Highlights
-
•
For avipoxvirus 282E4 strain, the avipoxvirus 282E4 was shown to be divergent from known fowlpox virus isolates.
-
•
For the genome avipoxvirus 282E4, the linear genome of avipoxvirus 282E4 was 308,826 bp in length, containing 313 open reading frames (ORFs), and 21 unique predicted ORFs.
-
•
Phylogenetic analysis based on p4b and DNA polymerase gene showed that avipoxvirus 282E4 and SWPV-1 belong to the same evolutionary branch.
Keywords: Avipoxvirus 282E4, Fowlpox virus, Genome-wide analysis, Phylogenetic analysis
Abstract
Avipoxvirus 282E4 strain was extensively applied into recombinant vaccine vector to prevent other infectious diseases. However, little information on the genomic background, functional and genetic evolutionary of the isolate 282E4 strain was clarified. The results showed that the linear genome of avipoxvirus 282E4 was 308,826 bp, containing 313 open reading frames (ORFs) and 12 new predicted ORFs. The 282E4 strain appears to encode two novel thymidine kinase proteins and two TGF-beta-like proteins that may be associated with the suppression of the host's antiviral response. Avipoxvirus 282E4 also encodes 57 ankyrin repeat proteins and 5 variola B22R-like proteins, which composed 7% of the avipoxvirus 282E4 genome. GO and KEGG analysis further revealed that 12 ORFs participate in viral transcription process, 7 ORFs may function during DNA repair, replication and biological synthesis, and ORF 208 is involved in the process of virus life cycle. Interestingly, phylogenetic analysis based on concatenated sequences p4b and DNA polymerase of avipoxviruses gene demonstrates that avipoxvirus 282E4 strain is divergent from known FWPV isolates and is similar to shearwater poxvirus (SWPV-1) that belongs to the CNPV-like virus. Sequencing avipoxvirus 282E4 is a significant step to judge the genetic position of avipoxviruses within the larger Poxviridae phylogenetic tree and provide a new insight into the genetic background of avipoxvirus 282E4 and interspecies transmission of poxviruses, meanwhile, explanation of gene function provides theoretical foundation for vaccine design with 282E4 strain as skeleton.
1. Introduction
Avipoxviruses, a member of the order Chitovirales, family Poxviridae, subfamily Chordopoxvirinae, are double-stranded DNA envelope viruses, ranging in size from 260 kbp to 365 kbp. Poxviruses replicate in the cytoplasm of the cell and bud through cellular endocytosis. (Mosad et al., 2020; Deng et al., 2022; Walker et al., 2021). Avipoxviruses extensively infect nonmammalian hosts including more than 300 species of birds (Bolte et al., 1999). Based on the characteristics of host species, Avipoxviruses roughly contained twelve species such as Fowlpox virus (FWPV), turkeypox virus, pigeonpox virus, canarypox virus, parrotpox virus, Flamingopox virus, Juncopox virus, Mynahpox virus, Penguinpox virus, Psittacinepox virus, Quailpox virus, Sparrowpox virus, and Starlingpox virus. (Ha et al., 2011, https://ictv.global/). Furthermore, recently researches suggested that the avipoxvirus was identified in others birds, for instance, Gymnorhina tibicen, Grallina cyanoleuca, Corvus bennetti, Pterodroma cookii and more (Sarker et al., 2020; Sarker et al., 2021(a); Sarker et al., 2022; Sarker et al., 2023). CNPV causes a high fatality rate in infected birds, both in the wild and in commercial aviaries, associating with serious economic losses (Zarifi et al., 2019). Compared with that of CNPV, FWPV infection is a disease with a low mortality proportion, which diminishes egg production and damages growth and fertility. But sometimes, the fatality of fowlpox reaches 50%∼100% without vaccine protection (Mosad et al., 2020; Zhao et al., 2014). Therefore, the vaccine is a necessary method for preventing avipoxviruses.
Currently, avipoxviruses have been used as the vectors for the development of recombinant vaccines to prevent infection with homologous or heterologous pathogens. For example, Fowlpox virus has received considerable attentions since it can be used as gene therapy vectors. Especially in vaccinal research field, the FWPV-based recombinant vaccines carrying the exogenous genes, which are inserted into or substitute for the nonessential region of the FWPV genome, were not only developed for the prevention of the fowlpox virus, but also used to prevent some infectious diseases caused by avian influenza virus (AIV), newcastle disease virus (NDV), infectious bronchitis virus (IBV), and infectious bursal disease virus (IBDV) (Hein et al., 2021; Skinner et al., 2005). In addition, an avipoxvirus 282E4 strain (previously believed to be FWPV) is significantly characterized by the limitation on viral replication in mammalian cells, so the recombinants are safe in mammals including those from foot-and-mouth disease virus (FMDV), porcine reproductive and respiratory syndrome virus (PRRSV) and bluetongue virus (BTV) (Zheng et al., 2006; Shen et al., 2007; Li et al., 2015). Interestingly, the vaccine, constructed by recombinant fowlpox virus (rFWPV) coexpressing HIV-1 gp120, produces specific cellular immunity against HIV-1 (Jiang et al., 2005). Importantly, in the field of gene therapy, FWPV is regarded as a non-oncolytic vector targeting to the treatment of melanoma and colon cancer in a clinical trial (Kaufman et al., 2014). Although vaccine and genetic engineering studies of the avipoxvirus 282E4 strain have already been performed (Zhu et al., 2018), little information on its genetic background and genomic information are known.
