Abstract
Polycystic ovary syndrome (PCOS) is the most prevalent endocrine disorder of women in reproductive age with significant effects on reproductive and metabolic functions. Many molecular players may be involved in PCOS pathology; however, miRNAs possess great ability in gene expression control in normal ovarian function and folliculogenesis. We appraised the relative expression of miR-146a, miR-222, miR-9, and miR-224 in serum and follicular fluid (FF) of PCOS patients compared to control subjects. PCOS (n = 35) and control (n = 30) subjects were recruited in the study during their enrolment in IVF cycles. Serum and FF of human subjects were collected and stored. Total RNA was isolated from samples and cDNA was synthesized using miRNA-specific stem-loop RT primers. Quantitative real-time PCR was used to evaluate the expression of miRNAs relative to U6 expression. The predictive value of miRNAs’ expression for discrimination of PCOS patients from control subjects was evaluated by receiver-operating characteristic (ROC) curve analysis. miR-224 was not detected in serum and FF samples. Significantly, higher levels of miR-146a and miR-9 in serum of PCOS group were detected. In contrast, relative expression of miR-146a and miR-9 significantly decreased in FF. In PCOS group, relative expression of all detected miRNAs was elevated in serum in comparison to FF, whereas in control group no change was noticed. Combination of FF miRNAs showed improved predictive value with area under the ROC curve (AUC) of 0.84, 93.8% sensitivity, and 83.3% specificity. Contradicting alternations of miRNAs in serum and FF are indicative of different sources of miRNAs in body fluids. Presumptive target genes of studied miRNAs in signalling pathways may show the potential role of these miRNA in folliculogenesis.
Key Words: miRNA, follicular fluid, Polycystic ovary syndrome, serum
Introduction
Polycystic ovary syndrome (PCOS) is a complex multifactorial endocrine disorder affecting 6-20% of women of childbearing age, based on the different criteria for diagnosis (1). PCOS disturbs hypothalamic–pituitary–ovarian axis and reveals diverse symptoms, including androgen excess, ovulatory dysfunction, polycystic ovaries, and metabolic difficulties (2). Affected patients manifest higher risk of insulin resistance, obesity, and diabetes (3). Impaired folliculogenesis of PCOS is described with the higher number of follicles in the early developmental stages and failure of selection of dominant follicle that ends in folliculogenesis blockade at pre-antral stage and development of polycystic ovary morphology and anovulation (3-5). The main cause of PCOS has not been discovered yet. Hence, countless studies have been carrying out to discover gene expression alternations and regulators of gene expression in different tissues and body fluids of PCOS patients to unravel complex molecular event contributing in PCOS pathophysiology and find novel biomarker for more accurate diagnosis of PCOS.
MicroRNAs (miRNAs) are small non-coding molecules with a length of about 20-25 nucleotides that play integral roles in disease and development through post-transcriptional regulation of gene expression (6). MiRNAs’ binding to 3´-untranslated regions of mRNAs creates a complex network of molecular interactions. The multifaceted function of miRNAs in gene expression control justifies their investigation in multifactorial and heterogamous disorders such as PCOS. In this regard, several groups have reported dysregulated expression of miRNAs in granulosa cells (GCs) (7), follicular fluid (FF) (8), blood (9), ovarian tissue (10), and adipose tissue of PCOS (11). Furthermore, hormone responsiveness of miRNAs has been confirmed (7, 12), which highlights the importance of miRNAs in PCOS as the disorder is associated with a dramatic imbalance of hormones.
Chen et al. reported increased levels of miR-93 in adipose tissue of PCOS patients, and subjects with insulin resistance showed significantly higher levels compared to non-insulin-resistant ones. They also demonstrated that miR-93 effectively targeted and regulated the expression of GLUT4 (glucose transporter), which suggested a significant role of miR-93 in insulin resistance of PCOS conditions (11). Increased proliferation of granulosa cells is a significant contributor to impaired folliculogenesis of PCOS. Elevated expression of miR-3940-5p in GCs of PCOS was reported. Furthermore, overexpression of miR-3940-5p stimulated the proliferation of GCs, which was revealed to be mediated by targeting voltage-gated channel subfamily A member 5 (KCNA5) (13). In addition, another study by Zhou et al. showed that KCNA5 could be the target of miR-3188 in regulating the viability and apoptosis of GCs (14). Up-regulated expression of miR-135a was observed in GCs of PCOS patients. It was also indicated that overexpression of miR-135a could promote apoptosis of GCs by targeting and suppressing vascular endothelial growth factor C (VEGFC) (15). Hormonal imbalance is a hallmark of PCOS, and miRNAs showed effective regulating effects in this context. Zhang et al. demonstrated significantly down-regulated expression of miR-320a in cumulus granulosa cells of PCOS patients. They further indicated that a dysregulated cascade of miR-320a/RUNX2/CYP11A1 contributed to compromised PCOS steroidogenesis (16).
