Summary
Sulfoquinovose (6-deoxy-6-sulfo-D-glucose, SQ), the polar head group of sulfolipids in plants, is abundant in nature. Many bacteria degrade SQ through pathways termed sulfoglycolysis producing C3 or C2 sulfonates, while certain bacteria degrade SQ through direct oxygenolytic cleavage of the SQ C-S bond, catalyzed by a flavin-dependent alkanesulfonate monooxygenase (sulfo-ASMO pathway). Here we report bioinformatics and biochemical studies revealing an alternative mechanism for oxygenolytic cleavage of the SQ C-S bond, catalyzed by an iron and α-ketoglutarate-dependent alkanesulfonate dioxygenase (SqoD, sulfo-ASDO pathway). In both the ASMO and ASDO pathways, the product 6-dehydroglucose is reduced to glucose by NAD(P)H-dependent SquF. Marinomonas ushuaiensis, a marine bacterium, which harbors the sulfo-ASDO gene cluster is shown utilizing SQ as a carbon source for growth, accompanied by increased transcription of SqoD. The sulfo-ASDO pathway highlights the range of microbial strategies for degradation of this ubiquitous sulfo-sugar, with potential implications for sulfur recycling in different biological environments.
Subject areas: Biochemistry, Microbial metabolism, Bioinformatics
Graphical abstract
Highlights
-
•
SqoD, an Fe/α-KG-dependent dioxygenase catalyzes the cleavage of the SQ C-S bond
-
•
NADPH-dependent SquF reduces 6-dehydroglucose formed by SqoD to glucose
-
•
This SQ degradation pathway is termed sulfo-ASDO pathway
-
•
Bacteria grow on SQ as a carbon source with increased SqoD-SquF transcription
Biochemistry; Microbial metabolism; Bioinformatics
Introduction
Sulfoquinovose (SQ) is a sulfonated homolog of glucose that acts as the polar head group of sulfoquinovosyl diacylglycerol (SQDG), a sulfolipid present in the thylakoid membranes of chloroplasts in plants, algae, and other photosynthetic eukaryotes, as well as in the photosynthetic membranes of most phototrophic bacteria.1,2,3 SQ is one of the most abundant organosulfur compounds on the planet, with an annual production estimated at 1010 tons.4 In photosynthetic organisms, inorganic sulfate obtained from the environment is first reduced to sulfite, which is then combined with UDP-glucose by UDP-SQ synthase to form UDP-SQ, the precursor to SQDG.5 SQ is broken down by heterotrophic bacteria (including decomposers, pathogens, and symbionts), which consume it as a source of carbon and energy for growth, returning most of the sulfonate sulfur to the environments in its inorganic forms and completing the sulfur cycle.6,7,8
The structural similarity between SQ and glucose-6-phosphate has led to the proposal that bacterial degradation of SQ occurs through processes analogous to glycolysis, referred to as sulfoglycolysis.6,9 To date, four distinct sulfoglycolytic mechanisms have been reported, of which one is analogous to the Embden-Meyerhof-Parnas pathway (sulfo-EMP),10,11,12,13 one is analogous to the Entner-Doudoroff pathway (sulfo-ED),14,15 and the remaining two sulfoglycolytic pathways rely on either a transaldolase (sulfo-TAL)16,17,18 or a transketolase (sulfo-TK)11 closely related to enzymes in the pentose phosphate pathway. While the former three pathways enable the use of three of the SQ carbons and yield a C3 sulfonate byproduct, the sulfo-TK pathway utilizes four of the SQ carbons and generates a C2 sulfonate byproduct. The resulting C3 and C2 sulfonates are further degraded by other bacteria, which release most of the sulfonate sulfur as either SO32−, SO42−, or H2S.7,19
Recent studies by our laboratory and other researchers have demonstrated that certain aerobic bacteria degrade SQ through a non-sulfoglycolytic pathway involving direct oxygenolytic cleavage of the SQ C-S bond, catalyzed by the sulfoquinovolytic AlkaneSulfonate MonoOxygenase SquD (sulfo-ASMO) pathway.11,20,21 SquD is closely related to other two-component flavin-dependent alkanesulfonate monooxygenases, such as E. coli SsuD, which require a reduced flavin cofactor provided by a separate NAD(P)H-dependent flavin reductase SsuE.22 These enzymes are believed to employ a reactive peroxyflavin intermediate, formed via the reaction of reduced flavin (FMN or FAD) with O2, to cleave the inert alkanesulfonate C-S bond, generating sulfite and an aldehyde as products.23 In the case of SquD, the resulting product 6-deoxyglucose is reduced by the NADP+-dependent glucose-6-dehydrogenase SquF to form glucose, which is then degraded through standard glycolytic pathways.11,21 Thus, in contrast to the sulfoglycolytic pathways, the sulfo-ASMO pathway permits utilization of all six SQ carbons, and results in direct organosulfur mineralization. Sulfo-ASMO gene clusters are commonly found in aerobic Alphaproteobacteria residing in soil,11,21 suggesting a possible role in the recycling of SQ from plant matter. In a recent study by Liu et al.,20 it was discovered that sulfo-ASMO genes are prevalent among marine bacteria belonging to the Roseobacter clade, where they may contribute to degradation of SQ from algae and cyanobacteria.20
Bacterial alkanesulfonate monooxygenases are typically involved in allowing sulfonates to be utilized as a sulfur source for growth, with the resulting sulfite being reduced by the assimilatory sulfite reductase.24 By contrast, the sulfo-ASMO pathway primarily functions to enable SQ to be used as a carbon and energy source, with the sulfite being directly exported.21 Apart from flavin-dependent monooxygenases, another class of enzyme known to catalyze C-S cleavage of unactivated alkanesulfonates is non-heme iron, α-ketoglutarate-dependent (Fe/α-KG) dioxygenases. Members of this functionally diverse superfamily of enzymes contain a mononuclear Fe(II) cofactor, coordinated by three protein-based ligands (two His and one Asp/Glu) and α-KG (α-ketoglutarate).25 The most extensively studied member of this superfamily is E. coli taurine dioxygenase TauD, which utilizes a highly reactive Fe(IV) oxo intermediate formed via the reaction of Fe(II) with α-KG and O2, to cleave the C-S bond of taurine, generating sulfite and 2-aminoacetaldehyde.26
Here we report a new pathway for SQ degradation, involving oxygenolytic cleavage of the SQ C-S bond, catalyzed by the Fe/α-KG-dependent sulfoquinovolytic AlkaneSulfonate DiOxygenase SqoD (sulfo-ASDO pathway). Similar to the sulfo-ASMO pathway, the resulting 6-deoxyglucose is reduced to glucose by a close homolog of SquF. Sulfo-ASDO gene clusters are present in marine Gammaproteobacteria including Marinobacterium aestuarii and Marinomonas ushuaiensis. We describe biochemical studies of recombinant enzymes from M. aestuarii, and growth experiments with M. ushuaiensis with SQ as a carbon source for growth.