Currently, safer and more available avipoxvirus 282E4 strain vaccines and/or vectors will profit from a fine virus genomic data. This study aimed to investigate the viral background, genomic function, and sequence similarity to other avipoxviruses usable in the GenBank database. The whole genome of avipoxvirus 282E4 strain was sequenced using Illumina HiSeq 2500 platform and raw data were assembled. Subsequently, the gene function of ORFs were annotated and predicted using Glimmer 3.0, and enriched with GO database or KEGG pathway. In addition, the genome collinearity analysis is performed using MUMmer 2.2, based on the reference genome sequence, and then P4b and DNA polymerase genes were used to further evaluate the phylogenetic relationship between the 282E4 strain and other avipoxviruses. We demonstrate that the isolated 282E4 strain (previously known as FWPV 282E4) is more closely related to canarypox-like virus, rather than fowlpox-like virus.
2. Materials and methods
2.1. Cells and virus
The primary chicken embryo fibroblasts (CEF) were prepared using 9 to 11 day-old specific pathogen-free (SPF) embryonated eggs and grown in Dulbecco's modified Eagle's medium (DMEM) (Hyclone, USA) supplemented with 5% fetal bovine serum (FBS, Hyclone, USA), 50 U/ml penicillin and 50 μg/ml streptomycin (Hyclone) at 37 °C and 5% CO2.
The Chinese attenuated avipoxvirus 282E4 strain (TCID50=10−7.3/0.1 ml) was kindly provided by the China Institute of Veterinary Drug Control.
2.2. Virion purification and identification
Avipoxvirus 282E4 strain was propagated in the primary CEF cells as described previously (Chung et al., 2006; Jensen et al., 1996). In brief, 1 × 106 cells/ml of CEF cells were seeded on the plate for 24 h and infected with avipoxvirus 282E4 with a multiplicity of infection (MOI) of 1 at 37 °C, 5% CO2. 2 hours post-infection (hpi), the infection media was removed and the cells were cultured in fresh DMEM supplemented with 5% FBS. the supernatant was harvested by low-speed centrifugation filtration with a 0.45 μm-filter, when the cytotoxic pathological effect was complete.
The virus was purified as described previously (Chung et al., 2006; Jensen et al., 1996). The collected supernatant was laid on sucrose cushion (20%) to centrifugate at 25,000 g for 80 min in a Beckman SW28 rotor. Thereafter, the virus pellet was resuspended in TM buffer, sonicated, and further purified by 25% to 50% sucrose density gradient centrifugation at 28,000 g for 80 min. The virions were diluted four-fold in TM buffer and centrifuged at 18,000 g for 45 min to remove the sucrose solution, followed by the resuspension of the virions with TM buffer. All stages of purification were performed at 4 °C and the virions were stored at -70 °C.Virion pellet was pipetted onto 200-mesh Formvar-coated copper grids and allowed to settle for 90 s. Following two washes in water, grids were negatively stained with 2% phosphotungstic acid (pH 6.8) for 60 s. TEM was carried with a JEM 1200-EXII transmitting electron microscope (JEOL-USA, Peabody, MA, USA).
2.3. Polymerase chain reaction (PCR) dectection, extracting and sequencing of viral genomic DNA
Viral genomic DNA was extracted from the virions using TIANamp Virus DNA/RNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer's protocol. The genomic DNA was dissolved in 50 μl of DNase-free ddH2O and stored at -20 °C until of use.
Primers (forward: 5′- GGACGCGTCAGCAGGTGCTAAACAACAA-3′; reverse: 5′- GGCTGCAGCGGTAGCTTAACGCCGAATA-3′) were designed and synthesized according to the highly conserved P4b gene sequence of FWPV (NC_002188.1). PCR was performed to detect the viral DNA using the primers and DNA polymerase with a Thermal Cycler (Thermo Electron Corporation, Milford, MA, USA). DNA concentration was measured by spectrophotometry and then whole-genome sequencing of the avipoxvirus 282E4 was performed by using the Illumina HiSeq 2500 platform (Illumina, Inc., San Diego, CA, USA).
2.4. Sequence assembly, analysis, and annotation
Raw data were filter, assembled into genomic sequences using SOAPdenovo software (Beijing Genomics Institute). and then the assembled genome or gene regions was mapped to FWPV US strain (NC_002188.1). The bioinformatic analyses were performed as described previously (Tu and Upton, 2019) and adjusted slightly. Briefly, the open reading frames (ORFs) were predicted and annotated using Glimmer 3.0 (The Center for Computational Biology at Johns Hopkins University) (Delcher et al., 2007). To ensure the biological meaning, only one high-quality information was treated as the annotation of each unigene. The functional annotation was accomplished by analysis of predicted ORFs. The Blastx program was used to assign annotations to all the unigenes combined with different databases, such as Clusters of Orthologous Groups (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and SwissProt and more. Of course, ORFs were annotated as putative genes and numbered from left to right based on FWPV US strain sequence to avoid any confusion. Protein identity analysis was performed using BLAST at NCBI database. The annotated sequence of avipoxvirus 282E4 strain was submitted to GenBank (Accession No. MG702259).
2.5. Genome alignment and phylogeny
Genome collinearity analysis is performed using MUMmer 2.2 (Kurtz et al., 2004), based on the reference genome sequence (NC_002188.1 and NC_005309.1). After the whole genome was scaled down according to the genome length, and the upper and lower coordinate axes in the linear collinearity map were constructed.
In addition, phylogenetic analyses based on the alignment of the P4b gene and DNA polymerase gene of avipoxvirus 282E4 strain and other Chordopoxvirus sequences obtained from NCBI were generated by using the Maximum-likelihood method in MEG5.0. Bootstrap analysis of 1000 replicates were applied to assess the reliability of the reconstructed phylogenies.
