Abstract
Epstein-Barr virus (EBV) represents one of the most important viral carcinogens. EBV nuclear antigen-1 (EBNA1) can induce the expression of different cellular and viral genes. In this study, we evaluated the EBNA1 effects on the expression patterns of human papillomavirus type 18 (HPV-18) E6 and E7 oncogenes and three cellular genes, including BIRC5, c-MYC, and STMN1, in a cervical adenocarcinoma cell line. HeLa cells were divided into three groups: one transfected with a plasmid containing the EBNA1 gene, one transfected with a control plasmid, and one without transfection. In all three groups, the expression levels of E6, E7, BIRC5, c-MYC, and STMN1 genes were checked using real-time PCR. Pathological staining was used to examine changes in cell morphology. Real-time PCR results showed that the expression level of HPV-18 E6 (P=0.02) and E7 (P=0.02) oncogenes significantly increased in HeLa cells transfected with the EBNA1 plasmid compared to cells transfected with control plasmid. Also, the presence of EBNA1 induced the expression of BIRC5 and c-MYC, which increased tenfold (P=0.03) and threefold (P=0.02), respectively. Regarding the STMN1 cellular gene, although the expression level in HeLa cells transfected with EBNA1 plasmid showed a twofold increase, this change was insignificant (P=0.11). Also, EBNA1 expression caused the creation of large HeLa cells with abundant cytoplasm and numerous nuclei. The EBV-EBNA1 could increase the expression levels of HPV-18 E6 and E7 viral oncogenes as well as c-MYC and BIRC5 cellular genes in the HeLa cell line. These findings indicate that the simultaneous infection of cervical cells with HPV-18 and EBV might accelerate the progression of cervical cancer.
Key Words: Cervical carcinoma, human papillomavirus, Epstein–Barr Virus, EBNA1
Introduction
Cancer is a global problem, and viruses have long been known as a strong risk factor for different types of cancer. About 15-20% of all cancer cases are related to viruses, including Epstein-Barr virus (EBV), Hepatitis B virus (HBV), Hepatitis C virus (HCV), Human Herpes virus type 8 (HHV8), Human T-cell lymphotropic virus type 1 (HTLV-1), Human Papillomavirus (HPV), and Merkel cell polyomavirus (MCPyV) (1).
Cervical carcinoma (CC) is the fourth most common cancer among women after breast, lung, and colon cancers, which accounts for 1.6% of cancer deaths in women (2). Economic-social and health status, number of births, type of births, race, circumcision factor in men, age of first sexual contact, smoking, use of birth control pills, and age of first childbirth are the risk factors for CC (3). Among these factors, the most important cause of CC is infection with high risk types of HPVs (HR-HPVs) (4). In terms of virology, HPV belongs to the Papillomaviridae family, which includes small, non-enveloped viruses, and their double-stranded circular DNA genome is between 5748 - 8607 bp long (5, 6). Most of the HPVs infecting the cervix and anogenital areas are high-risk types, especially two types, 16 and 18, which have a high ability to cause CC (7). Among the eight non-structural proteins of this virus, the role of E6 and E7 in causing CC has been well proven (5). E6 protein can bind to tumor suppressor protein P53 and destroy it by forming a complex with cell ubiquitinating enzyme E6AP, so the proliferation of infected cell increases and the cell moves towards becoming cancerous (5). E7 protein binds to the cell transcription factor E2F and causes its release from retinoblastoma (RB), resulting in transcription of genes related to the proliferative phase of the cell cycle and cell division (5).
Recently, many clinical studies have acknowledged the role of EBV/HPV co-infection in the development of cancers related to HR-HPVs, especially CC (8, 9). EBV can easily infect cervical epithelial origin cells that are even infected with HPVs due to the abundance of EBV/C3d receptors on the surface of uterine cells (10-12). It is important to note that as the severity of an HPV-infected epithelial cell lesion increases due to further integration of the HPV genome in the host's chromosome, the conditions for EBV entry into such a cell improve (13, 14). Also, EBV infection in an HPV-derived cells can have a wide range of effects on cell signaling pathways, hastening the development of CC (11, 12).
