirus genus), and Bunyavirales order. While extensive work has been done to study alphavirus and flavivirus biology, there is still much unknown about bunyavirus mechanisms of entry, pathogenesis, and evolution, despite their high epidemic potential.
La Crosse virus is an emerging neuroinvasive arbovirus that has been a major cause of pediatric encephalitis throughout the United States, with persistent high-risk clusters of cases in the Midwest and Appalachian regions (11). Here, we used the structural similarities of the CHIKV E1 and the LACV Gc class II fusion glycoproteins to hypothesize the importance of the ij loop for cholesterol-dependent entry and infectivity in vitro and in vivo. Like alphaviruses, we found that La Crosse virus entry was cholesterol dependent in both mammalian and mosquito cell lines. While CHIKV appears to be more sensitive, both CHIKV and LACV show a decrease in infected cells with depleting levels of cholesterol and lipids, signifying the importance of cholesterol for LACV entry. However, in the presence of cholesterol-modulating compounds, LACV is less sensitive to treatment, as only higher concentrations of the compounds decreased the percentage of infected cells in Vero cells. These results may be due to differences in host and cell type as previous works with imipramine and statins were completed in human cells (32, 33). Nonetheless, these results show that while LACV entry is cholesterol dependent, similar to CHIKV, replication in LACV is less affected by cholesterol-manipulating treatments, shown through no differences in infectious particle titers. Characterizing LACV’s cholesterol dependence is crucial, as a re-emergent CHIKV strain from the Indian Ocean islands outbreak shifted its dependency on cholesterol and enhanced transmissibility in Ae. albopictus (25). The increased transmission of CHIKV during this epidemic highlights the importance of monitoring LACV and other arboviruses for mechanisms of entry and replication that could be enhanced due to single or multiple point mutations.
The alphavirus E1 A226 residue resides on the ij loop, which has been characterized extensively as important for infectivity (18, 21), cholesterol dependence (23), and transmission (24, 25). The ij loop of orthobunyaviruses is highly conserved and contains a conserved histidine and arginine, as well as an analogous alanine to the alphavirus A226 residue. We found that all three ij loop variants were attenuated in BHK-21 cells with the A1195V mutant having reduced infectivity, potentially explaining its attenuated growth in vitro. It is interesting that the H1193A and R1197E variants had no impact on infectivity yet still showed reduced growth in vitro, suggesting they may play other roles in the viral life cycle. However, in neonatal mice, only the H1193A is attenuated, with one trial having no infectious particles found in the brain. A similar histidine in the alphavirus ij loop has been shown to play important roles in Semliki Forest virus fusion (18), suggesting a similar role in LACV. Future studies are under way to continue to further dissect the mechanisms of how the ij loop functions in vitro and in vivo.
Although the primary vector of La Crosse virus is the Ae. triseriatus mosquito, LACV has been found to infect other Aedes species, including Ae. albopictus, Ae. japonicus, and Ae. aeygpti (10, 43). Here, we demonstrate that LACV (Human/78 strain) can infect and disseminate in both Ae. aegypti and Ae. albopictus mosquitoes, but not to the same extent as CHIKV. Importantly, given the need for in vivo models to study arboviral evolution, we found that sequencing LACV from infected mice and mosquitoes identified multiple point mutations in the M segment, specifically in the Gc head domain. One mutation in particular at residue R618 has been found previously during mosquito passage, indicating that this residue could be important for vector infections (44). There have been few studies highlighting the importance of the Gc head domain, yet it has been shown to be highly immunogenic and a potential target as a vaccine against another orthobunyavirus, Schmallenberg virus (45). Studying the LACV Gc head domain as a potential hotspot for evolution is essential as these prominent residues in our studies reveal how LACV can evolve and emerge in different vector hosts.
Together, our studies highlight the importance of the cholesterol-dependent LACV Gc ij loop in the LACV life cycle in vitro and in vivo. The highly conserved residues found in the orthobunyavirus Gc ij loop are critical for entry, infectivity, and evolution of this emerging virus. Further studies are under way to evaluate these and other Gc residues and their importance in LACV pathogenesis and transmission in vitro and in vivo.
