Summary
Mitochondrial networks remodel their connectivity, content, and subcellular localization to support optimized energy production in conditions of increased environmental or cellular stress. Microglia rely on mitochondria to respond to these stressors, however our knowledge about mitochondrial networks and their adaptations in microglia in vivo is limited. Here, we generate a mouse model that selectively labels mitochondria in microglia. We identify that mitochondrial networks are more fragmented with increased content and perinuclear localization in vitro vs. in vivo. Mitochondrial networks adapt similarly in microglia closest to the injury site after optic nerve crush. Preventing microglial UCP2 increase after injury by selective knockout induces cellular stress. This results in mitochondrial hyperfusion in male microglia, a phenotype absent in females due to circulating estrogens. Our results establish the foundation for mitochondrial network analysis of microglia in vivo, emphasizing the importance of mitochondrial-based sex effects of microglia in other pathologies.
Subject areas: Natural sciences, Biological sciences, Physiology, Neuroscience, Systems neuroscience, Sensory neuroscience
Graphical abstract

Highlights
-
•
Mitochondrial connectivity, content, and location in microglia is environment-dependent
-
•
Mitochondrial networks adapt in microglia after optic nerve crush (ONC)
-
•
Microglial UCP2KO induces mitochondrial hyperfusion in males after ONC
-
•
Circulating estrogens prevent this phenotype in UCP2KO female microglia
Natural sciences; Biological sciences; Physiology; Neuroscience; Systems neuroscience; Sensory neuroscience
Introduction
Mitochondria have been classically defined as autonomous organelles that support eukaryotic cell function by providing energy through cellular respiration.1 In recent years, this traditional view has expanded to include several mechanisms of mitochondrial-mediated cellular signaling such as, inter-organelle contacts, mitochondrial-derived vesicle trafficking and release of metabolites or mitochondrial DNA.2,3,4,5 In line with these observations, changes in cellular metabolism and mitochondrial-associated gene signatures have been frequently associated to pathological conditions, highlighting mitochondria as key players in cell signaling and disease.2
In physiological conditions, mitochondrial networks constantly remodel through the process of organelle fission and fusion, termed mitochondrial dynamics.1 Mitochondrial fusion supports matrix content exchange and/or enhanced ATP production, while fission assists redistribution and elimination of dysfunctional organelles.6 Cellular stress, such as increased reactive oxygen species (ROS), leads to imbalanced mitochondrial dynamics often resulting in a more fragmented network. This is reflected in an increased number of small-volume, spherical mitochondria7 and expedites removal of damaged mitochondria in a process termed mitophagy. To compensate for mitophagy, mitochondrial biogenesis occurs which induces protein synthesis allowing the organelles to increase in size, leading to transient alterations in cellular mitochondrial content.8 Fragmented mitochondrial networks also facilitate organelle trafficking and redistribution to different subcellular compartments, which can influence cytoplasmic and nuclear calcium loads and transcriptional activity.9,10,11 Under certain stress conditions, mitochondria can also form highly connected, hyperfused networks, which increase membrane potential and ATP production helping mitigate stress and promotes cell survival.12,13,14 Therefore, the adaptations of mitochondrial network connectivity, content, and localization provide valuable insight into cellular stress and energy demands.
Degenerative disease environments increase cellular stress, and failure to mitigate this stress results in its accumulation and cellular damage.15 This is particularly relevant in the context of immune cells, whose primary role is to detect and rapidly react to perturbations in their environment.2,16,17 Microglia, the resident immune cells of the brain, respond to environmental cues through morphological, transcriptional, and metabolic adaptations.18,19,20 Several studies have shown that mitochondria are essential to microglial surveillance and response. Mitochondrial network fragmentation is required for the metabolic shift to glycolysis in responsive microglia in vitro.21 This was corroborated in cell isolation studies where aberrant metabolic reprogramming was identified as the cause for microglial dysfunction in Alzheimer’s mouse models.20 Microglia are able to adapt to available energy sources to maintain their surveillance functions,22 however in germ-free mice where fuel sources are scarce, their mitochondrial function diminishes which is reflected in increased mitochondrial mass, ROS and reduced numbers.23 Therefore, mitochondrial network adaptations are necessary for effective microglial responses, and provide important insights into cellular stress and metabolic changes that could predict disease-related phenotypes in the neuronal environment.