EBV is classified as an oncogenic double-stranded DNA virus (length of roughly 170 kb) (12). It belongs to the family of herpesviridae and is found in more than 90% of adult humans, the majority of whom do not exhibit any symptoms (15). Saliva and genital secretions are the primary means of transmission for EBV (12, 16). EBV nuclear antigen-1 (EBNA1) is the sole viral protein present in all EBV latency types (17). EBNA1 has a regulatory role in the transcription of both viral and cellular promoters (17). The promoters of the genes whose transcription is controlled by EBNA1 are among the DNA sequences in the cellular genome that this protein can bind to as a viral transcription factor (18, 19).
The first cellular gene investigated in this study was BIRC5. The product of this gene inhibits cell apoptosis by interfering with the function of caspases. So the increase in the expression of this gene has been reported in many malignancies (20, 21). MYC, as a transcription factor, has a significant impact on a number of biological functions, including cell growth, proliferation, apoptosis, and cellular metabolism (22). Stathmin-1 (also known as oncoprotein 18) is a 17-kDa protein that plays an important role in cytoskeleton regulation (23). Numerous cellular processes, such as cytoplasmic organization, cell division, and cell motility, depend on the cytoskeleton, which serves as a scaffold (24).
Our understanding of cancer and its molecular underpinnings can be improved by identifying host-virus interactions, which can also help us in the discovery of prognostic indicators, novel antiviral therapies, and therapeutic interventions. In this study, we aimed to evaluate the EBV-EBNA1 effects on the expression patterns of HPV-18 E6 and E7 oncogenes as well as three cellular genes, including BIRC5, c-MYC, and STMN1 in a cervical adenocarcinoma cell line.
Materials and methods
Plasmids, bacterial transformation, and plasmid extraction
In this study, the Invitrogen pCEP4 plasmid (an EBV-based plasmid containing the EBNA1 gene and the hygromycin B resistance gene) as well as pcDNA3.1/hygro+ as control plasmid that contains the hygromycin B resistance gene but lacked the EBNA1 gene were used. The plasmids were transformed into Escherichia coli (the Top10 strain) and multiplied. To confirm the presence of the EBNA1 gene in the plasmid, enzyme digestion and colony PCR methods were used. Then, these plasmids were extracted, and their quality and concentrations were determined using gel electrophoresis and spectrophotometry.
Cell culture, transfection, and clonal selection by hygromycin B
HeLa cells (cervical adenocarcinoma cell line containing HPV-18) were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS) at 37°C and 5% CO2 in a six-well plate. After reaching the cell confluency of about 70%, one group of cells was transfected with the pCEP4 plasmid (containing the EBNA1 gene) and the other group with the control plasmid (lacking the EBNA1 gene) using an optimized concentration of DNA fectamine (BioBasic, Canada). All experiments were performed in duplicate, i.e. we have two well for each group (two well for EBNA1 transfected group, two well for control plasmid transfected group). After 24 hours of transfection, both EBNA1- and control-plasmid transfected cell groups were treated with 350 μg/mL hygromycin B to select EBNA1 transfected cells (cells with stable EBNA1 expression) and cells containing control plasmid. These cell groups were cultured for 16 days in the presence of hygromycin B during several passages to achieve 100% selection.
RNA isolation and cDNA synthesis
An RNA Isolation Kit (Dena Zist, Iran) was used to extract the total RNAs. To assess the quality and quantity of the extracted RNAs, gel electrophoresis and spectrophotometry (NanodropTM Spectrophotometer, Thermo Scientific, USA) were used. Following the manufacturer's instructions, isolated RNA (1000 ng/μL from each sample) was reverse transcribed into cDNA using an EasycDNA Synthesis Kit (AddScript RT-PCR SYBR Master, AddBio, Sweden).