MATERIALS AND METHODS
Cell lines
BHK-21 cells (ATCC CCL-10) were grown in complete media, consisting of Dulbecco’s Modified Eagle Medium (Corning) with 10% fetal bovine serum (FBS; Atlanta Biologics), 1% nonessential amino acids (NEAA; Corning), and 1% HEPES (Invitrogen). BHK-BSR/T7 cells were a gift from Dr. Steven Whitehead [National Institutes of Health (NIH)] and maintained in the same media as the BHK-21 cells above with 1-mg/mL gentamicin (Gibco) added every other passage (46). Vero cells (ATCC CCL-81) were grown in media, composed of DMEM with 10% newborn calf serum (Sigma). Mammalian cells were maintained at 37°C with 5% CO2. C6/36 Ae. albopictus cells (ATCC CRL-1660) were maintained in L-15 Lebovitz Medium with 10% FBS, 1% NEAA, and 1% tryptose phosphate broth at 28°C with 5% CO2. All cell lines were confirmed to be mycoplasma free.
Viruses
La Crosse virus strain Human/78 was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH (NR-540). The LACV strain LACV/78/NC-cl infectious clone was a gift from Dr. Steven Whitehead (NIH) (46). LACV Gc ij loop variants were generated by overlapping Phusion PCR using the primers in Table 1. In brief, a ~2.4-kb overlapping PCR product containing 5′ EcoRI and 3′ NotI restriction sites was generated and subcloned into the same restriction sites of the wild-type plasmid. All plasmids were sequenced in full at Plasmidsaurus. To generate infectious virus, BHK BSR/T7 cells were transfected with 2 µg of each T7-driven plasmid encoding S, M, and L segments using TransIT-LT1 reagent (Mirus) following the manufacturer’s instructions (46). Twenty-four hours post-transfection, the media was replaced with fresh media, and cells were incubated at 37°C for 4 days. To generate a working stock of all viruses, LACV was amplified on Vero CCL-81 cells; the supernatant was clarified by centrifugation; and the virus was aliquoted and stored at −80°C. Infectious viral titers were quantified by plaque assay as described below. The entire coding region of the LACV genome (S, M, and L segments) was Sanger sequenced at Genewiz. Sanger sequences were used to generate reference genomes for variant detection.
TABLE 1.
Site-directed mutagenesis primers used in this study a
| LACV strain | Primer name | Sequence | ||
|---|---|---|---|---|
| 78/NC-cl | LACV Gc H1192A For | GATTTGACTACCTATGCgctTTAGCTAGCAGAAAG | ||
| 78/NC-cl | LACV Gc H1192A Rev | CTTTCTGCTAGCTAAagcGCATAGGTAGTCAAATC | ||
| 78/NC-cl | LACV Gc A1195V For | CTACCTATGCCATTTAgtcAGCAGAAAGGAAGTC | ||
| 78/NC-cl | LACV Gc A1195V Rev | GACTTCCTTTCTGCTgacTAAATGGCATAGGTAG | ||
| 78/NC-cl | LACV Gc R1197E For | GCCATTTAGCTAGCgagAAGGAAGTCATTG | ||
| 78/NC-cl | LACV Gc R1197E Rev | CAATGACTTCCTTctcGCTAGCTAAATGGC | ||
| 78/NC-cl | LACV EcoRI For | CAAATATGATGAATTCTGGGATG | ||
| 78/NC-cl | LACV NotI Rev | GCTACGCGGCCGCCTTAGCC | ||
Lowercase = mutant codon.
The chikungunya virus strain 06-049 (accession no. AM258994) infectious clone encoding a ZsGreen reporter has been previously described (27). Ten micrograms of the infectious clone plasmid was linearized overnight with NotI, purified by phenol:chloroform extraction and ethanol precipitation, and resuspended in nuclease free water. CHIKV-ZsGreen RNA was in vitro transcribed using the mMessage mMachine SP6 kit (Invitrogen) following the manufacturer’s instructions. RNA was purified by phenol:chloroform extraction and ethanol precipitation, diluted to 1 µg/µL, aliquoted, and stored at −80°C. To generate infectious virus, BHK-21 cells (107/mL) were electroporated with 10 µg of in vitro transcribed RNA by 1 pulse of 1,200 V, 25 Ω, and infinite resistance. Electroporated cells were added to a T25 flask in BHK-21 media (DMEM, 10% FBS, 1% NEAA, and 1% HEPES) and incubated at 37°C for 72 h. After incubation, the supernatant was centrifuged at 1,200 rpm for 5 min, and the virus was aliquoted and stored at −80°C. To generate a working stock, virus from the initial electroporation was amplified once on BHK-21 cells, the supernatant was clarified by centrifugation at 1,200 rpm for 5 min and virus was aliquoted and stored at −80°C. All work with CHIKV and LACV variants was completed under ABSL3 conditions at the NYU Grossman School of Medicine.
