Abstract
Molecular methods are increasingly used to identify microbes in clinical samples. A common technical problem with PCR is failed amplification due to the presence of PCR inhibitors. Initial attempts at amplification of the bacterial 16S rRNA gene from inoculated blood culture media failed for this reason. The inhibitor persisted, despite numerous attempts to purify the DNA, and was identified as sodium polyanetholesulfonate (SPS), a common additive to blood culture media. Like DNA, SPS is a high-molecular-weight polyanion that is soluble in water but insoluble in alcohol. Accordingly, SPS tends to copurify with DNA. An extraction method was designed for purification of DNA from blood culture media and removal of SPS. Blood culture media containing human blood and spiked with Escherichia coli was subjected to an organic extraction procedure with benzyl alcohol, and removal of SPS was documented spectrophotometrically. Successful amplification of the extracted E. coli 16S rRNA gene was achieved by adding 5 μl of undiluted processed sample DNA to a 50-μl PCR mixture. When using other purification methods, the inhibitory effect of SPS could be overcome only by dilution of these samples. By our extraction technique, even uninoculated blood culture media were found to contain bacterial DNA when they were subjected to broad-range 16S rRNA gene consensus PCR. We conclude that the blood culture additive SPS is a potent inhibitor of PCR, is resistant to removal by traditional DNA purification methods, but can be removed by a benzyl alcohol extraction protocol that results in improved PCR performance.
The role of nucleic acid amplification technology in diagnostic microbiology continues to expand. Amplification of microbial sequences from clinical samples offers the potential for rapid detection and specific identification of pathogens, either directly from tissues or body fluids or after culture of such samples. The use of PCR for the detection and identification of microbes in blood cultures has several theoretical advantages over existing technology. First, phylogenetically informative microbial sequences, such as bacterial 16S rRNA genes, can be used to identify microbes that can be cultivated but that defy classification by traditional phenotypic tests. Second, with automation, sequence-based microbial identification may reduce the time between the detection of positive blood cultures and definitive microbial identification. Third, even noncultivated microbes may be detected by highly sensitive PCR assays. For instance, microbes may not grow in the laboratory if patients are receiving antibiotics at the time that the sample for culture is obtained or are bacteremic or fungemic because of infection with microbes that resist propagation by standard culture techniques (e.g., Ehrlichia).
A common limitation to PCR-based methods is failed amplification due to the presence of inhibitory substances in the sample. PCR inhibitors include heme compounds found in blood (2), aqueous and vitreous humors (26), heparin (15), EDTA (12), urine (16), polyamines (1), and plant polysaccharides (6). To deal with this problem, PCR inhibitors must be diluted, inactivated, or removed from the sample. In a series of initial experiments, bacteria were inoculated into blood culture media, and DNA was purified for use as target in a broad-range 16S rRNA gene PCR assay. These assays repeatedly failed due to the presence of a substance inhibitory to the PCR. Multiple attempts to purify the microbial DNA failed to remove the inhibitor. We describe the identity of the PCR inhibitor detected in blood culture media and a method for removing the inhibitor so as to allow amplification of microbial DNA from blood culture systems without dilution of the sample containing the DNA target.
MATERIALS AND METHODS
Preparation of inoculated blood culture media.
Ten milliliters of human blood obtained in a sterile fashion from one of the authors was added to a bottle of commercial blood culture medium (BacT Alert anaerobic media; Organon Teknica). Escherichia coli DH5α (Bethesda Research Laboratories) was grown in Luria-Bertani (LB) broth in the logarithmic phase, and then 0.129 ml was inoculated into 0.871 ml of this blood culture medium to produce a final concentration of 2 × 107 CFU/ml. The numbers of CFU were determined by plating serial dilutions of E. coli in LB broth onto LB agar plates and counting the colonies after an overnight incubation.
DNA purification methods.
Blood culture medium containing human blood and spiked with E. coli was prepared as noted above and was subjected to the following digestion and DNA purification methods. Starting volumes of medium and final volumes of DNA target were 0.1 ml unless otherwise noted.
(i) Phenol-chloroform extraction (method A).
A total of 0.1 ml of the inoculated medium was added to 0.1 ml of digestion buffer consisting of 10 mM Tris, 1 mM EDTA (pH 8.0), 0.4 mg of proteinase K (Sigma Chemical, St. Louis, Mo.) per ml, and 1% Laureth-12 detergent (PPG Industries Inc., Gurnee, Ill.). The sample was digested for 2 h at 55°C and was then heated to 95°C for 10 min to inactivate the proteinase K. The sample volume was adjusted to 0.5 ml with 0.3 ml of 10 mM Tris–1 mM EDTA buffer (pH 8.5), and 0.5 ml of phenol-chloroform (1:1; vol/vol) was added to the sample; the components were mixed by vortexing, and then the mixture was centrifuged at 7,000 × g for 5 min. The aqueous layer was removed and was subjected to two more rounds of phenol-chloroform extraction, followed by a chloroform extraction. The aqueous layer was harvested, and a 1/10 volume of 3.0 M sodium acetate was added, followed by the addition of 2 volumes of 100% ethanol. The sample was placed at −20°C for 10 min. After centrifugation at 13,000 × g for 15 min at 4°C, the supernatant was decanted and the pellet was washed with 1.0 ml of 70% ethanol. The pellet was air dried and was resuspended in 0.1 ml of 10 mM Tris–0.1 mM EDTA buffer at pH 8.5.