Primer design and real-time PCR
The NCBI gene database and AlleleID software (version 7) were used to design primers. Table 1 displays the sequences of designed primers. SYBR green-based real-time PCR was used to assess the expression of the E6 and E7 viral oncogenes and cellular genes, including BIRC5, c-MYC, and STMN1. The qRT-PCR ABI QuantStudioTM3 instrument (Applied Biosystems, Grand Island, NY, USA) was employed for this purpose. The beta-actin gene was used as the reference gene. 2x Master Mix Green (Ampliqon Inc., Denmark), cDNA, primers (at a concentration of 10 pmol), and water made up the final volume of each reaction, which was 15 µL. For the PCR program, a denaturation phase of 95 °C lasting 15 minutes was followed by 40 cycles of 95 °C for 15 seconds and annealing/extension at 62 °C for 1 minute.
Table 1.
Primers used for evaluation of gene expression by relative quantitative real-time PCR
| Gene Name | Sequence | Product size | Primer position |
|---|---|---|---|
| E6 | 5'-TTAATAAGGTGCCTGCGGTG-3' 5'-GCGTCGTTGGAGTCGTTC-3' |
156 bp | F: 304-323 R: 459-442 |
| E7 | 5'-TCACGAGCAATTAAGCGACT-3' 5'-CACGGACACACAAAGGACAG-3' |
219 bp | F: 81-100 R: 299-280 |
| c-MYC | 5'-TCACACCCTTCTCCCTT-3' 5'-CGCTCCACATACAGTCC-3' |
180 bp | F: 1432-1448 R: 1611-1595 |
| BIRC5 | 5'-AGTTGGAGTGGAGTCTGG-3' 5'-CTTGCTGGTCTCTTCTGG-3' |
144 bp | F: 2346-2363 R: 2489-2472 |
| STMN1 | 5'-GCTTGTCTTCTATTCACCAT-3' 5'-TTGCGTCTTTCTTCTGC-3' |
203 bp | F: 239-258 R: 441-425 |
| EBNA1 | 5'-GGGTGGTTTGGAAAGCATCG-3' 5'-CTTACTACCTCCATATACGAACACA-3' |
156 bp | F: 1257-1276 R: 1413-1387 |
| Beta-actin | 5'-GCCTTTGCCGATCCGC-3' 5'-GCCGTAGCCGTTGTCG-3' |
90 bp | F: 135-150 R: 240-224 |
Verification of EBNA1 gene expression
To remove plasmid contamination, total RNA extracted from transfected cells was treated with DNase (Sinaclon, Tehran, Iran) according to the manufacturer's instructions. The expression of EBNA1 was verified using real-time PCR. Total RNA that had been subjected to DNase was used as the negative control.
Pathology staining
At the end of the 16th day, cells with stable EBNA1 expression and cells containing control plasmid were harvested. To examine morphological changes, hematoxylin and eosin (H&E) staining was used.
Data analysis
All experiments were performed in duplicate. The final results were calculated based on the average of corresponding experiments for each group. The CtNorm algorithm, which can be accessed online at http://ctnorm.sums.ac.ir, was used to equalize the Ct values of the qRT-PCR runs (25). The data were computed in Microsoft Excel after Ct normalization. Mann –Whitney U test in GraphPad Prism software was applied to compare means. Statistical significance was defined as a P-value less than 0.05.
Results
Real-time PCR results analysis of the expression of HPV-18 E6, and E7 oncogenes following EBNA1 transfection
The expression levels of two HPV-18 oncogenes, E6 and E7, were compared between EBNA1 transfected cells and mock plasmid transfected cells (Figure 1A). Real-time PCR results showed that the expression of E6 (P=0.02) and E7 (P=0.02) genes was considerably increased in HeLa cells containing EBNA1 gene compared to mock plasmid transfected controls (E6 gene expression increased twofold, and E7 gene expression increased thrice).
Fig.1.
Real-time PCR results analysis of the expression of E6, E7, BIRC5, STMN1, and c-MYC genes: A) Comparison of expression changes of E6, and E7 viral oncogenes between EBNA1 transfected HeLa cells and control cells B) Comparison of expression changes of BIRC5, STMN1, and c-MYC genes between EBNA1 transfected HeLa cells and control cells
Real-time PCR results analysis of BIRC5, c-MYC, and STMN1 gene expression following EBNA1 transfection
The expression levels of BIRC5, c-MYC, and STMN1 cellular genes were compared between cells transfected with the EBNA1 plasmid and controls (Figure 1B). According to real-time PCR results, in the presence of EBNA1, the expression levels of BIRC5 and c-MYC genes was increased tenfold (P = 0.03) and threefold (P=0.02), respectively. Regarding the STMN1 cellular gene, although the expression of this gene in EBNA1 transfected HeLa cells showed a twofold increase, this change was not significant (P = 0.11).