LACV Sanger sequencing
The genome of all LACV working stocks and mouse and mosquito samples were Sanger sequenced at Genewiz. In brief, viral RNA was extracted with Trizol following the manufacturers’ instructions. cDNA was generated using the Maxima H First Strand cDNA kit (Thermo Scientific) and the S, M, and L segments were amplified by Phusion PCR (Thermo Scientific) using the primers in Table 2. PCR products were visualized on a 1% agarose gel, PCR column purified (Macherey-Nagel), and sent for Sanger sequencing at Genewiz with the primers in Table 2.
TABLE 2.
Sequencing primers used in this study
| LACV strain(s) | Genome segment | Sequence |
|---|---|---|
| Human/78 | S | gtagtgtaccccacttgaatac |
| Human/78 | S | ccccacttgaatactttg |
| Human/78 | S | ctatctatcattctttccagg |
| Human/78 | S | gggtagaatttgaaagtttc |
| Human/78/78/NC-cl | S | gatttatagttttttgctgtcccc |
| Human/78/78/NC-cl | S | gtagtgtgccccactgaatac |
| 78/NC-cl | S | gttgttgactgttttttacc |
| 78/NC-cl | S | gtcggatttggtgttttatg |
| 78/NC-cl | S | ccagagatctatctatc |
| Human/78/78/NC-cl | M | gtagtgtactaccaagtatag |
| Human/78/78/NC-cl | M | gttttgaatcaaagccaaag |
| Human/78/78/NC-cl | M | cctgcggcccaaaatctgtac |
| Human/78 | M | gatcttccagacgacatgttg |
| Human/78 | M | gaaggaagatttccatgtccagc |
| Human/78/78/NC-cl | M | gcaagaatgttaaggatgttg |
| Human/78 | M | ccaaagcacatctctaactg |
| Human/78 | M | gttgggtggctgacatttgc |
| Human/78/78/NC-cl | M | ggcaattttaggagatgtc |
| Human/78 | M | cacatttattatatctatagtag |
| Human/78/78/NC-cl | M | catcatatttgaaatttgcc |
| Human/78/78/NC-cl | M | gcaaaataaatttacgctg |
| Human/78/78/NC-cl | M | gtagtgtgctaccaag |
| Human/78/78/NC-cl | M | CTGCAATCAGCACCAGTATGCG |
| Human/78/78/NC-cl | M | GTTAAGTGGGGGGTGGGGAAC |
| Human/78/78/NC-cl | M | cctatttgaggtacagttc |
| 78/NC-cl | M | gcttatctctatttaagtatg |
| 78/NC-cl | M | gcgaagagtttggttgcctg |
| Human/78 | L | gtagtgtacccctatctacaaaac |
| Human/78/78/NC-cl | L | cagaaaattcagtcatatcac |
| Human/78 | L | gatagatttaaagaactttac |
| Human/78/78/NC-cl | L | ctcaaaatctctccatgacc |
| Human/78/78/NC-cl | L | gtatctagctagcctaaccatg |
| Human/78 | L | gtgccagagaattgtttcctc |
| Human/78 | L | ctaaaggactacatgagaag |
| Human/78/78/NC-cl | L | cagattatatatcaactaaag |
| Human/78 | L | ctaaagatggaaataaatgc |
| Human/78/78/NC-cl | L | cccttttcaatatatggcag |
| Human/78/78/NC-cl | L | gatatttagttctagatgcag |
| Human/78 | L | gatgggcatgtcctgttcaac |
| Human/78/78/NC-cl | L | ccataatgcagcataatgccac |
| Human/78 | L | gctaggtgcaaggcatggac |
| Human/78 | L | ctcaagatttgcttaacc |
| Human/78/78/NC-cl | L | gtatcattaataaaattgttg |
| Human/78/78/NC-cl | L | cccttggagccaatttctc |
| Human/78/78/NC-cl | L | cacagacattgcgcagctatgg |
| Human/78/78/NC-cl | L | cagttgctgttaatccccctc |
| Human/78 | L | gtagtgtgcccctatctac |
| Human/78/78/NC-cl | L | gccaatttccaaaaccctcc |
Plaque assay
Infectious virus was quantified using a plaque assay where 10-fold serial dilutions of each sample with the virus in DMEM were added to a monolayer of Vero cells. After incubating the virus and cells for 1 h at 37°C, a medium containing 0.8% agarose and DMEM with 2% FBS was added for 3 days at 37°C. The cells were then fixed with 4% formalin for 1 h, after which the agarose was removed and the plaques were visualized using crystal violet. Viral titers were determined using the lowest countable dilution.