Results
SQ degradation gene clusters containing a Fe/αKG-dependent dioxygenase
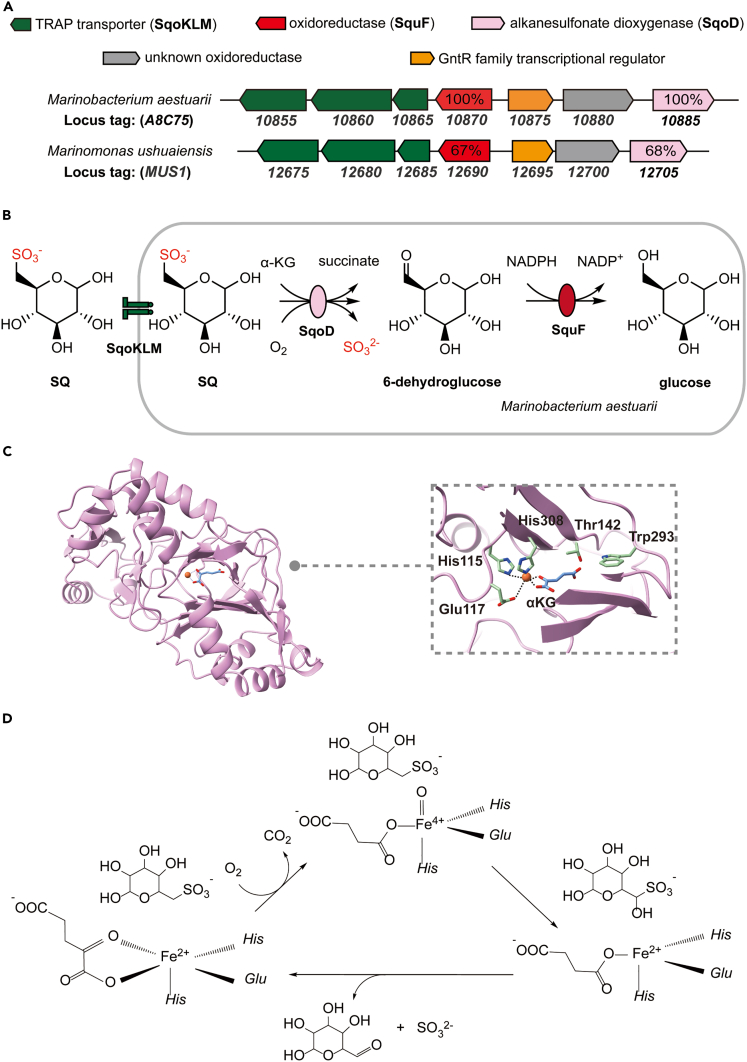
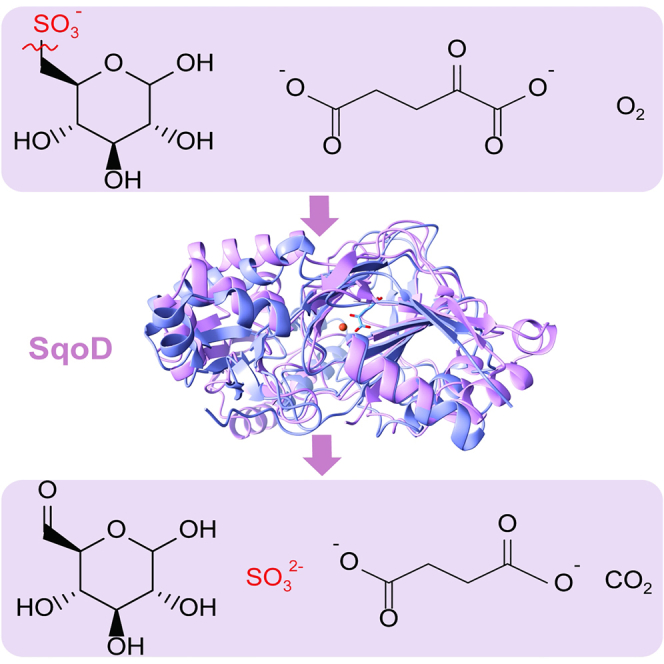
While examining the genome neighborhood of close homologs of SquF (also known as SmoB) using the Enzyme Function Initiative Genome Neighborhood Tool (EFI-GNT),27 we noticed that some of the sequences were not associated with homologs of SquD (luciferase-like monooxygenase family, PF00296), but rather with Fe/α-KG-dependent dioxygenases (TauD family, PF02668) (Figure 1A), implying an alternative mechanism for cleavage of SQ into 6-deoxyglucose via an SQ dioxygenase (SqoD) (Figure 1B). For instance, in M. aestuarii, SquF (UniProt A0A1A9EZ66, 52% sequence identity to A9CEY6, Agrobacterium tumefaciens SmoB) is associated with the putative SqoD (A0A1A9EZ58), as well as a GntR family transcription factor, and a TRipartite ATP-independent Periplasmic (TRAP) transporter (Figure 1A), which typically facilitates import of carboxylic and sulfonic acids.28
Figure 1.
Reaction scheme and gene cluster for the proposed sulfo-ASDO pathway
(A) Gene cluster for the sulfo-ASDO pathway in M. aestuarii and M. ushuaiensis.
(B) The proposed sulfo-ASDO pathway diagram.
(C) AlphaFold model of the active site of SqoD (UniProt A0A1A9EZ58), in which the positions of the Fe and α-ketoglutarate were estimated by overlaying with the crystal structure of E. coli TauD (PDB 1OS7).
(D) Reaction scheme for SqoD. See also Figures S2, S6, and S7 and Table S1.
Comparison of the AlphaFold model of M. aestuarii SquF (available from the UniProt page of A0A1A9EZ66),29,30 with the crystal structure of A. tumefaciens SmoB in complex with NADPH and glucose (PDB 7BC1)21 confirmed that the substrate-binding residues are conserved (See Figure S1; Table S2). In addition, comparison of the AlphaFold model of M. aestuarii SqoD (available from the UniProt page of A0A1A9EZ58) with the crystal structure of E. coli TauD (PDB 1OS7, 18% sequence identity with M. aestuarii SqoD)31 confirmed the presence of Fe-coordinating residues in SqoD (Figure 1C), which is consistent with an analogous mechanism for SQ C-S cleavage involving Fe(II), O2, and α-KG (Figure 1D).
Detection of reaction products of M. aestuarii SqoD and SquF
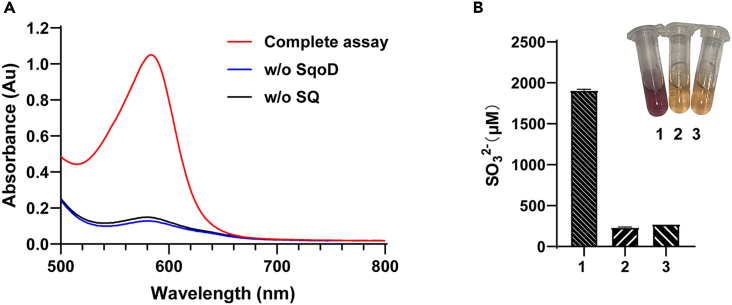
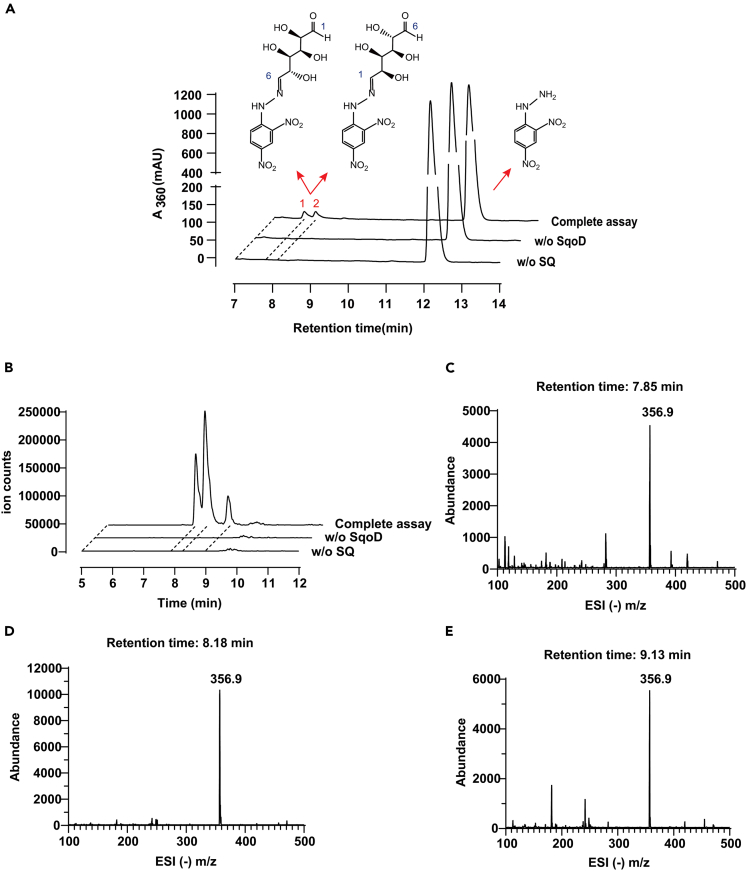
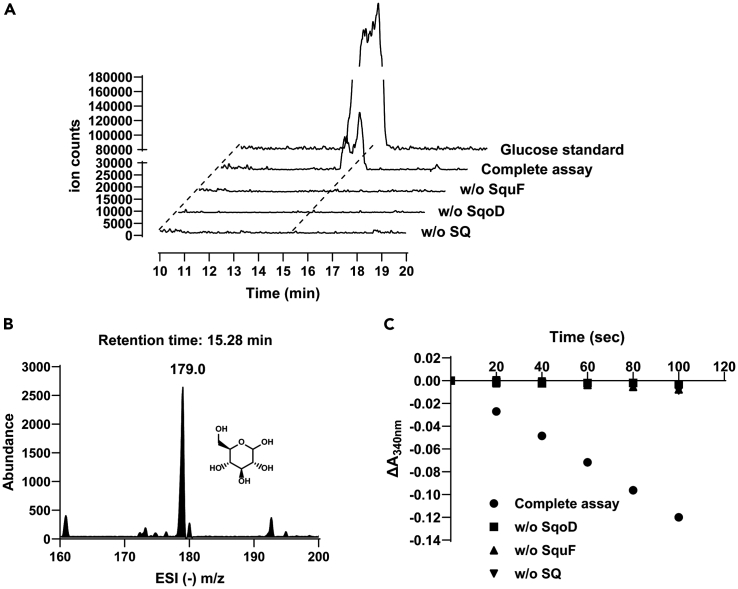
To test our hypothesis, we recombinantly produced SqoD and SquF from M. aestuarii, and the proteins were purified to homogeneity (See Figure S2). To supply the Fe(II) cofactor, 25 μM FeSO4 and 50 μM ascorbic acid were included in all SqoD assays. Incubation of SQ with SqoD and -KG led to the release of sulfite as detected by a colorimetric Fuchsin assay (Figure 2). No sulfite release was detected when SQ was replaced with taurine as the sulfonate substrate (See Figure S4). Derivatization with 2,4-dinitrophenylhydrazine (DNPH) followed by LC-MS analysis revealed two peaks with negative ion m/z (−) 356.9, corresponding to two isomers of DNPH-6-dehydroglucose (Figure 3), as previously reported for assays of SquD.11 Inclusion of SquF and NADPH in the assay followed by LC-MS analysis led to the appearance of a peak with m/z (−) 179.0, identical to a commercial