3. Results
3.1. Identification and purification of avipoxvirus 282E4 particles
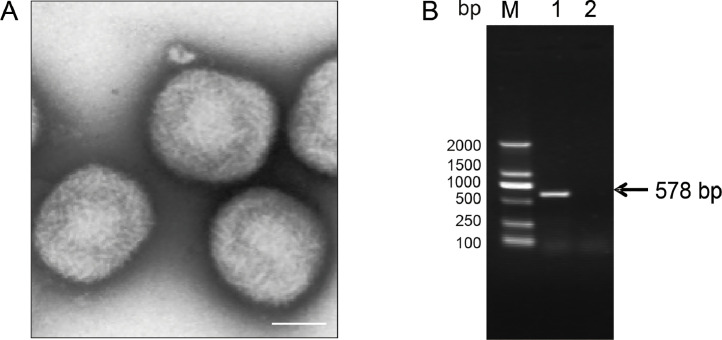
Collecting cell specimans infected with avipoxvirus 282E4 strain and performed virus purification after freeze-thaw. To observe viral particles, electron microscopic analyses were performed to visualize virions. The results showed that oval or brick shape virions with a diameter of about 200 nm were observed in the purified sample (Fig. 1A) in agreement with previous research (Delcher et al., 2007). To further confirm the presence of avipoxvirus 282E4 strain genomic DNA, conventional PCR using primers of the P4b gene was performed to detect avipoxvirus 282E4 strain DNA. The results showed that avipoxvirus 282E4 strain -specific products of the expected size, a 578 bp product, were amplified from the sample (Fig. 1B), and no specific PCR products were detected in the negative control indicating the sample of DNA could be used for sequencing.
Fig. 1.
Purification and identification of the avipoxvirus 282E4. (A) Electron microscopy of purified virions using negative phosphotungstic acid staining, the bar is 200 nm; (b) PCR amplification of avipoxvirus 282E4. M-DL2000 DNA Marker; Lane 1, PCR products of FWPV using P4b primers; Lane 2, negative control.
3.2. Genomic characteristics of avipoxvirus 282E4 strain
The complete genome of avipoxvirus 282E4 strain was identified as a contiguous sequence of 308,829 bp in length, and presented to GeneBank under accession number MG702259, which is larger than the primarily FWPV US strain and attenuated FP9 strain (Afonso et al., 2000; Laidlaw et al., 2004). Because of the absence of hairpin loop sequences, base number one was arbitrarily assigned to the left-most nucleotide of the assembled sequence. The avipoxvirus 282E4 strain genome is composed of a central coding region that is surrounded by two identical inverted terminal repeats (ITRs) as same like other avipoxviruses (Afonso et al., 2000; Tulman et al., 2004). The A+T content of the whole genome of avipoxvirus 282E4 strain was evaluated as 72%, with partial genome regions being higher/lower than that of the whole genome. We annotated 313 ORFs as putative genes from the whole genome of the avipoxvirus 282E4 strain. These genes covered a 90.16% coding density and encode structural and functional proteins of 39 to 1862 amino acids (Table S1).
To explore the homology of avipoxvirus 282E4 strain predicted genes, which were blasted in the NCBI database. Previous study showed that the ENV and LTR gene sequences of avian reticuloendothelium virus (REV) were recombined with the fowlpox virus genome, and even the whole sequence of REV was recombined with FWPV genome (Tadese et al., 2008; Hertig et al., 1997; García et al., 2003). However, our results showed that no one gene of REV was blasted in avipoxvirus 282E4 strain genome (data no shown). Gene annotation analysis showed that the homology of 126 ORFs were above 80% between avipoxvirus 282E4 strain and ChePV-1 in the best matching group (Table S1). In addition, compared with FWPV US and CNPV, the average amino acid homology of the whole genome is 44.26% and 63.42%, respectively. Surprisingly, the linear genome of avipoxvirus 282E4 strain is comparable to FWPV US, the terminal region genome of avipoxvirus 282E4 strain performed gene insertion, deletion, replacement, and rearrangement, expanding to the central genome (Table S1). These results indicated that homology of genome is a significant difference between avipoxvirus 282E4 strain and FWPV US.
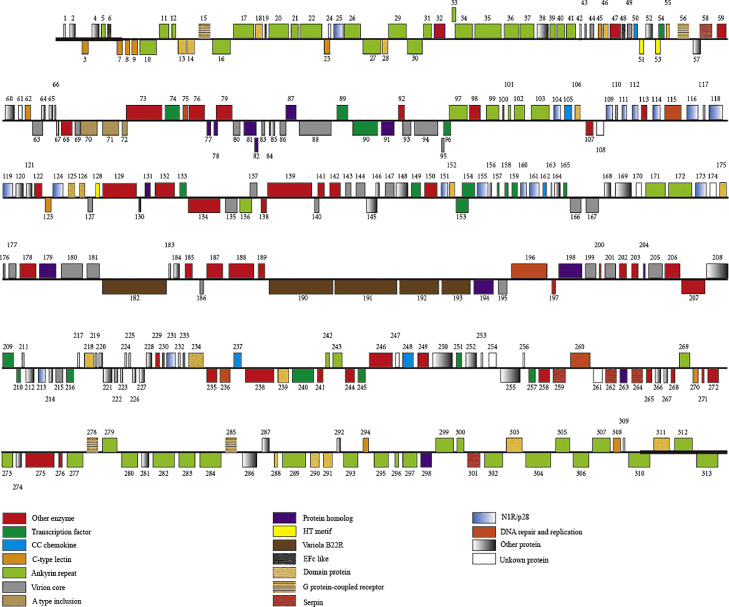
A total of 86 conserved genes from subfamily chordopoxvirinae was identified under the genome central region of avipoxvirus 282E4 strain, involving to the viral replication process, containing viral transcription, viral DNA replication, and the structure and assembly of intracellular mature virus particles and extracellular envelope virus particles (Table S1). 86 conserved ChPV genes were present in both avipoxvirus 282E4 strain and Cheloniid poxvirus (ChePV-1), sharing on average 91% amino acid identity, The most conserved protein was ORF 83 and 149, (Virion envelope protein and late transcription factor, 99% amino acid identity), while the most divergent was ORF 92 (immunodominant virion protein,52% amino acid identity) (Table S1). However, the conserved genes of both avipoxvirus 282E4 strain and CNPV shared on average 83% amino acid identity, anticipately, compared with FWPV US genome, the average amino acid homology of 86 conserved genes is only 72%. The research demonstrated that these conserved genes are mainly located in the central region of avipoxvirus 282E4 strain (ORF 62 to ORF 96, ORF 127 to ORF 149, ORF 167 to ORF 222, ORF 236 to ORF 251). Moreover, five variola B22R-like proteins were predicted from avipoxvirus 282E4 strain, containing the most numbers of amino acids, composed 7% of the avipoxvirus 282E4 strain. And some ankyrin repeat proteins were annotated at the two terminal regions of the genome (Fig. 2 and Table S4). Among them, 12 predicted ORFs are unique to avipoxvirus 282E4 strain, and none of them match any sequence in NCBI database of NR proteins using BLASTP. These ORFs were predicted to generally encode 30 to 100 amino acid residues apart from ORF 254.