However, nearly all of the evidence investigating mitochondrial adaptations are from in vitro or cell isolation studies.20,21,23,24,25 Microglia isolated from their environment quickly transition to a responsive phenotype, preventing us from deciphering whether novel insights result from the microglia within the tissue environment or from artifacts arising due to the additional stress of tissue dissociation, processing, or the primary culture environment.26,27,28 Furthermore, spatial information and environmental factors such as sex are lost in cell isolation studies, which are important contributing factors known to influence microglia.29 A major limitation in addressing mitochondria in microglia in vivo is the lack of tools to effectively visualize mitochondrial networks specifically in microglia.
Here, we generated a mouse model that selectively labels mitochondria in microglia. First, we established the differences between in vitro and in vivo mitochondrial networks. Then, we analyzed spatially isolated microglial populations in the brain using the retina as a model. Here, microglia predominately reside in inner- and outer plexiform layers (IPL and OPL, respectively), and respond to optic nerve crush (ONC) which induces axonal injury of retinal neurons distant from the retinal tissue.30,31,32 We show that after ONC, microglia have more fragmented mitochondrial networks with increased content per cell and altered localization in IPL microglia, while OPL microglia further from the dying neurons only minimally respond. Next, we manipulated cellular stress in microglia by selectively depleting uncoupling protein 2 (UCP2), a mitochondria-localized gene associated with stress mitigation and microglial function.33,34 UCP2-depleted IPL microglia responded similarly to wildtype after ONC with the exception of mitochondrial number. We uncovered a sex-specific difference in stress mitigation, where male UCP2KO microglia exhibited hyperfused mitochondrial networks after ONC. Ovariectomized UCP2KO females represented a similar mitochondrial phenotype suggesting that circulating estrogens influence this effect, and highlighting that there are sex-based differences in microglial stress mitigation.
Results
Mitochondrial networks of microglia differ between in vitro and in vivo environments
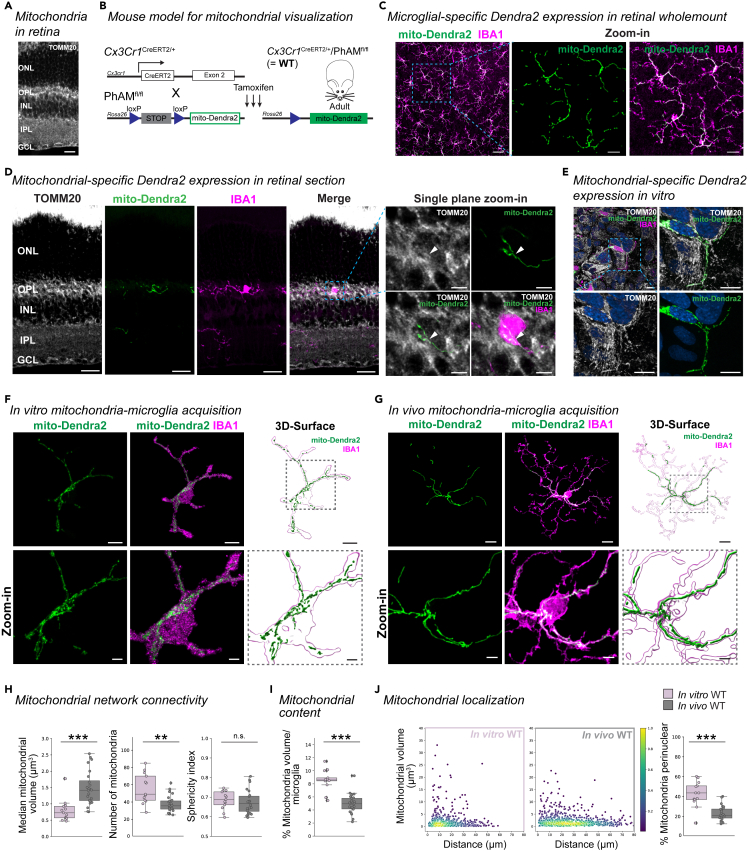
Visualizing mitochondrial networks of retinal microglia by immunostaining for markers such as TOMM20 (translocase of outer mitochondria membrane 20) results in a dense and ill-defined labeling in the OPL and IPL, where microglia predominantly reside (Figure 1A). Thus, we took advantage of the PhAMfl/fl mouse, which carries a floxed transgene encoding a mitochondrial-localized Dendra2 fluorophore.35 We crossed this mouse with the Cx3cr1CreERT2 mouse expressing tamoxifen-inducible Cre recombinase in cells of myeloid origin.36 After tamoxifen administration, Cx3cr1CreERT2/+/PhAMfl/fl mice, hereafter referred to as wildtype (WT) mice (Figure 1B), microglia labeled with IBA1 (ionized calcium-binding adaptor molecule 1) showed selective expression of the mito-Dendra2 fluorophore (Figure 1C). We verified mito-Dendra2 localization at the mitochondria of microglia by co-labeling with TOMM20 in retinal sections (Figure 1D), as well as in 4-hydroxytamoxifen treated primary mixed glial cultures prepared from WT pups (Figure 1E).