D-glucose standard, demonstrating the SquF-catalyzed reduction of 6-dehydroglucose to glucose (Figures 4A–4C). SquF also catalyzed the reverse reaction, oxidation of glucose to 6-dehydoglucose (See Figure S5B). Incubation of SquF with NADP+ and D-glucose followed by DNPH derivatization and LC-MS revealed two peaks with m/z (−) 356.9, consistent with two isomers of DNPH-6-dehydroglucose (See Figures S5A, S5C, and S5D). Together, these results demonstrate that SqoD is an iron-dependent SQ dioxygenase that catalyzes SQ cleavage into sulfite and 6-dehydro-D-glucose, while SquF is a NADP+-dependent D-glucose 6-dehydrogenase.
Figure 2.
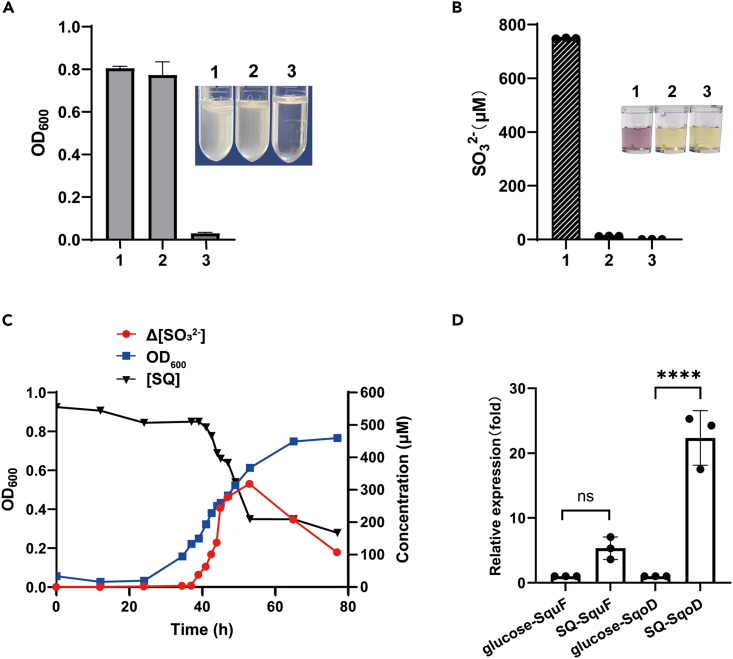
Detection of sulfite formation in the MaSqoD -catalyzed SQ cleavage by a colorimetric Fuchsin assay
(A) UV-vis absorption spectra of the complete assay and the negative controls omitting MaSqoD or SQ are shown in red, blue, and black, respectively.
(B) Absorbance at 580 nm of each assay. Inset: Photographs of reaction mixtures. The complete assay 1, the negative control omitting SQ 2, and the negative control omitting SqoD 3. Data are represented as mean ± SD. See also Figures S3 and S4.
Figure 3.
LC-MS detection of the SqoD reaction product 6-dehydroglucose
(A) HPLC elution profiles of the DNPH derivatized products of the SqoD reaction, monitoring the absorbance at 360 nm, showing product peaks for 1 and 2 in the full assay, corresponding to two stereoisomers of DNPH-6-dehydroglucose.
(B) Extracted ion chromatograms (m/z (−) 356.9, the predicted mass of the DNPH-6-dehydroglucose monoanion), showing the same two product peaks 1 and 2.
(C–E) The ESI m/z (−) spectrum of the region spanned by three peaks in Figure 3B, showing a species with m/z (−) 356.9 corresponding to different isomers of DNPH-6-dehydroglucose (two possible isomers are depicted). See also Figure S5.
Figure 4.
In vitro reconstitution of the sulfo-ASDO pathway showing the formation of glucose
(A) Extracted ion chromatograms (m/z (−) 179.0, the predicted mass of glucose monoanion), showing product peaks in the full assay coeluting with glucose standard.
(B) ESI (−) m/z spectrum of the product peak showing a species with m/z (−) 179.0 corresponding to glucose.
(C) Enzyme activity assay monitoring NADPH consumption accompanying 6-dehydroglucose reduction by MaSquF. See also Figure S1.
Reaction kinetics of SqoD
SqoD reaction kinetics were measured using a coupled spectrophotometric assay with excess SquF and NADPH, monitoring the decrease of A340nm corresponding to NADPH consumption accompanying 6-dehydroglucose reduction (See Figure S6A). The optimal reaction pH for SqoD was determined to be pH 7.0 (See Figure S6B). Under the specified reaction conditions, activity was directly proportional to the amount of SqoD added (See Figure S6C). Apparent Michaelis-Menten kinetic parameters of SqoD for SQ (kcat = 0.16 ± 0.01 s−1, KM = 1.0 ± 0.2 mM, kcat/KM = (1.6 ± 0.3) × 102 M−1 s−1) and α-KG (kcat = 0.73 ± 0.03 s−1, KM = 26 ± 4 μM, kcat/KM = (2.8 ± 0.4) × 104 M−1 s−1) were determined by keeping the concentration of one substrate constant at 5 mM while varying the concentration of the other substrate (0.25–8.0 mM for SQ, or 3–200 μM for α-KG) (See Figures S6D and S6E).