Intracellular LACV staining and quantification of infected cells
Cells were fixed with 4% paraformaldehyde (PFA), incubated with 0.25% TritonX-100 for 10 min followed by blocking buffer (0.2% bovine serum albumin and 0.05% saponin) for 1 h. Cells were then incubated with LACV antisera (a gift from Dr. Karin Peterson, NIH) diluted in blocking buffer for 2 h. After three PBS washes, cells were then stained with a goat anti-rabbit IgG-Alexa488 secondary antibody (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI) for 1 h at room temperature. To quantify infected cells, we used a Cell-Insight CX7 high-content microscope (Thermo Scientific). In brief, the percentage of infected cells was quantified by dividing the number of ZsGreen-positive (CHIK) or Alexa488 (LACV)-positive cells by the total number of DAPI stained cells.
Cholesterol depletion and complementation assays
BHK-21 cells (20,000 cells/well) were seeded in a Costar 96-well plates and incubated for 24 h at 37°C. The cells were pretreated with increasing concentrations of methyl-β-cyclodextrin for 1 h at 37°C. Following incubation, cells were washed once with phosphate-buffered saline and then incubated with each virus at an MOI of 1 for 1 h at 37°C. After incubation, media containing ammonium chloride was added to a final concentration of 20 mM and the cells were incubated at 37°C for 24 h. The cells were then fixed with 4% paraformaldehyde and stained with DAPI (Thermo Scientific) and LACV antibody as described below, and infected cells were quantified using a Cell-Insight CX7 high-content microscope as stated above. Cholesterol complementation assay was completed as described above with 10 mM MβCD, and the additional step of adding 200 µg/mL of water-soluble cholesterol (Sigma) for 1 h at 37°C before infection.
MTT cell proliferation assay
To measure the cytotoxicity of various treatments, BHK-21 or Vero cells (20,000 cells/well) were seeded in a 96-well plate in DMEM supplemented with either complete media or Vero media and incubated for 24 h at 37°C. Cells were then incubated with increasing concentrations of DMSO or each compound for 24 h at 37°C. An MTT assay was performed following the manufacturer’s guidelines with the CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega). For the proliferation readout, 15 µL of the dye solution was added to each well and incubated for 1 h at 37°C. Solubilization solution (100 µL) was added to stop the reaction, and absorbance was read at 570 nm using a PerkinElmer EnVision plate reader.
Delipidated insect cell assay
Delipidated C6/36 cells were generated as previously described (20). In brief, FBS was delipidated by continuous mixing with 3% (wt/vol) Cab-O-Sil (ACROS Organics) at 4°C for 48 h. The FBS was sterile filtered, and C6/36 cells were passaged four times in the presence of delipidated serum. C6/36 insect cells were seeded (100,000 cells/well) in a 96-well plate with either complete media or delipidated media and were incubated for 24 h at 28°C. The cells were infected with either LACV WT (MOI of 0.1) or CHIKV WT (MOI of 0.01) for 1 h at 28°C. After washing the cells three times with PBS, either complete media or delipidated media was added back to the cells and left incubating at 28°C. At 24 h post-infection, the supernatants from each well were harvested, and infectious titers were determined via plaque assay.
Drug assays
Vero cells (20,000 cells/well) were pretreated with increasing concentrations of either imipramine (TCI America), atorvastatin (TCI America), lovastatin (TCI America) or DMSO diluted in Vero media. After a 1-h incubation at 37°C, cells were infected with either LACV or CHIKV-ZsGreen diluted in DMEM at an MOI of 0.1 mixed with each respective compound for an additional hour. The cells were then washed with PBS twice, and each compound diluted in Vero media was given to the cells for an additional 24 incubation. Supernatants after 24 h were collected and used to calculate infectious titers via plaque assay. The cells were then fixed with 8% PFA and stained. Infected cells were quantified using a Cell-Insight CX7 high-content microscope as stated above.