It was reported that miR-146a in human granulosa cells regulated the apoptosis process by targeting interleukin‑1 receptor‑associated kinase and tumor necrosis factor receptor‑associated factor 6 (17). Furthermore, the importance of miR-146a in granulosa cells’ function was highlighted by Cho et al., which reported the association of miR-143a polymorphism with broad gene expression alternation in granulosa cells (18). MiR-222 could play significant roles in the pathogenesis of PCOS by affecting proliferation and apoptosis of granulosa cells through targeting p27 Kip1 (19). More supporting findings for the role of miR-222 in the pathogenesis of PCOS was reported by Ortega et al. that showed its potential relevance with insulin sensitivity of PCOS (20). During PCOS condition, miR-9 can regulate the expression of vitamin D receptor in granulosa cells resulted in significant effects on the proliferation and apoptosis of these cells (21). miR-224 is a TGF-beta-responsive miRNA, which several reports indicated its potential roles in cumulus expansion, oocyte competency, and embryo development (22, 23). In this work, the expression of miR-146a-5p, miR-222-3p, miR-9-5p, and miR-224-5p was evaluated in serum and FF of PCOS patients to study their role in PCOS condition and evaluate their function as biomarkers of PCOS.
Materials and methods
Participants and sample collection
This study was approved by the Research Deputy and Ethics Committee of Tehran University of Medical Sciences (IR.TUMS.MEDICINE.REC.1396.4213). All participants were enrolled at the Infertility Department of Shariati Hospital (Tehran, Iran) after giving their informed consent. PCOS patients (n = 35) were selected according to the revised Rotterdam European Society of Human Reproduction and Embryology/American Society for Reproductive Medicine Criteria (24) after exclusion of other etiologies including non-classic congenital adrenal hyperplasia. Control subjects (n=30) were IVF candidates for male factor etiologies and sex selection without any previous record of menstrual irregularities. All human subjects did not receive any medication affecting steroids and glucose metabolism for at least three months before the study and were scheduled for the first cycle of IVF treatment. Exclusion criteria were background and affection of other endocrinopathies, endometriosis, and premature ovarian failure. Basic clinical features of human subjects were assessed by measurement of age, body mass index (BMI), Ferriman-Gallwey score, total testosterone (TT), free testosterone (FT), 17-Hydroxyprogesterone (17-OHP), follicle-stimulating hormone (FSH), luteinizing hormone (LH), LH/FSH ratio, estradiol (E2), prolactin (ng/mL), thyroid-stimulating hormone (TSH), fasting plasma glucose, fasting serum insulin, and homeostatic model assessment for insulin resistance (HOMA-IR).
Gonadotropin hormone-releasing hormone (GnRH) antagonist protocol was used for all human subjects. Stimulation protocol was initiated by daily administration of recombinant FSH (Gonal-F, Merck Serono SA, Switzerland; 150-300 IU) at day 3 of the menstrual cycle and sustained until two follicles (14–15 mm) were observed. Afterwards, the dosages of gonadotropins were customized according to the patient’s age, estradiol levels, and transvaginal ultrasonic measurements of the follicles. Administration of GnRH antagonist (cetrorelix; Merck Serono SA, Switzerland; 0.25 mg/day) began and continued till the observation of at least two follicles with a diameter of 18 mm. Subsequently, 10,000 IU human chorionic gonadotropin (hCG) (Ovitrelle, Merck Serono SA, Switzerland) was administered. Finally, oocyte retrieval was performed after 36 h, and FF was collected. Serum samples were collected on the day of FF collection.
RNA isolation, cDNA synthesis and quantitative real-time PCR
RNA isolation from 200 µl of serum and FF was carried out by Trizol reagent (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instruction. RNA concentration was quantified using a spectrophotometer (WPA, Biochrom). Genomic DNA contamination was removed by RNase-free DNase I (Thermo Scientific, Waltham, MA, USA) treatment. RNA samples were reversely transcribed to cDNA by first-strand cDNA synthesis kit (Sinaclon, Iran) primed with stem-loop miRNA specific primers (100 ng template RNA for each miRNA) or random hexamer (500 ng RNA for U6).
Primers were designed using AlleleID 6 software and listed in Table 1. Quantitative RT-PCR reactions were composed of 10 μl 2X RealQ Plus ProbeMix (Ampliqon, Denmark), 0.8 μL of each primer, 0.5 μL probe, 2 μL first-strand cDNA template (1:2 in distilled water) and 5.9 μL distilled water. Cycling factors were 95 °C for 15 min to activate the enzyme, 40 cycles of 95 °C for 25 s followed by 60 °C for 60 s. Thermocycling was performed by Rotor-Gene Q instrument (Qiagen). To normalize the expression of miRNAs, RNU6-1 RNA (U6) expression level was parallelly measured (25, 26). The mean threshold cycles of the duplicate analysis of each sample were considered and used for quantification of miRNAs expression levels using the 2-∆Ct formula.
Table 1.
Sequence of oligonucleotides for quantification of miRNAs
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| RT Primer miR-146a | GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCAACCCA |
| Forward Primer miR-146a | GCGTGAGAACTGAATTCCA |
| RT Primer miR-222 | GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCACCCAG |
| Forward Primer miR-222 | GCGAGCTACATCTGGCTA |
| RT Primer miR-9 | GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCTCATAC |
| Forward Primer miR-9 | CGCCTCTTTGGTTATCTAGC |
| RT Primer miR-224 | GGTCGTATGCAAAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCCTAAAC |
| Forward Primer miR-224 | GTCAAGTCACTAGTGGTTCC |
| Universal Reverse Primer | AAGCAGGGTCCGAGGT |
| Universal Probe | FAM-TCCATCGCACGCATCGCACT-BHQ1 |
| Forward U6 | GCTTCGGCAGCACATATAC |
| Reverse U6 | ATTTGCGTGTCATCCTTGC |
| U6 Probe | FAM-CAGGGGCCATGCTAATCTTCTCT-BHQ1 |
Bioinformatic analysis
To estimate the potential effects of miRNAs’ dysregulation (miR-146a, miR-222, and miR-9) on biological processes and signalling pathways, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed by DIANA mirPath v.3 tool using Targetscan algorithm (27). A network of miRNAs with the members of important signalling pathways from KEGG analysis was made by Cytoscape (version 3.9.1) and ClueGO plugin.