Pathological staining for morphological examination
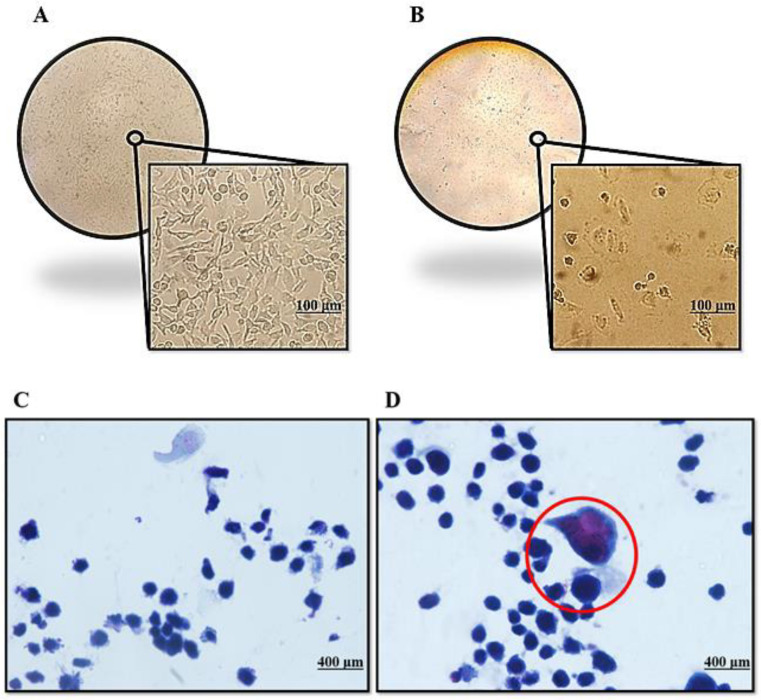
We observed alterations in the morphology of EBNA1 transfected cells after selecting hygromycin B-resistant clones. Pathological staining revealed, as depicted in Fig. 2, that EBNA1 expression changed the shape of HeLa cells in comparison to the control (mock plasmid-transfected) cells. HeLa cells transfected with the EBNA1 plasmid became abnormally large cells with numerous nuclei and abundant cytoplasm
Fig.2.
Morphological changes of HeLa cell line transfected with EBNA1 and control plasmid: A) HeLa cells transfected with the control plasmid; B) abnormal morphology in the culture of the HeLa cells transfected with EBNA1; C) pathological staining of control plasmid transfected cell; D) pathological staining of EBNA1 plasmid transfected cell
Discussion
Today, the study of the connection between viruses and cancer is of interest to many researchers. Numerous cancers and EBV have been proven to be related thus far. However, the effect of this virus on some cancers, including CC, remains unknown. Recently some studies have reported the frequency of EBV in epithelial tissues isolated from CC patients (9, 26, 27). Based on a study conducted in 2019 by Guidry et al., it is stated that when EBV enters the cell infected with HR-HPV, the prevailing conditions of the cell cause EBV not to replicate, and enter to its latent phase (28). The only viral protein presents in all EBV latency types is EBNA1 (17). EBNA1 can affect the cellular and viral genes expression by binding to their promoters (17, 29).