Fig. 2.
Linear map of the avipoxvirus 282E4 genome. The avipoxvirus 282E4 ORFs are numbered from left to right based on the position of the methionine initiation codon. The ORFs transcribed from 5′-3′ forward are located above horizontal lines, Conversely, ORFs transcribed from 3′ -5′forward are located below horizontal lines. Genes with similar functions and gene family members are marked with the same color. Unknow protein indicates that there is no gene information record in NCBI database. ITRs are delegated to thick line on the ORF map, respectively.
Compared with the FWPV US strain, avipoxvirus 282E4 strain encodes 65 predicted genes for which any homologue is deficient from or highly fragmented in FWPV US, involving in multiple gene family, such as C-type lectin, ankyrin repeat protein, TGF-beta-like protein, nucleotide metabolism genes and 14 hypothetical proteins and more.
3.3. Functional annotation and clustering of genomic encoded proteins
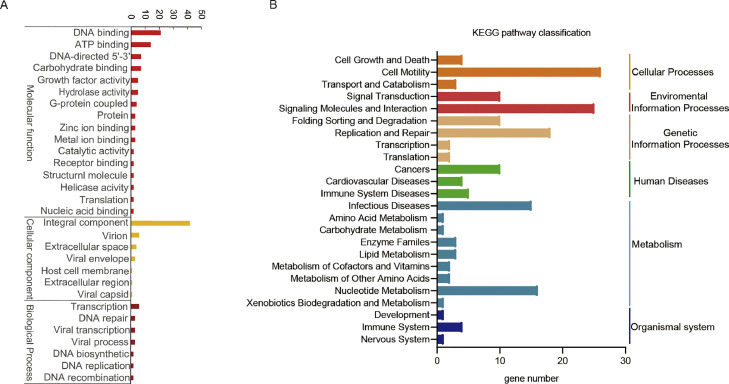
To investigate the function of avipoxvirus 282E4 strain ORFs-encoded proteins, GO analysis was performed and the result showed that a total of 276 ORFs were annotated. Of these, 119 ORFs were grouped into molecular function, 58 ORFs were grouped into cellular component, and 40 ORFs were grouped into biological process (Fig. 3A). In the molecular function group, a large proportion of ORFs were related to DNA binding (21/119, 17.6 %) and ATP binding (14/119, 11.7 %). Within cellular component group, unigenes related to the integral component of membrane represented the largest group (42/58, 72.4 %). In the category of biological process group, a high percentage of ORFs were involved in transcription, DNA-templated activity (6/40, 15 %).
Fig. 3.
GO and KEGG pathway enrichment analysis of avipoxvirus 282E4 genome. (A) GO terms are on the x axis. Enrichment number of genes encode ORFs shown as GO terms for molecular function, cellular component and biological process. (B) KEGG pathways are on the y axis. Enrichment number of genes encode ORFs displayed with the name of KEGG pathway for six categories, organismal systems, metabolism, humane diseases, genetic information processing, environmental information processing, cellular processes.
Meanwhile, KEGG analysis was performed to the biological pathways in avipoxvirus 282E4 strain encoding proteins, we mapped the predicted ORFs to the reference of typical pathways in KEGG database. Among 313 ORFs, 169 matched genes were classified into 24 different functional KEGG pathways (Fig. 3B): cell motility pathway was the predominant pathway, with 26 unigenes (26/169, 15.4 %), followed by signaling molecules and interaction (25/169, 14.8 %), replication and repair (18/169, 10.7 %), nucleotide metabolism (16/169, 9.5 %), and infectious diseases (15/169, 8.9 %). Other pathways included relatively fewer unigenes, such as nervous system, development, xenobiotics biodegradation and metabolism, carbohydrate metabolism, and amino acid metabolism.
Further analysis revealed 12 ORFs were involved in viral transcription process, containing RNA polymerase (ORF 68, ORF 73, ORF 92, ORF 139, ORF 141, ORF 189 and ORF 241), nucleoside triphosphate phosphohydrolase I (ORF 246) and poly (A) polymerase (ORF 187). Early transcription factor (ORF 90) and late transcription factor (ORF 96 and 194) also were identified in avipoxvirus 282E4 strain genome. In addition, ORF 75, ORF 98, ORF 115, ORF 196, ORF 203, ORF 236 and ORF 260 were participated in DNA repair, replication and biological synthesis. Interestingly, avipoxvirus 282E4 strain including Thymidine kinase (ORF 200, ORF203 and ORF 271), deoxycytidine kinases (ORF 107, ORF 122 and ORF 202), dUTP (ORF 265) and DNase II (ORF 272) contributed in nucleotide metabolism. While the coding gene for glutaredoxin was not found in the avipoxvirus 282E4 strain genome. Furthermore, ORF 208 was involved in the process of virus life cycle, and ORF 263 may play a significant role in inhibiting host cell apoptosis the same as ORF50 was employed in immune response.