LACV growth curves
BHK-21 (50,000 cells/well in a 24-well plate) were infected with wild-type LACV or each Gc ij loop variant at an MOI of 0.1 for 1 h followed by two washes with PBS, and complete media were added. Samples of the supernatant were collected at various time points, and viral titers were quantified by plaque assay as described above.
Mouse infections
Individual C57BL/6J (Jackson Laboratory) breeding pairs were set up to use each liter as a biological replicate. Within 1 L, male and female 7- to 8-day-old mice were infected with 50 PFU of wild-type LACV or each ij loop variant subcutaneously in the back. Each virus was represented at least one within each liter. Mice were monitored daily then euthanized 3 days post-infection. The brain was harvested, collected in DMEM with 2% serum and homogenized using a Tissue-Lyser II (QIAGEN). Samples were centrifuged at 8,000 rpm for 8 min, and viral titers were quantified using a plaque assay as described above. All animal experiments were completed in accordance with the NYU Grossman School of Medicine Institutional Animal Care and Use Committee guidelines (protocol no. IA16-01783).
Mosquito infections
Aedes albopictus (New York City, USA) and Aedes aegypti (Poza Rica, Mexico) mosquitoes were previously described (27, 37, 47). In brief, female mosquitoes were fed each virus (106 PFU/mL) diluted in PBS-washed sheep blood containing 5 mM ATP. Mosquitoes were fed for 30 min, anesthetized at 4°C, and engorged females sorted into cups. Mosquitoes were incubated at 28°C with 70% relative humidity and 12 h:12 h diurnal light cycle with 10% sucrose applied ad libitum. At 7 days post- infection, mosquitoes were cold anesthetized, and legs and wings were removed. The bodies and legs and wings were placed in 200 µL PBS, and both were ground in a Tissue-Lyser II and clarified by centrifugation. Viral titers were quantified by plaque assay as described above.
Protein structures and sequence alignments
The CHIKV E1 glycoprotein (PDB: 3N42), LACV Gc glycoprotein (PDB: 7A57), and LACV Gc head domain (PDB: 6H3W) were rendered in PyMol (Version 2.5.2). The bunyavirus Gc ij loop sequence alignment was generated using MegAlign (DNA Star) using the following accession numbers: La Crosse virus (AF528166.1), Bunyamwera virus (NC_001926), California encephalitis virus (NC_055118), Jamestown Canyon virus (U88058), Oropouche virus (KP691604), Schmallenberg virus (NC_043584), and Snowshoe hare virus (NC_055196). LACV nucleotide sequencing alignments were generated with SeqMan Ultra (DNA Star).
Statistics and data analysis
All data were analyzed using GraphPad Prism (Version 9.3.1). All in vitro experiments were completed with at least two biological replicates and internal technical duplicates or triplicates (exact details can be found in the figure legends). All in vivo experiments were completed with at least n = 6 for mice and n = 10 for mosquito infections. A P value of <0.05 is considered statistically significant.
ACKNOWLEDGMENTS
We thank all members of the Stapleford Lab for their helpful comments on this project. We thank Dr. Meike Dittmann at the New York University Grossman School of Medicine for the use of the CX7 high-content microscope and Drs. Ludovic Desvignes and Dominick Papandrea for their support with the NYU ABSL3 high-containment laboratory.
This work was supported by funding from the NYUGSoM Start up and NIAID/NIH R01 AI162774-01A1.
Contributor Information
Kenneth A. Stapleford, Email: kenneth.stapleford@nyulangone.org.