Growth of Marinomonas ushuaiensis DSM 15871 with SQ as a carbon source
To examine the physiological role of SqoD, growth experiments were carried out on the commercially available strain M. ushuaiensis DSM 15871,32 which harbors the sulfo-ASDO gene cluster, and lacks the sulfo-ASMO and other known pathways for SQ degradation. M. ushuaiensis also lacks homologs of sulfoacetaldehyde acetyltransferase (Xsc),33 (R)-sulfolactate sulfolyase (SuyAB),34 or L-cysteate sulfolyase (CuyA),35 involved in dissimilation of C2 and C3 sulfonates.36,37 The bacterial strain exhibited robust growth in minimal medium containing either 10 mM glucose or SQ as a carbon source (growth also required addition of 2 g/L peptone in the media), but not in media where glucose or SQ were omitted (Figure 5A). Production of sulfite was observed in the medium with SQ but not with glucose, as detected by the Fuchsin assay (See Figures S3 and5B). Growth in the SQ medium was accompanied by depletion of SQ and corresponding production of sulfite (Figure 5C). The ∼2-fold difference of sulfite level between Figures 5B and 5C is probably due to excessive oxygen exposure through multiple samplings in the growth curve assay and consequently more sulfite oxidized into sulfate. We also noted that the sulfite content decreased lately during the growth indicating sulfite oxidation, which was also observed by a related study (Figure 5C).21 qPCR assays demonstrated the increased transcription of both SqoD (UniProt X7E6N4, 22-fold) and SquF (X7E6P9, 5-fold) in cells grown on SQ relative to cells grown on glucose (Figure 5D).
Figure 5.
Growth of M. ushuaiensis in minimal medium containing different carbon sources
(A) Tubes 1, 2, and 3 show cells grown in media with SQ, glucose or no additional carbon source. Detect the optical density at 600 nm. Data are represented as mean ± SD.
(B) Quantification of sulfite formed in cultures 1, 2, and 3 by a colorimetric fuchsin assay. Inset: Photographs of the respective assays.
(C) Optical density of M. ushuaiensis culture (blue) and concentration of SQ ([SQ]) (black) and change in concentration of sulfite (Δ[sulfite]) (red), with respect to time.
(D) qPCR analyses of the transcription levels of SqoD and SquF. The transcriptional levels of genes of interest were normalized by that of the 16S rRNA. The induction by SQ was displayed in comparison with the transcriptional data from glucose-grown cells. The error bars represent the standard deviation of three individual experiments. Data are represented as mean ± SD. Significance was assessed by an ordinary one-way ANOVA test (∗∗∗∗p value <0.0001). See also Figures S3 and S8 and Table S2.
Occurrence of sulfo-ASMO and sulfo-ASDO gene clusters in different bacteria
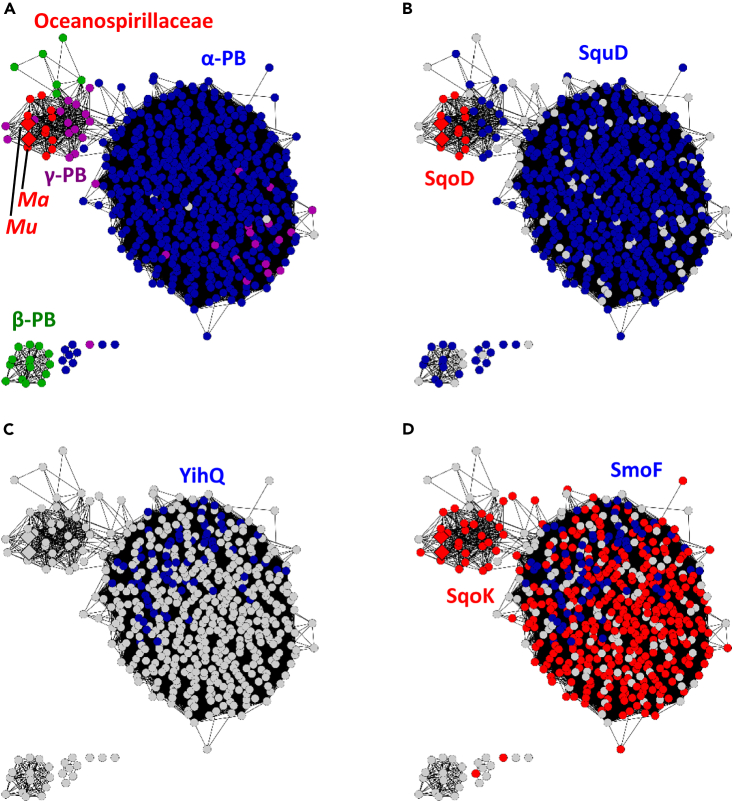
The glucose-6-dehydrogenase SquF serves as a marker for the identification of both sulfo-ASMO and sulfo-ASDO gene clusters. The EFI-GNT tool was used to examine the genome neighborhoods of SquF homologs within a 10 open reading frame (10-ORF) window, which showed that close homologs of SquF within the clusters UniRef50_A0A0K0Y4B2 are associated with other sulfo-ASMO and sulfo-ASDO genes (members of a UniRef50 cluster share >50% sequence identity with the reference sequence of the cluster).38 To obtain an overview of these sequences, a Sequence Similarity Network (SSN) of the 584 SquF sequences within UniRef50_A0A0K0Y4B2 was constructed using the EFI Enzyme Similarity Tool (EFI-EST),39 with an edge E-value cutoff of ≥10−110 (the cutoff value was chosen to fractionate the sequences by taxonomic class), and plotted using Cytoscape (Figure 6).40 The analysis revealed that the majority of SquF sequences are present in α-Proteobacteria, with a minority of sequences present in β- and γ-Proteobacteria (Figure 6A). In addition, the majority of SquF sequences are associated with SquD (PF00296), corresponding to the sulfo-ASMO pathway, with a minority associated with SqoD (PF02668), corresponding to the sulfo-ASDO pathway (Figure 6B). The sulfo-ASDO gene clusters localize in γ-Proteobacteria in the family Oceanospirillaceae.
Figure 6.
SSN of close homologs of glucose-6-dehydrogenase SquF within UniRef50_A0A0K0Y4B2
(A) Sequences present in α-proteobacteria (blue), β-proteobacteria (green), γ-proteobacteria in the family Oceanospirillaceae (red, Mu = Marinomonas ushuaiensis, Ma = Marinobacterium aestuarii), and other γ-proteobacteria (purple).
(B) Sequences associated with SquD (blue, sulfo-ASMO pathway) or SqoD (red, sulfo-ASDO pathway), within a 10-ORF window.
(C) Sequences associated with sulfoquinovosidase YihQ.
(D) Sequences associated with the sulfoquinovoside transporter substrate-binding subunit SmoF (UniRef50_I9XG35), or the putative sulfoquinovose transporter substrate-binding subunit SqoK (UniRef50_V9WJD9).
A fraction of the gene clusters also harbor sulfoquinovosidase YihQ (InterPro family IPR044112) (Figure 6C). Recent research by Sharma et al. on Agrobacterium tumefaciens C58 indicates that SQ, SQDG, SQDG-derived sulfoquinovosyl glycerol (SQGro), and other sulfoquinovosides are imported by an ATP-binding cassette (ABC) transporter SmoEFGH, followed by cleavage of the latter by YihQ to release SQ for further degradation.21,41 The authors also reported crystal structures of the periplasmic substrate-binding subunit SmoF in complex with SQ and various sulfoquinovosides.21,41 Our genome neighborhood analysis shows that close homologs of SmoF (within UniRef50_I9XG35) often co-occur with YihQ, supporting the conclusion by Sharma et al. that SmoEFGH transports sulfoquinovosides. In contrast, the sulfo-ASDO gene clusters in the Marinomonas sp. contain a TRAP transporter, which we refer to as SqoKLM (Figure 6D). Close homologs of the periplasmic substrate-binding subunit SqoK (within UniRef50_V9WJD9) do not co-occur with YihQ, suggesting that SqoKLM likely transports for SQ rather than sulfoquinovosides. These observations are consistent with recent research by Liu et al., on Roseobacter clade marine bacteria that degrade SQ via the sulfo-ASMO pathway.20 Similar to Marinomonas sp., these bacteria lack YihQ, but contain a TRAP transporter that may import free SQ, which has been suggested to originate from marine algae and cyanobacteria.20 Growth of M. ushuaiensis on other SQ-glycosides, of which biologically relevant examples are sulfoquinovosyl glycerol and sulfoquinovosyl diacylglycerol, is currently unknown.