3.4. Avipoxvirus 282E4 strain has a close genetic relationship with CNPV strain
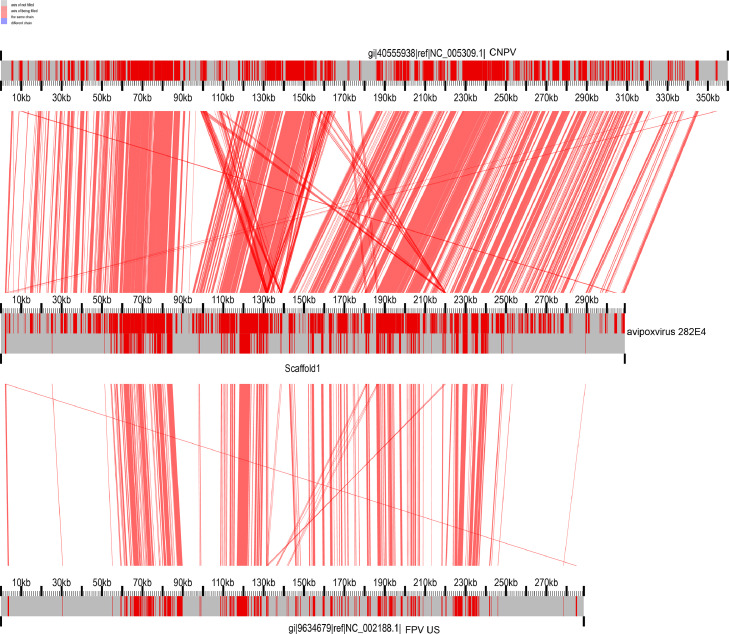
To verify the accuracy of the avipoxvirus 282E4 strain genome map, the genome-wide collinearity comparison was performed between the avipoxvirus 282E4 strain and the FWPV US strain (NC_002188.1) or CNPV strain (NC_005309.1) deposited in GenBank. As shown in the Fig. 4, 64,917 bp (21.02 %) of avipoxvirus 282E4 strain was identical to that of FWPV US strain, the genome collinearity region is the lowest between the range of 1 kbp to 60 kbp and 240 kbp to 280 kbp (Fig. 4), involving in ankyrin protein and enzyme predicted ORFs, while 176,888 bp (57.28 %) of avipoxvirus 282E4 strain was covered by CNPV strain (Fig. 4). Especially, there is a high level of gene synteny in the central region of genome (60 kbp to 85 kbp, 105 kbp to 135 kbp and 190 kbp to 210 kbp). Altogether, the results indicated that avipoxvirus 282E4 strain more closed to the CNPV strain.
Fig. 4.
Analysis of gene collinearity. The genome sequence of avipoxvirus 282E4 strain is compared to that of FWPV-US (NC_002188.1) and CNPV (NC_005309.1). Genome is represented by coordinate axis, genome with collinear regions is marked with the red color, genome with noncollinear regions is marked with the gray color.
3.5. Avipoxvirus 282E4 may have originated from a Canarypox-like viruses
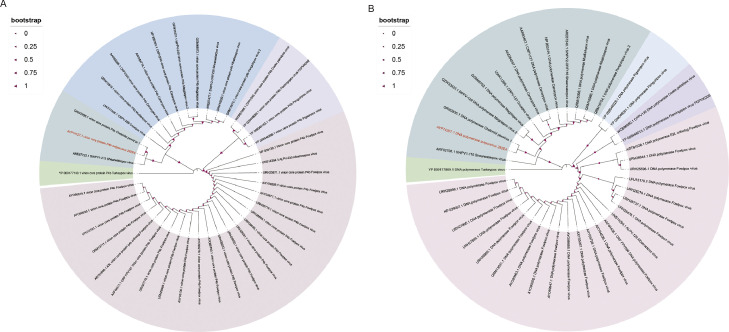
To further evaluate the relationship between avipoxvirus 282E4 strain and other avipoxviruses. The viral core protein gene P4b and DNA polymerase were used to compare with avipoxviruses obtained from GenBank (Table S2). Currently, there are three clades of Avipoxviruses, A (Fowlpox-like virus), B (Canarypox-like virus) and C (Parrotpox-like virus), as well as two proposed clades, D (Quailpox virus (QPV)) and E (Turkeypox virus (TKPV)) (Carulei et al., 2017). As shown Fig. 5, phylogenetic analysis suggested that avipoxvirus 282E4 strain clustered in global clade B (Canarypox-like virus), indicating that the SWPV and ChePV-1 identified recently are closely related to the isolated 282E4 strain, which diverged from fowlpox viruses (Fig. 5A and 5B).
Fig. 5.
Phylogenetic and homology relationships between avipoxvirus 282E4 and other avipoxvirus. Phylogenetic tree and homology were constructed from nucleotide sequence of the P4b core protein and DNA polymerase gene in NCBI database using Mega 5.0 version software. The numbers on the northwest map show bootstrap values as percentages. The labels at branch tips refer to original avipoxvirus GenBank accession numbers followed by species names. Branch with similar clades of avipoxvirus is marked with the same colors, avipoxvirus 282E4 strain was highlighted with red. (A) and (B) phylogenetic analysis of p4b and DNA polymerase gene.
3.6. Nucleotide sequence accession numbers
The complete genomic sequence of avipoxvirus 282E4 strain has been deposited in the GenBank database under accession number MG702259.