In this study, we found that HPV-18 E6 oncogene, was upregulated at the mRNA level in HeLa cells that were transfected with a plasmid containing EBNA1 gene compared to control plasmid transfected cells. E6 protein, by forming a complex with the cellular ubiquitination enzyme E6AP, can bind to the tumor suppressor protein P53 and cause its destruction through the proteasomal degradation pathway (5). Following the reduction of P53 protein in the cell, E6 can induce the BIRC5 gene promoter. Therefore, this drop in P53 level and increase in the BIRC5 gene product, both can inhibit apoptosis of the infected cell (5, 30). Also, the E6/E6AP complex, by binding to another cellular protein called NFX1-91, which plays a role in inhibiting the expression of the human telomerase reverse transcriptase (hTERT) enzyme, causes its destruction (31). Therefore, the NFX1-91 inhibitory effect on the expression of hTERT is removed (31). On the other hand, E6 protein of HPV-18 by forming a complex with c-MYC cellular protein (E6/c-MYC), directly affects the hTERT gene promoter and increases its transcription. So this enzyme is produced in a high amount, which results in the immortalization of the host cell (31). The E6/E6AP complex also interacts with other cellular proteins, including the pro-apoptotic protein BAK1, the Fas-associated protein with death domain (FADD), and procaspase 8, to inhibit apoptosis in infected cells (32). In 2018, Al-Thawadi et al. revealed that the co-expression of the E6 gene of HR-HPVs and the EBV-LMP1 gene is associated with a phenotype of poorly differentiated squamous cell carcinoma (26). So, the expression of the DNA-binding protein inhibitor 1 (Id-1) increases greatly and, in combination with nuclear NF κB p65, can be associated with aggressive CC behavior and poorer clinical outcomes (33, 34). As a result, increased E6 expression by EBNA1 may hasten the progression of cervical lesions to cancerous status in HPV/EBV coinfection compared to HPV mono-infection.
Also we found that the expression level of HPV-18 E7 gene was significantly increased in EBNA1 transfected HeLa cells. The product of this viral oncogene binds to the cell transcription factor E2F, causing it to be released from RB. In this way, E2F enters the cell nucleus and transcribes genes related to the proliferative phase of the cell cycle, so the infected cell goes from G1 to the S-phase and continues to multiply (5). E7 protein also interacts with cellular histone deacetylase 1 and 2 (HDAC 1, 2) enzymes and thus leads to activation of transcription (35). Also, E7 can bind to the TMEM173/STING protein complex in the infected cell and prevent the identification of the viral genome in the cytoplasm. So, the innate immune responses of interferon alpha and beta are inhibited (36). It has also been reported that in the case of HPV/EBV co-infection, the HPV-E7 protein prevents the replication of EBV and drives this virus to its latent phase, which might contribute to the progression of the cell to become cancerous (28). Hence, the expression of EBNA1 in HPV/EBV co-infection could increase the potency of viral carcinogenesis more than in HPV mono-infection. To the best of our knowledge, for the first time, we showed that EBV-EBNA1 could increase the expression level of HPV-18 E6 and E7 oncogenes in HeLa cell line.
According to our study, the expression level of BIRC5 gene was about tenfold higher in EBNA1 transfected HeLa cells (compared to control cells). In agreement with our study, Lu et al. stated that in Burkitt's lymphoma, EBNA1 increases the expression of the BIRC5 gene by forming a complex and binding to the promoter of this gene (37). BIRC5 (also named survivin) is a member of the inhibitors of apoptosis proteins (IAPs) family, which by interfering in the functions of caspases 3, 7 and also inhibiting Bax and Fas-induced apoptotic pathways, causes abnormal cells to survive (38). So, increased expression of BIRC5 by EBNA1 might contribute to the pathogenesis of CC in HPV/EBV co-infected cervical cells.
Our study indicated that the expression level of the c-MYC gene increased threefold in HeLa cells transfected with EBNA1 plasmid compared to control plasmid transfected cells. Based on the study by Ferber et al., it was found that in CC, the HPV-18 genome is integrated in a region near the c-MYC gene and can cause the activation of this proto-oncogene (39). Also, it is stated that the product of this gene in the presence of HR-HPVs-E6 protein, can induce the cell hTERT enzyme and ultimately lead to the immortality of the infected cell (31, 39). According to studies conducted in Burkitt lymphoma cases, it is clear that EBV is involved in 90% of c-MYC gene translocations, which can cause the activation of this proto-oncogene, and subsequently, be involved in the development and/or progression of cancers (40). Also, c-MYC protein is able to bind to the promoter region of vascular endothelial growth factor A (VEGFA) to stimulate more production of this growth factor and then cause angiogenesis (41). Therefore, as a multi-functional proto-oncogene, increased expression of c-MYC by EBNA1 in cervical cells co-infected with HR-HPVs and EBV might push cervical lesions to CC more than in HPV infection alone.