Statistical analysis
The normality of data distribution was assessed by D'Agostino & Pearson omnibus normality test. Kruskal–Wallis test was used for comparison of miRNAs’ expression. Pairwise comparisons were performed by Mann–Whitney U test, and Bonferroni correction was considered for the estimation of the significance threshold. Clinical and demographic features were analysed by t-test or Mann–Whitney U test. Correlation analysis was performed by Spearman’s rank correlation test. Diagnostic values for the relative expression level of miRNAs in follicular fluid and serum were evaluated by receiver-operating characteristic (ROC) analysis using SPSS 22 (SPSS Inc., Chicago, IL). To evaluate cumulative diagnostic efficacy, miRNAs-based scores were obtained using the logistic regression model. All data were presented as mean ± standard error of the mean. GraphPad Prism 6.01 was used for other statistical analysis and presentations of data.
Results
Basic clinical and demographic features of participants
Basic and demographic characteristics of human subjects are listed in Table 2. Mean Ferriman-Gallwey score (P = 0.03), total testosterone (<0.0001), free testosterone (<0.0001), LH (P = 0.003), and LH/FSH ratio (P=0.02) in serum of PCOS patients were significantly higher than the control group. However, no significant differences were noticed in age, BMI, 17-OHP, FSH, E2, Prolactin, TSH, fasting plasma glucose, fasting serum insulin, and HOMA-IR between study groups.
Table 2.
Basic clinical features of participants
| Control (n = 30) | PCOS (n =35) | P-value | |
|---|---|---|---|
| Age (year) | 31.8 ± 0.67 | 28.7 ± 1.08 | 0.06 |
| BMI (kg/m2) | 25.4 ± 0.88 | 25.8 ± 0.60 | 0.69 |
| Ferriman-Gallwey Score | 6.05 ± 0.80 | 8.79 ± 1.07 | 0.03 |
| Total T (nmol/liter) | 1.18 ± 0.16 | 2.65 ± 0.30 | < 0.0001 |
| Free T (nmol/liter) | 0.015 ± 0.001 | 0.034 ± 0.004 | < 0.0001 |
| 17-OHP (ng/mL) | 0.53 ± 0.07 | 0.77 ± 0.10 | 0.1 |
| FSH (mIU/mL) | 6.02 ± 0.61 | 5.27 ± 0.49 | 0.34 |
| LH (mIU/mL) | 6.66 ± 0.89 | 9.80 ± 0.92 | 0.003 |
| LH/FSH | 1.33 ± 0.22 | 2.00 ± 0.21 | 0.02 |
| E2 (nmol/liter) | 0.17 ± 0.01 | 0.18 ± 0.01 | 0.55 |
| Prolactin (ng/mL) | 16.21 ± 1.82 | 15.75 ± 2.48 | 0.84 |
| TSH (μIU/mL) | 2.10 ± 0.25 | 1.96 ± 0.17 | 0.65 |
| Fasting plasma glucose (mmol/liter) | 5.26 ± 0.18 | 5.41 ± 0.16 | 0.53 |
| Fasting serum insulin (mIU/liter) | 5.21 ± 0.33 | 6.17 ± 0.51 | 0.13 |
| HOMA-IR | 1.23 ± 0.1 | 1.48 ± 0.13 | 0.15 |
Relative expression levels of miRNAs in follicular fluid, and serum
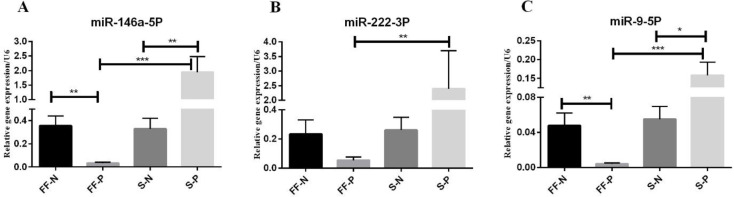
We detected significantly reduced levels of miR-146a in FF of the PCOS compared to the control group (P = 0.0005); however, serum levels of miR-146a were significantly elevated in the PCOS group versus the control group (P=0.0041). In PCOS group, serum levels of miR-146a were significantly increased compared to FF levels in this group (P < 0.0001) (Figure 1A).
Fig.1.
Relative expression of miR-146a, miR-222, and miR-9 in follicular fluid and serum of POCS patients and control group subjects. FF-N, follicular fluid of control (normal) group; FF-P, follicular fluid of PCOS group; S-N, seru
Regarding miR-222, the only significant difference was the elevated relative expression of miR-222 in the serum of PCOS versus FF of this group (P < 0.0001) (Figure 1B).