4. Discussion
In our research, the morphology of avipoxvirus 282E4 strain virion was observed and its genomic DNA was identified, assembled and analyzed. The whole length of the avipoxvirus 282E4 strain genome was 308,829 bp, which was larger than FP9 and FWPV US genomes, but smaller than the CNPV genome. In addition, compared with the FWPV US genome, a total of 65 ORFs with a failed blast and unknown functions were certified in the avipoxvirus 282E4 strain (Table S1). Some research shows that the Deoxycytidine kinase (dCK) protein is unique to the fowlpox virus (Afonso et al., 2000). Indeed, the avipoxvirus 282E4 strain genome contains ORF107 encoding deoxycytidine kinase (dCK) protein. However, the later study confirmed that genes encoding dCK and DNase II were identified in CNPV, ALPV, SWPV-1, and ChePV-1 (Tulman et al., 2004; Sarker et al., 2017; Sarker et al., 2021(b); Sarker et al., 2021(c)), dCK and DNase II are consequently not unique to FWPV. Although, the structure of the avipoxvirus 282E4 linear genome is greatly different from that of FWPV US, avipoxvirus 282E4 strain still could enter mammalian cells to express its own or exogenous proteins (Du et al., 2015). Ankyrin repeats protein composed of two α-helices linking with β-turns and short loops, which can interact with host protein (Herbert et al., 2015). The rearrangement and abundance of ANK repeats have a potential effect on the host range of poxvirus, meanwhile, they may prohibit virus-induced apoptosis (Afonso et al., 2000) Furthermore, ORF 106, 121, and 135 connected with the host range of avipoxvirus 282E4 strain, the homology of these genes is related to the virus titer and host range of CNPV (Tulman et al., 2004). Little is known that avipoxvirus 282E4 strain can replicate in other cells and hosts.
Avipoxvirus 282E4 strain plays an important role in the development of recombinant vaccines and the application of gene therapy vectors. The thymidine kinase (TK) gene is a non-essential gene of the fowlpox virus and also a common insertion site of an exogenous gene (Boyle and Coupar, 1988). ORF 200 and 203 of avipoxvirus 282E4 strain can encode TK protein, which may be the target of the exogenous insertion gene. Moreover, our previous research demonstrated that avipoxvirus 282E4 strain with ORF150 mutation, ORF193 deletion, and exogenous gene insertion did not affect the morphogenesis, replication, and proliferation of intracellular recombinant viruses (Du et al., 2015). Significantly, totals of 12 ORFs unannotated in the Genebank database may also be targeted for new exogenous gene insertions, but further validation is required. The avipoxvirus 282E4 strain linear genome contains 86 conserved ORFs of the poxvirus genus, as these ORFs play an important role in viral replication and assembly (Afonso et al., 2000), the disfunction of conserved ORFs should be avoided during exogenous gene insertion. Furthermore, 276 ORFs were analyzed for GO enrichment, involving multiple biological processes of virus activity. 169 ORFs were enriched in the relevant KEGG pathway, especially 26 ORFs in the signal pathway of cell motility (Fig. 4B).
Genomic homology, which represents the range of similarities and differences between the genomes of two different species, is important for species evolution. The analysis of gene collinearity could explain the homology relationship between species. currently, gene collinearity analysis is used for the evolution of mammals, homology matching of bacterial genomes, and analysis of the evolution of the same gene among species (Li et al., 2021; Zeng et al., 2022). In this study, gene collinearity analysis of the avipoxvirus 282E4 strain, FWPV US, and CNPV revealed that the genomic homology of avipoxvirus 282E4 strain and CNPV reached 57.2%, implying that avipoxvirus 282E4 strain is more closely related to CNPV. Interestingly, the P4b gene, a viral core protein conserved in the avipoxvirus, is homologous to Vaccinia virus A3L and is conserved in avipoxviruses. The evolutionary analysis of the P4b gene showed that avipoxvirus 282E4 strain was a closely evolutionary branch with SWPV-1, but apart from the evolutionary branch of FWPV in the Genebank database. And the results of DNA polymerase as the second locus for phylogenetic analysis of the genus avipoxvirus were generally consistent with the results of evolutionary analysis of the P4b gene. DNA polymerase gene was first reported as a universal evolutionary marker of avipoxvirus in 2013, used in the epidemiological investigation of some avian poxviruses and the analysis of newly discovered poxviruses (Gyuranecz et al., 2013; Sahu et al., 2020). The above results indicated that avipoxvirus 282E4 strain is significantly divergent from FWPV.
5. Conclusion
We primarily sequenced the genome of avipoxvirus 282E4 strain, and characterized its genome structure and annotated gene functions, providing a reference for the virus to be used as a skeleton for the preparation of recombinant vaccines. An evolutionary analysis of the P4b and DNA polymerase indicated that avipoxvirus 282E4 strain is significantly divergent from the reported FWPV viruses and more closely related to ChePV-1 and SWPV-1, suggesting that the avipoxvirus 282E4 strain, previously thought to be a fowlpox virus, actually represents a Canarypox-like virus.
Consent for publication
Not applicable.
Author statement
Lingcong Deng has made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; Cunxia Liu has drafted the work or revised it critically for important intellectual content; Letian Li has made devotion of the investigation of the work; Pengfei Hao has made devotion of the visualization of the work; Maopeng Wang has made contributions to revising the paper; Ningyi Jin has made contributions to the development of the work; Ronglan Yin has contributed to data analysis; Shouwen Du made important contributions to Writing-Review & Editing of the work. I have approved the final version to be published.
I agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons who have made substantial contributions to the work reported in the manuscript, including those who provided editing and writing assistance but who are not authors, are named in the Acknowledgments section of the manuscript and have given their written permission to be named.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (31702210, 31972719, 31502093) and the CAMS Innovation Fund for Medical Sciences (2020-12M-5-001).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199218.
Contributor Information
Ronglan Yin, Email: yinronglan@163.com.
Shouwen Du, Email: du-guhong@163.com.