(ii) QIAmp silica column purification (method B).
A total of 0.1 ml of inoculated medium was digested, and the DNA was purified according to the manufacturer’s directions by using the QIAmp blood kit (Qiagen Corporation, Chatsworth, Calif.). In this method, DNA adsorbs to silica in the presence of a chaotrope, is washed with buffer, and is eluted from the column in 0.1 ml of 10 mM Tris–0.1 mM EDTA buffer at pH 8.5.
(iii) Isoquick organic extraction (method C).
A total of 0.1 ml of inoculated medium was digested and DNA was extracted according to the manufacturer’s directions by using the Isoquick kit (ORCA Research Inc., Bothell, Wash.). In this method, DNA is purified by an alternative organic extraction procedure. After alcohol precipitation, the target DNA was resuspended in 0.1 ml of 10 mM Tris–0.1 mM EDTA buffer at pH 8.5.
(iv) Benzyl alcohol-guanidine hydrochloride organic extraction (method D).
A total of 0.1 ml of the inoculated medium was added to 0.1 ml of lysis buffer and was briefly mixed with a vortex mixer. Lysis buffer consisted of 5.0 M guanidine hydrochloride–100 mM Tris (pH 8.0) in sterile water. A total of 0.4 ml of water was added, followed by the addition of 0.8 ml of 99% benzyl alcohol (Sigma Chemical Co.), and the sample was mixed again by vortexing. The sample was centrifuged at 7,000 × g for 5 min. A total of 0.4 ml of the aqueous supernatant was removed and was placed in a new microcentrifuge tube. A total of 0.040 ml of 3.0 M sodium acetate was added, followed by the addition of 0.44 ml of isopropranol, and the sample was centrifuged at 16,000 × g for 15 min at 4°C. The precipitated DNA was washed with 70% ethanol and the pellet was air dried. The DNA was resuspended in 0.1 ml of 10 mM Tris–0.1 mM EDTA buffer at pH 8.5.
(v) Chelex digestion (method E).
A total of 0.1 ml of blood culture medium was added to 0.5 ml of a 50% (vol/vol) solution of Chelex-100 resin (Bio-Rad Laboratories, Hercules, Calif.) in water, and the mixture was boiled for 10 min. After centrifugation at 7,000 × g for 5 min, about 0.25 ml of supernatant was removed and was used for PCR.
(vi) Washing by centrifugation (method F).
A total of 0.1 ml of inoculated blood culture medium was added to 1.0 ml of water, and the mixture was centrifuged at 7,000 × g for 5 min. The supernatant was removed, and the pellet was resuspended in 1.0 ml of water. This washing step was repeated two more times, and the pellet was finally resuspended in 0.1 ml of water. This method was used to process the blood culture medium prior to other digestion and extraction procedures, where noted.
(vii) Ultrafiltration (method G).
Ultrafiltration was used to process DNA that had already been liberated and purified by other digestion and purification methods, where noted. DNA was added to 1.0 ml of TE (Tris-EDTA) buffer in a Centricon 100 concentrator (Amicon Inc., Beverly, Mass.), and the mixture was centrifuged at 2,000 × g for 20 min. An additional 1.0 ml of TE buffer was added to the reservoir, and the centrifugation was repeated. The concentrator was then inverted and the ultrafiltered fraction was spun into a cap by centrifugation at 500 × g for 2 min.
PCR amplification of 16S rRNA gene.
PCR was performed with broadly conserved bacterial 16S rRNA gene primers (Table 1). For most experiments primer pair 516F-13R was used for amplification of E. coli or Bordetella pertussis DNA. For experiments in which a bacterial 16S rRNA gene was amplified from uninoculated BACTEC medium (Becton Dickinson, Sparks, Md.), primer pairs 516F-806R and 516F-911R were used for PCR. For experiments in which a bacterial 16S rRNA gene was amplified from uninoculated BacT Alert blood culture media, primer pairs 516F-806R and 806F-13R were used for PCR. One unit of Amplitaq DNA polymerase (Perkin-Elmer, Foster City, Calif.) was used in each 50-μl reaction mixture, which also contained 20 pmol of each primer, each deoxynucleoside triphosphate at a concentration of 200 μM, 2 mM MgCl2, and 1× PCR buffer II (Perkin-Elmer Applied Biosystems Inc. [ABI], Foster City, Calif.). The water used for PCR reactions and controls was sterile water for injection (Abbott Laboratories, North Chicago, Ill.) and was irradiated for 30 min on a UV transilluminator. One or 5 μl of target was used in each 50-μl PCR mixture. PCR consisted of 35 or 50 cycles of amplification on a Perkin-Elmer GeneAmp 2400 thermal cycler. Each cycle consisted of 30 s of melting at 94°C, 30 s of annealing at 55°C, and 30 s of extension at 72°C. Prior to the first cycle, the samples were heated to 94°C for 3 min. The last cycle was followed by a final extension at 72°C for 7 min. Amplification products were detected by electrophoresis on 2% agarose gels that were stained with ethidium bromide and visualized with a UV transilluminator. On some occasions (e.g., with the use of the 516F-806R primer pair for 50 cycles), the master mixture was irradiated for 2 to 5 min on a UV transilluminator (shortwave; UVP Inc.) prior to the addition of the DNA target in order to eliminate false-positive background amplification.