Mark T. Heise, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
REFERENCES
- 1. Luethy D. 2023. Eastern, Western, and Venezuelan equine encephalitis and West Nile viruses: clinical and public health considerations. Vet Clin North Am Equine Pract 39:99–113. doi: 10.1016/j.cveq.2022.11.007 [DOI] [PubMed] [Google Scholar]
- 2. Hollidge BS, González-Scarano F, Soldan SS. 2010. Arboviral encephalitides: transmission, emergence, and pathogenesis. J Neuroimmune Pharmacol 5:428–442. doi: 10.1007/s11481-010-9234-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weaver SC, Charlier C, Vasilakis N, Lecuit M. 2018. Zika, Chikungunya, and other emerging vector-borne viral diseases. Annu Rev Med 69:395–408. doi: 10.1146/annurev-med-050715-105122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malet H, Williams HM, Cusack S, Rosenthal M. 2023. The mechanism of genome replication and transcription in bunyaviruses. PLoS Pathog 19:e1011060. doi: 10.1371/journal.ppat.1011060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boshra H. 2022. An overview of the infectious cycle of bunyaviruses. Viruses 14:2139. doi: 10.3390/v14102139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eifan S, Schnettler E, Dietrich I, Kohl A, Blomström A-L. 2013. Non-structural proteins of arthropod-borne bunyaviruses: roles and functions. Viruses 5:2447–2468. doi: 10.3390/v5102447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Windhaber S, Xin Q, Lozach PY. 2021. Orthobunyaviruses: from virus binding to penetration into mammalian host cells. Viruses 13:872. doi: 10.3390/v13050872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plassmeyer ML, Soldan SS, Stachelek KM, Martín-García J, González-Scarano F. 2005. California serogroup Gc (G1) glycoprotein is the principal determinant of pH-dependent cell fusion and entry. Virology 338:121–132. doi: 10.1016/j.virol.2005.04.026 [DOI] [PubMed] [Google Scholar]
- 9. Rowe RD, Odoi A, Paulsen D, Moncayo AC, Trout Fryxell RT, Samy AM. 2020. Spatial-temporal clusters of host-seeking Aedes albopictus, Aedes japonicus, and Aedes triseriatus collections in a La Crosse virus endemic county. PLoS ONE 15:e0237322. doi: 10.1371/journal.pone.0237322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Westby KM, Fritzen C, Paulsen D, Poindexter S, Moncayo AC. 2015. La Crosse encephalitis virus infection in field-collected Aedes albopictus, Aedes japonicus, and Aedes triseriatus in Tennessee. J Am Mosq Control Assoc 31:233–241. doi: 10.2987/moco-31-03-233-241.1 [DOI] [PubMed] [Google Scholar]
- 11. Day CA, Odoi A, Trout Fryxell R. 2023. Geographically persistent clusters of La Crosse virus disease in the Appalachian region of the United States from 2003 to 2021. PLoS Negl Trop Dis 17:e0011065. doi: 10.1371/journal.pntd.0011065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McJunkin JE, de los Reyes EC, Irazuzta JE, Caceres MJ, Khan RR, Minnich LL, Fu KD, Lovett GD, Tsai T, Thompson A. 2001. La Crosse encephalitis in children. N Engl J Med 344:801–807. doi: 10.1056/NEJM200103153441103 [DOI] [PubMed] [Google Scholar]
- 13. Edridge AWD, van der Hoek L. 2020. Emerging orthobunyaviruses associated with CNS disease. PLoS Negl Trop Dis 14:e0008856. doi: 10.1371/journal.pntd.0008856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teleron ALA, Rose BK, Williams DM, Kemper SE, McJunkin JE. 2016. La Crosse encephalitis: an adult case series. Am J Med 129:881–884. doi: 10.1016/j.amjmed.2016.03.021 [DOI] [PubMed] [Google Scholar]
- 15. Guardado-Calvo P, Rey FA. 2017. The envelope proteins of the bunyavirales. Adv Virus Res 98:83–118. doi: 10.1016/bs.aivir.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 16. Vaney MC, Rey FA. 2011. Class II enveloped viruses. Cell Microbiol 13:1451–1459. doi: 10.1111/j.1462-5822.2011.01653.x [DOI] [PubMed] [Google Scholar]
- 17. Guardado-Calvo P, Atkovska K, Jeffers SA, Grau N, Backovic M, Pérez-Vargas J, de Boer SM, Tortorici MA, Pehau-Arnaudet G, Lepault J, England P, Rottier PJ, Bosch BJ, Hub JS, Rey FA. 2017. A glycerophospholipid-specific pocket in the RVFV class II fusion protein drives target membrane insertion. Science 358:663–667. doi: 10.1126/science.aal2712 [DOI] [PubMed] [Google Scholar]
