Abstract
Advances in long-read sequencing technologies have broadened our understanding of genetic variation in the human population, uncovered new complex structural variants and offered an opportunity to elucidate new variant associations with disease.
Recent sequencing innovations in both read length and accuracy have led to the emergence of complete, telomere-to-telomere chromosome assemblies1, and uncovered new complex structural variants2. Additionally, as long-read sequencing becomes more economical and available for large-scale production, our understanding of rare and common variants across diverse human haplotypes will become more comprehensive. A complete assessment of variation, small and large, offers an opportunity to explore sources of hidden heritability. Here we explore the promise that long-read sequencing holds for uncovering complex variants through access to more-complete human reference genomes, and how such resources can support large-scale disease-association studies in the future.
Since its initial release, the human reference genome3,4 has served as a critical map for variant discovery5,6. Resulting catalogs have described variant frequency within the population5, defined shared haplotypes6 and broadened our understanding of the functional role of a number of variants in human health and disease7. For decades these surveys have been limited by technology: efforts have largely focused on single-nucleotide-variant calls in regions where short, paired read sequences could be confidently mapped to a single human reference genome. By contrast, structural variation — defined by genomic events that involve 50 or more base pairs — is difficult to predict in traditional whole-genome-sequencing studies confidently. As a result, larger variants in the genome in the form of deletions or insertions, inversions, translocations and complex rearrangements (for example, involving segmental duplications and satellite DNA) are not readily available in biomedical research. It is important to include these events to understand genome biology and function, as structural variations are commonly associated with cancer, developmental disorders and complex disease8-10. It has previously been estimated that roughly 70% of structural variations have remained undetected owing to mapping limitations of short-read data and inherent reference biases11. With long-read data and the development of new reference resources, we are able to more confidently identify structural variations. For example, advanced long-read studies of complex, clinically relevant structural variations involving the LPA gene — which has shown clinical utility as a predictor of vascular-related diseases12 — have offered a richer understanding of coding and haplotype-level sequence diversity13. Overall, it is understood that our catalogs of sequence variation are biased for small events, and that larger events may cumulatively have a greater effect on phenotype by affecting a larger genomic region. Therefore, as long-read data become more economical and scalable, we expect the standards of variant reporting to broaden and become more comprehensive.
Advances in sequencing technologies have increased the accuracy (to more than 99.9%) and length of sequencing reads, which has enabled the production of highly continuous genome assemblies and has increased the capacity for large structural-variation detection. High-fidelity reads from Pacific Biosciences are extremely accurate at the base level (over 99.9%) and typically within the range of 18–25 kilobases (kb) in length14. By comparison, Oxford Nanopore Technologies has released long-read duplex data (median of 25–35 kb), in which the template and complement strand of a single molecule of DNA are sequenced in succession to achieve sequencing results of very high accuracy (over 99.9%). Additionally, using a separate Oxford Nanopore Technologies ‘ultra-long’ (ONT-UL) protocol, nanopore sequencing supports read data with median lengths of 50–150 kb with slightly lower accuracy15 (R10.4.1, kit 14, with median sequence identities in the range of 98–99%). Moreover, long reads (from both PacBio and Oxford Nanopore Technologies) inherently encompass information about epigenetic patterns16,17 such as CpG methylation (but also 5hmC and others, including potentially novel ones), adding yet another layer to the variant characterization that we anticipate becoming routinely incorporated into future studies18. Therefore, the future holds rich datasets of conjoint genetic and epigenetic variation.
Genome assembly methods using ONT-UL and highly accurate reads (high-fidelity or duplex) result in highly continuous reference assemblies that have markedly increased the representation of complex, highly repetitive sequences of the genome19. Notably, the release of the first complete human reference genome (T2T-CHM13)1 revealed nearly 200 million bases that were missing from the previous human reference genome (Fig. 1). These new sequences represent pericentromeric and subtelomeric regions, recent segmental duplications, duplicated gene families and ribosomal DNA arrays that are distributed in regions of the genome that are known to be important for fundamental cellular processes. Further, these long-read complete assemblies provide a more-accurate reconstruction of regions harboring medically relevant genes that were either collapsed or incorrectly characterized using previous references2,20. Efforts to automate the assembly of complete, telomere-to-telomere diploid reference genomes have benefited from the combination of highly accurate and ultra-long reads21. It is now possible to routinely reach fully phased diploid telomere-to-telomere chromosomes with the addition of haplotype information (from parental data (familial trios), chromatin capture (Hi-C) or the Strand-seq method). Thus, we are entering into an exciting new era in which complete and phased genome assemblies are expected to be routinely available to the research community. As a result, variants in complex regions can be more confidently mapped and identified. Additionally, generating multiple assembly-to-assembly alignments of a collection of complete, diverse human diploid genomes (or a human ‘pangenome’) offers a new opportunity to study common variants and their associated haplotype structure.