The results of our study indicated that STMN1 gene transcript level was doubled in HeLa cells transfected with EBNA1 plasmid, although this change in STMN1 gene expression was not significant. According to Miceli et al., STMN1 gene expression is increased in breast cancer and is directly related to tumor size and degree of damage (42). Also, in agreement with our results, a study performed in 2012 on nasopharyngeal carcinoma (NPC) determined that the expression level of the STMN1 gene increased threefold in the presence of EBV-EBNA1 (43). Also, in another study, it is stated that in cancers related to HPVs, such as oropharyngeal cancer, the expression level of this cellular gene increases significantly (44).
Based on the results of our study, pathological staining revealed that the expression of EBNA1 had an impact on the morphology of HeLa cells. Similar results were obtained by Wang et al., who found that EBNA1 affected NPC cell shape as well as the expression of markers for the epithelial-mesenchymal transition (EMT) (45). They also showed that the EBNA1 was highly expressed in NPC tissue samples, tying this expression to NPC lymph node metastasis (45). To the best of our knowledge, for the first time, we showed that the expression of EBNA1 can lead to significant morphological changes in HeLa cells. Other studies did not report such morphological changes in epithelial cells, which could be attributed to their method of transiently transfecting the cells, whereas we looked at stabilized long-term EBNA1 effects. All of these findings point to a link between EBNA1 expression and the expression of viral and cellular genes in the HeLa cell line, and we believe that such a relationship, in the case of simultaneous infection with HR-HPVs and EBV, causes the development and progression of CC more than HPV infection alone.
In conclusion, our results indicated that the EBV-EBNA1 could increase the expression levels of HPV-18 E6 and E7 oncogenes as well as cellular genes, including BIRC5, and c-MYC in the HeLa cell line. Therefore, simultaneous EBV and HPV-18 infection of cervical cells might accelerate the progression of CC to a higher degree not only by HPV carcinogenesis mechanisms but also by inducing and over-expression of HPV-18 E6 and E7 oncogenes, as well as BIRC5, and c-MYC cellular genes through EBV-EBNA1. Further studies are recommended to clarify these results.
Acknowledgments
This study was extracted from Amir Hossein Alipour's M.Sc. thesis. Shiraz University of Medical Sciences provided financial assistance for this work (Grant No. 25219).
References
- 1.McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta. 2008;1782:127–50. doi: 10.1016/j.bbadis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzmaurice C, Allen C, et al. Global Burden of Disease Cancer C. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kashyap N, Krishnan N, Kaur S, et al. Risk Factors of Cervical Cancer: A Case-Control Study. Asia Pac J Oncol Nurs. 2019;6:308–14. doi: 10.4103/apjon.apjon_73_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrestha AD, Neupane D, Vedsted P, et al. Cervical Cancer Prevalence, Incidence and Mortality in Low and Middle Income Countries: A Systematic Review. Asian Pac J Cancer Prev. 2018;19:319–24. doi: 10.22034/APJCP.2018.19.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasi Bonab F, Baghbanzadeh A, Ghaseminia M, et al. Molecular pathways in the development of HPV-induced cervical cancer. EXCLI J. 2021;20:320–37. doi: 10.17179/excli2021-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vashisht S, Mishra H, Mishra PK, et al. Structure, Genome, Infection Cycle and Clinical Manifestations Associated with Human Papillomavirus. Curr Pharm Biotechnol. 2019;20:1260–80. doi: 10.2174/1389201020666190802115722. [DOI] [PubMed] [Google Scholar]
- 7.Weaver BA. Epidemiology and natural history of genital human papillomavirus infection. J Am Osteopath Assoc. 2006;106:S2–8. [PubMed] [Google Scholar]