Chang Li, Email: lichang78@163.com.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Afonso C.L., Tulman E.R., Lu Z., Zsak L., Kutish G.F., Rock D.L. The genome of fowlpox virus. J. Virol. 2000;74(8):3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte A.L., Meurer J., Kaleta E.F. Avian host spectrum of avipoxviruses. Avian Pathol.: J. W.V.P.A. 1999;28(5):415–432. doi: 10.1080/03079459994434. [DOI] [PubMed] [Google Scholar]
- Boyle D.B., Coupar B.E. Construction of recombinant fowlpox viruses as vectors for poultry vaccines. Virus Res. 1988;10(4):343–356. doi: 10.1016/0168-1702(88)90075-5. [DOI] [PubMed] [Google Scholar]
- Carulei O., Douglass N., Williamson A.L. Comparative analysis of avian poxvirus genomes, including a novel poxvirus from lesser flamingos (Phoenicopterus minor), highlights the lack of conservation of the central region. Bmc Genomics. 2017;18(1):947. doi: 10.1186/s12864-017-4315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.S., Chen C.H., Ho M.Y., Huang C.Y., Liao C.L., Chang W. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 2006;80(5):2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Xia X., Deng Y., Zhao M., Gu C., Geng Y., Wang J., Yang Q., He M., Xiao Q., Xiao W., He L., Liang S., Xu H., Lü M., Yu Z. ANI analysis of poxvirus genomes reveals its potential application to viral species rank demarcation. Virus Evol. 2022;8(1):veac031. doi: 10.1093/ve/veac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Liu C., Zhu Y., Wang Y., Ren D., Wang M., Tan P., Li X., Tian M., Zhang Y., Li J., Zhao F., Li C., Jin N. Construction and characterization of novel fowlpox virus shuttle vectors. Virus Res. 2015;197:59–66. doi: 10.1016/j.virusres.2014.12.015. [DOI] [PubMed] [Google Scholar]
- García M., Narang N., Reed W.M., Fadly A.M. Molecular characterization of reticuloendotheliosis virus insertions in the genome of field and vaccine strains of fowl poxvirus. Avian Dis. 2003;47(2):343–354. doi: 10.1637/0005-2086. [DOI] [PubMed] [Google Scholar]
- Gyuranecz M., Foster J.T., Dán Á., Ip H.S., Egstad K.F., Parker P.G., Higashiguchi J.M., Skinner M.A., Höfle U., Kreizinger Z., Dorrestein G.M., Solt S., Sós E., Kim Y.J., Uhart M., Pereda A., González-Hein G., Hidalgo H., Blanco J.M., Erdélyi K. Worldwide phylogenetic relationship of avian poxviruses. J. Virol. 2013;87(9):4938–4951. doi: 10.1128/JVI.03183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H.J., Howe L., Alley M., Gartrell B. The phylogenetic analysis of avipoxvirus in New Zealand. Vet. Microbiol. 2011;150(1-2):80–87. doi: 10.1016/j.vetmic.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Hein R., Koopman R., García M., Armour N., Dunn J.R., Barbosa T., Martinez A. Review of Poultry Recombinant Vector Vaccines. Avian Dis. 2021;65(3):438–452. doi: 10.1637/0005-2086-65.3.438. [DOI] [PubMed] [Google Scholar]
- Herbert M.H., Squire C.J., Mercer A.A. Poxviral ankyrin proteins. Viruses. 2015;7(2):709–738. doi: 10.3390/v7020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig C., Coupar B.E., Gould A.R., Boyle D.B. Field and vaccine strains of fowlpox virus carry integrated sequences from the avian retrovirus, reticuloendotheliosis virus. Virology. 1997;235(2):367–376. doi: 10.1006/viro.1997.8691. [DOI] [PubMed] [Google Scholar]
- Jensen O.N., Houthaeve T., Shevchenko A., Cudmore S., Ashford T., Mann M., Griffiths G., Krijnse Locker J. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 1996;70(11):7485–7497. doi: 10.1128/JVI.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Jin N., Cui S., Li Z., Zhang L., Zhang H., Wang H., Han W. Construction and characterization of recombinant fowlpox virus coexpressing HIV-1(CN) gp120 and IL-2. J. Virol. Methods. 2005;130(1-2):95–101. doi: 10.1016/j.jviromet.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Kaufman H.L., Kim D.W., Kim-Schulze S., DeRaffele G., Jagoda M.C., Broucek J.R., Zloza A. Results of a randomized phase I gene therapy clinical trial of nononcolytic fowlpox viruses encoding T cell costimulatory molecules. Hum. Gene Ther. 2014;25(5):452–460. doi: 10.1089/hum.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw S.M., Skinner M.A. Comparison of the genome sequence of FP9, an attenuated, tissue culture-adapted European strain of Fowlpox virus, with those of virulent American and European viruses. J. Gen. Virol. 2004;85(Pt 2):305–322. doi: 10.1099/vir.0.19568-0. [DOI] [PubMed] [Google Scholar]
- Li J., Yang T., Xu Q., Sun E., Feng Y., Lv S., Zhang Q., Wang H., Wu D. DNA vaccine prime and recombinant FPV vaccine boost: an important candidate immunization strategy to control bluetongue virus type 1. Appl. Microbiol. Biotechnol. 2015;99(20):8643–8652. doi: 10.1007/s00253-015-6697-8. [DOI] [PubMed] [Google Scholar]
- Li S., Zhao G., Han H., Li Y., Li J., Wang J., Cao G., Li X. Genome collinearity analysis illuminates the evolution of donkey chromosome 1 and horse chromosome 5 in perissodactyls: A comparative study. Bmc Genomics. 2021;22(1):665. doi: 10.1186/s12864-021-07984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosad S.M., El-Tholoth M., El-Kenawy A.A., Abdel-Hafez L.J.M., El-Gohary F.A., El-Sharkawy H., Elsayed M.M., Saleh A.A., Elmahallawy E.K. Molecular Detection of Reticuloendotheliosis Virus 5′ Long Terminal Repeat Integration in the Genome of Avipoxvirus Field Strains from Different Avian Species in Egypt. Biology. 2020;9(9):257. doi: 10.3390/biology9090257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B.P., Majee P., Mishra C., Dash M., Biswal S., Sahoo N., Nayak D. The emergence of subclades A1 and A3 avipoxviruses in India. Transbound. Emerg. Dis. 2020;67(2):510–517. doi: 10.1111/tbed.13413. [DOI] [PubMed] [Google Scholar]