Discussion
Our experiments provide evidence for yet another mechanism for SQ degradation, namely the sulfo-ASDO pathway, involving oxygenolytic C-S cleavage by a Fe/α-KG-dependent SQ dioxygenase SqoD. This pathway shares some similarities with the previously described sulfo-ASMO pathway, where C-S cleavage is catalyzed by a flavoenzyme monooxygenase SquD.11,21 Both enzyme families have long been known to participate in utilization of sulfonates as a sulfur source for growth,24 but are now known to facilitate utilization of the sulfo-sugar SQ as a carbon and energy source, with the sulfonate-derived sulfur being exported as a waste product. In both cases, C-S cleavage produces 6-deoxyglucose, which is reduced to glucose by SquF, providing a more direct path for entry into glycolysis relative to the sulfoglycolytic pathways, while utilizing all six SQ carbons. The O2-dependency of the C-S cleavage reactions means that both pathways are limited to aerobic organisms. However, a key difference between the two pathways is that while the sulfo-ASMO pathway requires a supply of NAD(P)H to generate the reduced flavin cofactor of SquD, the sulfo-ASDO pathway requires a supply of α-KG, which is oxidized to succinate on every turnover of SqoD.
Fe/α-KG-dependent dioxygenases encompass a broad range of enzymes with various functions, such as post-translational modifications (e.g., collagen prolyl 4-hydroxylase),42 signaling (e.g., hypoxia inducible factor prolyl hydroxylase),43 epigenetics (e.g., TET methylcytosine dioxygenases),44 sulfur assimilation (e.g., TauD),45 biosynthesis of primary metabolites (e.g., carnitine)46 and a variety of secondary metabolites.25 However, there are limited examples of Fe/α-KG-dependent dioxygenases catalyzing reactions in carbon and energy metabolism, which involve significantly greater metabolic flux in the cell. One complication in employing Fe/α-KG-dependent enzymes in carbon metabolism is that the complete metabolic pathway must support the replenishment of the co-substrate α-KG. A case in point is the E. coli aerobic lysine degradation pathway,47 in which the Fe/α-KG-dependent glutarate-2-hydroxylase CsiD catalyzes the formation of L-2-hydroxyglutarate, which is then oxidized to α-KG to replenish this co-substrate (See Figure S7). In the case of SqoD, the necessary α-KG is most likely provided by the TCA cycle, which generates two molecules of α-KG for every molecule of glucose. Consequently, utilizing SQ as a carbon source for growth necessitates that at least half of the carbon flux is processed via the TCA cycle.
The presence of SQ in various natural environments is consistent with the wide range of bacterial taxa capable of metabolizing SQ, and the diversity of biochemical mechanisms for SQ degradation.7,8 While the sulfo-ASMO pathway is present in different classes of Gram negative isolated from various terrestrial and marine environments, the sulfo-ASDO pathway is only present in halophilic marine γ-proteobacteria in the family Oceanospirillaceae. These include strains isolated from seawater, marine, and estuarine sediments, as well as from seagrass (Marinomonas posidonica and Marinomonas aquiplantarum), salt marsh cordgrass (Marinomonas spartinae), and coral (Marinomonas fungiae) suggesting a possible role in degradation of SQ from marine and coastal plants, or phytoplankton. However, any advantage of employing the sulfo-ASDO pathway over the sulfo-ASMO pathway in these organisms remains unknown. Nevertheless, identification of the sulfo-ASDO enzymes and associated transporters will aid future bioinformatics studies of SQ degradation and provide a deeper understanding of bacterial recycling of this prevalent organosulfur compound.
In brief
Zhang et al. reported a novel α-KG/Fe2+ dependent dioxygenase, SqoD responsible for an alternative mechanism of the C-S cleavage of SQ. SQ is thus converted to glucose in a two-step pathway catalyzed by SqoD followed by glucose 6-dehydrogenase (SquF).
Limitations of the study
In qPCR assay, the growth on SQ is accompanied by transcription of SqoD. Despite our efforts, we were unable to quantify the levels of SqoD and SquF proteins as they were not expressed at sufficiently high levels for accurate quantitation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Marinomonas ushuaiensis | DSMZ | DSM 15871 |
| E. coli DH5α | TransGen Biotech | Cat#CD501 |
| E. coli BL21 | Biomed | Cat#zc0380-m0243 |
| Chemicals, peptides, and recombinant proteins | ||
| Sulfoquinovose | ABCR | CAS: 3458-06-8 |
| α-Ketoglutaric acid | Solarbio | CAS: 328-50-7 |
| Ascorbic acid | Solarbio | CAS: 50-81-7 |
| SspI | NEB | Cat#B0132S |
| Critical commercial assays | ||
| Q5 High-Fidelity 2× Master Mix | NEB | Cat#M0492L |
| GoScript™ Reverse Transcription System | Promega | Cat#A5000 |
| GoTaq® qPCR Master Mix | Promega | Cat#A6001 |
| RNAprep Pure Cell/Bacteria Kit | TIANGEN | Cat#4992235 |
| Deposited data | ||
| Source data obtained in this study | This paper | https://data.mendeley.com/datasets/z4f53mdy6t/draft?a=a3eee788-5c4b-4f8b-a0b6-e30a24b1648b |
| Oligonucleotides | ||
| Primers for qPCR: see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| HT-SqoD | General Biosystems | N/A |
| HT-SquF | General Biosystems | N/A |
| Software and algorithms | ||
| Graphpad Prism 9 | GraphPad | https://www.graphpad.com/ |
| Adobe Illustrator 2021 | Adobe Illustrator | https://www.adobe.com/cn/products/illustrator.html |
| Chimera 1.11.2 | Chimera | https://www.cgl.ucsf.edu/chimera/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yan Zhang (yan.zhang@tju.edu.cn).
Materials availability
All of the reagents reported in this study are available from the lead or correspondence contact with Materials Transfer Agreement as long as stocks remain available.
Experimental model and study participant details
Bacterial strains
Plasmids used in this study were synthesized and inserted into the SspI site of the modified pET28 vector HT by General Biosystems, Inc (Anhui, China). The proteins were expressed in E. coli BL21 (DE3) cells.
Method details
Materials and general methods
Sulfoquinovose (SQ) was purchased from ABCR (Germany). Marinomonas ushuaiensis strain U1 (DSM 15871) was purchased from DSMZ (Germany). Methanol and acetonitrile used for liquid chromatography-mass spectrometry (LC-MS) were high-purity solvents from Concord Technology (Minnesota, USA). Formic acid was purchased from Merck (New Jersey, USA). Water used in this work was ultrapure deionized water from Millipore Direct-Q. Protein purification chromatographic experiments were performed on an “ÄKTA pure” FPLC machine equipped with appropriate columns in a 4°C cold cabinet. Protein concentrations were calculated from their absorption at 280 nm measured with Nanodrop One (Thermo Fisher Scientific). The extinction coefficient for each protein at 280 nm was obtained using the ExPASy ProtParam tool. Data was analyzed and plotted using GraphPad Prism 9.
Gene synthesis
E. coli codon-optimized DNA fragments encoding Marinobacterium aestuarii SqoD (MaSqoD, A0A1A9EZ58) and SquF (MaSquF, A0A1A9EZ66) were synthesized and inserted into the SspI site of the modified pET28 vector HT by General Biosystems, Inc (Anhui, China). The resulting plasmids contain an N-terminal His6-tag and a Tobacco Etch Virus (TEV) protease cleavage site, followed by the gene of interest. Both plasmids were confirmed by DNA sequencing.
Expression and purification of MaSqoD, MaSquF
MaSqoD and MaSquF were heterologously expressed in Escherichia coli BL21 (DE3) cells harboring the corresponding plasmids. The transformants containing the respective plasmids were grown in LB supplemented with 50 μg/mL kanamycin at 37°C while being shaken at 220 rpm. When OD600 reached ∼0.8, the temperature was decreased to 16°C and isopropy β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3 mM to induce the production of the proteins. After 18 h, cells were harvested by centrifugation (5,000 × g for 15 min at 4°C).