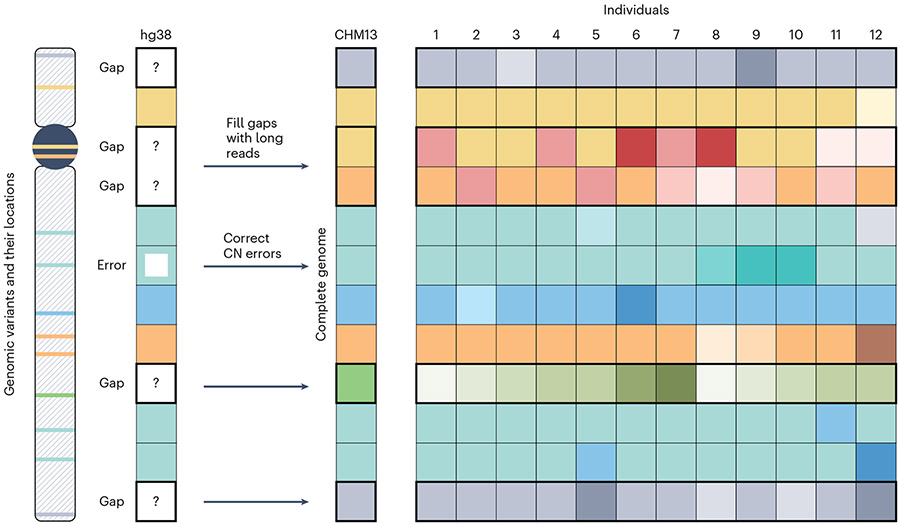
Fig. 1 ∣. Genomic variants and schematics of their location on the chromosome.
Locations on the chromosome include previously inaccessible regions such as telomeres, centromeres and pericentromeres, satellite DNA, and segmental duplications. A complete genome, such as T2T-CHM13, closes gaps in the assembly and corrects misassemblies, including copy number (CN) errors. Diverse samples of the human population are needed to differentiate between common and rare variants, to comprehensively catalog this variation and to find new disease associations.
The Human Pangenome Reference Consortium22 aims to ‘reboot’ the previous, linear human reference genome to represent a collection of complete, telomere-to-telomere assemblies that represent global genomic diversity. The pangenome represents the combination, or alignment, of these complete references and can be defined as a variation graph23. Ultimately, this reference represents a comprehensive catalog of common variants that will serve as a critical genomic resource for biomedical research and precision medicine. Long-read data will broaden our understanding of common single nucleotide variants and structural variations in the human population; however, arguably once this improved reference is available, efforts to identify and characterize variants using short-read datasets will markedly improve. Alignments of short-read or long-read data to the draft human pangenome24 have revealed clear improvements in structural variation genotyping and discovery. Methods such as PanGenie25 leverage short and longer linkage disequilibrium structures inherent in the pangenome assemblies to infer the genome of a new sample for which only short reads are available, and thereby enable the inclusion of tens of thousands of additional structural-variation alleles into genome-wide association studies. In the future, as large-scale long-read sequencing projects become more economical, it will be possible to formally explore the role of structural variation and rare variants as a source of missing heritability in disease-association studies. Long reads also face current limitations as we shift our studies to genetic and epigenetic variants within single cells, low-abundant cell-free DNA or intact tissues. Other than the cost, the current limitations of applying long-read sequencing data include baseline error rate and biases in sequencing that may influence predictions of rare somatic variants. In summary, long reads have brought us to the era of complete genomes and present an opportunity to expand our knowledge of variation in the human population, including the most repeat-rich sequences, which are the most dynamic in terms of copy number in the human genome.
Acknowledgements
K.H.M. and M.C. are supported by NIH/NHGRI U01HG010971.
Footnotes
Competing interests
K.H.M. is a science advisory board member of Centaura, Inc. and has received travel funds to speak at events hosted by Oxford Nanopore Technologies.
References
- 1.Nurk S. et al. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aganezov S. et al. Science 376, eabl3533 (2022).35357935 [Google Scholar]
- 3.Lander ES et al. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Venter JC et al. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 5.The 1000 Genomes Project Consortium. Nature 526, 68–74 (2015).26432245 [Google Scholar]
- 6.The International HapMap Consortium. Nature 426, 789–796 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Manolio TA Nat. Rev. Genet 14, 549–558 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Thibodeau ML et al. Genet. Med 22, 1892–1897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurles ME, Dermitzakis ET & Tyler-Smith C Trends Genet. 24, 238–245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weischenfeldt J, Symmons O, Spitz F & Korbel JO Nat. Rev. Genet 14, 125–138 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Ebert P. et al. Science 372, eabf7117 (2021).33632895 [Google Scholar]
- 12.Trinder M, Uddin MM, Finneran P, Aragam KG & Natarajan P JAMA Cardiol. 6, 287–295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin C-S et al. Preprint at bioRxiv 10.1101/2022.06.08.495395 (2022). [DOI] [Google Scholar]
- 14.Wenger AM et al. Nat. Biotechnol 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M. et al. Nat. Biotechnol 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flusberg BA et al. Nat. Methods 7, 461–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson JT et al. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Gershman A. et al. Science 376, eabj5089 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis ED et al. Nature 611, 519–531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner J. et al. Nat. Biotechnol 40, 672–680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rautiainen M. et al. Preprint at bioRxiv 10.1101/2022.06.24.497523 (2022). [DOI] [Google Scholar]
- 22.Wang T. et al. Nature 604, 437–446 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eizenga JM et al. Annu. Rev. Genomics Hum. Genet 21, 139–162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao W-W et al. Preprint at bioRxiv 10.1101/2022.07.09.499321 (2022). [DOI] [Google Scholar]
- 25.Ebler J. et al. Nat. Genet 54, 518–525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]