- 8.Shi Y, Peng SL, Yang LF, et al. Co-infection of Epstein-Barr virus and human papillomavirus in human tumorigenesis. Chin J Cancer. 2016;35 doi: 10.1186/s40880-016-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lima MAP, Neto PJN, Lima LPM, et al. Association between Epstein-Barr virus (EBV) and cervical carcinoma: A meta-analysis. Gynecol Oncol. 2018;148:317–28. doi: 10.1016/j.ygyno.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Joharinia N, Faghihinejad S, Seyedi K, et al. Co-existing of HSV1/2 or EBV Infection with the Presence of High-Risk HPV DNA in Cervical Lesions in the Southwest of Iran. Asian Pac J Cancer Prev. 2020;21:1459–64. doi: 10.31557/APJCP.2020.21.5.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui X, Cao Z, Ishikawa Y, et al. Immunization with Epstein-Barr Virus Core Fusion Machinery Envelope Proteins Elicit High Titers of Neutralizing Activities and Protect Humanized Mice from Lethal Dose EBV Challenge. Vaccines (Basel) 2021:9. doi: 10.3390/vaccines9030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanco R, Carrillo-Beltran D, Osorio JC, et al. Role of Epstein-Barr Virus and Human Papillomavirus Coinfection in Cervical Cancer: Epidemiology, Mechanisms and Perspectives. Pathogens. 2020:9. doi: 10.3390/pathogens9090685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oyervides-Munoz MA, Perez-Maya AA, Rodriguez-Gutierrez HF, et al. Understanding the HPV integration and its progression to cervical cancer. Infect Genet Evol. 2018;61:134–44. doi: 10.1016/j.meegid.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Makielski KR, Lee D, Lorenz LD, et al. Human papillomavirus promotes Epstein-Barr virus maintenance and lytic reactivation in immortalized oral keratinocytes. Virology. 2016;495:52–62. doi: 10.1016/j.virol.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Yu Z, Shen G, et al. Association between Epstein-Barr virus and Thymic epithelial tumors: a systematic review. Infect Agent Cancer. 2019;14:32. doi: 10.1186/s13027-019-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mui UN, Haley CT, Tyring SK. Viral Oncology: Molecular Biology and Pathogenesis. J Clin Med. 2017:6. doi: 10.3390/jcm6120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, Xie C, Lung HL, et al. EBNA1-targeted inhibitors: Novel approaches for the treatment of Epstein-Barr virus-associated cancers. Theranostics. 2018;8:5307–19. doi: 10.7150/thno.26823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sompallae R, Callegari S, Kamranvar SA, et al. Transcription profiling of Epstein-Barr virus nuclear antigen (EBNA)-1 expressing cells suggests targeting of chromatin remodeling complexes. PLoS One. 2010;5:e12052. doi: 10.1371/journal.pone.0012052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood VH, O'Neil JD, Wei W, et al. Epstein-Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFbeta signaling pathways. Oncogene. 2007;26:4135–47. doi: 10.1038/sj.onc.1210496. [DOI] [PubMed] [Google Scholar]
- 20.Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res. 2019;38 doi: 10.1186/s13046-019-1362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sah NK, Khan Z, Khan GJ, et al. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–71. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, et al. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2022;19:23–36. doi: 10.1038/s41571-021-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai T, Yokobori T, Altan B, et al. High STMN1 level is associated with chemo-resistance and poor prognosis in gastric cancer patients. Br J Cancer. 2017;116:1177–85. doi: 10.1038/bjc.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–3. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramezani A. CtNorm: Real time PCR cycle of threshold (Ct) normalization algorithm. J Microbiol Methods. 2021;187:106267. doi: 10.1016/j.mimet.2021.106267. [DOI] [PubMed] [Google Scholar]