Cells (∼1 g wet weight) were suspended in 5 mL of lysis buffer [50 mM Tris-HCl, pH 8.0, 1 mM phenylmethanesulfonyl fluoride (PMSF), 0.4 mg/mL lysozyme, 0.03% Triton X-100, and 0.03 mg/mL of DNase I (Roche, Germany)]. The cell suspension was frozen in a −80°C freezer, and then thawed and incubated at 25°C for 40 min to allow cell lysis. A 6% solution of streptomycin sulfate in water was added to a final concentration of 1% to precipitate the DNA. The precipitate was removed by centrifugation (12,000 × g for 10 min at 4°C). The supernatant was then filtered through a 0.45 μm filter. β-mercaptoethanol (BME) was added to a final concentration of 5 mM, and the sample was loaded onto a 10 mL TALON Co2+-affinity column, pre-equilibrated with buffer A [20 mM Tris/HCl, pH 7.5, 0.2 M KCl and 5 mM BME]. The column was washed with 10 column volumes (CV) of buffer A, and then the protein was eluted with 5 CV of buffer A containing 150 mM imidazole. Eluted protein (∼20 mL) was dialyzed against 2 L of buffer A to remove imidazole. The dialyzed protein was frozen in aliquots with liquid nitrogen, and stored at −80°C until further use. The purified SqoD (ε280nm = 42,065 M−1 cm−1) and SquF (ε280nm = 46,200 M−1 cm−1) were examined by SDS-PAGE on a commercial gel (SurePAGE, Bis-Tris, 8–16%).
LC-MS assay for 6-dehydroglucose formation from SQ by MaSqoD
To detect the product 6-dehydroglucose of the MaSqoD reaction, a 200 μL reaction mixture containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM SQ, 5 mM -KG, 25 μM FeSO4, 50 μM ascorbic acid and 4 μM MaSqoD was incubated at RT for 2 h. Two negative controls omitting either MaSqoD or SQ were also performed. The product 6-dehydroglucose was detected by derivatization with 2,4-dinitrophenylhydrazine (DNPH) (MACKLIN, Shanghai, China). After the enzyme reaction, 50 μL of reaction solution was mixed with 550 μL of 0.73 M sodium acetate buffer, pH 5.0, followed by 400 μL of freshly prepared DNPH solution (10 mg dissolved in 25 mL methanol). The mixture was incubated at 50°C for 1 h and then was filtered through a 0.22 μm nylon membrane filter prior to LC-MS analysis. LC-MS analysis was performed on an Agilent 6420 Triple Quadrupole LC/MS instrument (Agilent Technologies), equipped with an Agilent ZORBAX SB-C18 column (4.6 × 250 mm). The drying gas temperature was maintained at 300°C with a flow rate of 10 L/min and a nebulizer pressure of 15 psi. The column was equilibrated with 95% of solvent A (0.1% formic acid in H2O), 5% of solvent B (0.1% formic acid in CH3CN) for 5 min, and developed at a flow rate of 1.0 mL/min from 25% B to 85% B in 25 min. UV detection was set at 360 nm.
Fuchsin assays for sulfite release from SQ by MaSqoD
Sulfite release upon cleavage of SQ by MaSqoD was detected using a colorimetric assay involving the formation of a colored complex between sulfite and Fuchsin dye in acidic solution.48 The Fuchsin reagent (0.8 M H2SO4, 0.08% Fuchsin and 1.6% formaldehyde, mixed with 7: 2: 1) was freshly prepared. Serial dilutions of sodium sulfite (200, 100, 50, 25, 12.5, 0 μM) were used to establish a standard curve (See Figure S3). A 200 μL reaction mixture containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM SQ, 5 mM α-KG, 25 μM FeSO4, 50 μM ascorbic acid and 4 μM MaSqoD was incubated at RT for 1 h. 50 μL aliquots of the reaction mixtures were then mixed with the acidic fuchsin reagent solution for color development. The absorbance at 580 nm was recorded, and the concentration of sulfite was calculated using the standard curve.
LC-MS assays for glucose formation from SQ by MaSqoD and MaSquF
To detect 6-dehydroglucose reductase activity of MaSquF, SQ was reacted with MaSqoD as described above, and then MaSqoD was removed from the reaction mixture using a centrifugal filter unit (1.5 mL YM-30 Amicon, Millipore). To the 100 μL of the flow-through, 10 μM MaSquF and 0.5 mM NADPH were added, mixed and incubated at RT for 30 min. Negative controls were prepared by omitting SQ or MaSquF/MaSqoD. After the reaction, an equal volume of acetonitrile was added, followed by incubation for 10 min at 4°C, and the precipitated protein was removed by centrifugation (10,000 × g for 5 min at 4°C). The supernatant was filtered through a 0.22 μm nylon membrane filter prior to LC-MS analysis. LC-MS was performed on an Agilent 6420 Triple Quadrupole LC/MS instrument (Agilent Technologies). LC-MS analysis was carried out on a ZIC-HILIC column (5 mm, 200 Å, 150 × 4.6 mm; Merck). The HPLC conditions were as follows: from 90% B to 70% B in 10 min, 70% B to 50% B in 20 min, and 90% B for 5 min. Solvent A was 90% 0.1 M ammonium acetate and 10% acetonitrile, and solvent B was acetonitrile. The flow rate was set to 0.5 mL/min. The mass spectrometer was run in ESI negative mode.
LC-MS assay for 6-dehydroglucose formation from glucose by MaSquF
To detect glucose-6-dehydrogenase activity of MaSquF, a 200 μL reaction mixture containing 50 mM CAPSO, pH 10.0, 100 mM NaCl, 10 mM NADP+, 100 mM glucose and 10 μM MaSquF was incubated at RT for 10 min. Reaction mixtures were derivatized with DNPH followed by LC-MS analysis as described for the MaSqoD assay.
Growth of M. ushuaiensis in minimal medium with SQ as a carbon source
The rich growth medium for M. ushuaiensis was prepared by dissolving 37.4 g 2216E Liquid Medium (Hope Bio, Qingdao, China, Catalog number: HB0132-1) in 1 L distilled water. To prepare the minimal medium for the growth of M. ushuaiensis DSM 15871, peptone 2 g, SQ 2.4 g (or glucose 2 g), ferric citrate 0.1 g, NaCl 19.45 g, MgCl2 5.9 g, Na2SO4 3.24 g, CaCl2 1.8 g, KCl 0.55 g, NaHCO3 0.16 g, KBr 0.08 g, SrCl2 34 mg, H3BO3 22 mg, Na2SiO3 4 mg, NaF 2.4 mg, NH4Cl 1.1 mg, NaNO2 1.4 mg, Na2HPO4 8 mg were dissolved in 1 L distilled water and autoclaved. M. ushuaiensis cells were inoculated into rich medium and grown overnight in a 22°C incubator with shaking at 220 rpm. Then 100 μL portions of the starter culture were transferred into 20 mL of minimal medium containing different carbon sources.
Sulfite produced during growth of M. ushuaiensis was detected using the fuchsin assay. The media were sampled prior to cell inoculation and after cells had grown to a density of OD600 ∼1, centrifuged at 8,000 × g for 10 min. The supernatant was used for the fuchsin assay. A 10 μL portion of sample was mixed with 190 μL of Fuchsin reagent, incubated for 10 min at RT, and the absorbance at 580 nm was recorded. Concentrations of sulfite were determined by reference to a standard curve constructed using Na2SO3 (200, 100, 50, 25, 12.5, 0 μM).
