Summary
Background
Severe community-acquired pneumonia (SCAP) results in high mortality as well as massive economic burden worldwide, yet limited knowledge of the bio-signatures related to prognosis has hindered the improvement of clinical outcomes. Pathogen, microbes and host are three vital elements in inflammations and infections. This study aims to discover the specific and sensitive biomarkers to predict outcomes of SCAP patients.
Methods
In this study, we applied a combined metagenomic and transcriptomic screening approach to clinical specimens gathered from 275 SCAP patients of a multicentre, prospective study.
Findings
We found that 30-day mortality might be independent of pathogen category or microbial diversity, while significant difference in host gene expression pattern presented between 30-day mortality group and the survival group. Twelve outcome–related clinical characteristics were identified in our study. The underlying host response was evaluated and enrichment of genes related to cell activation, immune modulation, inflammatory and metabolism were identified. Notably, omics data, clinical features and parameters were integrated to develop a model with six signatures for predicting 30-day mortality, showing an AUC of 0.953 (95% CI: 0.92–0.98).
Interpretation
In summary, our study linked clinical characteristics and underlying multi-omics bio-signatures to the differential outcomes of patients with SCAP. The establishment of a comprehensive predictive model will be helpful for future improvement of treatment strategies and prognosis with SCAP.
Funding
National Natural Science Foundation of China (No. 82161138018), Shanghai Municipal Key Clinical Specialty (shslczdzk02202), Shanghai Top-Priority Clinical Key Disciplines Construction Project (2017ZZ02014), Shanghai Key Laboratory of Emergency Prevention, Diagnosis and Treatment of Respiratory Infectious Diseases (20dz2261100).
Keywords: Severe community-acquired pneumonia (SCAP), Metagenomic, Transcriptomic, Signatures, Outcome
Research in context.
Evidence before this study
We searched PubMed database, for published studies evaluating the metagenomic and transcriptomic signatures for predicting outcomes of patients with severe Community-acquired Pneumonia (CAP) in the past five years. The search terms used were (“Community-acquired pneumonia” OR “Community-acquired infection”) AND (“genomics” OR “gene expression profiling”) AND (“transcriptomic” OR “transcriptome”) AND (“outcomes” OR “prognosis”). We identified only 2 studies of CAP in children.
Added value of this study
Our study comprehensively investigated metagenomic and transcriptomic signatures in severe CAP and their associations with prognosis. We found a significant difference in host gene expression pattern presented between 30-day death group and the survival group. Most importantly, we integrated omics data, clinical features and parameters to develop a model with six signatures for predicting 30-day mortality, showing an AUC of 0.953.
Implications of all the available evidence
This study suggested a potentially critical connection between host response and prognosis of SCAP. The comprehensive predicting model could develop precision therapeutics of clinical practices.
Introduction
Community-acquired pneumonia (CAP) is a major infectious disease worldwide and contributes to high mortality and massive economic burden.1, 2, 3 Hospital mortality among the severe CAP (SCAP) remains high, ranging from 25% to more than 50%.4 Disease progression and prognosis in SCAP, from first symptoms to the need for supportive care such as mechanical ventilation, can vary widely among patients. It is believed that pulmonary microbiota—host interaction plays fundamental roles in development and severity of lung infectious diseases.5,6 However, studies about microbiota-host interactions on outcomes of SCAP are still in their infancy.
Many microorganisms including bacteria, viruses and fungi, coexist in the lungs of healthy individuals to constitute the lung microbiome. Published articles demonstrated the diversity of sputum microbiota predicted mortality in patients with chronic obstructive pulmonary disease, and the community composition of respiratory microbiota predicted exacerbations in bronchiectasis and respiratory infections in infants.7, 8, 9 Moreover, among mechanically ventilated critically ill patients, variation in lung microbiota predicts ICU outcomes.10 In particular, microorganisms residing in the upper respiratory tract can alter the susceptibility to the outcomes of infectious diseases.11 The underlying mechanisms may be the induction of the immune response in the host and colonization resistance.12 To date, no study has determined whether altered lung microbiota predict disease outcomes in the population of SCAP.
It has been observed that in many cases, death is not solely attributable to the direct effect of the pathogen or any toxin it produces. Rather, it is often the consequence of the systemic response in the host.13,14 There are at least two distinct biological components to the mortality risk: susceptibility to infection and propensity to develop harmful pulmonary inflammation. For example, susceptibility to respiratory viruses such as influenza and SARS-COV-2 is heritable and known to be associated with specific genetic variants.15,16 Another example is tuberculosis, in which outcomes vary greatly among different individuals infected. Studies show that TLR2, CCL2 and SLC11A1 genes may associate with susceptibility to tuberculosis and relate to its outcomes.17 Understanding the host response could be an invaluable tool guiding clinical treatment, as well as in understanding of the infectious disease development, progression and outcomes. However, a comprehensive assessment of the heterogeneity in the adult host response to varied outcomes and severity of community-acquired pneumonia has not previously been reported.
Massive attempts have been tried to find clinical factors and biomarkers to assess the severity and predict the risk of poor outcome of CAP patients. Previous studies indicated age and sex were related to the incidence and outcomes of CAP patients.18,19 It was also pointed out that some scoring system, such as pneumonia severity index (PSI), CURB-65 (acronym for confusion, urea, respiratory rate, blood pressure and 65 years of age) and quick sepsis-related organ failure assessment (qSOFA), were associated with prognosis of CAP.20,21 Meanwhile, C-reactive protein (CRP), procalcitonin (PCT), neutrophil-to-lymphocyte ratio (NLR), lactic dehydrogenase (LDH) and some cytokines were used as biomarkers for the prediction of outcomes.4,22,23 However, the variations of host response underlying these characteristics are still ambiguous. The determinations of these variations may help to uncover the mechanism underlying the progression of CAP, and discover signatures or clinical-genetic combination of new models to improve the accuracy of prediction contributing to the decrease of CAP mortality.
In the current study, we enrolled SCAP patients to explore the pulmonary microbiota and host responses of different outcomes, since the genetic differences could be detected more easily in the most severe patients as opposed to mildly severe ones.24 We performed DNA and RNA-based metagenomic next generation sequencing of bronchoalveolar lavage fluid (BALF), sputum and whole blood samples from 275 SCAP patients with varied characteristics and outcomes, to analyze the differences in the microbes and host responses between them. Besides, we tried to establish a comprehensive risk prediction model with those microbes, host genes and clinical characteristics correlated with outcomes, aiming to enable physicians to more accurately predict prognosis and provide appropriate treatments for SCAP patients.
Methods
Study design
All adult patients in this study were from our previous work.25 Blood samples, sputum samples and bronchoalveolar lavage fluid (BALF) were chosen for further DNA or RNA sequencing or both. The DNA-seq and RNA-seq data were applied for the analysis of microbial diversity and host gene expression profiles respectively. Infectious pathogens, microbial diversity and host response were compared between 30-day survivals and 30-day deaths. Clinical characteristics (such as age and sex), scoring systems (including PSI and CURB-65), oxygenation and laboratory parameters (blood routine and biochemical tests) were collected, and associations of host gene expressions with them were analyzed.
Sample collection and nucleic acid extraction
Three types of samples including 54 BALF (5 mL), 211 blood (3 mL) and 113 sputum (5 mL) samples were collected from 275 SCAP patients and were pretreated before further nucleic acid extraction. For blood sample, volume of 3 mL blood was drawn from patient, placed in cell-free DNA storage tube, and stored at room temperature. Plasma was separated within 96 h by centrifugation at 1600g for 10 min at 4 °C, then transferred to new sterile tube for next step. For sputum sample, 5 mL sputum sample from patient was collected according to standard procedures and then placed in a sterile container and inactivated at 65 °C for 30 min. Then sputum sample was liquefied by using 0.1% dithiothreitol (DTT) for 30 min at room temperature and then used for nucleic acid extraction. For BALF sample, 5 mL BALF was collected based on the standard clinical procedure and then placed in a sterile container and inactivated at 65 °C for 30 min before nucleic acid extraction. BALF collection was standardized according to the guideline issued by the Chinese Medical Association, which was performed by sequentially instilling two 10-mL aliquots in the pulmonary lobe directed by abnormal imaging. Aspiration was performed immediately after instilling each aliquot.
DNA of some pretreated samples (BALF, blood and sputum) from SCAP patients was extracted respectively and used for further DNA library construction. For blood sample, 300 μL plasma, spiked with 0.05 ng DNA as internal control, which was a nucleic acid fragment of known sequence, was used for DNA extraction. For sputum and BALF sample, 1.5 mL microcentrifuge tube with 500 μL sample and 1g 0.5 mm glass beads were attached to a horizontal platform on a vortex mixer and agitated vigorously at 2800–3200 rpm for 30 min. And then 300 μL of the supernatant was transferred to 1.5 mL microcentrifuge tube and mixed with 0.2 ng of internal DNA control. DNA was extracted using the TIANamp Micro DNA Kit (DP316, TIANGEN BIOTECH) according to the manufacturer’s recommendation. The extracted DNA was quantified by Qubit and 100 ng DNA was used for the following library construction, and if the DNA yield was less than 100 ng, then all the DNA from the sample was used for the following library construction.