TABLE 1.
16S rRNA gene primers used for PCR and sequencing
| Primer | Sequence (5′→3′) | E. coli positiona |
|---|---|---|
| 516F | TGCCAGCAGCCGCGGTAA | 516–533 |
| 806F | ATTAGATACCCTGGTAGTCC | 787–806 |
| 806R | GGACTACCAGGGTATCTAAT | 806–787 |
| 911R | GCCCCCGTCAATTCMTTTGA | 930–911 |
| 1175F | GAGGAAGGTGGGGATGACGT | 1175–1194 |
| 1175R | ACGTCATCCCCACCTTCCTC | 1194–1175 |
| 13R | AGGCCCGGGAACGTATTCAC | 1390–1371 |
E. coli position numbers are based on reference 5.
In some experiments, positive control DNA for PCR was made by digesting the pellet from a 5-ml Stainer-Sholte broth culture of B. pertussis and purifying the DNA with the QIAmp kit (Qiagen Corporation). One microliter of this positive control DNA at a concentration of 79 ng/μl was added to a 50-μl PCR mixture. The presence of activity inhibitory to the PCR in a target sample was assayed for by adding 1 μl of the positive control DNA to a PCR mixture along with the target in question. A PCR inhibitor was deemed to be present when a reaction mixture containing positive control DNA alone produced an amplification product of the appropriate size, but a reaction mixture containing positive control DNA plus target DNA (e.g., E. coli from a blood culture) produced no band. (This type of experiment that involves spiking the target sample with a known amount of amplifiable DNA is referred to as a “spike back” experiment.) In other experiments, positive control DNA was made by growing E. coli DH5α in LB broth. Bacteria in the logarithmic phase were diluted in TE buffer and were frozen for use in PCR or were diluted in LB broth and plated onto LB agar to determine the numbers of CFU after an overnight incubation. To determine the approximate number of gene copies of the E. coli 16S rRNA gene added to a PCR mixture, the numbers of CFU of bacteria at a given dilution were multiplied by seven gene copies per E. coli cell.
Spectrophotometry.
Sodium polyanetholesulfonate (SPS) was detected in samples by performing a scan of the optical density from 200 to 700 nm with a Beckman DU-64 spectrophotometer and looking for the characteristic absorption peak at 284 nm. SPS was obtained from Sigma Chemical Co. Spectrophotometry was performed with quartz cuvettes.
Sequencing and phylogenetic analysis.
Amplification products from uninoculated blood culture media were purified with Wizard PCR Preps (Promega, Madison, Wis.) and were directly sequenced with the fluorescent Dye Terminator Cycle Sequencing kit with Amplitaq FS polymerase (ABI). Sequencing products were electrophoresed and analyzed on a 373 automated sequencer (ABI). Electropherograms were processed with Factura software (ABI), and overlapping sequences from forward and reverse strands were assembled into a consensus sequence with Autoassembler software (ABI). The sequencing primers used in these experiments were designed from conserved bacterial 16S rRNA gene sequences and are listed in Table 1.
Preliminary phylogenetic associations of directly sequenced 16S rRNA genes were determined by using the Similarity Rank software and services of the Ribosomal Database Project (20) and the BLAST search algorithm of GenBank (National Center for Biotechnology Information) (3, 4).
The initial alignment of the amplified sequences was done with the automated 16S rRNA sequence aligner of the ARB software package (Technical University of Munich, Munich, Germany) against a database of more than 4,000 complete and partial sequences. Ambiguously and incorrectly aligned positions were aligned manually on the basis of conserved primary sequence and secondary structure.
The phylogenetic relationships of amplified sequences were determined from unambiguously aligned (masked) positions with a maximum-likelihood algorithm (9, 21). The sequence amplified from uninoculated BACTEC blood culture medium contained 370 masked positions, while that from uninoculated BacT Alert blood culture medium contained 554 masked positions. These relationships were confirmed with least-squares and parsimony algorithms. Phylogenetic trees were created with the ARB software package and were manually pruned. Bootstrap values were obtained from 500 resamplings (10).
Nucleotide sequence accession numbers. The nucleotide sequences of the BacT Alert (streptococcus) and the BACTEC (bacillus) cultures have been deposited in the GenBank database under accession no. AF078908 and AF078909, respectively.
RESULTS
Blood culture medium inhibits PCR.
In preliminary experiments, attempts to amplify the 16S rRNA gene of E. coli (2 × 107 CFU/ml) from inoculated blood culture medium (BacT Alert anaerobic medium) by PCR failed. Spike-back experiments (see Materials and Methods) confirmed that an inhibitor was present in the DNA obtained from blood culture medium. Attempts to remove the PCR inhibitor(s) by standard DNA purification methods failed (see methods A to C and E to G). Even combinations of DNA purification methods failed to remove the PCR inhibitor. For instance, washing of inoculated blood culture medium by centrifugation (method F) followed by QIAmp column purification (method B) failed to remove the inhibitor. Similarly, phenol-chloroform extraction (method A) followed by ultrafiltration (method G) also failed. The PCR inhibitor persisted after each of these purification methods whether blood culture medium that had been inoculated with blood, bacteria, blood and bacteria, or none of these was used. Blood culture media from different manufacturers were tested, including BACTEC aerobic medium, anaerobic medium, and mycobacterial media 13A and 12B (Becton Dickinson) and BacT Alert aerobic and anaerobic media (Organon Teknica). The only product free of activity that was inhibitory to the PCR, the BACTEC 12B mycobacterial culture medium, was also the only product without the additive SPS.