Relative expression of miR-9 was significantly down-regulated in the FF of the PCOS group compared to the control group (P =0.0034). On the other hand, serum levels of miR-9 were significantly up-regulated in the PCOS group versus the control group (P = 0.0118). In the PCOS group, serum levels of miR-9 were significantly increased compared to FF levels in this group (P < 0.0001) (Figure 1C). We did not observe a stable and reproducible signal for the relative expression of miR-224; therefore, it was undetectable in serum and FF.
Correlation analysis and Predictive value of miRNAs in follicular fluid and serum
The correlation of clinical and demographic features with the relative expression of miRNAs and the correlation of miRNAs’ relative expression levels in serum and FF are summarized in Table 3. In the control group, serum miR-222 was positively correlated with FF levels of miR-222 (r = 0.54, P = 0.029). Serum level of miR-9 was negatively correlated with BMI of the control group (r = -0.53, P = 0.027). In PCOS group, serum level of miR-9 was positively correlated with age (r = 0.49, P = 0.046). By unifying all studied subjects in a single group, Follicular level of miR-146a was negatively correlated with the Ferriman-Gallwey score and LH (r = -0.38, P = 0.019; r = -0.39, P = 0.015, respectively).
Table 3.
Correlation analysis of clinical features with expression level of miRNAs
| miRNA | r | P value | |
|---|---|---|---|
| Control Subjects | |||
| S-miR-222 | FF-miR-222 | 0.54 | 0.029 |
| BMI | S-miR-9 | -0.53 | 0.027 |
| PCOS Subjects | |||
| Age | S-miR-9 | 0.49 | 0.046 |
| All Subjects | |||
| Ferriman-Gallwey Score | FF-miR-146 | -0.38 | 0.019 |
| LH | FF-miR-146 | -0.39 | 0.015 |
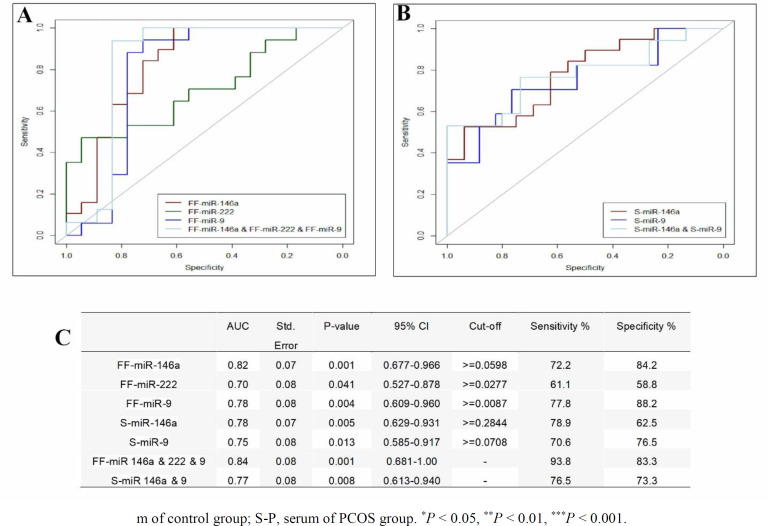
To determine the applicability of serum and FF differential relative expression of miRNAs for discrimination of PCOS patient from control subjects, ROC curve analysis was conducted, and the area under the ROC curve (AUC) was calculated. FF levels of miR-146a, miR-222, and miR-9 showed AUC of 0.82, 0.7, and 0.78, respectively. Accordingly, sensitivity and specificity of FF miRNAs were 72.2% and 84.2% for miR-146a; 61.1% and 58.8% for miR-222; 77.8% and 88.2% for miR-9, respectively (Figure 2 A and C). Serum levels of miR-146a and miR-9 showed AUC of 0.78 and 0.75, respectively. The sensitivity and specificity of serum miRNAs were 78.9% and 62.5% for miR-146a; 70.6% and 76.5% for miR-9, respectively (Figure 2 B and C). The combination of FF miRNAs improved the AUC (93.8) and revealed 93.8% sensitivity and 83.3% specificity (Figure 2 A and C). By combining serum miRNAs, AUC reached to 0.76 with 76.5% sensitivity and 73.3% specificity.
Fig.2.