RNA of some pretreated samples (BALF, blood and sputum) from SCAP patients was extracted respectively and further applied for the synthesis of cDNA before DNA library generation. For plasma sample, 140 μL sample was used for extraction of RNA. For sputum and BALF sample, 200 μL sample was first centrifuged at 1500g for 30 min at 4 °C to reduce human derived nucleic acid before extraction, and then 140 μL supernatant was used for RNA extraction. The sputum and BALF samples were handled the same way for the RNA extraction, library construction and RNA sequencing steps. RNA was extracted by using QIAAMP VIRALRNA MINI KIT (52904#, QIAGEN) according to the manufacturer’s recommendation. Then complementary DNA (cDNA) was generated from an RNA template by reverse transcription, followed by second strand synthesis of cDNA. Then the double strand DNA was used for the construction of DNA library.
Library preparation and sequencing
DNA library was constructed through DNA-fragmentation, end-repair, adapter-ligation and PCR amplification. Constructed library was qualified by Agilent 2100 (Agilent Technologies, Santa Clara, CA) and Qubit 2.0 (Invitrogen, USA). The concentration required for the constructed library was ≥1 ng/μL. Qualified double strand DNA library was transformed into single-stranded circular DNA library through DNA-denaturation and circularization. DNA nanoballs (DNBs) were generated from single-stranded circular DNA using rolling circle amplification (RCA). The DNBs were qualified using Qubit 2.0. Qualified DNBs were loaded on the flow cell and sequenced on MGISEQ-2000 platform (MGI, China). The requirement for the amount of data obtained from sequencing must be more than 15 M reads.
Metagenomic analysis
The DNA-seq data were applied for the analysis of microbial diversity. Firstly, the high-throughput sequencing raw data were filtered by fastp to remove low quality reads and adapters, and the quality of filtered data were further controlled using FastQC (Version 0.11.9).26,27 Then the filtered reads were mapped to the human genome (GRCh38) using HISAT2 (2.2.1 release) to remove human sequences.28 The identifications of microbial species were performed on the clean reads by Kraken2.29 The DNA-seq data were then normalized to RPM before further analysis. Shannon Wiener index were calculated by the known formula.30 PCA was performed by ade4 R package.31 All the PCA plots, box plots and heatmap figures were generated by utilizing the ggplot2 package in R v4.0.3.32
Transcriptome analysis
The RNA-seq data were used for the analysis of host transcriptomic profiles. Firstly, the RNA-seq raw data were filtered to remove low quality reads and adapters by fastp.26 Subsequently, the rRNA sequences of filtered RNA-seq data were removed using SortMeRNA (version 4.3.3) with rRNA databases including RFAM 5S and 5.8S databases from the ARB package.33,34 Then, the quality of RNA-seq filtered reads were further controlled using FastQC (Version 0.11.9).27 Later, the RNA-seq filtered reads were mapped to the human genome (GRCh38) used hisat2 (2.2.1 release),29 and then the RPKM values of host gene expressions were calculated by the R package “edgeR”.35 Host differentially expressed genes (DEGs) between two different groups were analyzed by RStudio (Version 1.3.959) with R software package DEseq2.36 The genes with absolute foldchange more than 1.2 between two groups and with adjusted p value <0.05 were presented as DEGs.37, 38, 39, 40 The p-value were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (<0.05). Genes with an adjusted p-value <0.05 (detected by DESeq2) were considered to be significantly differentially expressed. DEGs were further showed in volcano plot as well as applied for gene functional enrichment analysis. The gene functional enrichment analysis of DEGs in host was performed in Metascape website (https://metascape.org/gp/index.html#/main/step1).41
The establishment of predictive model
We tried to apply transcriptomic, microbial and clinical signatures to develop models for predicting outcomes (30-day mortality or 30-day survival) of SCAP patients. Previous studies showed that random forests gave outstanding binary predictions on similar data types, which were a mix of numeric, characters and Boolean values.42 Therefore, our models in this study were built in random forest. Models were trained using data from the host genes, microbes and clinical characteristics. Microbes and genes correlated with death (spearman correlation coefficient |rho| > 0.3) were selected and clinical characteristics including gender, age, CURB65 (acronym for confusion, urea, respiratory rate, blood pressure and 65 years of age), CRB65, pneumonia severity index score, PSI class, Neutrophil-Lymphocyte Ratio (NLR), LDH, ratio of the partial pressure of oxygen (PaO2) to the fraction of inspired oxygen (FiO2), blood urea nitrogen (BUN), albumin, arterial blood pH and sodium, were utilized to further screen high effective markers for generating predictive models. RPKM values of genes, RNA/DNA ratio of microbes and the detection values of clinical characteristics were used respectively for the screen of signatures building the models.
In total, 9 models were built based on different sample types and markers. Among them, three models were developed only with host gene signatures from BALF, sputum and blood samples respectively, namely BALF gene-based model (BALF-G), sputum gene-based model (SPU-G) and blood gene-based model (BLO-G). Besides, two models were generated with microbial signatures from BALF (model BALF-M) and sputum (model SPU-M) samples. Since there were no microbes which showed correlation (|rho| > 0.3) with outcomes of SCAP patients in blood samples, no microbe-based model was constructed for blood samples. In addition, one model only consisting of clinical signatures (model C) was developed to examine the performance of clinical features in predicting SCAP outcomes. Moreover, we hypothesized that comprehensive signatures should improve the precision and specificity of predicting outcomes of SCAP patients, and hence three different types of signatures (genes, microbes, and clinical characteristics) were combined to develop combined models for sputum (model SPU-GMC) and blood (model BLO-GC) samples. No combined model was built for BALF samples due to insufficient death cases (only 3 cases) for testing the generated model, which limits its ability in making practical predictions. Additionally, to take advantage of information from the three types of samples simultaneously to further improve prediction performance, one integrated model (ALL-GMC) was developed. The model ALL-GMC was built based on all the markers obtained from sputum, blood and the patients’ clinical characteristics.
For the model building, numeric values were scaled so that the mean of each numeric marker was zero and fell between −1 and 1. Models SPU-G, SPU-M, BLO-G, SPU-GMC, and BLO-GC included only the complete datasets, but ALL-GMC would assign “NA” with values imputed from random forest proximity matrix if the patient had done either one of the NGS sample tests. Data were split into 80% for training and 20% for testing. As more patients survived in the 30-day window, our datasets were imbalanced. Therefore, a specific number of death cases were forced to be included in the testing set to ensure sufficient death cases for model testing.
To optimize the model parameters, ten trees per grid search was applied to determine the optimal number of trees, which was 200–300 in this study. In total, two rounds of modelling were made. In the first round 100 training iterations using 80% of all data selected randomly were done. During each round, the combinations of the most critical markers which led to the least model errors were noted. After 100 iterations, markers were selected if they were considered important more than 20 times. In the second round of training, the markers selected from the previous step were used and RF models were built.
To validate the prediction power of the markers selected, 10 rounds of random forest training were done on each type of model, and during each round, data was randomly split into training (80%) and testing (20%) datasets. To ensure that a sufficient amount of death cases can be tested, around 35% of total death cases were included in the testing dataset. Ten-fold cross-validation was not used here, because there were much more survival cases than death cases, and ten-fold cross validation would inevitably result in a training or testing dataset without any death cases. Including death cases in the testing dataset is a must, since if we would like to implement predictive markers clinically, correctly predicting patients at high risk of mortality is crucial to promptly provide medical care or intervention before their condition worsens.