Identification of the PCR inhibitor in blood culture media: SPS.
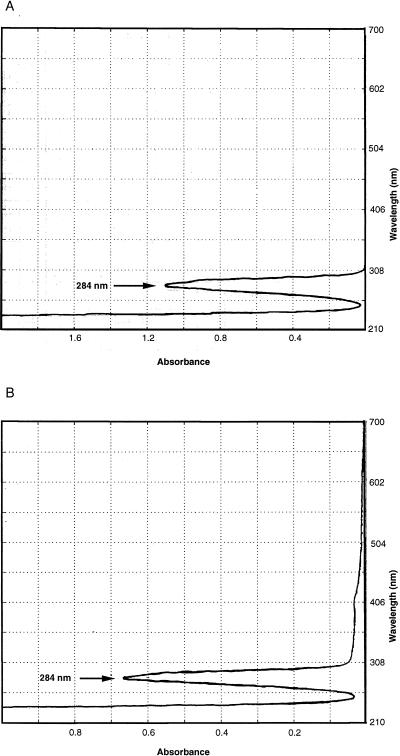
To test the hypothesis that SPS is the PCR inhibitor present in blood culture media, a solution of 0.0135% SPS in water was made and was analyzed spectrophotometrically. A narrow absorption peak at 284 nm was noted (Fig. 1A). When DNA samples from blood cultures that were purified by the above standard methods mentioned above were scanned, the samples all had a single absorption peak at 284 nm, suggesting the presence of SPS. Figure 1B shows the spectrophotometric scan from one attempt to purify DNA from blood culture medium plus blood in which a combination of method F (centrifugation wash) followed by method B (QIAmp column) was used. Note that no DNA absorption peak is visible at 260 nm because most of the leukocyte DNA from the blood was lost with the washing step. To confirm the ability of SPS to copurify with DNA and inhibit the PCR, 0.1 ml of a 0.05% solution of SPS was subjected to several of the standard DNA purification methods described above (methods A, B, and G) to produce 0.1 ml of “purified” product. For each method, the fraction that normally contains purified DNA was scanned, revealing an absorption peak at 284 nm. When 1 μl of the processed sample was added to a 50-μl positive control PCR mixture with primers 516F and 13R, the sample was inhibitory. When water controls or BACTEC 12B medium was subjected to the same methods, no peaks were detected on scanning and no PCR inhibition occurred (data not shown).
FIG. 1.
Spectrophotometric scan. (A) SPS at 0.0135% dissolved in water. (B) BacT Alert blood culture medium (0.1 ml) inoculated with blood was washed three times by centrifugation in 10 volumes of water (method F) and was then processed by the QIAmp blood digestion and DNA purification protocol (method B). Water eluate from the silica column, which normally contains purified DNA, was scanned.
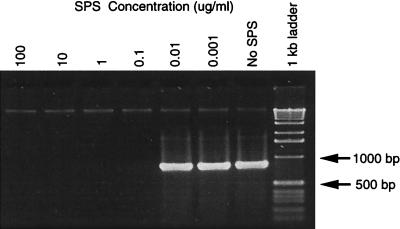
Figure 2 demonstrates the PCR-inhibitory activity of SPS at various concentrations when it was added to 79 ng of purified B. pertussis DNA and subjected to PCR with conserved 16S rRNA gene primers. An SPS concentration of 100 ng/ml in the 50-μl reaction mixture was inhibitory, whereas use of a mixture with SPS at 10 ng/ml resulted in successful amplification of the target. The concentration of SPS in BACTEC aerobic and anaerobic blood culture media is 0.05%, or 500 μg/ml. Thus, unprocessed blood culture media would have to be diluted more than 5,000-fold in order to amplify microbial DNA in this assay.
FIG. 2.
Effect of SPS concentration on PCR. Agarose gel electrophoresis of PCR products amplified from B. pertussis target DNA with conserved bacterial 16S rRNA gene primers 516F and 13R in the presence of various concentrations of SPS in each PCR reaction, as listed, including no SPS (positive control). The amplification product size is about 874 bp.
A comparison of four DNA purification methods for blood cultures: removal of SPS is the key.
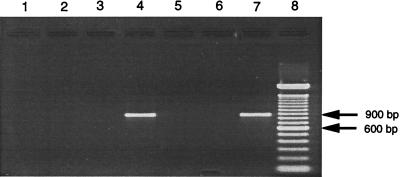
Removal of SPS and purification of DNA for PCR amplification was achieved by the modified organic extraction method described above with lysis buffer consisting of guanidine hydrochloride in Tris buffer as the aqueous phase and benzyl alcohol as the organic phase. This SPS extraction method (method D) was tested head to head against three other DNA purification methods: phenol-chloroform extraction (method A), QIAmp silica column adsorption (method B), and Isoquick extraction (method C). Blood culture medium inoculated with human blood and E. coli as described above was subjected to each of the four DNA purification methods. The purified DNA was then resuspended in 10 mM Tris–0.1 mM EDTA buffer at the original volume digested (0.1 ml), and 5 μl was used as the target in a 50-μl 16S rRNA gene PCR assay mixture. Figure 3 shows the PCR products produced with the target from the various purification procedures. Only the SPS extraction protocol produced a visible product from an inoculated blood culture (lane 4). Spectrophotometric analysis was performed with negative control samples of blood culture medium (without E. coli) processed by the four purification methods. Scanning showed that the 284-nm absorption peak characteristic of SPS was absent from the products from samples processed by methods D and C but was present for products from samples processed by methods A and B (data not shown).