ROC curve analysis to evaluate the predictive significance of miRNAs. (A) miR-146a, miR-222, and miR-9 in FF and their combinatory curve. (B) miR-146a, and miR-9 in serum and their combinatory curve. (C) Combination of miRNAs in FF reveled modest increase in AUC
In silico analysis
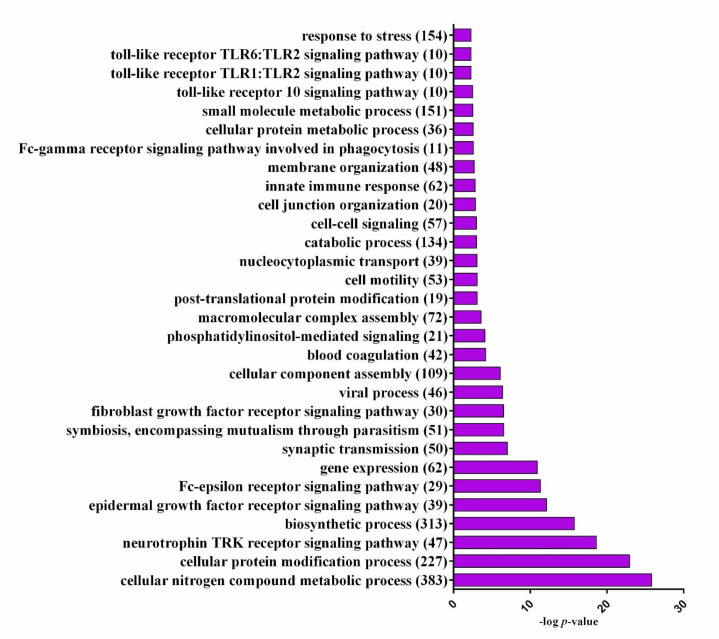
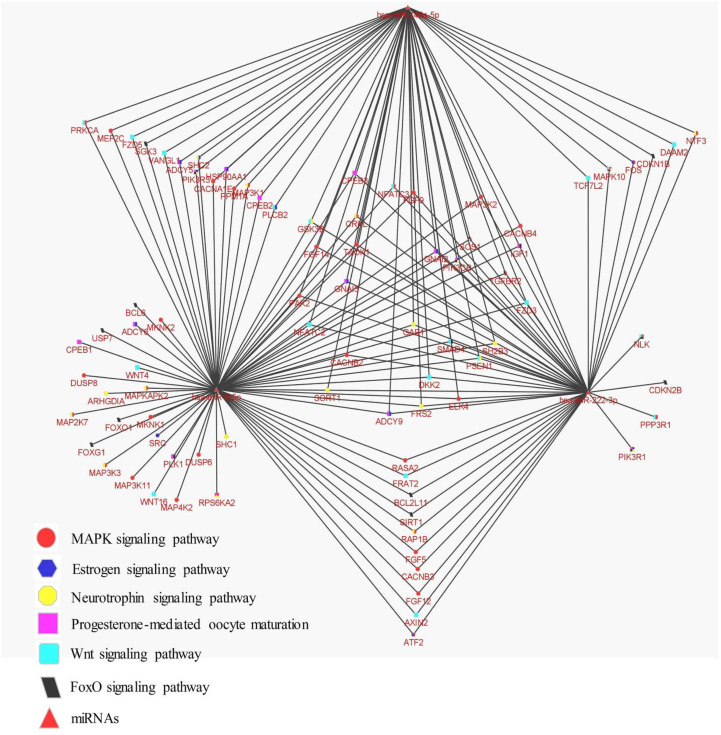
DIANA mirPath v.3 (recruited Targetscan data) was utilized to study the biological processes and signalling pathways that may be influenced by up/down-regulation of miRNAs expression, which in turn may cause vast alternations in their target genes. The most significant biological processes were found by GO analysis and illustrated in Figure 3. KEGG analysis was used to find the most significant signalling pathways that may be modulated by studied miRNAs. After the removal of cancer-related pathways and pathways that were affected by only a single miRNA, the most important pathways were listed in Table 4. KEGG analysis revealed significant pathways with important roles in folliculogenesis and reproductive function, including MAPK signalling pathway, Estrogen signalling pathway, Neurotrophin signalling pathway, FoxO signalling pathway, Progesterone-mediated oocyte maturation, and Wnt signalling pathway. The vast potential of a limited number of miRNAs for targeting and regulating different signalling pathways was illustrated by a network of miRNAs with presumptive targets in signalling pathways from KEGG analysis (Figure 4).
Fig.3.
Significant biological processes were identified by DIANA mirPath. P-value was presented after -log10 transformation. The number of contributor genes in each term was demonstrated in parenthesis
Table 4.
KEGG pathway analysis of potential target genes for miR-146a, miR-222, and miR-9
| KEGG pathways | Genes number | P -value |
|---|---|---|
| Prolactin signaling pathway | 17 | 4.87 |
| Signaling pathways regulating pluripotency of stem cells | 27 | 4.23 |
| ErbB signaling pathway | 20 | 3.03 |
| MAPK signaling pathway | 38 | 2.35 |
| Axon guidance | 19 | 2.35 |
| Hippo signaling pathway | 16 | 2.35 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 13 | 2.35 |
| Gap junction | 15 | 2.34 |
| Estrogen signaling pathway | 16 | 2.08 |
| Neurotrophin signaling pathway | 22 | 1.91 |
| Type II diabetes mellitus | 11 | 1.81 |
| Oxytocin signaling pathway | 25 | 1.68 |
| Ras signaling pathway | 32 | 1.59 |
| FoxO signaling pathway | 19 | 1.59 |
| Sphingolipid signaling pathway | 15 | 1.59 |
| Progesterone-mediated oocyte maturation | 16 | 1.59 |
| Chronic myeloid leukemia | 14 | 1.59 |
| Adherens junction | 11 | 1.59 |
| Thyroid hormone signaling pathway | 16 | 1.40 |
| Bacterial invasion of epithelial cells | 14 | 1.40 |
| Wnt signaling pathway | 20 | 1.33 |
| Focal adhesion | 28 | 1.33 |
Fig.4.
Network of miR-146a, miR-222, and miR-9 with predicted targets of important signaling pathways from KEGG analysis was generated by the Cytoscape software
Discussion
The multifaceted nature of PCOS has motivated many investigators to study the alternation of numerous molecular factors in the tissues and biological fluids of these patients to unravel the pathophysiology of the disease and discover potential diagnostic and therapeutic methods. In the current work, we have investigated the relative expression of miR-146a, miR-222, and miR-9 in serum and FF of PCOS patients and compared them with control subjects. ROC curve analysis was utilized to assess the applicability of the relative expression of miRNAs for the discrimination of PCOS patients from control subjects. Finally, we appraised the potential influence of dysregulated miRNAs on the main signalling pathways of folliculogenesis through gene ontology and KEGG enrichment analysis.