To evaluate the performances of the models, the area under the receiver operating characteristic curve (AUC-ROC) was calculated for each model. AUC-ROC was calculated from the values predicted by the random forests on the test datasets, after 10 rounds of model training. F1 scores, calculated as below, were also listed as indicators of model performances at a cut-off value of 0.5:
Statistical analysis
Principal component analysis (PCA) is a method to extract and keep the most important information from variables with many dimensions/features in a large dataset to simplify the description of data set and then make it easier for analyzing.43 Comparisons of differences between groups in PCA plot were tested by pairwise permutational multivariate analysis of variation (PERMANOVA). Comparisons of microbial diversity, microbe abundance and gene expression level between two groups were assessed by Wilcoxon rank sum test,44 and between three groups were examined by Kruskal–Wallis test.45 Correlations between microbial taxa or host genes and clinical characteristics were tested using Spearman correlation analysis or multiple linear regression analysis. The Spearman correlation coefficients between 0.3 and 0.6 was regarded as moderate correlation, and more than 0.6 indicated high correlation.46, 47, 48, 49 Spearman correlation analysis was performed with the R package “Hmisc”.50 Multiple linear regression analysis was performed using R base function glm() and the boot.lasso.proj() from the hdi package.51,52 The associations of outcome with clinical characteristics were measured by multiple logistic regression analysis using R base function glm().51 The function was used with argument “family” of “binomial”. For incomplete clinical data, missing values were imputed using the rfImpute() function from the randomForest R package. All clinical features and outcomes (survival/morality) were included to build the multiple logistic regression models. All genes and ten outcome-related clinical features were imported to build the multiple linear regression model. We used Least Absolute Shrinkage and Selection Operator (LASSO) logistic or linear regression as a feature selection strategy. As p-values were calculated using bootstrapping to test whether the corresponding regression coefficient were significant, features with insignificant impact on the output were therefore omitted. The model performances were assessed using the area under the receiver operating characteristic curve (AUC-ROC), as well as F1 scores to compensate for the possible misleading AUC-ROC information due to our imbalanced data. F1 scores were calculated at a cut-off value of 0.5, regardless of which model was tested.
Ethics statement
The current study was approved by the Ruijin Hospital Ethics Committee, Shanghai Jiaotong University School of Medicine (reference number: 2017-186). Informed consent was signed by all patients.
Role of funders
The funding sources had no role in the design of this study, data collection, analyses, interpretation of the data or writing the manuscript.
Results
Patient characteristics and sequence data assessment
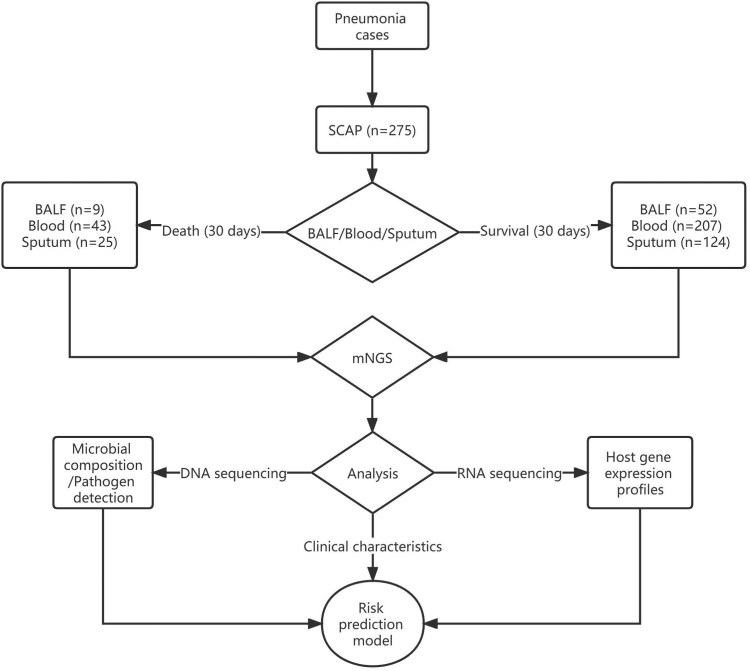
From 1 June 2018 to 31 December 2019, 275 adult patients with SCAP from 17 hospitals were included for the final analysis. The detailed characteristics of these patients were shown in the previous study.25 Fifty-two out of the 275 patients eventually died within 30 days after admission to the hospital. Three types of samples including BALF, blood and sputum were collected from non-survival (n = 9, 43, 25, respectively) and survival (n = 52, 207, 124, respectively) patients as soon as possible after the presentation, which were further sequenced by DNA or RNA or both-based metagenomic next generation sequencing (mNGS) method. The median (interquartile range, IQR) of clean data for samples with DNA-based sequencing was 24.43 (18.07–35.09) M reads, and for samples with RNA-based sequencing was 24.37 (16.34–32.42) M reads. The DNA data were applied to detect pathogens as well as characterize microbial compositions, and the RNA data were used for the analysis of host gene expression profiles. Furthermore, the microbes, the differentially expressed genes (DEGs) and clinical characteristics were exploited to develop a risk prediction model for 30-day death (Fig. 1).
Fig. 1.
Study design. mNGS: metagenomic next generation sequencing. BALF: bronchoalveolar lavage fluid. Pathogen detection is an integrated diagnosis including routine bacterial/fungal cultures, polymerase chain reaction (PCR) for virus detection, urine—antigen test, pathogen—specific antibody titre, metagenomic next generation sequencing (mNGS) method, as well as features (laboratory tests and imaging findings).
The impact of different pathogens on outcomes of SCAP patients
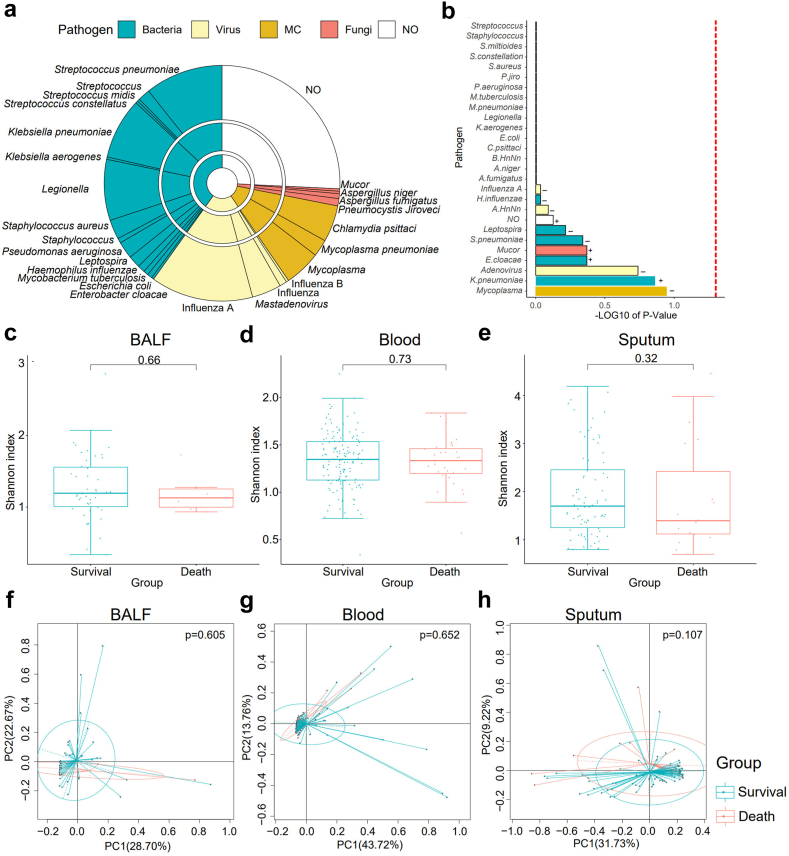
Different pathogens, including bacteria, virus, fungi, mycoplasma and chlamydia, were finally identified by integrated diagnosis methods in 198 (72.0%) of 275 recruited SCAP patients, while no pathogens were identified in the other 77 patients. Among the 198 cases, 119 patients (60.1%) were infected by bacterial pathogens. Fifty-seven (28.8%), 35 (17.7%) and 7 (3.5%) patients were confirmed to be infected by virus, Mycoplasma and Chlamydia, and fungi, respectively (Fig. 2a). Some cases were simultaneously infected by two or more pathogens according to the final diagnosis (Supplementary Table S1). Then we investigated associations of pathogens with outcomes of SCAP patients. We didn’t find obvious evidence that the outcome of 30-day mortality was dependent on bacteria (p = 0.636, Fisher's exact test), virus (p = 0.836, Fisher's exact test), fungi (p = 0.343, Fisher's exact test), mycoplasma or chlamydia infections (p = 0.586, Fisher's exact test) and co-infection (p = 0.138, Fisher's exact test) in this study. Moreover, we further investigated associations of specific pathogens with outcomes of SCAP patients by Chi-square test. The results indicated that 30-day mortality might be independent of these identified pathogens (Fig. 2b).
Fig. 2.
The identified pathogen profiles and associations of microbial diversity with outcomes of SCAP patients. (a) Pathogen profiles identified in SCAP patients. MC: Mycoplasm and Chlamydia. NO: unknown etiologies. The innermost pie indicates the main categories of pathogens: bacteria (blue), virus (yellow), MC (orange), fungi (red), and NO (white). The second pie shows the taxa of pathogens at genera level. The outside pie shows the species or genera of pathogens identified. (b) The associations of different pathogens with 30-day death of SCAP patients by Chi-square test. The vertical red line indicating the p = 0.05. (c) The associations of microbial alpha-diversity with outcomes of SCAP in BALF, blood (d) and sputum (e), respectively. The statistical significances between the 30-day death and survival groups were tested by Wilcoxon rank sum test. (f) The associations of microbial beta-diversity with outcomes of SCAP in BALF, blood (g) and sputum (h), respectively. The statistical significances between the 30-day death and survival groups were tested by pairwise permutational multivariate analysis of variation (PERMANOVA). p < 0.05 means that the difference was significant.