Figure 1.
Mitochondria in microglia in vivo establish networks distinct from in vitro
(A) Retinal section immunostained for TOMM20 (white). Scale bar: 20 μm. ONL, outer nuclear layer. OPL, outer plexiform layer. INL, inner nuclear layer. IPL, inner plexiform layer. GCL, ganglion cell layer.
(B) Schematic of mouse model for mitochondrial visualization with floxed mito-Dendra2 at the Rosa26 locus (PhAMfl/fl). Phamfl/fl are crossed to Cx3cr1CreERT2 for microglia-selective mito-Dendra2 expression (green) after receiving tamoxifen injection for three consecutive days. Referred to as WT throughout the study.
(C) Overview image of immunostained microglia (IBA1, magenta) in the IPL expressing the mitochondrial label (Dendra2, green) in retinal wholemounts for WT. Scale bar: 50 μm, zoom-in: 20 μm.
(D) TOMM20-immunostained retinal cross sections from WT mouse expressing mito-Dendra2 (green) and co-labeled with IBA1 (magenta). Scale bar: 20 μm. Single plane zoom-in (dashed outline) show overlap (white arrow) for mito-Dendra2 and TOMM20 (white). Scale bar: 5 μm.
(E) TOMM20 (white) and IBA1 (magenta)-immunostained primary mixed glia culture with mito-Dendra2 (green) expression from WT mouse after 4-hydroxytamoxifen treatment. Scale bar: 15 μm. Dashed outline, zoom-in single plane. Blue: Hoechst labeled nuclei. Scale bar: 15 μm.
(F and G) Immunostained microglia (IBA1, magenta) expressing mito-Dendra2 (green) in microglia in vitro from WT primary mixed glial cultures (E) or WT IPL in vivo (F). Scale bar: 10 μm. Next, 3D-surface rendering of IBA1 microglia (magenta outline) and Dendra2 mitochondria (green) with zoom-in (dashed line). Scale bar: 10 μm, zoom-in: 3 μm.
(H–J) Mitochondrial parameters. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. Each overlaid point represents data point of a single microglia. In vitro: dusty pink. In vivo: gray. (H) Mitochondrial network connectivity determined by median mitochondrial volume (left, Wilcoxon rank-sum test: p < 0.0001), number of organelles within a single cell (center, Welch’s t-test: p = 0.0033) and mean mitochondrial sphericity (right, t-test, p = 0.551017). (I) Percentage of mitochondrial volume per microglial volume. T-test: p < 0.0001.(J) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) in vitro (left, dusty pink) or in vivo (right, gray) with pseudo-colored point density. Right: Percentage of total mitochondrial volume localized within the perinuclear region (0–10 μm). Wilcoxon rank-sum test: p < 0.0001. ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. p > 0.05: not significant. See Table S1 for experimental, retina and cell numbers, statistical tests and corresponding data.