The integral role of miRNAs in mammalian folliculogenesis has been emphasized by the studies that utilized conditional knockout models of Dicer (a key regulatory molecule in miRNA signalling) in oocytes and granulosa cells, which reported an array of reproductive disorders, including impaired folliculogenesis, premature ovarian failure, increased early follicle recruitment, higher follicle degeneration, and female infertility (28, 29). A growing body of evidence has been reporting the role of miRNAs in early embryonic development (30, 31), different aspects of folliculogenesis (32), and female reproductive disorders such as PCOS (33), recurrent pregnancy loss (34), endometriosis (35), and premature ovarian insufficiency (36). In this regard, several studies in the last decade have described the differential expression of miRNAs in granulosa cells (7), FF (8), blood (9), and adipose tissue (11) of PCOS patients with mechanistic connections with androgen responsiveness (7), insulin resistance (11, 37), proliferation and apoptosis of granulosa cells (13, 14), and steroidogenesis (38). It has been demonstrated that miR-146a plays critical roles in granulosa cells’ apoptosis and gene expression regulation (17, 18). Similar to our findings, decreased expression of miR-146a in FF and elevated levels of this miRNA in the blood of PCOS patients have been recently reported by different groups (39, 40). MiR-222 roles in the pathophysiology of PCOS have been documented by several studies that reported its contribution to insulin sensitivity (20) and folliculogenesis by modulating steroidogenesis (26) and granulosa cells’ apoptosis and proliferation (19). We observed increased and diminished levels of miR-222 in serum and FF of PCOS patients, respectively; however, these findings did not reach the significant threshold. Accordingly, elevated levels of miR-222 in serum of PCOS patients have been indicated (9), while insignificant changes were detected in FF of PCOS patients (26). Our result of increased expression of miR-9 in serum of PCOS group was in conformity of previous study by Kong et al.(21). We found diminished levels of miR-9 in FF of PCOS group in accordance with the report by Butler and colleagues (41). However, contradicting results have been published which revealed significantly increased levels of miR-9 in FF of PCOS patients (8). Contradicting findings of miRNAs’ expression in PCOS patients may originate from heterogamous nature of PCOS, recruiting patients with different inclusion criteria, diverse genetic background of patients, and difference of detection methods.
FF is a complex mixture of locally released molecules from follicular cells and exudates of blood after being excreted over the blood-follicle barrier (42). Although the exact source of each individual component of FF is not clear, the close proximity of FF with follicular elements offers an outstanding window for scrutinizing the process of folliculogenesis, specifically in disorders with noticeable irregularities of follicular development, such as PCOS. The small size of miRNAs renders superior stability to these RNA species compared to larger ones (43), which provides a promising possibility for focusing on them as biomarkers of PCOS (44). FF profile of miRNAs can be representative of intra-follicular events participated by different cell types (42). However, the miRNAs in serum can originate from all body organs. Therefore, concurrent evaluation of miRNAs in various body fluids may disclose more detailed information regarding their role and potential to be used as biomarkers. In this study, in addition to the comparison of the control group with PCOS patients, we performed an intragroup comparison of miRNAs in serum and FF. In the control group, no significant differences were observed between serum and FF. However, in PCOS patients, significantly higher levels for all three miRNAs were detected in serum. These differences may originate from concurrent up-regulation in serum and down-regulation in FF. The decreased levels of miRNAs in FF reflect the overall alternations in ovarian follicles. However, PCOS is a systemic disease affecting many organs other than the reproductive system (1-3), so the increased blood levels of miRNAs can be attributed to the up-regulation of studied miRNAs in tissues other than the ovary. The tissue-specific function and expression of miRNAs may implicate this intriguing finding (45) and foster future studies for evaluating miRNA alterations and roles in different tissues of PCOS.
KEGG analysis of putative targets of miR-146a, miR-222, and miR-9 suggested signalling pathways with well-documented roles in follicular development, including MAPK signalling pathway (46), estrogen signalling pathway (47), neurotrophin signalling pathway (48), FoxO signalling pathway (49), and WNT signalling pathway (50). This highlights the noticeable roles of miRNAs in the pathophysiology of PCOS as dysregulation of a small number of miRNAs, due to their mode of action, can provoke vast downstream cellular consequences.
In sum, a combination of three miRNAs can be recommended for the discrimination of PCOS from control subjects. However, due to the small number of subjects studied in the present study, a larger-scale investigation is required to validate this data. The contradicting pattern of dysregulation for miR-146a and miR-9 in serum versus FF may originate from tissue-specific expression of miRNAs and different cellular sources of expression in FF and serum. Future studies in this context should consider that concurrent evaluation of miRNAs in different tissues might create a more accurate view concerning the miRNAs’ function in PCOS.
Acknowledgments
We greatly appreciate the patients who participated in the study, as well as the collaboration of staff at the embryology laboratory of Shariati Hospital. This work was supported by Tehran University of Medical Sciences (Grant number: 96-03-30-35981).