- 26.Al-Thawadi H, Ghabreau L, Aboulkassim T, et al. Co-Incidence of Epstein-Barr Virus and High-Risk Human Papillomaviruses in Cervical Cancer of Syrian Women. Front Oncol. 2018;8:250. doi: 10.3389/fonc.2018.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khenchouche A, Sadouki N, Boudriche A, et al. Human papillomavirus and Epstein-Barr virus co-infection in cervical carcinoma in Algerian women. Virol J. 2013;10:340. doi: 10.1186/1743-422X-10-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guidry JT, Myers JE, Bienkowska-Haba M, et al. Inhibition of Epstein-Barr Virus Replication in Human Papillomavirus-Immortalized Keratinocytes. J Virol. 2019:93. doi: 10.1128/JVI.01216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canaan A, Haviv I, Urban AE, et al. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc Natl Acad Sci U S A. 2009;106:22421–6. doi: 10.1073/pnas.0911676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borbely AA, Murvai M, Konya J, et al. Effects of human papillomavirus type 16 oncoproteins on survivin gene expression. J Gen Virol. 2006;87:287–94. doi: 10.1099/vir.0.81067-0. [DOI] [PubMed] [Google Scholar]
- 31.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–17. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- 32.Yuan CH, Filippova M, Tungteakkhun SS, et al. Small molecule inhibitors of the HPV16-E6 interaction with caspase 8. Bioorg Med Chem Lett. 2012;22:2125–9. doi: 10.1016/j.bmcl.2011.12.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darnel AD, Wang D, Ghabreau L, et al. Correlation between the presence of high-risk human papillomaviruses and Id gene expression in Syrian women with cervical cancer. Clin Microbiol Infect. 2010;16:262–6. doi: 10.1111/j.1469-0691.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- 34.Wu Z, Li J, Zhang Y, et al. Synchronous co expression of Id 1 and nuclear NF kappaB p65 promotes cervical cancer progression and malignancy, and is associated with a poor prognosis and chemosensitivity. Oncol Rep. 2019;42:2075–86. doi: 10.3892/or.2019.7301. [DOI] [PubMed] [Google Scholar]
- 35.Lau L, Gray EE, Brunette RL, et al. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–71. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Garcia E, Olague C, Rius-Rocabert S, et al. TMEM173 Alternative Spliced Isoforms Modulate Viral Replication through the STING Pathway. Immunohorizons. 2018;2:363–76. doi: 10.4049/immunohorizons.1800068. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, Murakami M, Verma SC, et al. Epstein-Barr Virus nuclear antigen 1 (EBNA1) confers resistance to apoptosis in EBV-positive B-lymphoma cells through up-regulation of survivin. Virology. 2011;410:64–75. doi: 10.1016/j.virol.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Duan N, Zhang C, et al. Survivin and Tumorigenesis: Molecular Mechanisms and Therapeutic Strategies. J Cancer. 2016;7:314–23. doi: 10.7150/jca.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferber MJ, Thorland EC, Brink AA, et al. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233–42. doi: 10.1038/sj.onc.1207006. [DOI] [PubMed] [Google Scholar]
- 40.De Falco G, Ambrosio MR, Fuligni F, et al. Burkitt lymphoma beyond MYC translocation: N-MYC and DNA methyltransferases dysregulation. BMC Cancer. 2015;15:668. doi: 10.1186/s12885-015-1661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, Xu X, Zhang Q, et al. tRNA synthetase counteracts c-Myc to develop functional vasculature. Elife. 2014;3:e02349. doi: 10.7554/eLife.02349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miceli C, Tejada A, Castaneda A, et al. Cell cycle inhibition therapy that targets stathmin in in vitro and in vivo models of breast cancer. Cancer Gene Ther. 2013;20:298–307. doi: 10.1038/cgt.2013.21. [DOI] [PubMed] [Google Scholar]
- 43.Cao JY, Mansouri S, Frappier L. Changes in the nasopharyngeal carcinoma nuclear proteome induced by the EBNA1 protein of Epstein-Barr virus reveal potential roles for EBNA1 in metastasis and oxidative stress responses. J Virol. 2012;86:382–94. doi: 10.1128/JVI.05648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohavanichbutr P, Houck J, Fan W, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–8. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Tian WD, Xu X, et al. Epstein-Barr virus nuclear antigen 1 (EBNA1) protein induction of epithelial-mesenchymal transition in nasopharyngeal carcinoma cells. Cancer. 2014;120:363–72. doi: 10.1002/cncr.28418. [DOI] [PubMed] [Google Scholar]