Primary microglia in mixed glial culture differ in their morphological shape compared to microglia in vivo (Figures 1F and 1G). To evaluate the mitochondrial network in both conditions, we generated 3-dimensional (3D) surfaces of both mitochondria and microglia and performed volumetric analyses (Figures 1F, 1G, and S1A, Videos S1, and S2). First, we quantified mitochondrial network connectivity for single cells in which we determined the median mitochondrial volume amongst all organelles of a cell (Figure S1B), the number of mitochondria, and the mean sphericity of mitochondria (Figure S1C). Microglia in vitro exhibited a reduced median mitochondrial volume and increased total number of mitochondria compared to in vivo (Figure 1H), indicating a more fragmented mitochondrial network, even though the sphericity did not differ. When we analyzed the mitochondrial content per cell (Figure S1D), we found that microglia in vitro had a significantly greater content compared to in vivo (Figure 1I), suggesting an imbalance of biogenesis and turnover between conditions, which can be associated with greater oxidative damage.37 To assess subcellular localization of mitochondria, we measured the distance from the centroid of each mitochondrion to the cell soma (Figure S1E) and represented the data in a scatterplot (Figure S1F). We found a greater percentage of a cell’s total mitochondrial volume was localized in the perinuclear region closer to the soma in vitro (Figures 1J and S1G). Collectively, these mitochondrial parameters demonstrate a difference between in vitro and in vivo conditions that align with a greater stress response in microglia in vitro. To substantiate this, we analyzed the expression of the endosomal-lysosomal marker CD68 (cluster of differentiation 68) in microglia (Figures S2A and S2B),which is upregulated in reactive microglia.38,39 The CD68 volume was significantly greater in vitro and coincided with increased vesicle volumes distributed along the length of processes when compared to microglia in vivo (Figures S2C and S2D). Together, this data emphasizes that microglia in vitro are distinct from microglia in vivo, not only morphologically, but also at the mitochondrial level.
Mitochondrial network alterations occur prominently in microglia proximal to dying neurons
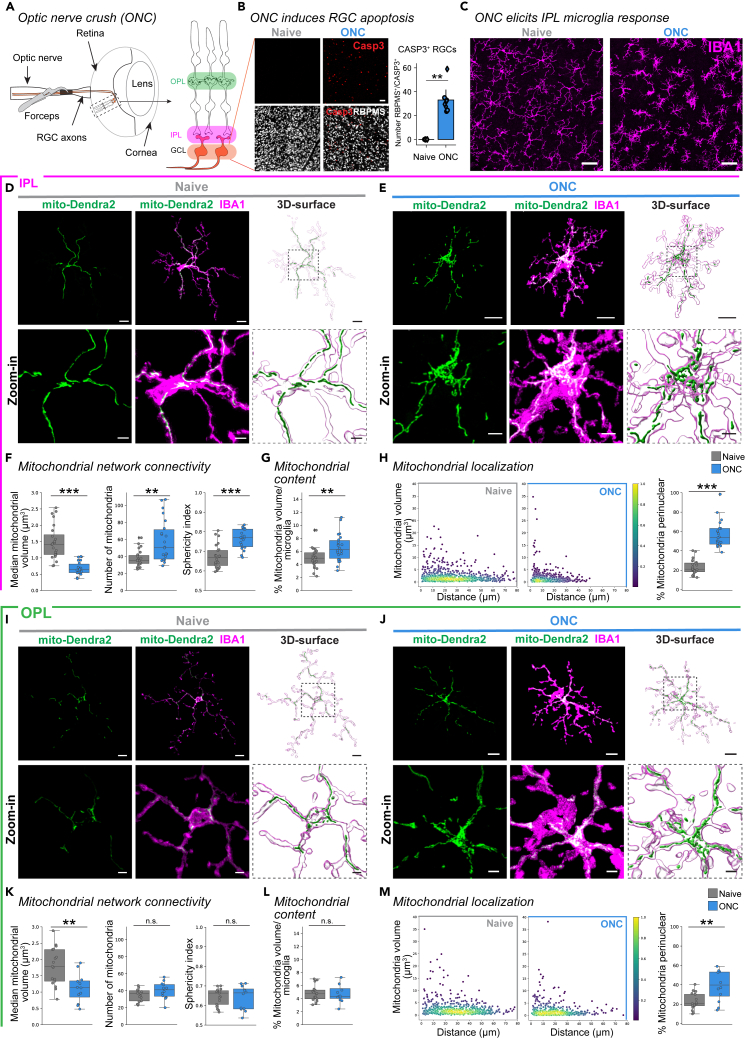
To identify how mitochondrial networks of retinal microglia in vivo adapt to stress conditions such as neuronal death, we took advantage of the optic nerve crush (ONC) model (Figure 2A).30,40 Here, a mechanical injury at the retinal ganglion cell axons posterior to the optic disc induces their apoptosis,30,41 which we confirmed by an increased number of cleaved-caspase 3+/RBPMS+-retinal ganglion cells (Figure 2B). Microglia showed morphological adaptation to these environmental changes throughout the IPL after ONC (Figure 2C). At the single cell level, microglia increased expression of CD68 (Figures S3A–S3C), where vesicles were distributed throughout the length of the processes (Figure S3D). This increase was corroborated by increased Cd68 transcript expression in FACS-sorted retinal microglia after ONC (Figure S3E). Accompanying this phenotype, microglia reduced their branching complexity, as verified by the increased number of Sholl intersections closer to the soma (Figure S3F). Next, we aligned mitochondrial network adaptations to this responsive microglial phenotype (Figures 2D and 2E). Relative to the naive environment, microglia showed significantly reduced median mitochondrial volume and an increased number of mitochondria with greater sphericity after ONC (Figure 2F). Together with increased mitochondrial content (Figure 2G), and the subcellular localization of the mitochondrial population closer to the soma (Figure 2H), this data indicates a more fragmented mitochondrial network with altered content and localization in responsive IPL microglia.