LC-MS assays for SQ consumption during the growth of M. ushuaiensis
The minimal medium (3 mL) containing 10 mM SQ was inoculated with M. ushuaiensis. Cultures were incubated for 77 h at 22 °C (220 rpm) with daily observations of optical density at 600 nm. 20 μL samples of culture supernatant were diluted with 380 μL of H2O and then was filtered through a 0.22 μm nylon membrane filter prior to LC-MS analysis. LC-MS was performed on an Agilent 6420 Triple Quadrupole LC/MS instrument (Agilent Technologies). LC-MS analysis was carried out on a ZIC-HILIC column (5 mm, 200 Å, 150 × 4.6 mm; Merck). The HPLC conditions were as follows: from 90% B to 70% B in 10 min, 70% B to 50% B in 20 min, and 90% B for 5 min. Solvent A was 90% 20 mM ammonium acetate and 10% acetonitrile, and solvent B was acetonitrile. The flow rate was set to 0.5 mL/min. The mass spectrometer was run in ESI negative mode. Extracted ion chromatograms (m/z (−) 242.9, the predicted mass of the SQ), exhibited the SQ peak. To monitor the consumption of SQ, serial dilutions of SQ (250, 125, 62.5, 31.2, 15.6, 0 μM) were used to establish a standard curve as a reference (See Figure S8). Peak integration was performed for SQ quantitation.
Real-time quantitative PCR analyses
RNAs were extracted from cultures 1 and 2 (∼1010 cells from each culture) using RNAprep pure Cell/Bacteria Kit (TIANGEN) and diluted to 500 ng/μL 1 μg RNA was revere transcribed in a 20 μL of reaction mixture containing 0.5 μg random primers and the GoScript Reverse Transcription System (Promega), according to the manufacturer’s recommended protocol. Obtained cDNA were frozen at −80°C for long-term storage. Prior to real-time quantitative PCR (qPCR), the reverse transcriptase was inactivated at 42°C for 15 min and then at 70°C for 15 min. In a typical qPCR reaction, 20 μL of reaction mixture contained 4 μL of 6 × diluted cDNA, 10 μM gene-specific forward and reverse primers and 1 × SYBR Green GoTaq qPCR Master Mix (Promega). qPCRs were performed on a LongGene Q2000. The primers were designed using primer3plus (http://www.primer3plus.com) (See Table S1). All reagents and consumables for the experiments were RNase-free. The data was analyzed by means of the ΔΔCt method and the values were normalized using 16S rRNA as an internal control.
MaSquF-coupled spectrophotometric activity assays for MaSqoD
In a typical assay, a 200 μL reaction mixture containing 2 μM SqoD and excess (10 μM) SquF, 0.5 mM SQ, 5 mM α-KG, 25 μM FeSO4, 50 μM ascorbic acid and 0.5 mM NADPH in 50 mM Tris-HCl pH 7.5 and 100 mM NaCl was prepared. For enzyme dose-dependent assays, the concentration of SqoD was varied from 0.25 μM to 4 μM while a fixed, saturating substrate concentration of 5 mM SQ was used. The concentration of SqoD in the following experiments was 2 μM. To determine the optimal reaction pH, the reaction buffer was replaced with 50 mM of different buffers: 2-(N-morpholino) ethanesulfonic acid (MES), pH 5.5, 6.5; Tris-HCl, pH 7.0, 7.5, 8.0; 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (CAPSO), pH 9.0, 10.0. To determine the apparent Michaelis-Menten kinetic parameters of SqoD, assays were carried out at optimal pH with varying concentrations of the substrates. The KM values for SQ was determined by keeping the concentration of α-ketoglutarate (α-KG) at 5 mM while varying the concentration of SQ from 0.25 mM to 8 mM. The KM value for α-KG was determined by keeping the concentration of SQ at 5 mM while varying the concentration of αKG from 3 μM to 200 μM). The absorbance of each assay mixture was monitored at 340 nm, at 10 s intervals. GraphPad Prism 9 was used for data analysis.
Quantification and statistical analysis
The data analysis was performed by using GraphPad Prism 9 software. The biochemistry experiments were repeated at least twice and the data were expressed as the mean ± SEM. Bar graphs and scatter blots represent the mean ± standard deviation (SD), when indicated. ANOVA was used followed by Tukey’s multiple comparison post-test comparing all pairs of conditions when multiple conditions were compared. Significant differences were indicated according to the p-values by asterisks with ∗∗∗∗ for p < 0.0001, ∗∗∗ for 0.0001 < p < 0.001, ∗∗ for 0.001 < p < 0.01, and ∗ for 0.01 < p < 0.05. Non-significant differences are indicated by n.s.
Acknowledgments
We thank the instrument analytical center of the School of Pharmaceutical Science and Technology at Tianjin University for providing the LC-MS analysis and Zhi Li for helpful discussion. This work was supported by the National Key R&D Program of China 2020YFA0907900 (Y.Z.) and the National Natural Science Foundation of China (NSFC) Distinguished Young Scholar of China Program 32125002 (Y.Z).
Author contributions
Z.Y. and L.J. conducted biochemical and molecular biology experiments; Z.Y. and Y.W. conducted the bioinformatic analyses. Z.Y., Y.W., and Y.Z. designed the experiments and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 31, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107803.
Supplemental information
Data and code availability
-
•
Source data underlying Figures 2, 3, 4, 5, 6, S3–S6, and S8 have been deposited at Mendeley Data and are publicly available as of the date of publication. Accession link is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- 2.Goddard-Borger E.D., Williams S.J. Sulfoquinovose in the biosphere: occurrence, metabolism and functions. Biochem. J. 2017;474:827–849. doi: 10.1042/bcj20160508. [DOI] [PubMed] [Google Scholar]
- 3.Benson A.A. In: Advances in Lipid Research. Paoletti R., Kritchevsky D., editors. Elsevier; 1963. The Plant Sulfolipid; pp. 387–394. [DOI] [Google Scholar]
- 4.Harwood J.L., Nicholls R.G. The plant sulpholipid-- a major component of the sulphur cycle. Biochem. Soc. Trans. 1979;7:440–447. doi: 10.1042/bst0070440. [DOI] [PubMed] [Google Scholar]
- 5.Sanda S., Leustek T., Theisen M.J., Garavito R.M., Benning C. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 2001;276:3941–3946. doi: 10.1074/jbc.M008200200. [DOI] [PubMed] [Google Scholar]
- 6.Roy A.B., Hewlins M.J.E., Ellis A.J., Harwood J.L., White G.F. Glycolytic breakdown of sulfoquinovose in bacteria: a missing link in the sulfur cycle. Appl. Environ. Microbiol. 2003;69:6434–6441. doi: 10.1128/aem.69.11.6434-6441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Y., Tong Y., Zhang Y. New mechanisms for bacterial degradation of sulfoquinovose. Biosci. Rep. 2022;42 doi: 10.1042/bsr20220314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snow A.J.D., Burchill L., Sharma M., Davies G.J., Williams S.J. Sulfoglycolysis: catabolic pathways for metabolism of sulfoquinovose. Chem. Soc. Rev. 2021;50:13628–13645. doi: 10.1039/D1CS00846C. [DOI] [PubMed] [Google Scholar]
- 9.Benson A.A., Lee R.F. The sulphoglycolytic pathway in plants. Biochem. J. 1972;128:29P–30P. doi: 10.1042/bj1280029Pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denger K., Weiss M., Felux A.K., Schneider A., Mayer C., Spiteller D., Huhn T., Cook A.M., Schleheck D. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature. 2014;507:114–117. doi: 10.1038/nature12947. [DOI] [PubMed] [Google Scholar]
- 11.Liu J., Wei Y., Ma K., An J., Liu X., Liu Y., Ang E.L., Zhao H., Zhang Y. Mechanistically diverse pathways for sulfoquinovose degradation in bacteria. ACS Catal. 2021;11:14740–14750. doi: 10.1021/acscatal.1c04321. [DOI] [Google Scholar]
- 12.Sharma M., Abayakoon P., Epa R., Jin Y., Lingford J.P., Shimada T., Nakano M., Mui J.W.Y., Ishihama A., Goddard-Borger E.D., et al. Molecular basis of sulfosugar selectivity in sulfoglycolysis. ACS Cent. Sci. 2021;7:476–487. doi: 10.1021/acscentsci.0c01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma M., Abayakoon P., Lingford J.P., Epa R., John A., Jin Y., Goddard-Borger E.D., Davies G.J., Williams S.J. Dynamic structural changes accompany the production of dihydroxypropanesulfonate by sulfolactaldehyde reductase. ACS Catal. 2020;10:2826–2836. doi: 10.1021/acscatal.9b04427. [DOI] [Google Scholar]