The correlations of microbes in BALF, blood and sputum with outcomes of SCAP patients
Various studies have shown that human diseases can be related with alterations of microbiota in human body. To test the associations of microbes with outcomes of SCAP patients, three hundred and seventy-eight samples including 54 BALF, 211 blood and 113 sputum samples from the 275 SCAP patients were first sequenced using mNGS for the analysis of microbial diversity. Significant differences in microbial diversity between BALF, blood and sputum were observed (Supplementary Fig. S1a and S1b). Hence, we next compared the microbiome diversity in BALF, blood and sputum between patients who died within 30 days and those who survived over 30 days, respectively. The results showed that there was no significant difference in microbial alpha- and beta-diversity between the two groups (Fig. 2c–h), indicating that the outcomes of SCAP patients were not distinctly dependent on BALF, blood, and sputum microbial diversity. We next calculated the correlations of specific microbial taxa (at species level) with the outcomes of SCAP patients by spearman's rank correlation analysis. Consistently, no strong associations (spearman's rank correlation coefficient |rho| > 0.6) between microbes and outcomes of SCAP patients were found. Only a few microbial species showed moderate negative or positive associations (|rho| > 0.3) with 30-day death (Supplementary Table S2).
The impact of differential host response on outcomes of SCAP patients
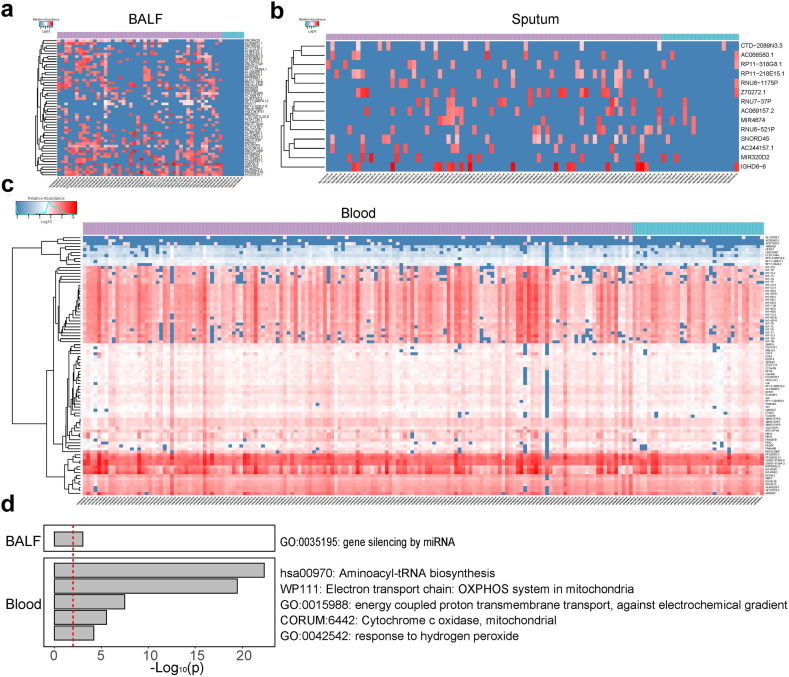
The principal component analysis (PCA) based on RNA sequencing data was performed to compare the dissimilarity of host response among different types of samples, which showed obvious differences in host responses between blood and BALF or blood and sputum, indicating that gene expression pattern was body site-dependent (Supplementary Fig. S2). To unravel whether host gene expressions affected outcomes of SCAP patients, we conducted analysis of differentially expressed genes (DEGs) in BALF, blood and sputum between SCAP patients who died within 30 days and those who survived over 30 days. The results showed that 52 genes were down-regulated in BALF in the 30-day death group compared to the survival group (Fig. 3a, Supplementary Table S3). These genes were mainly enriched in one pathway: gene silencing by miRNA. Among them, two immune-related genes including IFNA17 and IGHD3-3 (immunoglobulin heavy diversity 3-3) were found, and IFNA17 was supposed to be associated with antiviral infection. Compared to BALF samples, a smaller number of DEGs (14 genes) were found in sputum between the 30-day death group and the patients who survived over 30 days (Fig. 3b, Supplementary Table S4). The 14 genes were not found to be enriched in certain functional pathways, including IGHD6-6 (immunoglobulin heavy diversity 6-6), RP11-218E15.1, SNORD45 (small nucleolar RNA SNORD45), Z70272.1, as well as AC069157.2. Besides, 87 genes were distinctly downregulated in blood in the non-survivors compared to the survivors (Fig. 3c, Supplementary Table S5). These genes were mainly involved in GO terms: aminoacyl-tRNA biosynthesis, electron transport chain, energy coupled proton transmembrane transport against electrochemical gradient, mitochondrial cytochrome c oxidase and response to hydrogen peroxide (Fig. 3d).
Fig. 3.
Comparison of host gene expression profiles in BALF, sputum and blood between different outcome groups. (a) The clustered heatmap shows the DEGs in BALF between SCAP patients who died within 30 days and those who survived over 30 days. The color scale indicating different values of log10(RPM). RPM: reads per million. The top horizontal bar indicating the survival (purple) and death (blue) outcomes. (b) The DEGs in sputum between the 30-day death and survival groups. (c) The DEGs in blood between the 30-day death and survival groups. (d) The GO and KEGG pathway enrichment analysis of DEGs in BALF and blood between the 30-day death and survival groups.
The associations of age and gender with outcomes of SCAP patients and the potential mechanisms
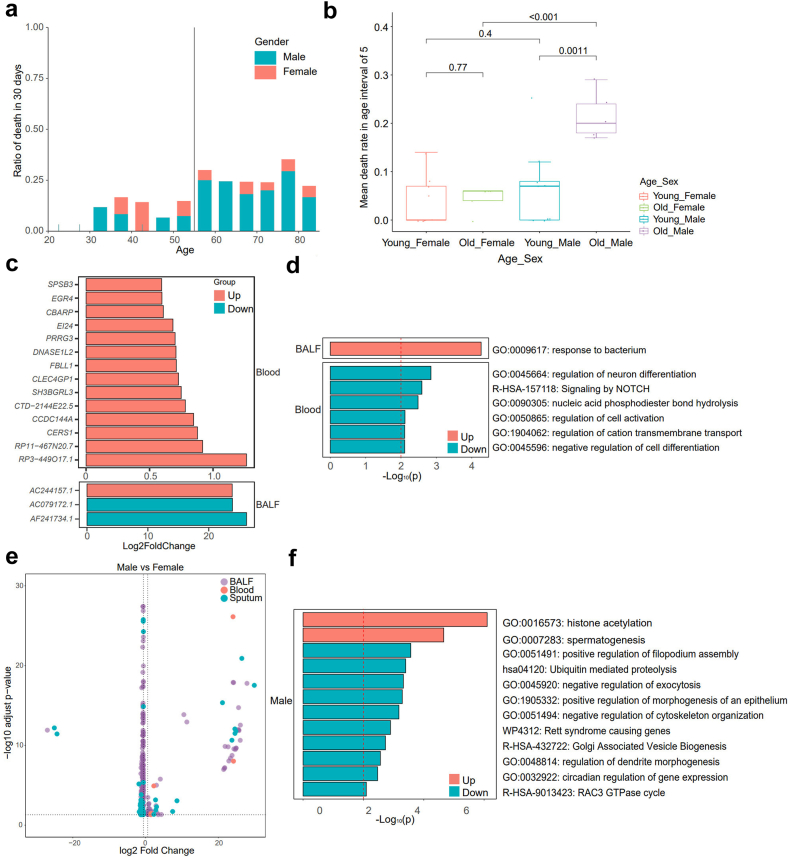
Previous studies indicated that the outcomes of SCAP patients were usually related with age and sex, thus we investigated the variations of 30-day mortality with the increase of age and compared their differences between males and females. The distribution of age and sex for the SCAP patients was shown in Supplementary Table S6. The results showed that there was a clear cut-off age for enrolled patients to be at the most significant risk of 30-day death, which was 55 years old (Fig. 4a). The patients who were 55 or under 55 years old displayed a significantly lower 30-day death rate than those aged over 55 (0.1 versus 0.260, p < 0.001, Welch's t-test). However, this age-dependent mortality was only observed in male patients not in female patients (Supplementary Fig. S3). Besides, males had distinctly higher 30-day mortality than females in the cohort of patients aged over 55 (mean 0.340 [95% CI: 0.02–0.66] versus 0.044 [95% CI: 0.01–0.08], p = 0.00043, Wilcoxon rank sum test), which was not significant between the male and female patients aged 55 or under 55 (mean 0.066 [95% CI: 0.00–0.13] versus 0.038 (95% CI: 0.00–0.08), p = 0.4, Wilcoxon rank sum test, Fig. 4b). These results may suggest that age factor played a more important role in outcomes of male versus female SCAP patients.