Figure 2.
Regional differences in mitochondrial network connectivity, content and localization of ONC-responsive IPL and OPL microglia
(A) Schematic of optic nerve crush (ONC). Forceps induce unilateral damage (red) to axonal projections (orange) of retinal ganglion cells (RGCs) posterior to the optic disc. Right, retinal side-view schematic. OPL, outer plexiform layer. IPL, inner plexiform layer. GCL, ganglion cell layer.
(B) Confocal images of RBPMS-immunostained naive WT retinas or 5 days after ONC WT co-labeled with cleaved caspase-3 (CASP3, red) to identify dying retinal ganglion cells (RGCs, white). Right, bar plot indicate number of caspase-3+/RBPMS+ cells. Wilcoxon rank-sum test: p = 0.0016. Scale bar: 50 μm.
(C) Overview confocal image of immunostained IPL microglia (IBA1, magenta) in naive WT (left) and 5 days after ONC WT (right) condition. Scale bar: 30 μm.
(D–M) Comparison of mitochondrial response in naive (magenta border) microglia WT (gray) and 5 days after ONC WT (blue). Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. (D–H) Mitochondrial parameters for IPL microglia. (D–E) Mito-Dendra2 (green) expression in the IBA1-immunostained IPL microglia for naive WT microglia (D) or ONC WT microglia (E). Next, corresponding 3D-surfaces. Below: Zoom-in of region of interest (dashed line) from image and 3D-surface. Scale bar: 10 μm, zoom-in: 3 μm. (F) Mitochondrial network connectivity determined by median mitochondrial volume (left, Welch’s t-test: p < 0.0001), number of organelles within a single cell (center, Wilcoxon rank-sum test: p = 0.0018) and mean mitochondrial sphericity (right, T-test: p < 0.0001). (G) Percentage of mitochondrial volume per microglial volume. T-test: p = 0.0040. (H) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in naive WT (left) or ONC WT (right) microglia. Point density, pseudo-colored. Percentage of total mitochondrial volume localized within the perinuclear region. Wilcoxon rank-sum test: p < 0.0001. (I–M) Mitochondrial parameters for OPL microglia. (I–J) Representative IBA1-immunostained OPL microglia (magenta) expressing mito-Dendra2 (green) from naive WT (I) or ONC WT microglia (J). Next, corresponding 3D-surfaces. Below: Zoom-in of region of interest (dashed line) from image and 3D-surface. Scale bar: 10 μm, zoom-in: 3 μm. (K) Mitochondrial network connectivity determined by median mitochondrial volume (left, Student’s t test: p = 0.0026), number of organelles within a single cell (center, Student’s t test: p = 0.0900) and mean mitochondrial sphericity (Student’s t test, p = 0.79389). (L) Percentage of mitochondrial volume per microglial volume. Student’s t test: p = 0.8441. (M) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in naive WT (left) or ONC WT (right) microglia. Point density, pseudo-colored. Percentage of total mitochondrial volume localized within the perinuclear region. Welch’s t-test: p = 0.0045. ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. p > 0.05: not significant. See Table S2 for retina and cell numbers, statistical tests and corresponding data.