- Sarker S., Athukorala A., Nyandowe T., Bowden T.R., Boyle D.B. Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi) Pathogens. 2021(a);10(5):575. doi: 10.3390/pathogens10050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker S., Athukorala A., Raidal S.R. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian passerine bird, mudlark (Grallina cyanoleuca) Virology. 2021(b);554:66–74. doi: 10.1016/j.virol.2020.12.011. [DOI] [PubMed] [Google Scholar]
- Sarker S., Batinovic S., Talukder S., Das S., Park F., Petrovski S., Forwood J.K., Helbig K.J., Raidal S.R. Molecular characterisation of a novel pathogenic avipoxvirus from the Australian magpie (Gymnorhina tibicen) Virology. 2020;540:1–16. doi: 10.1016/j.virol.2019.11.005. [DOI] [PubMed] [Google Scholar]
- Sarker S., Das S., Lavers J.L., Hutton I., Helbig K., Imbery J., Upton C., Raidal S.R. Genomic characterization of two novel pathogenic avipoxviruses isolated from pacific shearwaters (Ardenna spp.) Bmc Genomics. 2017;18(1):298. doi: 10.1186/s12864-017-3680-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker S., Hannon C., Athukorala A., Bielefeldt-Ohmann H. Emergence of a Novel Pathogenic Poxvirus Infection in the Endangered Green Sea Turtle (Chelonia mydas) Highlights a Key Threatening Process. Viruses. 2021(c);13(2):219. doi: 10.3390/v13020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker S., Raidal S.R. A Novel Pathogenic Avipoxvirus Infecting Vulnerable Cook's Petrel (Pterodroma cookii) in Australia Demonstrates a High Genomic and Evolutionary Proximity with South African Avipoxviruses. Microbiol. Spectr. 2023;11(2) doi: 10.1128/spectrum.04610-22. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker S., Sutherland M. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian little crow (Corvus bennetti) directly from the clinical sample. Sci. Rep. 2022;12(1):15053. doi: 10.1038/s41598-022-19480-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Jin N., Ma M., Jin K., Zheng M., Zhuang T., Lu H., Zhu G., Jin H., Jin M., Huo X., Qin X., Yin R., Li C., Li H., Li Y., Han Z., Chen Y., Jin M. Immune responses of pigs inoculated with a recombinant fowlpox virus coexpressing GP5/GP3 of porcine reproductive and respiratory syndrome virus and swine IL-18. Vaccine. 2007;25(21):4193–4202. doi: 10.1016/j.vaccine.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Skinner M.A., Laidlaw S.M., Eldaghayes I., Kaiser P., Cottingham M.G. Fowlpox virus as a recombinant vaccine vector for use in mammals and poultry. Expert Rev. Vaccines. 2005;4(1):63–76. doi: 10.1586/14760584.4.1.63. [DOI] [PubMed] [Google Scholar]
- Tadese T., Fitzgerald S., Reed W.M. Detection and differentiation of re-emerging fowlpox virus (FWPV) strains carrying integrated reticuloendotheliosis virus (FWPV-REV) by real-time PCR. Vet. Microbiol. 2008;127(1-2):39–49. doi: 10.1016/j.vetmic.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Tu S.L., Upton C. Bioinformatics for Analysis of Poxvirus Genomes. Methods Mol. Biol. 2019;2023:29–62. doi: 10.1007/978-1-4939-9593-6_2. [DOI] [PubMed] [Google Scholar]
- Tulman E.R., Afonso C.L., Lu Z., Zsak L., Kutish G.F., Rock D.L. The genome of canarypox virus. J. Virol. 2004;78(1):353–366. doi: 10.1128/jvi.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., Davison A.J., Dempsey D.M., Dutilh B.E., García M.L., Harrach B., Harrison R.L., Hendrickson R.C., Junglen S., Knowles N.J., Krupovic M., Kuhn J.H., Lambert A.J., Łobocka M., Nibert M.L., Zerbini F.M. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2021;166(9):2633–2648. doi: 10.1007/s00705-021-05156-1. [DOI] [PubMed] [Google Scholar]
- Zarifi F., Nakhaei P., Nourani H., Mirshokraei P., Razmyar J. Characterization of Iranian canarypox and pigeonpox virus strains. Arch. Virol. 2019;164(8):2049–2059. doi: 10.1007/s00705-019-04277-y. [DOI] [PubMed] [Google Scholar]
- Zeng X., Xiong L., Wang W., Zhao Y., Xie Y., Wang Q., Zhang Q., Li L., Jia C., Liao Y., Zhou J. Whole-genome sequencing and comparative analysis of Helicobacter pylori GZ7 strain isolated from China. Folia Microbiol. 2022;67(6):923–934. doi: 10.1007/s12223-022-00989-y. [DOI] [PubMed] [Google Scholar]
- Zhao K., He W., Xie S., Song D., Lu H., Pan W., Zhou P., Liu W., Lu R., Zhou J., Gao F. Highly pathogenic fowlpox virus in cutaneously infected chickens, China. Emerg. Infect. Dis. 2014;20(7):1208–1210. doi: 10.3201/eid2007.131118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Jin N., Zhang H., Jin M., Lu H., Ma M., Li C., Yin G., Wang R., Liu Q. Construction and immunogenicity of a recombinant fowlpox virus containing the capsid and 3C protease coding regions of foot-and-mouth disease virus. J. Virol. Methods. 2006;136(1-2):230–237. doi: 10.1016/j.jviromet.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Du S., Zhang Y., Liu J., Guo Y., Liu C., Bai J., Wang M., Zhao F., Cao T., Xu W., Bai B., Zhang K., Ma Y., Li C., Jin N. SIV-Specific Antibodies are Elicited by a Recombinant Fowlpox Virus Co-expressing SIV Gag and envT. Indian J. Microbiol. 2018;58(3):345–352. doi: 10.1007/s12088-018-0728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.