- 18. Chanel-Vos C, Kielian M. 2004. A conserved histidine in the ij loop of the Semliki forest virus E1 protein plays an important role in membrane fusion. J Virol 78:13543–13552. doi: 10.1128/JVI.78.24.13543-13552.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Umashankar M, Sánchez-San Martín C, Liao M, Reilly B, Guo A, Taylor G, Kielian M. 2008. Differential cholesterol binding by class II fusion proteins determines membrane fusion properties. J Virol 82:9245–9253. doi: 10.1128/JVI.00975-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phalen T, Kielian M. 1991. Cholesterol is required for infection by Semliki forest virus. J Cell Biol 112:615–623. doi: 10.1083/jcb.112.4.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chatterjee PK, Vashishtha M, Kielian M. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki forest virus fusion. J Virol 74:1623–1631. doi: 10.1128/jvi.74.4.1623-1631.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsetsarkin KA, McGee CE, Higgs S. 2011. Chikungunya virus adaptation to Aedes albopictus mosquitoes does not correlate with acquisition of cholesterol dependence or decreased pH threshold for fusion reaction. Virol J 8:376. doi: 10.1186/1743-422X-8-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vashishtha M, Phalen T, Marquardt MT, Ryu JS, Ng AC, Kielian M. 1998. A single point mutation controls the cholesterol dependence of semliki forest virus entry and exit. J Cell Biol 140:91–99. doi: 10.1083/jcb.140.1.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsetsarkin KA, Chen R, Weaver SC. 2016. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol 16:143–150. doi: 10.1016/j.coviro.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3:e201. doi: 10.1371/journal.ppat.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stapleford KA, Coffey LL, Lay S, Bordería AV, Duong V, Isakov O, Rozen-Gagnon K, Arias-Goeta C, Blanc H, Beaucourt S, Haliloğlu T, Schmitt C, Bonne I, Ben-Tal N, Shomron N, Failloux A-B, Buchy P, Vignuzzi M. 2014. Emergence and transmission of arbovirus evolutionary intermediates with epidemic potential. Cell Host Microbe 15:706–716. doi: 10.1016/j.chom.2014.05.008 [DOI] [PubMed] [Google Scholar]
- 27. Noval MG, Rodriguez-Rodriguez BA, Rangel MV, Stapleford KA. 2019. Evolution-driven attenuation of alphaviruses highlights key glycoprotein determinants regulating viral infectivity and dissemination. Cell Rep 28:460–471. doi: 10.1016/j.celrep.2019.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McIver LA, Siddique MS. 2022. Atorvastatin. StatPearls, Treasure Island (FL). [Google Scholar]
- 29. Duong H, Bajaj T. 2022. Lovastatin. StatPearls, Treasure Island (FL). [Google Scholar]
- 30. Osuna-Ramos JF, Reyes-Ruiz JM, Del Ángel RM. 2018. The role of host cholesterol during flavivirus infection. Front Cell Infect Microbiol 8:388. doi: 10.3389/fcimb.2018.00388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Españo E, Nam J-H, Song E-J, Song D, Lee C-K, Kim J-K. 2019. Lipophilic statins inhibit zika virus production in vero cells. Sci Rep 9:11461. doi: 10.1038/s41598-019-47956-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wichit S, Hamel R, Bernard E, Talignani L, Diop F, Ferraris P, Liegeois F, Ekchariyawat P, Luplertlop N, Surasombatpattana P, Thomas F, Merits A, Choumet V, Roques P, Yssel H, Briant L, Missé D. 2017. Imipramine inhibits chikungunya virus replication in human skin fibroblasts through interference with intracellular cholesterol trafficking. Sci Rep 7:3145. doi: 10.1038/s41598-017-03316-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bakhache W, Neyret A, Bernard E, Merits A, Briant L, Dutch RE. 2020. Palmitoylated cysteines in chikungunya virus nsP1 are critical for targeting to cholesterol-rich plasma membrane microdomains with functional consequences for viral genome replication. J Virol 94. doi: 10.1128/JVI.02183-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plassmeyer ML, Soldan SS, Stachelek KM, Roth SM, Martín-García J, González-Scarano F. 2007. Mutagenesis of the La Crosse virus glycoprotein supports a role for Gc (1066-1087) as the fusion peptide. Virology 358:273–282. doi: 10.1016/j.virol.2006.