To determine whether microglia residing more distant from the apoptotic retinal ganglion cells show a similar effect, we repeated the above analysis in microglia of the OPL. OPL microglia showed mild morphological adaptations (Figure S3G). Upon single cell analysis (Figures 2I and 2J), OPL microglia had significantly reduced median mitochondrial volume, whereas the number and the sphericity of mitochondria, and total content remained unaltered (Figures 2K and L). Similar to IPL microglia, the subcellular mitochondrial localization shifted toward the soma (Figure 2M), CD68 volume increased (Figures S3H–S3K), and the microglial branching complexity was moderately altered (Figure S3L). This data indicates that proximity to apoptotic neurons is a key determinant in the robustness of the mitochondrial-microglial response, where more distant microglia also exhibit some mitochondrial network alterations.
Selective microglial UCP2KO increases cellular and mitochondrial stress with no effect on the mitochondrial network
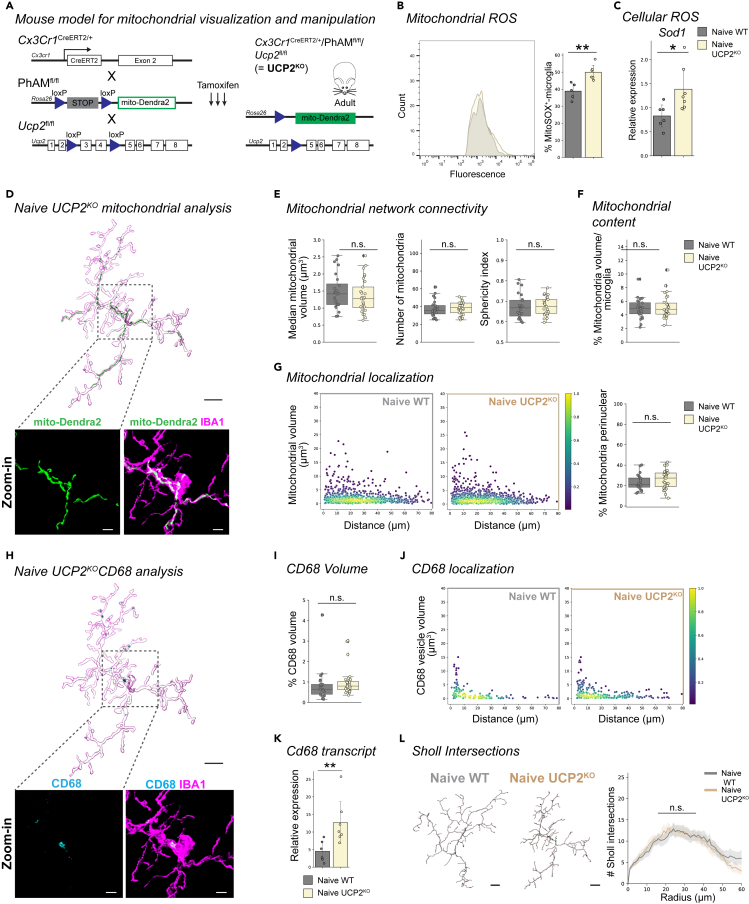
Electron leakage during oxidative phosphorylation leads to the formation of mitochondrial superoxides and reactive oxygen species (ROS). Strategies to mitigate ROS include the induction of mild uncoupling to increase the respiration rate and reduce electron leak, or detoxification of superoxides with cellular enzymes such as superoxide dismutase 1 (SOD1).42 However, these mitigation strategies are less effective in conditions of cellular damage or mitochondrial dysfunction, and can lead to ROS accumulation.43 One negative regulator of mitochondria ROS generation is uncoupling protein 2 (UCP2, Slc25a8), a mitochondrial-associated gene highly enriched in immune cells, namely retinal microglia (Figures S4A and S4B).42,44,45,46,47,48 Upregulation of Ucp2 transcript has been reported in microglia within disease environments,49,50 and we confirmed increased Ucp2 transcript expression in microglia five days after ONC (Figures S4C and S4D), making Ucp2 a candidate gene to disrupt mitochondrial function and alter ROS accumulation.