FIG. 3.
Amplification of bacterial DNA from blood cultures by four digestion and extraction methods. Agarose gel electrophoresis of PCR products amplified with 16S rRNA gene primers 516F and 13R. In the first four lanes, E. coli was inoculated into blood culture medium containing human blood and was processed by various digestion and purification protocols to produce target DNA for PCR. Lane 1, proteinase K digestion with phenol-chloroform extractions (method A); lane 2, Isoquick protocol (method C); lane 3, protocol with the QIAmp blood kit; (method B); lane 4, benzyl alcohol-guanidine hydrochloride extraction protocol (method D); lane 5, method D performed with sterile LB broth inoculated into blood culture medium (negative control); lane 6, method D performed with sterile LB broth added to TE buffer (negative control); lane 7, bacterial DNA-positive control; lane 8, 100-bp DNA ladder. The amplification product size is about 874 bp.
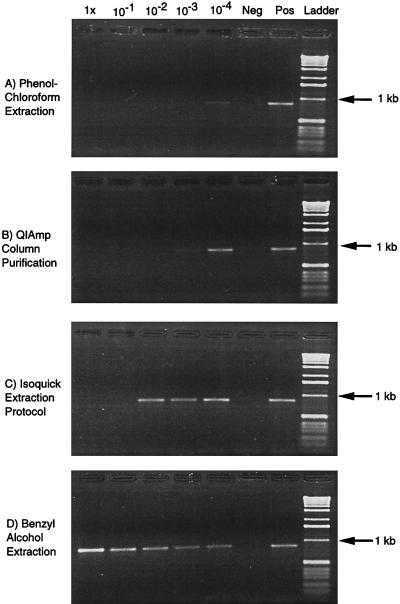
To confirm that a PCR inhibitor was present in samples prepared by the other methods, assays with amplification controls were performed. One microliter (79 ng) of B. pertussis DNA was added to each PCR mixture, as was 5 μl of E. coli DNA (at dilutions of 1 to 10−4) purified from the inoculated blood culture by one of the four purification methods. Accordingly, each reaction should produce a product if it is free of PCR inhibitors. Serial dilutions of each processed target sample in water were made in order to measure the dilution required for the restoration of normal amplification activity (Fig. 4). For methods A, B, and C higher residual concentrations of PCR inhibitor necessitate larger dilutions before amplification is seen.
FIG. 4.
Amplification controls. Agarose gel electrophoresis of PCR products amplified with 16S rRNA gene primers 516F and 13R (about 874 bp). E. coli was inoculated into blood culture medium containing human blood and processed by one of the four listed digestion and purification protocols. In the first five lanes, the processed DNA was diluted in sterile, UV-irradiated water as indicated, and 5 μl of DNA target were added to a 50-μl PCR mixture along with 1 μl of additional B. pertussis DNA. Lane Neg, unprocessed, sterile water (negative control); and Lane Pos, 1 μl (79 ng) of B. pertussis DNA (positive control) subjected to PCR. A 1-kb DNA ladder is present in the last lane.
Method D yielded a target that produced a strong PCR band even with undiluted sample. Amplification products were detectable throughout the dilution series. In comparison, method C (Isoquick extraction) was the next best method, but it still demonstrated the presence of inhibitor at dilutions of 1 and 10−1. Method B (QIAmp silica column absorption) produced a faint band at a 10−3 dilution and a strong band at a 10−4 dilution, whereas method A (phenol-chloroform extraction) was the worst performer, producing only a faint band at a 10−4 dilution. The product band intensity was greater for the undiluted (dilution of 1) target in the sample processed by method D than in the positive control sample because amplifiable bacterial DNA from both the inoculated blood culture (E. coli) and the positive control DNA (B. pertussis) were present.
Uninoculated blood culture medium contains bacterial DNA.
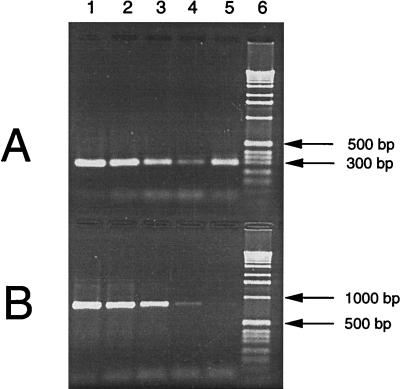
When 0.1 ml of uninoculated blood culture medium (no blood, no added bacteria) was extracted twice by method D and 1 or 5 μl of the DNA fraction was added to a 50-μl PCR mixture with broadly conserved 16S rRNA gene primers 516F and 806R and a low DNA Taq polymerase concentration (Taq LD; ABI), small amplification products in the 300-bp size range were consistently produced. Negative controls containing water subjected to the same purification method did not produce a product. These results were reproducible in more than 20 experiments. Direct sequencing of these small amplification products and comparison to known 16S rRNA gene sequences in the Ribosomal Database Project revealed that each commercial blood culture medium has a unique contaminating bacterial sequence. PCR with primers directed against larger target sequences, such as the approximately 874-bp product of primers 516F and 13R, failed to produce a visible product with DNA from uninoculated blood culture medium. Figure 5 shows that primer pairs 516F-806R and 516F-13R have similar sensitivities of about 20 to 200 gene copies when they are used to detect E. coli DNA added to a PCR mixture subjected to 35 cycles. Yet, when DNA from uninoculated BacT Alert anaerobic blood culture medium purified by method D is the target, primer pair 516F-806R produces an intense band with ethidium bromide staining, whereas primer pair 516F-13R does not.