Fig. 4.
Impact of age and gender on outcomes of SCAP patients and its potential mechanisms. (a) The column chart showing differential 30-day morality of SCAP patients at different ages. The vertical line shows the cut-off age for patients to be at higher risk of 30-day death. The males were indicated in blue. The females were marked in red. (b) The box plot showing the 30-day death rate of four groups including the females aged ≤55 (red), the females >55 (light green), the males aged ≤55 (blue), the males >55 (purple). The statistical analysis was conducted by Wilcoxon rank sum test. (c) The column chart shows DEGs in BALF and blood in the elder (patients aged >55) group compared to the young (patients aged ≤55) group. (d) The GO and KEGG pathway enrichment analysis of the outcomes-related DEGs between the elder male patients and the young male patients. (e) The volcano plot showing outcomes-related DEGs in BALF, blood and sputum between the males and females aged over 55. The outcomes-related DEGs: the rest DEGs between the males and females aged over 55 after the removal of the overlapped DEGs (present not only in DEGs between the male and female SCAP patients aged >55 years but also in DEGs between the male and female SCAP patients aged ≤55 years) except those associated with age. (f) The GO and KEGG pathway enrichment analysis of the outcomes-related DEGs between the males and females aged over 55.
To explore the potential mechanisms of the impact of age on outcomes of SCAP, the host DEGs between the elder (>55) and the young (≤55) patients were first analyzed. Three and 14 genes were differentially expressed in BALF and blood between the two groups respectively, as shown in Fig. 4c. No DEG was found in sputum between the elder and the young groups. Since age showed no significant impact on outcomes of female SCAP patients, we further compared the DEGs between the elder and young patients by sex to find the exact mechanisms responsible for the higher mortality in the elder male patients. In order to remove the genes related with age but uncorrelated with outcomes, among the DEGs between the elder and young male patients, the DEGs which were also present between the elder and young female patients were removed except for those with different (2 folds) foldchanges in male patients and female patients. After the removal of genes unrelated with outcomes, 20 genes were significantly upregulated in BALF in the elder male patients versus the young male patients (Supplementary Table S7). Interestingly, three of the 20 genes including CXCL8 (IL-8), CXCL5 (IL-5), and DEFB114 (β-Defensin 114) were related to response to bacterium (Fig. 4d). There were 14 genes significantly downregulated in the elder male patients and interestingly 5 of them belonged to immunoglobulin kappa variable 1 (pseudogene) including IGKV1OR22-5, IGKV1OR2-1, IGKV1OR2-2, IGKV1OR1-1, and IGKV1OR2-118. Fifty-three genes were significantly upregulated in blood in the elder male patients (Supplementary Table S8), such as PI4K2B (Type II phosphatidylinositol 4-kinase β), PIGC (phosphatidylinositol glycan anchor biosynthesis class C), and (gap junction protein epsilon 1). PI4K2B played an important role in early T cell activation.53 Fifty-four genes were distinctly downregulated in blood (Supplementary Table S8) in the elder male patients and enriched in 6 terms including regulation of neuron differentiation, signaling by NOTCH, nucleic acid phosphodiester bond hydrolysis, regulation of cell activation, regulation of cation transmembrane transport, and negative regulation of cell differentiation (Fig. 4d). No DEG was found in sputum in the elder versus the young male patients.
We further analyzed the role of sex in the influence of age on outcomes. We separately investigated host DEGs between male and female SCAP patients aged >55 years and those aged ≤55 years. The overlapped DEGs except those associated with age were removed, so that genes correlated to sex not to outcomes were eliminated. The results showed that 53 genes were specifically upregulated and 176 genes downregulated in BALF of males compared to females (Fig. 4e, Supplementary Table S9). Most of genes upregulated in BALF of males were involved in histone acetylation and spermatogenesis, while those downregulated were correlated with pathways such as promotion of filopodium assembly, ubiquitin mediated proteolysis, morphogenesis of epithelium, dendrite development, and inhibition of exocytosis, cytoskeleton organization as well as cellular component organization (Fig. 4f). Besides, there were 6 genes (such as MIR320D2, HBA2 and RNU6-1175P) upregulated in blood, and 17 genes upregulated (such as VTRNA2-1, IGHD6-6 and HIST1H1PS1) as well as 95 genes downregulated (such as POLA1, IL1RAPL2, XACT, and RRAGB) in sputum in male versus female population (Supplementary Tables S10 and S11). VTRNA2-1 produces a non-coding RNA (lncRNA), inhibiting the activation of protein kinase R (PKR) which plays a critical role in host innate immune response to defend against viral infections by suppressing protein synthesis.54,55 This suggests that the upregulation of VTRNA2-1 in sputum (log2FoldChange = 26, p = 2.11 × 10−42, Wald test) in the male patients (age >55) may increase the patients’ susceptibility to viral infection leading to poor outcomes.
The associations of host gene expressions with other outcome-related clinical characteristics in SCAP patients
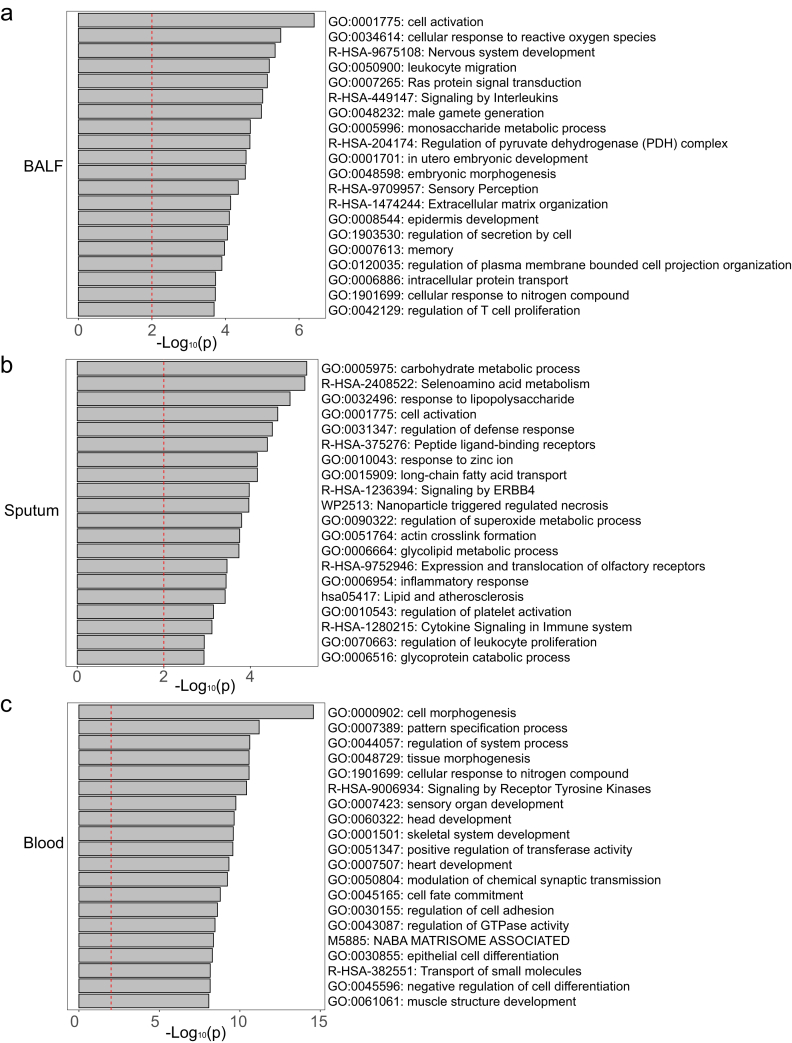
To unravel clinical characteristics synergistically associated with outcome, multiple logistic regression analysis between clinical characteristics (Supplementary Table S12) and outcome was performed. The results showed that besides age, characteristics including prothrombin time, neutrophil count, LDH, CD4/CD3 ratio, base excess (BE), direct bilirubin, lactic acid, glycated hemoglobin, neutrophil/lymphocyte ratio (NLR), fibrinogen and kidney disease were significantly associated with 30-day morality of SCAP patients (p < 0.05; Bootstrapping, Supplementary Table S12). Further, to understand the host response underlying these outcome-related clinical characteristics, we investigated the associations of host gene expressions with them by multiple linear regression analysis (age had been discussed above). The results indicated that these outcome-related characteristics were associated with BALF genes mainly involved in leukocyte and lymphocyte activation (such as IRF8, IFNA2, IFNA4), response to reactive oxygen species, carbohydrate metabolism, signaling by interleukins, and male gamete generation (Fig. 5a and Supplementary Table S13). Similarly, in sputum, genes associated with these characteristics were enriched in carbohydrate metabolism, leukocyte and lymphocyte activation, defense against infection, superoxide metabolism, and inflammatory response (Fig. 5b and Supplementary Table S14). In contrast, blood genes associated with these features were mainly referred to cell morphogenesis, organ development, cellular response to nitrogen compound and cell differentiation (Fig. 5c and Supplementary Table S15).