Loss of UCP2 has been reported to elicit cellular ROS accumulation.51,52 Thus, to evaluate this consequence on the mitochondrial network in retinal microglia, we generated microglia-specific UCP2KO mice by crossing the Ucp2fl/fl mouse with our mitochondrial-labeled WT mice (Cx3cr1CreERT2/+/PhAMfl/fl/Ucp2fl/fl, Figure 3A). This model, hereafter referred to as UCP2KO mice, showed microglia-selective mitochondrial labeling (Figure S4E). We confirmed successful Ucp2 knockout at both the transcript (Figure S4F) and protein level (Figures S4G–S4I) in which we performed qRT-PCR of FACS-isolated Dendra2+-retinal microglia three weeks after tamoxifen induction and Western blot analysis of microglia after 4-OHT (4-hydroxytamoxifen) treatment in primary mixed glial cultures, respectively. To determine the consequences of UCP2-knockout on microglial ROS levels, we first compared mitochondrial superoxide in retinal microglia from naive WT and UCP2KO mice using FACS-sorted MitoSOX stained cells. UCP2KO mice exhibited a significantly increased percentage of MitoSOX+-microglia (Figure 3B). This coincided with significantly enhanced gene transcript expression of the detoxifying enzyme Sod1 (Figure 3C), together indicating elevated mitochondrial and cellular ROS in UCP2KO microglia. When we compared the mitochondrial networks of UCP2KO to WT in the naive condition, we did not detect differences in mitochondrial network connectivity, content, or localization (Figures 3D–3G). Interestingly, the overall expression of CD68 was slightly elevated in UCP2KO microglia (Figures 3H–J) and aligned with increased CD68 transcript (Figure 3K), whereas the microglial morphology remained unaffected (Figure 3L). Together, this data indicates increased cellular stress in UCP2KO microglia with no direct effect on the mitochondrial network in the naive environment.
Figure 3.
Mitochondrial networks are unaffected by increased stress from microglia-selective knockout of UCP2
(A) Schematic of mouse model. Cx3cr1CreERT2/+/Phamfl/fl/Ucp2fl/fl for selective mitochondria-labeling in parallel with UCP2-knockout. Upon tamoxifen injection for three consecutive days in adult mice, Cx3cr1CreERT2-expressing microglia excise the stop cassette in Phamfl/fl and exon 3 and 4 of Ucp2 to induce selective mito-Dendra2 (green) expression and UCP2-knockout (here after referred as UCP2KO).
(B) Frequency plot of MitoSOX fluorescence from FACSed mito-Dendra2+ retinal microglia for naive WT (gray) and naive UCP2KO (tan). Corresponding bar plot of percentage of microglia that are MitoSOX+. Student’s t test: p = 0.0033.
(C) Bar plot depicting relative Sod1 transcript expression from FACSed mito-Dendra2+ microglia in naive WT or UCP2KO. Student’s t test: p = 0.0141.
(D) 3D-surface of representative IBA1-immunostained microglia (magenta) expressing mito-Dendra2 (green) from naive UCP2KO. Below: Zoom-in of region of interest (dashed line) showing confocal images of mito-Dendra2 (green) expression or co-labeled IBA1-immunostaining (magenta). Scale bar: 10 μm, zoom-in: 3 μm.
(E–G) Mitochondrial parameters. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. Each overlaid point represents data point of a single microglia. Naive WT: gray. Naive UCP2KO: tan. (E) Mitochondrial network connectivity determined by median mitochondrial volume (left, Student’s t test: p = 0.4131), number of organelles within a single cell (center, Student’s t test: p = 0.8337) and mean mitochondrial sphericity (right, Student’s t test, p = 0.8732). (F) Percentage of mitochondrial volume per microglial volume. Wilcoxon rank-sum test: p = 0.7410. (G) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in naive WT (left) or naive UCP2KO (right) microglia. Point density, pseudo-colored. Percentage of total mitochondrial volume localized within the perinuclear region. Student’s t test: p = 0.1941.