References
- 1.Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. 2018;14:270–84. doi: 10.1038/nrendo.2018.24. [DOI] [PubMed] [Google Scholar]
- 2.Witchel SF, Oberfield SE, Pena AS. Polycystic Ovary Syndrome: Pathophysiology, Presentation, and Treatment With Emphasis on Adolescent Girls. J Endocr Soc. 2019;3:1545–73. doi: 10.1210/js.2019-00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodarzi MO, Dumesic DA, Chazenbalk G, et al. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7:219–31. doi: 10.1038/nrendo.2010.217. [DOI] [PubMed] [Google Scholar]
- 4.Webber LJ, Stubbs S, Stark J, et al. Formation and early development of follicles in the polycystic ovary. Lancet. 2003;362:1017–21. doi: 10.1016/s0140-6736(03)14410-8. [DOI] [PubMed] [Google Scholar]
- 5.Hughesdon PE. Morphology and morphogenesis of the Stein-Leventhal ovary and of so-called "hyperthecosis". Obstet Gynecol Surv. 1982;37:59–77. doi: 10.1097/00006254-198202000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naji M, Aleyasin A, Nekoonam S, et al. Differential Expression of miR-93 and miR-21 in Granulosa Cells and Follicular Fluid of Polycystic Ovary Syndrome Associating with Different Phenotypes. Sci Rep. 2017;7:14671. doi: 10.1038/s41598-017-13250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth LW, McCallie B, Alvero R, et al. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J Assist Reprod Genet. 2014;31:355–62. doi: 10.1007/s10815-013-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long W, Zhao C, Ji C, et al. Characterization of serum microRNAs profile of PCOS and identification of novel non-invasive biomarkers. Cell Physiol Biochem. 2014;33:1304–15. doi: 10.1159/000358698. [DOI] [PubMed] [Google Scholar]
- 10.Jiang L, Huang J, Li L, et al. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. J Clin Endocrinol Metab. 2015;100:E729–38. doi: 10.1210/jc.2014-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YH, Heneidi S, Lee JM, et al. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62:2278–86. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song J, Luo S, Li SW. miRNA-592 is downregulated and may target LHCGR in polycystic ovary syndrome patients. Reprod Biol. 2015;15:229–37. doi: 10.1016/j.repbio.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Gao L, Wu D, Wu Y, et al. MiR-3940-5p promotes granulosa cell proliferation through targeting KCNA5 in polycystic ovarian syndrome. Biochem Biophys Res Commun. 2020;524:791–7. doi: 10.1016/j.bbrc.2020.01.046. [DOI] [PubMed] [Google Scholar]
- 14.Zhou S, Xia L, Chen Y, et al. miR-3188 Regulates proliferation and apoptosis of granulosa cells by targeting KCNA5 in the polycystic ovary syndrome. Acta Biochim Pol. 2021;68:83–9. doi: 10.18388/abp.2020_5441. [DOI] [PubMed] [Google Scholar]
- 15.Wei Y, Lu S, Hu Y, et al. MicroRNA-135a Regulates VEGFC Expression and Promotes Luteinized Granulosa Cell Apoptosis in Polycystic Ovary Syndrome. Reprod Sci. 2020;27:1436–42. doi: 10.1007/s43032-020-00155-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang CL, Wang H, Yan CY, et al. Deregulation of RUNX2 by miR-320a deficiency impairs steroidogenesis in cumulus granulosa cells from polycystic ovary syndrome (PCOS) patients. Biochem Biophys Res Commun. 2017;482:1469–76. doi: 10.1016/j.bbrc.2016.12.059. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Xie M, Liu D, et al. Downregulation of microRNA‑146a inhibits ovarian granulosa cell apoptosis by simultaneously targeting interleukin‑1 receptor‑associated kinase and tumor necrosis factor receptor‑associated factor 6. Mol Med Rep. 2015;12:5155–62. doi: 10.3892/mmr.2015.4036. [DOI] [PubMed] [Google Scholar]
- 18.Cho SH, An HJ, Kim KA, et al. Single nucleotide polymorphisms at miR-146a/196a2 and their primary ovarian insufficiency-related target gene regulation in granulosa cells. PLoS One. 2017;12:e0183479. doi: 10.1371/journal.pone.0183479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, She L, Luo X, et al. MiR-222 promotes the progression of polycystic ovary syndrome by targeting p27 Kip1. Pathol Res Pract. 2019;215:918–23. doi: 10.1016/j.prp.2019.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–83. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 21.Kong F, Du C, Wang Y. MicroRNA-9 affects isolated ovarian granulosa cells proliferation and apoptosis via targeting vitamin D receptor. Mol Cell Endocrinol. 2019;486:18–24. doi: 10.1016/j.mce.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Yao G, Liang M, Liang N, et al. MicroRNA-224 is involved in the regulation of mouse cumulus expansion by targeting Ptx3. Mol Cell Endocrinol. 2014;382:244–53. doi: 10.1016/j.mce.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Vahdat-Lasemi M, Hosseini S, Jajarmi V, et al. Intraovarian injection of miR-224 as a marker of polycystic ovarian syndrome declines oocyte competency and embryo development. J Cell Physiol. 2019;234:13858–66. doi: 10.1002/jcp.28067. [DOI] [PubMed] [Google Scholar]
- 24.Rotterdam EA-SPCWG. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Naji M, Nekoonam S, Aleyasin A, et al. Expression of miR-15a, miR-145, and miR-182 in granulosa-lutein cells, follicular fluid, and serum of women with polycystic ovary syndrome (PCOS) Arch Gynecol Obstet. 2018;297:221–31. doi: 10.1007/s00404-017-4570-y. [DOI] [PubMed] [Google Scholar]