Fig. 5.
GO and KEGG pathway enrichment analysis of host genes associated with outcome-related clinical characteristics. The GO and KEGG pathway enrichment of associated genes in BALF (a), sputum (b), and blood (c).
Combined signatures for predicting outcomes of SCAP patients
To compare the performances of transcriptomic and metagenomic signatures for predicting outcomes (30-day death or survival) of SCAP patients, we used microbes and genes correlated with 30-day death (|rho| > 0.3) to develop models respectively (Table 1, Supplementary Table S16). Notably, we combined signatures from genetic, microbial and clinical perspectives to build comprehensive models with regard to different types of samples to see the precision and specificity of predicting outcomes of SCAP patients. The combined model for sputum samples (SPU-GMC) exhibited excellent performance with an AUC of 0.953 (95% CI: 0.92–0.98) and an F1 of 0.87, which consisted of 6 signatures including four genes (RP11.513M16.8, RPL23AP48, TECTB, RNU1.36P) from sputum sample and two clinical characteristic neutrophil/lymphocyte ratio (NLR) and age. RP11.513M16.8 produces a non-coding lncRNA, and RPL23AP48 is ribosomal protein L23a pseudogene 48 (NCBI database), however, the functions of them were not discovered yet. TECTB encoding a non-collagenous glycoprotein (β-tectorin), was involved in the formation of tectorial membrane which was important for hearing.56 RNU1.36P is a pseudogene with unknown functions, which produces RNA, U1 Small Nuclear 36, Pseudogene (NCBI database).
Table 1.
The developed risk prediction models for SCAP.
| Model content | Model abbreviation | Number of markers | Markers | ROC (%) (test) | ROC 95% CI |
|---|---|---|---|---|---|
| BALF gene | BALF-G | 3 | RNU7.141P, AC009502.1, RP11.80H5.6 | 91.38 | 85.28–97.47 |
| BALF microbes | BALF-M | 10 | Corynebacterium.diphtheriae, Waddlia.chondrophila, Fonsecaea.monophora, Xanthobacter.tagetidis, Bipolaris.oryzae, Paucibacter.toxinivorans, Aspergillus.brunneoviolaceus, Streptococcus.pneumoniae, Pneumocystis.jirovecii, X.Candida.glabrata | 60.00 | 43.83–76.17 |
| Sputum gene | SPU-G | 16 | RPL23AP48, PDSS1P2, RP11.513M16.8, CTC.499B15.7, RAC1P3, OR52W1, RNU1.36P, TECTB, DNAJC3, AC018890.4, MINOS1P2, RP11.983C2.3, SEC61B, UBE2V1P10, AC007204.3, RNA5SP285 | 91.36 | 87.27–95.45 |
| Sputum microbes | SPU-M | 7 | Clostridiaceae.bacterium, Acanthamoeba.castellanii, Corynebacterium.matruchotii, Chryseobacterium.contaminans, Schaalia.vaccimaxillae, Bacteroidales.bacterium.KA00251, Fusobacterium.necrophorum | 74.91 | 66.47–83.34 |
| Sputum combined | SPU-GMC | 6 | RP11.513M16.8, RPL23AP48, TECTB, RNU1.36P, NLR, age | 95.32 | 92.19–98.45 |
| Blood gene | BLO-G | 3 | RP11.325O24.1, UGT3A2, ANKRD66 | 74.76 | 69.28–80.23 |
| Blood combined | BLO-GC | 7 | ldh, albumin, RP11.325O24.1, UGT3A2, ANKRD66, psi_score, age | 85.05 | 80.90–89.20 |
| Clinic | C | 5 | Age, ldh, pH, albumin, psi_score | 73.47 | 67.56–79.38 |
| All markers combined | ALL-GMC | 9 | BUN, RPL23AP48_sputum, TECTB_sputum, psi_level, RP11.513M16.8_sputum, OR52W1_sputum, RNU1.36P_sputum, AC007204.3_sputum, RP11.325O24.1_blood, age | 79.58 | 73.62–85.53 |
Discussion
In the current study, we found that 30-day mortality was independent of pathogen category, microbial diversity or specific microbial taxa, while significant differences in host gene expression patterns were suggested to be responsible for different outcomes. Clinical characteristics analysis showed that male sex with age over 55 years was a risk factor for poor prognosis, and specific enrichment of genes and signaling pathways were found in omics data. Besides, in addition to age and sex, characteristics including prothrombin time, neutrophil count, LDH, CD4/CD3 ratio and neutrophil/lymphocyte ratio (NLR), were also related to outcomes, and associated genes underlying were mainly involved in leukocyte and lymphocyte activation, carbohydrate metabolism, defense against infection, and response to reactive oxygen species. Furthermore, the sputum model SPU-GMC showed the best performance for predicting 30-day mortality, indicating the significance of exploring markers from different perspectives.
Association analysis to clinical outcomes of SCAP indicated tremendous changes in transcriptomic profiles in relation to the host response to the disease. We observed that the identified DEGs in BALF between the 30-day death and the survival group were mainly enriched in gene silencing by miRNA. This may suggest that the host response differences in BALF mainly present in protein level since only few functional genes such as IFNA17 and IGHD3-3 were found. In blood, genes enriched in aminoacyl-tRNA biosynthesis were negatively modulated in the 30-day death group versus the surviving group, which was similar to the observation that genes related to aminoacyl-tRNA biosynthesis such as TRNS1 and TRNM were downregulated in patients with severe pneumonia in comparison with healthy individuals.57 Besides, genes related to mitochondrial functions were also downregulated in non-survivors in our study, including electron transport chain: OXPHOS system in mitochondria, energy coupled proton transmembrane transport against electrochemical gradient, and mitochondrial cytochrome c oxidase. Previous findings demonstrated that genes associated with oxidative phosphorylation such as electron transport chain complex I, IV and V were downregulated in blood of non-surviving patients with sepsis caused by CAP.58 The downregulation of energy coupled proton transmembrane transport, which is essential for ATP synthesis, may contribute to poor outcome. This is supported by evidence that decreased ATP concentrations in skeletal muscle are related with multiorgan failure and eventual death in septic patients.59 Genes related to response to hydrogen peroxide were also downregulated in the 30-day death group. Hydrogen peroxide belongs to reactive oxygen species (ROS), which can induce oxidative stress and cytotoxicity to cells. The reduced transcription of genes involved in response to hydrogen peroxide suggested that the capacity of anti-cytotoxicity was declined in the 30-day death patients, which may be also responsible for the poor outcome. The differential host responses in different body sites were understandable since the composition of cells, microbiota and microenvironment were divergent.
A variety of previous studies had shown that mortality of SCAP was associated with age and sex, since elder or male patients had higher mortality, which were consistent with our finding. Clinical characteristics were the macroscopical reflection of host-related element, while the microscopical mechanism lay in the variation of host gene expressions.18,60, 61, 62 To date, there is still lack of studies to investigate the differential host gene expressions between varied age and sex from transcriptomic profiles. In our study, we found that three genes related with host immune response were significantly upregulated in the elder male patients compared to the young male patients, including CXCL8 (IL-8), CXCL5 (IL-5), and DEFB114 (β-Defensin 114). IL-8 is a key proinflammatory cytokine for the recruitment of neutrophils to infection sites. A meta-analysis from 13 studies found that IL-8 was in higher level in SCAP patients than in patients with non-severe pneumonia.63 It was also reported that IL-5 concentration was increased in severe Coronavirus Disease 2019 (COVID-19) patients in comparison with those with moderate COVID-19 and it displayed an upward trend during the progress of the disease in severe COVID-19 patients.64 Defensins are antimicrobial peptides which are important components of host innate immunity. Influenza virus infection could induce the expression of some murine β-defensins in respiratory tract mucosa of mice.65 Human β-defensin 114 was found to be able to bind Lipopolysaccharide (LPS) in vitro and modulate LPS-mediated Inflammation.66 As a whole, CXCL8, CXCL5, and DEFB114 may serve as the important modulators in the poor outcome of the elder male SCAP patients.