FIG. 5.
Comparison of two primer pairs used in PCR amplification. PCR was performed with primer pair 516F-806R (A) or 516F-13R (B), and the products were subjected to gel electrophoresis. One microliter of target was added to each 50-μl PCR mixture, as follows: 20,000 gene copies of E. coli 16S rDNA (lane 1), 2,000 gene copies (lane 2), 200 gene copies (lane 3), 20 gene copies (lane 4), DNA purified from uninoculated BacT Alert anaerobic blood culture medium by method D (lane 5), and 1-kb DNA ladder (lane 6).
Since PCR amplification of large segments of the 16S rRNA gene from uninoculated blood culture medium was not successful, smaller products were made for direct sequencing. PCR products were produced with primer pairs 516F-806R, 516F-911R, and 806F-13R (Table 1). Initial analysis of the sequences by a similarity search revealed that each product from a particular blood culture medium produced the same similarity scores, even with different primer pairs. In addition, the products shared overlapping regions of sequence identity. Sequence data for some of these small products were combined to generate sequence blocks for phylogenetic analysis. For the BACTEC media, the sequence used to make the phylogenetic tree was assembled from products produced with primer pairs 516F-806R and 516F-911R and consisted of 375 bp prior to alignment and creation of a mask. For the BacT Alert media, the sequence used to make the phylogenetic tree was assembled from products produced with the 806F-13R primer pair and consisted of 562 bp prior to alignment and the creation of a mask.
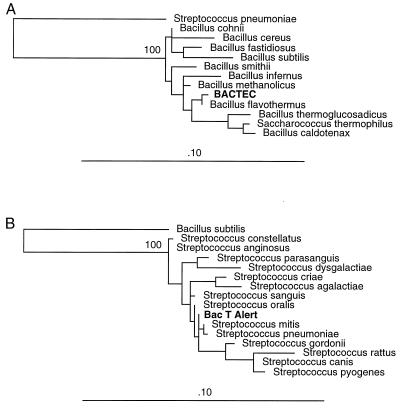
Sequencing and phylogenetic analysis of amplification products revealed that uninoculated aerobic and anaerobic BACTEC blood culture media contain 16S rRNA genes from the Bacillus genus (Fig. 6A). Bacillus flavothermus is the closest phylogenetic neighbor of the organism whose 16S rRNA gene was amplified from BACTEC blood culture media. B. flavothermus is a facultative aerobe and a facultative thermophile which can grow at temperatures ranging from 30 to 70°C (14). Given the limited size of the 16S rRNA gene that could be amplified and submitted for phylogenetic comparison, we are hesitant to make any firm conclusions about the identity of the organism represented, other than to say that the organism is most closely related to the Bacillus genus and distinct from the Streptococcus genus, as supported by bootstrap analysis.
FIG. 6.
Phylogenetic trees. The 16S rRNA gene sequence amplified from uninoculated blood culture medium was aligned with other known 16S rRNA gene sequences, and phylogenetic relationships were inferred by using a maximum likelihood algorithm. The branch length is proportional to the evolutionary distance, and the bar labeled .10 represents 0.1 estimated base changes per position. (A) Sequence in BACTEC medium with 370 masked positions. S. pneumoniae was used as an outgroup. (B) Sequence in BacT Alert medium with 554 masked positions. B. subtilis was used as an outgroup.
Uninoculated BacT Alert blood culture media (aerobic and anaerobic) also contain a bacterial 16S rRNA gene, but the organism represented is most closely related to organisms from the Streptococcus genus (Fig. 6B). Streptococcus mitis, Streptococcus pneumoniae, and Streptococcus oralis are the closest phylogenetic neighbors to the organism whose 16S rRNA gene was amplified from BacT Alert blood culture media. Although more sequence data were obtained for this organism, we remain cautious about definitively identifying the organism since only a third of the 16S rRNA gene was analyzed in the present investigation. Nevertheless, bootstrap analysis again confirms that the sequence is distinct from sequences present in the Bacillus genus. Thus, BACTEC and BacT Alert blood culture media are both contaminated with a bacterial 16S rRNA gene, but the genes have distinct sequence types that appear to be specific for each manufacturer. This specificity was confirmed by sequencing multiple amplification products from each type of blood culture medium with different primer pairs on different occasions with different lots of media. The same 16S rRNA gene sequence was present in aerobic and anaerobic culture media from the same manufacturer.