- 26.Sang Q, Yao Z, Wang H, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98:3068–79. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 27.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–6. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan S, Ortogero N, Wu Q, et al. Murine follicular development requires oocyte DICER, but not DROSHA. Biol Reprod. 2014;91 doi: 10.1095/biolreprod.114.119370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei L, Jin S, Gonzalez G, et al. The regulatory role of Dicer in folliculogenesis in mice. Mol Cell Endocrinol. 2010;315:63–73. doi: 10.1016/j.mce.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross N, Kropp J, Khatib H. MicroRNA Signaling in Embryo Development. Biology (Basel) 2017:6. doi: 10.3390/biology6030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azizi E, Ghaffari Novin M, Naji M, et al. Effect of vitrification on biogenesis pathway and expression of development-related microRNAs in preimplantation mouse embryos. Cell Tissue Bank. 2021;22:103–14. doi: 10.1007/s10561-020-09870-z. [DOI] [PubMed] [Google Scholar]
- 32.Tu J, Cheung AH, Chan CL, et al. The Role of microRNAs in Ovarian Granulosa Cells in Health and Disease. Front Endocrinol (Lausanne) 2019;10:174. doi: 10.3389/fendo.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deswal R, Dang AS. Dissecting the role of micro-RNAs as a diagnostic marker for polycystic ovary syndrome: a systematic review and meta-analysis. Fertil Steril. 2020;113:661–9 e2. doi: 10.1016/j.fertnstert.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Jairajpuri DS, Malalla ZH, Mahmood N, et al. Differentially expressed circulating microRNAs associated with idiopathic recurrent pregnancy loss. Gene. 2021;768:145334. doi: 10.1016/j.gene.2020.145334. [DOI] [PubMed] [Google Scholar]
- 35.Bjorkman S, Taylor HS. MicroRNAs in endometriosis: biological function and emerging biomarker candidatesdagger. Biol Reprod. 2019;100:1135–46. doi: 10.1093/biolre/ioz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Sun J, Lai D. Role of microRNAs in premature ovarian insufficiency. Reprod Biol Endocrinol. 2017;15:38. doi: 10.1186/s12958-017-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cirillo F, Catellani C, Lazzeroni P, et al. MiRNAs Regulating Insulin Sensitivity Are Dysregulated in Polycystic Ovary Syndrome (PCOS) Ovaries and Are Associated With Markers of Inflammation and Insulin Sensitivity. Front Endocrinol (Lausanne) 2019;10:879. doi: 10.3389/fendo.2019.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Liu Y, Lv M, et al. miR-323-3p regulates the steroidogenesis and cell apoptosis in polycystic ovary syndrome (PCOS) by targeting IGF-1. Gene. 2019;683:87–100. doi: 10.1016/j.gene.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Jiang X, Li J, Zhang B, et al. Differential expression profile of plasma exosomal microRNAs in women with polycystic ovary syndrome. Fertil Steril. 2021;115:782–92. doi: 10.1016/j.fertnstert.2020.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Rooda I, Hasan MM, Roos K, et al. Cellular, Extracellular and Extracellular Vesicular miRNA Profiles of Pre-Ovulatory Follicles Indicate Signaling Disturbances in Polycystic Ovaries. Int J Mol Sci. 2020:21. doi: 10.3390/ijms21249550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler AE, Ramachandran V, Hayat S, et al. Expression of microRNA in follicular fluid in women with and without PCOS. Sci Rep. 2019;9:16306. doi: 10.1038/s41598-019-52856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamah AM, Hassis ME, Albertolle ME, et al. Proteomic analysis of human follicular fluid from fertile women. Clin Proteomics. 2015;12:5. doi: 10.1186/s12014-015-9077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamaddon M, Azimzadeh M, Tavangar SM. microRNAs and long non-coding RNAs as biomarkers for polycystic ovary syndrome. J Cell Mol Med. 2022;26:654–70. doi: 10.1111/jcmm.17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sood P, Krek A, Zavolan M, et al. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hua G, George JW, Clark KL, et al. Hypo-glycosylated hFSH drives ovarian follicular development more efficiently than fully-glycosylated hFSH: enhanced transcription and PI3K and MAPK signaling. Hum Reprod. 2021;36:1891–906. doi: 10.1093/humrep/deab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang ZR, Zhang R, Lian ZX, et al. Estrogen-Receptor Expression and Function in Female Reproductive Disease. Cells. 2019:8. doi: 10.3390/cells8101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaves RN, Alves AM, Lima LF, et al. Role of nerve growth factor (NGF) and its receptors in folliculogenesis. Zygote. 2013;21:187–97. doi: 10.1017/S0967199412000111. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Lin F, Zhao J, et al. Expression Regulation and Physiological Role of Transcription Factor FOXO3a During Ovarian Follicular Development. Front Physiol. 2020;11:595086. doi: 10.3389/fphys.2020.595086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez Gifford JA. The role of WNT signaling in adult ovarian folliculogenesis. Reproduction. 2015;150:R137–48. doi: 10.1530/REP-14-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]