In the current study, we exploited microbes, host genes, clinical characteristics, and parameters as signatures, and developed 9 models with them respectively to predict 30-day death of SCAP patients. Among these, the model from sputum with host genes, age and NLR showed the best performance. This result was supported by previous findings, in addition to age mentioned above, NLR has also been reported to be correlated with poor outcomes and act as biomarker predicting prognosis.67,68 The predicting models in previous published studies were usually established according to the signatures of single level, while the comprehensive model further improved the predictive capacity and reliability. In addition, our comprehensive model was raised from multi-omics analysis, and genetic changes would better reflect the underlying mechanism of disease progress. Finally, this was an ideal and easy-to-use model for assessing 30-day mortality risk of SCAP, since sputum is much easier to collect than blood or BALF.
Some limitations of this study should also be acknowledged. There were some unavoidable biases in identifying and recruiting participants. For example, more male SCAP patients were enrolled than the female patients, and the number of surviving patients was much larger than that of non-survivors. Meanwhile, the conclusion that pathogens didn’t affect outcomes needs to be further evidenced since the specimens in each category were not quite enough and balanced. Besides, limited to the sample size, we failed to enroll a separate cohort of SCAP patients to further verify the predicting capacity of comprehensive model. Thus, a study with a larger sample size is needed to bring a more credible result. Notably, 77 (28%) patients were found pathogen-negative. To the best of our knowledge, the identification rate of responsible pathogens in our study was among the highest in similar studies, and the identification of responsible pathogen by the scientific committee made the result clinically reliable. We thought the initial empiric treatment before enrollment in part of patients covered the responsible pathogens may somehow lead to pathogen-negative findings. Moreover, the cell types and numbers were not analyzed in BALF, blood and sputum samples, and therefore the potential impacts of cell composition shift induced by pathogen infection on variance in gene expression were unclear, though there were centrifugation steps to remove most of host cells before RNA extraction. And the lack of evaluation on RNA integrity of all samples before RNA sequencing should be considered when we over-interpret these data, and further study with more strict and thorough RNA quality control is required to confirm these data.
In conclusion, the findings in our study suggest a potentially critical connection between host response and prognosis of SCAP. Elderly male patients were a risk factor for poor outcomes and a series of clinical parameters were also associated with outcomes, in which differential host gene enrichments lay behind. According to metagenomic as well as in transcriptomic data, a comprehensive predicting model based on six signatures was established and has a strong predictive ability for 30-day mortality in SCAP patients, which could provide guidance to make further clinical decisions.
Contributors
Conceptualization: Min Zhou, Jinmin Ma, Jing Zhang, and Jieming Qu.
Methodology: Jiumeng Min, Rosary Sin Yu Yao, Zhonglin Chen, Zhongyi Zhu, Yanping Gong, Weiwei Luo, Ao Shen, Ruixue Sun, Yang Lin, Lingling Yang, Jiali Zhou, Zhenzi Ma, Yongquan Lin, Xifang Nie, and Xiangyan He.
Investigation: Yu Chen, Yi Huang, Yusang Xie, Yuping Li, Dongwei Shi, Jinfu Xu, Qiuyue Wang, Bei He, Ning Shen, Bin Cao, Danyang She, Yi Shi, Xin Su, Hua Zhou, Hong Fan, Feng Ye, Qiao Zhang, Xinlun Tian, and Guoxiang Lai.
Visualization: Jiumeng Min, Rosary Sin Yu Yao, and Xiangyan He.
Validation: Jiumeng Min, Rosary Sin Yu Yao, Xiangyan He, and Jingya Zhao.
Funding acquisition: Ming Zhou, Jinmin Ma, Jing Zhang, and Jieming Qu.
Project administration: Yu Chen, Yi Huang, Yusang Xie, Yuping Li, Dongwei Shi, Jinfu Xu, Qiuyue Wang, Bei He, Ning Shen, Bin Cao, Danyang She, Yi Shi, Xin Su, Hua Zhou, Hong Fan, Feng Ye, Qiao Zhang, Xinlun Tian, and Guoxiang Lai.
Supervision: Jinmin Ma, Jing Zhang, and Jieming Qu.
Writing—original draft: Jingya Zhao and Xiangyan He.
Writing—review & editing: Ming Zhou and Jing Zhang.
All authors read and approved the final version of the manuscript, and ensure it is the case. Jiumeng Min and Rosary Sin Yu Yao have directly accessed and verified the underlying data.
Data sharing statement
All data are available in the main text or the Supplementary Materials. Correspondence and requests for materials should be addressed to Ming Zhou, Jinmin Ma, Jing Zhang and Jieming Qu. The non-human raw sequencing data were deposited in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0004448).
Declaration of interests
Authors declare that they have no competing interests.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 82161138018), Shanghai Municipal Key Clinical Specialty (shslczdzk02202), Shanghai Top-Priority Clinical Key Disciplines Construction Project (2017ZZ02014), Shanghai Key Laboratory of Emergency Prevention, Diagnosis and Treatment of Respiratory Infectious Diseases (20dz2261100). This work was also supported by Innovative research team of high-level local universities in Shanghai.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104790.
Contributor Information
Min Zhou, Email: doctor_zhou_99@163.com.
Jinmin Ma, Email: majinmin@genomics.cn.
Jing Zhang, Email: zhang.jing@zs-hospital.sh.cn.
Jieming Qu, Email: jmqu0906@163.com.
Appendix A. Supplementary data
References
- 1.Spoorenberg S., Bos W.J.W., Heijligenberg R., et al. Microbial aetiology, outcomes, and costs of hospitalisation for community-acquired pneumonia; an observational analysis. BMC Infect Dis. 2014;14(1):1–9. doi: 10.1186/1471-2334-14-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Troeger C., Forouzanfar M., Rao P.C., et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the global burden of disease study 2015. Lancet Infect Dis. 2017;17(11):1133–1161. doi: 10.1016/S1473-3099(17)30396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welte T., Torres A., Nathwani D. Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax. 2012;67(1):71–79. doi: 10.1136/thx.2009.129502. [DOI] [PubMed] [Google Scholar]
- 4.Cillóniz C., Dominedò C., Garcia-Vidal C., Torres A. Community-acquired pneumonia as an emergency condition. Curr Opin Crit Care. 2018;24(6):531–539. doi: 10.1097/MCC.0000000000000550. [DOI] [PubMed] [Google Scholar]
- 5.Siow W.T., Koay E.S.-C., Lee C.K., et al. The use of polymerase chain reaction amplification for the detection of viruses and bacteria in severe community-acquired pneumonia. Respiration. 2016;92(5):286–294. doi: 10.1159/000448555. [DOI] [PubMed] [Google Scholar]
- 6.Quah J., Jiang B., Tan P.C., Siau C., Tan T.Y. Impact of microbial aetiology on mortality in severe community-acquired pneumonia. BMC Infect Dis. 2018;18(1):1–9. doi: 10.1186/s12879-018-3366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leitao Filho F.S., Alotaibi N.M., Ngan D., et al. Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am J Respir Crit Care Med. 2019;199(10):1205–1213. doi: 10.1164/rccm.201806-1135OC. [DOI] [PubMed] [Google Scholar]
- 8.Bosch A.A., de Steenhuijsen Piters W.A.A., van Houten M.A., et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med. 2017;196(12):1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 9.Rogers G.B., Zain N.M.M., Bruce K.D., et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc. 2014;11(4):496–503. doi: 10.1513/AnnalsATS.201310-335OC. [DOI] [PubMed] [Google Scholar]
- 10.Dickson R.P., Schultz M.J., van der Poll T., et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med. 2020;201(5):555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Steenhuijsen Piters W.A., Binkowska J., Bogaert D. Early life microbiota and respiratory tract infections. Cell Host Microbe. 2020;28(2):223–232. doi: 10.1016/j.chom.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Libertucci J., Young V.B. The role of the microbiota in infectious diseases. Nat Microbiol. 2019;4(1):35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 13.Angus D.C., Van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 14.Stacy A., Andrade-Oliveira V., McCulloch J.A., et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell. 2021;184(3):615–627.e17. doi: 10.1016/j.cell.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pairo-Castineira E., Clohisey S., Klaric L., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 16.Clohisey S., Baillie J.K. Host susceptibility to severe influenza A virus infection. Crit Care. 2019;23(1):1–10. doi: 10.1186/s13054-019-2566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patarčić I., Gelemanović A., Kirin M., et al. The role of host genetic factors in respiratory tract infectious diseases: systematic review, meta-analyses and field synopsis. Sci Rep. 2015;5(1):1–10. doi: 10.1038/srep16119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbagelata E., Cilloniz C., Dominedo C., Torres A., Nicolini A., Solidoro P. Gender differences in community-acquired pneumonia. Minerva Med. 2020;111(2):153–165. doi: 10.23736/S0026-4806.20.06448-4. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez F., Masia M., Mirete C., et al. The influence of age and gender on the population-based incidence of community-acquired pneumonia caused by different microbial pathogens. J Infect. 2006;53(3):166–174. doi: 10.1016/j.jinf.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Tokioka F., Okamoto H., Yamazaki A., Itou A., Ishida T. The prognostic performance of qSOFA for community-acquired pneumonia. J Intensive Care. 2018;6(1):1–8. doi: 10.1186/s40560-018-0307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma C.-M., Wang N., Su Q.-W., Yan Y., Yin F.-Z. The performance of CURB-65 and PSI for predicting in-hospital mortality of community-acquired pneumonia in patients with type 2 diabetes compared with the non-diabetic population. Diabetes Metab Syndr Obes. 2021;14:1359. doi: 10.2147/DMSO.S303124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salazar M.G., Neugebauer S., Kacprowski T., et al. Association of proteome and metabolome signatures with severity in patients with community-acquired pneumonia. J Proteonomics. 2020;214 doi: 10.1016/j.jprot.2019.103627. [DOI] [PubMed] [Google Scholar]