DISCUSSION
Blood cultures are among the most important tests used for the diagnosis of infectious diseases; they are commonly ordered and detect a wide variety of microbes, and the results are usually clinically relevant. The use of PCR for the detection and identification of microbes in blood cultures has been rather limited to date (11, 13, 18, 19), despite certain theoretical advantages to sequence-based identification, such as the speed and the specificity of identification. In some of the studies from which successful bacterial DNA amplification was reported, SPS-free blood culture medium was used for PCR, such as noncommercial broth (13) or BACTEC 12B mycobacterial medium (11). In another study, BACTEC 13A medium containing SPS was used, and the presence of a PCR inhibitor was noted after phenol-chloroform extraction of the samples, but the inhibitor was not identified (18). Those investigators did have success in amplifying mycobacterial DNA from blood cultures by using a combination of washing of the cells by centrifugation, sodium iodide lysis, and alcohol precipitation to purify DNA and by using the polymerase Tth plus for amplification. Another approach with an alkaline wash step with heat lysis has also been used to prepare mycobacterial DNA from BACTEC 13A medium for PCR amplification (19). Technical problems related to the presence of PCR inhibitors may have hindered the wider application of amplification technology to the detection of microbes in blood cultures.
Although blood is known to possess substances inhibitory to PCR (2), many DNA purification methods are successful in eliminating these inhibitors. We detected persistent inhibitory activity from inoculated blood culture medium, despite the use of six standard DNA purification techniques. The PCR inhibitor present in commercial blood culture media tends to copurify with DNA and was identified as SPS. SPS is added to blood culture media for its anticoagulant and anticomplementary activities, which are believed to increase the level of growth of most microbes (8, 17, 22–25). Given the chemical similarities between DNA and SPS, it should not be surprising that they copurify. Both are high-molecular-weight polyanions that are soluble in water but insoluble in alcohols. Accordingly, we found that phenol-chloroform fails to extract SPS. Alcohol precipitation merely precipitates SPS along with the DNA. SPS binds to silica in the presence of chaotropes and elutes with water, just like DNA. Ultrafiltration in Centricon-100 columns concentrates SPS because it is too large to pass through the membrane. Washing of cell pellets by centrifugation and resuspension are variably successful in removing SPS. Because SPS may bind to hemoglobin (7, 23) and erythrocyte membranes (which also pellet in blood culture media) and is inhibitory at extremely low concentrations, multiple wash cycles are required for successful removal. In addition, we found that SPS fails to bind to Chelex resin and is stable to boiling in the presence of Chelex, like DNA.
This study demonstrates the superiority of an organic extraction method with benzyl alcohol and guanidine hydrochloride over several other methods for the removal of SPS. The standard DNA purification methods tested in the present investigation failed to eliminate SPS, resulting in PCR inhibition unless the SPS was diluted. The chaotrope guanidine hydrochloride plays a critical role in the extraction process. Extraction with benzyl alcohol and water alone fails to remove SPS. The addition of other salts such as sodium chloride or guanidine isothiocyanate to the extraction does not result in the complete removal of SPS (data not shown). However, the addition of guanidine hydrochloride to the extraction promotes the partition of SPS into benzyl alcohol, while the DNA remains associated with the aqueous phase. The guanidine may function as an organic compound-compatible counterion to the SPS. Method C (Isoquick extraction) also uses a benzyl alcohol extraction, but in conjunction with guanidine isothiocyanate. The use of guanidine isothiocyanate instead of guanidine hydrochloride appears to account for the inferior performance of method C on the basis of some preliminary experiments. Method D is less effective if blood is absent from the blood culture medium. Blood binds to SPS, as described above, and may help carry SPS into the lower organic phase. Bloodless culture medium requires a second extraction for the complete removal of SPS (data not shown).
Even after the successful removal of SPS, PCR of microbial DNA from blood culture medium remains problematic if broad-range primers are used. Commercial blood culture medium is sterile but is not free of microbial DNA. Microbial growth in medium components prior to sterilization probably accounts for the residual DNA detectable in uninoculated culture medium. One would have to analyze the individual medium components from each manufacturer to determine the source of the Bacillus DNA present in BACTEC media and the Streptococcus DNA present in BacT Alert media. This residual microbial DNA appears to be highly fragmented, since amplification occurs only with primers directed against small targets (e.g., 291 bp but not 874 bp). The fact that PCRs with primer pairs 516F-806R and 516F-13R have similar sensitivities when relatively unfragmented E. coli DNA is used confirms that this difference in amplification is not due to differences in assay sensitivity. An alternative explanation for these results is that the bacterial 16S rRNA gene present in blood culture medium does not contain a nucleotide sequence complementary to the 13R primer; hence, there is no amplification with the 516F-13R primer pair. This explanation is refuted by the successful amplification of the DNA target from blood culture medium with primer pair 806F-13R.
If patient-associated microbes grow to high copy numbers in blood culture or if larger DNA segments (e.g., 874 bp) are targeted by PCR, then the problem of background contamination may be overcome. Nevertheless, the presence of microbial DNA in commercial blood culture medium should prompt investigators to perform rigorous controls when analyzing clinical samples for sequence-based evidence of microbial growth. The presence of a streptococcal sequence related to S. pneumoniae in BacT Alert blood culture media is particularly disturbing since one could easily attach significance to this sequence where none is warranted. To avoid the misattribution of blame, uninoculated culture medium should be included in nucleic acid amplification controls after SPS is removed from these samples.