- 14.Felux A.-K., Spiteller D., Klebensberger J., Schleheck D. Entner–Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1. Proc. Natl. Acad. Sci. USA. 2015;112:E4298–E4305. doi: 10.1073/pnas.1507049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Epa R., Scott N.E., Skoneczny D., Sharma M., Snow A.J.D., Lingford J.P., Goddard-Borger E.D., Davies G.J., McConville M.J., Williams S.J. A sulfoglycolytic Entner-Doudoroff pathway in Rhizobium leguminosarum bv. trifolii SRDI565. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/aem.00750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frommeyer B., Fiedler A.W., Oehler S.R., Hanson B.T., Loy A., Franchini P., Spiteller D., Schleheck D. Environmental and intestinal phylum firmicutes bacteria metabolize the plant sugar sulfoquinovose via a 6-deoxy-6-sulfofructose transaldolase pathway. iScience. 2020;23 doi: 10.1016/j.isci.2020.101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Wei Y., Zhou Y., Ang E.L., Zhao H., Zhang Y. A transaldolase-dependent sulfoglycolysis pathway in Bacillus megaterium DSM 1804. Biochem. Biophys. Res. Commun. 2020;533:1109–1114. doi: 10.1016/j.bbrc.2020.09.124. [DOI] [PubMed] [Google Scholar]
- 18.Snow A.J.D., Sharma M., Abayakoon P., Williams S.J., Blaza J.N., Davies G.J. Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism. Structure. 2023;31:244–252.e4. doi: 10.1016/j.str.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Burrichter A., Denger K., Franchini P., Huhn T., Müller N., Spiteller D., Schleheck D. Anaerobic degradation of the plant sugar sulfoquinovose concomitant with H(2)S production: Escherichia coli K-12 and Desulfovibrio sp. strain DF1 as co-culture model. Front. Microbiol. 2018;9:2792. doi: 10.3389/fmicb.2018.02792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L., Chen X., Ye J., Ma X., Han Y., He Y., Tang K. Sulfoquinovose is a widespread organosulfur substrate for Roseobacter clade bacteria in the ocean. ISME J. 2023;17:393–405. doi: 10.1038/s41396-022-01353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma M., Lingford J.P., Petricevic M., Snow A.J.D., Zhang Y., Järvå M.A., Mui J.W.Y., Scott N.E., Saunders E.C., Mao R., et al. Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2116022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis H.R. Mechanism for sulfur acquisition by the alkanesulfonate monooxygenase system. Bioorg. Chem. 2011;39:178–184. doi: 10.1016/j.bioorg.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Robbins J.M., Ellis H.R. Identification of critical steps governing the two-component alkanesulfonate monooxygenase catalytic mechanism. Biochemistry. 2012;51:6378–6387. doi: 10.1021/bi300138d. [DOI] [PubMed] [Google Scholar]
- 24.Kertesz M.A. Riding the sulfur cycle – metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 2000;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 25.Gao S.-S., Naowarojna N., Cheng R., Liu X., Liu P. Recent examples of α-ketoglutarate-dependent mononuclear non-haem iron enzymes in natural product biosyntheses. Nat. Prod. Rep. 2018;35:792–837. doi: 10.1039/C7NP00067G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price J.C., Barr E.W., Hoffart L.M., Krebs C., Bollinger J.M., Jr. Kinetic dissection of the catalytic mechanism of taurine:alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry. 2005;44:8138–8147. doi: 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- 27.Zhao S., Sakai A., Zhang X., Vetting M.W., Kumar R., Hillerich B., San Francisco B., Solbiati J., Steves A., Brown S., et al. Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. Elife. 2014;3 doi: 10.7554/eLife.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan C., Fischer M., Thomas G.H. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 2011;35:68–86. doi: 10.1111/j.1574-6976.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 29.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien J.R., Schuller D.J., Yang V.S., Dillard B.D., Lanzilotta W.N. Substrate-induced conformational changes in Escherichia coli taurine/alpha-ketoglutarate dioxygenase and insight into the oligomeric structure. Biochemistry. 2003;42:5547–5554. doi: 10.1021/bi0341096. [DOI] [PubMed] [Google Scholar]
- 32.Prabagaran S.R., Suresh K., Manorama R., Delille D., Shivaji S. Marinomonas ushuaiensis sp. nov., isolated from coastal sea water in Ushuaia, Argentina, sub-Antarctica. Int. J. Syst. Evol. Microbiol. 2005;55:309–313. doi: 10.1099/ijs.0.64517-0. [DOI] [PubMed] [Google Scholar]
- 33.Ruff J., Denger K., Cook A.M. Sulphoacetaldehyde acetyltransferase yields acetyl phosphate: purification from Alcaligenes defragrans and gene clusters in taurine degradation. Biochem. J. 2003;369:275–285. doi: 10.1042/bj20021455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rein U., Gueta R., Denger K., Ruff J., Hollemeyer K., Cook A.M. Dissimilation of cysteate via 3-sulfolactate sulfo-lyase and a sulfate exporter in Paracoccus pantotrophus NKNCYSA. Microbiology. 2005;151:737–747. doi: 10.1099/mic.0.27548-0. [DOI] [PubMed] [Google Scholar]
- 35.Denger K., Smits T.H.M., Cook A.M. L-Cysteate sulpho-lyase, a widespread pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3T. Biochem. J. 2006;394:657–664. doi: 10.1042/BJ20051311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook A.M., Denger K. Dissimilation of the C2 sulfonates. Arch. Microbiol. 2002;179:1–6. doi: 10.1007/s00203-002-0497-0. [DOI] [PubMed] [Google Scholar]
- 37.Cook A.M., Denger K., Smits T.H.M. Dissimilation of C3-sulfonates. Arch. Microbiol. 2006;185:83–90. doi: 10.1007/s00203-005-0069-1. [DOI] [PubMed] [Google Scholar]
- 38.Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 39.Zallot R., Oberg N., Gerlt J.A. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry. 2019;58:4169–4182. doi: 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snow A.J.D., Sharma M., Lingford J.P., Zhang Y., Mui J.W.Y., Epa R., Goddard-Borger E.D., Williams S.J., Davies G.J. The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids. Curr. Res. Struct. Biol. 2022;4:51–58. doi: 10.1016/j.crstbi.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutton J.J., Tappel A.L., Udenfriend S. Cofactor and substrate requirements of collagen proline hydroxylase. Arch. Biochem. Biophys. 1967;118:231–240. doi: 10.1016/0003-9861(67)90302-5. [DOI] [Google Scholar]
- 43.Semenza G.L. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 44.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichhorn E., van der Ploeg J.R., Kertesz M.A., Leisinger T. Characterization of α-Ketoglutarate-dependent Taurine Dioxygenase from Escherichia coli. J. Biol. Chem. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- 46.Hulse J.D., Ellis S.R., Henderson L.M. Carnitine biosynthesis. beta-Hydroxylation of trimethyllysine by an alpha-ketoglutarate-dependent mitochondrial dioxygenase. J. Biol. Chem. 1978;253:1654–1659. doi: 10.1016/S0021-9258(17)34915-3. [DOI] [PubMed] [Google Scholar]
- 47.Knorr S., Sinn M., Galetskiy D., Williams R.M., Wang C., Müller N., Mayans O., Schleheck D., Hartig J.S. Widespread bacterial lysine degradation proceeding via glutarate and L-2-hydroxyglutarate. Nat. Commun. 2018;9:5071. doi: 10.1038/s41467-018-07563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denger K., Ruff J., Rein U., Cook A.M. Sulphoacetaldehyde sulpho-lyase (EC 4.4.1.12) from Desulfonispora thiosulfatigenes: purification, properties and primary sequence. Biochem. J. 2001;357:581–586. doi: 10.1042/bj3570581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Source data underlying Figures 2, 3, 4, 5, 6, S3–S6, and S8 have been deposited at Mendeley Data and are publicly available as of the date of publication. Accession link is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.