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soldan SS, Hollidge BS, Wagner V, Weber F, González-Scarano F. 2010. La Crosse virus (LACV) Gc fusion peptide mutants have impaired growth and fusion phenotypes, but remain neurotoxic. Virology 404:139–147. doi: 10.1016/j.virol.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hollidge BS, Salzano M-V, Ibrahim JM, Fraser JW, Wagner V, Leitner NE, Weiss SR, Weber F, González-Scarano F, Soldan SS. 2022. Targeted mutations in the fusion peptide region of La Crosse virus attenuate neuroinvasion and confer protection against encephalitis. Viruses 14:1464. doi: 10.3390/v14071464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rangel MV, Catanzaro N, Thannickal SA, Crotty KA, Noval MG, Johnson KEE, Ghedin E, Lazear HM, Stapleford KA, Parrish CR. 2022. Structurally conserved domains between flavivirus and alphavirus fusion glycoproteins contribute to replication and infectious-virion production. J Virol 96. doi: 10.1128/JVI.01774-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney M-C, Lavenir R, Pardigon N, Reynes J-M, Pettinelli F, Biscornet L, Diancourt L, Michel S, Duquerroy S, Guigon G, Frenkiel M-P, Bréhin A-C, Cubito N, Desprès P, Kunst F, Rey FA, Zeller H, Brisse S. 2006. Genome microevolution of chikungunya viruses causing the Indian ocean outbreak. PLoS Med 3:e263. doi: 10.1371/journal.pmed.0030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. 2009. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae.aegypti mosquitoes. PLoS One 4:e6835. doi: 10.1371/journal.pone.0006835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shan C, Xia H, Haller SL, Azar SR, Liu Y, Liu J, Muruato AE, Chen R, Rossi SL, Wakamiya M, Vasilakis N, Pei R, Fontes-Garfias CR, Singh SK, Xie X, Weaver SC, Shi P-Y. 2020. A zika virus envelope mutation preceding the 2015 epidemic enhances virulence and fitness for transmission. Proc Natl Acad Sci U S A 117:20190–20197. doi: 10.1073/pnas.2005722117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lambert AJ, Blair CD, D’Anton M, Ewing W, Harborth M, Seiferth R, Xiang J, Lanciotti RS. 2010. La Crosse virus in Aedes albopictus mosquitoes. Emerg Infect Dis 16:856–858. doi: 10.3201/eid1605.100170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hellert J, Aebischer A, Wernike K, Haouz A, Brocchi E, Reiche S, Guardado-Calvo P, Beer M, Rey FA. 2019. Orthobunyavirus spike architecture and recognition by neutralizing antibodies. Nat Commun 10:879. doi: 10.1038/s41467-019-08832-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bara JJ, Parker AT, Muturi EJ. 2016. Comparative susceptibility of Ochlerotatus japonicus, Ochlerotatus triseriatus, Aedes albopictus, and Aedes aegypti (diptera: culicidae) to La Crosse virus. J Med Entomol 53:1415–1421. doi: 10.1093/jme/tjw097 [DOI] [PubMed] [Google Scholar]
- 44. Borucki MK, Kempf BJ, Blair CD, Beaty BJ. 2001. The effect of mosquito passage on the La Crosse virus genotype. J Gen Virol 82:2919–2926. doi: 10.1099/0022-1317-82-12-2919 [DOI] [PubMed] [Google Scholar]
- 45. Wernike K, Aebischer A, Audonnet JC, Beer M. 2022. Vaccine development against schmallenberg virus: from classical Inactivated to modified-live to scaffold particle vaccines. One Health Outlook 4:13. doi: 10.1186/s42522-022-00069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bennett RS, Gresko AK, Nelson JT, Murphy BR, Whitehead SS. 2012. A recombinant chimeric La Crosse virus expressing the surface glycoproteins of Jamestown Canyon virus is immunogenic and protective against challenge with either parental virus in mice or monkeys. J Virol 86:420–426. doi: 10.1128/JVI.02327-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaczmarek ME, Herzog NL, Noval MG, Zuzworsky J, Shah Z, Bajwa WI, Stapleford KA. 2020. Distinct New York City Aedes albopictus mosquito populations display differences in salivary gland protein D7 diversity and chikungunya virus replication. Viruses 12:698. doi: 10.3390/v12070698 [DOI] [PMC free article] [PubMed] [Google Scholar]