In conclusion, SPS is a common component in commercially available blood culture medium and is a potent inhibitor of PCR. Several standard DNA purification methods failed to remove SPS, resulting in failed PCR amplification. An organic extraction procedure with benzyl alcohol and guanidine hydrochloride buffer successfully removes SPS, yielding DNA that can be amplified by PCR without further processing or dilution. PCR amplification technology is dogged by false-negative reactions (e.g., from the presence of PCR inhibitors such as SPS) as well as false-positive reactions (e.g., from the presence of contaminating target sequences). With the successful extraction of SPS-free DNA, the advantages and limitations of sequence-based identification of blood culture isolates can be further studied.
ACKNOWLEDGMENTS
David N. Fredricks is supported by a Physician Scientist Award from NIH (award K11-AI01360). David A. Relman is supported in part by a grant from the Donald E. and Delia B. Baxter Foundation.
We thank Mark Troll for sharing his expertise in organic chemistry and Doug Smith, Deborah Dodge, and Nicole Ellis from Perkin-Elmer Applied Biosystems Inc. for technical assistance and support. Thanks also go to Paul Lepp for advice on phylogenetic analysis.
REFERENCES
- 1.Ahokas H, Erkkila M J. Interference of PCR amplification by the polyamines, spermine and spermidine. PCR Methods Appl. 1993;3:65–68. doi: 10.1101/gr.3.1.65. [DOI] [PubMed] [Google Scholar]
- 2.Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Demeke T, Adams R P. The effects of plant polysaccharides and buffer additives on PCR. BioTechniques. 1992;12:332–334. [PubMed] [Google Scholar]
- 7.Edberg S C, Edberg M K. Inactivation of the polyanionic detergent sodium polyanetholsulfonate by hemoglobin. J Clin Microbiol. 1983;18:1047–1050. doi: 10.1128/jcm.18.5.1047-1050.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escamilla J, Santiago L T, Uylangco C V, Cross J H. Evaluation of sodium polyanethanol sulfonate as a blood culture additive for recovery of Salmonella typhi and Salmonella paratyphi A. J Clin Microbiol. 1983;18:380–383. doi: 10.1128/jcm.18.2.380-383.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Forbes B A, Hicks K E. Ability of PCR assay to identify Mycobacterium tuberculosis in BACTEC 12B vials. J Clin Microbiol. 1994;32:1725–1728. doi: 10.1128/jcm.32.7.1725-1728.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenfield L, White T J. Sample preparation methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: American Society for Microbiology; 1993. pp. 122–137. [Google Scholar]
- 13.Hassan K M, Baldeh I, Secka O, Falade A, Greenwood B. Detection of Streptococcus pneumoniae DNA in blood cultures by PCR. J Clin Microbiol. 1994;32:1721–1724. doi: 10.1128/jcm.32.7.1721-1724.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinen W, Lauwers A M, Mulders J W. Bacillus flavothermus, a newly isolated facultative thermophile. Antonie Leeuwenhoek. 1982;48:265–272. doi: 10.1007/BF00400386. [DOI] [PubMed] [Google Scholar]
- 15.Holodniy M, Kim S, Katzenstein D, Konrad M, Groves E, Merigan T C. Inhibition of human immunodeficiency virus gene amplification by heparin. J Clin Microbiol. 1991;29:676–679. doi: 10.1128/jcm.29.4.676-679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan G, Kangro H O, Coates P J, Heath R B. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J Clin Pathol. 1991;44:360–365. doi: 10.1136/jcp.44.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogstad D J, Murray P R, Granich G G, Niles A C, Ladenson J H, Davis J E. Sodium polyanethol sulfonate inactivation of aminoglycosides. Antimicrob Agents Chemother. 1981;20:272–274. doi: 10.1128/aac.20.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulski J K, Khinsoe C, Pryce T, Christiansen K. Use of a multiplex PCR to detect and identify Mycobacterium avium and M. intracellulare in blood culture fluids of AIDS patients. J Clin Microbiol. 1995;33:668–674. doi: 10.1128/jcm.33.3.668-674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulski J K, Pryce T. Preparation of mycobacterial DNA from blood culture fluids by simple alkali wash and heat lysis method for PCR detection. J Clin Microbiol. 1996;34:1985–1991. doi: 10.1128/jcm.34.8.1985-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 22.Tenney J H, Reller L B, Wang W L, Cox R L, Mirrett S. Comparative evaluation of supplemented peptone broth with sodium polyanetholesulfonate and Trypticase soy broth with sodium amylosulfate for detection of septicemia. J Clin Microbiol. 1982;16:107–110. doi: 10.1128/jcm.16.1.107-110.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traub W H, Fukushima P I. Neutralization of human serum lysozyme by sodium polyanethol sulfonate but not by sodium amylosulfate. J Clin Microbiol. 1978;8:306–312. doi: 10.1128/jcm.8.3.306-312.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traub W H, Fukushima P I. Variable neutralization of several nonspecific antibacterial systems in fresh, defibrinated human blood by sodium polyanetholsulfonate and sodium amylosulfate. J Clin Microbiol. 1979;10:27–31. doi: 10.1128/jcm.10.1.27-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traub W H, Kleber I. Inactivation of classical and alternative pathway-activated bactericidal activity of human serum by sodium polyanetholsulfonate. J Clin Microbiol. 1977;5:278–284. doi: 10.1128/jcm.5.3.278-284.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiedbrauk D L, Werner J C, Drevon A M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]