- 23.Bermejo-Martin J.F., Almansa R., Martin-Fernandez M., Menendez R., Torres A. Immunological profiling to assess disease severity and prognosis in community-acquired pneumonia. Lancet Respir Med. 2017;5(12):e35–e36. doi: 10.1016/S2213-2600(17)30444-7. [DOI] [PubMed] [Google Scholar]
- 24.Baillie J.K. Translational genomics. Targeting the host immune response to fight infection. Science. 2014;344(6186):807–808. doi: 10.1126/science.1255074. [DOI] [PubMed] [Google Scholar]
- 25.Qu J., Zhang J., Chen Y., et al. Aetiology of severe community acquired pneumonia in adults identified by combined detection methods: a multi-centre prospective study in China. Emerg Microbes Infect. 2022;11(1):556–566. doi: 10.1080/22221751.2022.2035194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews S. 2010. FASTQC. A quality control tool for high throughput sequence data. [Google Scholar]
- 28.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spellerberg I., Fedor P. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ index. Glob Ecol Biogeogr. 2003;12:177–179. [Google Scholar]
- 31.Dray S., Dufour A.-B. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 2007;22(4):1–20. [Google Scholar]
- 32.Wickham H.C.W., Wickham M.H. Package ‘ggplot2’. Create elegant data visualisations using the grammar of graphics. Version. 2016;2(1):1–189. [Google Scholar]
- 33.Burge S.W., Daub J., Eberhardt R., et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(D1):D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig W., Strunk O., Westram R., et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10) doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nassa G., Giurato G., Cimmino G., et al. Splicing of platelet resident pre-mRNAs upon activation by physiological stimuli results in functionally relevant proteome modifications. Sci Rep. 2018;8(1):498. doi: 10.1038/s41598-017-18985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan J., Kensler K.H., Hu Z., et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet. 2020;16(2) doi: 10.1371/journal.pgen.1008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harvey T.J., Davila R.A., Vidovic D., Sharmin S., Piper M., Simmons D.G. Genome-wide transcriptomic analysis of the forebrain of postnatal Slc13a4(+/-) mice. BMC Res Notes. 2021;14(1):269. doi: 10.1186/s13104-021-05687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan C., Chen K., Zhou J., et al. Systematic analysis to identify transcriptome-wide dysregulation of Alzheimer's disease in genes and isoforms. Hum Genet. 2021;140(4):609–623. doi: 10.1007/s00439-020-02230-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tin Kam H. Vol. 1. 1995. Random decision forests; pp. 278–282. (Proceedings of 3rd international conference on document analysis and recognition; 1995 14-16 Aug. 1995). [Google Scholar]
- 43.Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374(2065) doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datta S., Satten G.A. Rank-sum tests for clustered data. J Am Stat Assoc. 2005;100(471):908–915. [Google Scholar]
- 45.McKight P.E., Najab J. 2010. Kruskal-Wallis Test. 2010. [Google Scholar]
- 46.Worm-Smeitink M., Gielissen M., Bloot L., et al. The assessment of fatigue: psychometric qualities and norms for the checklist individual strength. J Psychosom Res. 2017;98:40–46. doi: 10.1016/j.jpsychores.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 47.Wiriyakijja P., Porter S., Fedele S., et al. Meaningful improvement thresholds in measures of pain and quality of life in oral lichen planus. Oral Dis. 2020;26(7):1464–1473. doi: 10.1111/odi.13379. [DOI] [PubMed] [Google Scholar]
- 48.Akoglu H. User's guide to correlation coefficients. Turk J Emerg Med. 2018;18(3):91–93. doi: 10.1016/j.tjem.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan Y.H. Biostatistics 104: correlational analysis. Singapore Med J. 2003;44(12):614–619. [PubMed] [Google Scholar]
- 50.Harrell F.E., Jr., Harrell M.F.E. 2019. Package ‘hmisc’. CRAN2018 2019; pp. 235–236. [Google Scholar]
- 51.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: a language and environment for statistical computing.http://www.R-project.org/ [Google Scholar]
- 52.Dezeure R., Bühlmann P., Meier L., Meinshausen N. High-dimensional inference: confidence intervals, $p$-values and R-Software hdi. Stat Sci. 2015;30(4):533–558. [Google Scholar]
- 53.Sinha R.K., Bojjireddy N., Kulkarni D., et al. Type II phosphatidylinositol 4-kinase β is an integral signaling component of early T cell activation mechanisms. Biochimie. 2013;95(8):1560–1566. doi: 10.1016/j.biochi.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Lee K., Kunkeaw N., Jeon S.H., et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17(6):1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balachandran S., Roberts P.C., Brown L.E., et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13(1):129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 56.Richardson G.P., Lukashkin A.N., Russell I.J. The tectorial membrane: one slice of a complex cochlear sandwich. Curr Opin Otolaryngol Head Neck Surg. 2008;16(5):458–464. doi: 10.1097/MOO.0b013e32830e20c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng C., Huang H., Huang S., et al. Identification of potential key genes associated with severe pneumonia using mRNA-seq. Exp Ther Med. 2018;16(2):758–766. doi: 10.3892/etm.2018.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nucci L.A., Santos S.S., Brunialti M.K., et al. Expression of genes belonging to the interacting TLR cascades, NADPH-oxidase and mitochondrial oxidative phosphorylation in septic patients. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0172024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brealey D., Brand M., Hargreaves I., et al. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet. 2002;360(9328):219–223. doi: 10.1016/S0140-6736(02)09459-X. [DOI] [PubMed] [Google Scholar]
- 60.Kothe H., Bauer T., Marre R., et al. Outcome of community-acquired pneumonia: influence of age, residence status and antimicrobial treatment. Eur Respir J. 2008;32(1):139–146. doi: 10.1183/09031936.00092507. [DOI] [PubMed] [Google Scholar]
- 61.Cillóniz C., Polverino E., Ewig S., et al. Impact of age and comorbidity on cause and outcome in community-acquired pneumonia. Chest. 2013;144(3):999–1007. doi: 10.1378/chest.13-0062. [DOI] [PubMed] [Google Scholar]
- 62.Ewig S., Schafer H., Torres A. Severity assessment in community-acquired pneumonia. Eur Respir J. 2000;16(6):1193–1201. doi: 10.1034/j.1399-3003.2000.16f27.x. [DOI] [PubMed] [Google Scholar]
- 63.Fernandes C.D., Arriaga M.B., Costa M.C.M., et al. Host inflammatory biomarkers of disease severity in pediatric community-acquired pneumonia: a systematic review and meta-analysis. Open Forum Infect Dis. 2019;6(12) doi: 10.1093/ofid/ofz520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lucas C., Wong P., Klein J., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong K.T., Thangavel R.R., Tang X. Enhanced expression of murine beta-defensins (MBD-1, -2,- 3, and -4) in upper and lower airway mucosa of influenza virus infected mice. Virology. 2008;380(1):136–143. doi: 10.1016/j.virol.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 66.Yu H., Dong J., Gu Y., et al. The novel human beta-defensin 114 regulates lipopolysaccharide (LPS)-mediated inflammation and protects sperm from motility loss. J Biol Chem. 2013;288(17):12270–12282. doi: 10.1074/jbc.M112.411884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cataudella E., Giraffa C.M., Di Marca S., et al. Neutrophil-to-lymphocyte ratio: an emerging marker predicting prognosis in elderly adults with community-acquired pneumonia. J Am Geriatr Soc. 2017;65(8):1796–1801. doi: 10.1111/jgs.14894. [DOI] [PubMed] [Google Scholar]
- 68.Ge Y.L., Zhang H.F., Zhang Q., et al. Neutrophil-to-lymphocyte ratio in adult community-acquired pneumonia patients correlates with unfavorable clinical outcomes. Clin Lab. 2019;65(5) doi: 10.7754/Clin.Lab.2018.181042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.