(H) 3D-surface of the IBA1-immunostained microglia (magenta) from Figure 3D co-labeled with CD68 (cyan). Below: Zoom-in of region of interest (dashed line) showing confocal images of CD68 (cyan) or co-labeled with IBA1-immunostaining (magenta). Scale bar: 10 μm, zoom-in: 3 μm.
(I) Percentage of total CD68 volume per microglia volume. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond points: outliers outside 1.5 IQRs. Wilcoxon rank-sum test: p = 0.0847.
(J) CD68 vesicle localization. Scatterplot depicting CD68 vesicle volume vs. distance from the cell soma (0, origin) of the population of vesicles in naive WT (left) and naive UCP2KO (right) conditions. Point density, pseudo-colored.
(K) Bar plot depicting relative Cd68 transcript expression from FACSed mito-Dendra2+ microglia in naive WT or naive UCP2KO. Student’s t test: 0.0062.
(L) 3D-filament tracings of microglia from naive WT (left) or naive UCP2KO (right). Scale bar: 10 μm. Line plot for mean number of Sholl intersections per radial distant from the soma (μm) with 95% confidence interval band. Linear mixed effects model: p = 0.9583. ∗p < 0.05, ∗∗p < 0.01, n.s. p > 0.05: not significant. See Table S3 for retina and cell numbers, statistical tests and corresponding data.
UCP2KO allows microglial response and mitochondrial adaptations after optic nerve crush
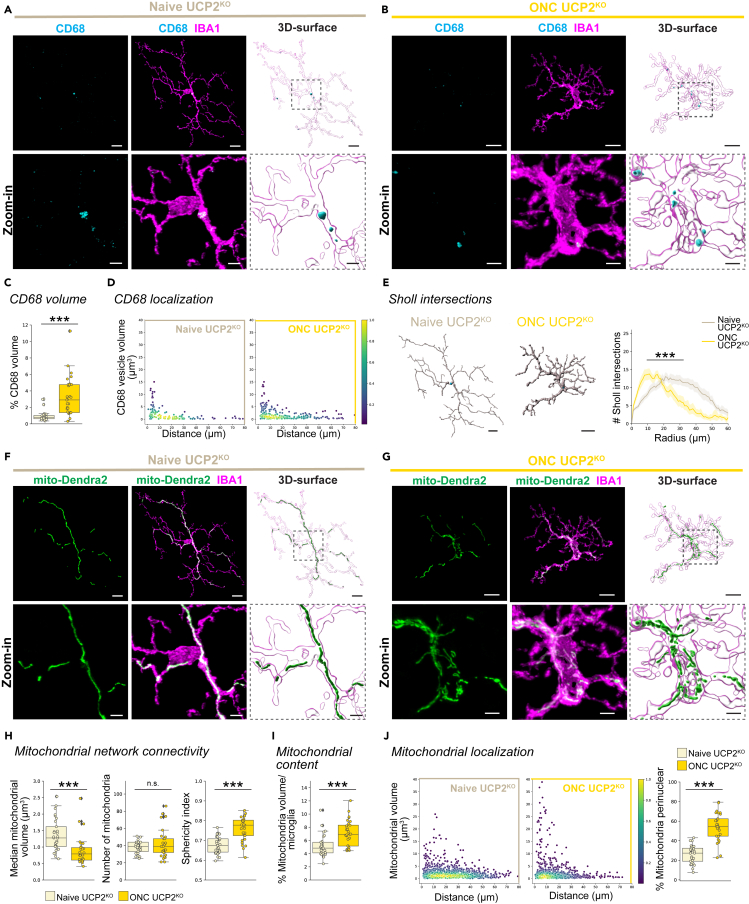
Based on previous studies reporting that UCP2KO prevents mitochondrial fission and microglial reactivity after high-fat diet,34,53 we anticipated no mitochondrial fragmentation or microglial response in the ONC injury model. Unexpectedly, ONC UCP2KO microglia showed increased CD68 volume, where vesicles were localized throughout the length of processes (Figures 4A–4D), and reduced branching complexity (Figure 4E) compared to naive UCP2KO. This aligned with the mitochondrial network becoming more fragmented as reflected in the reduced median mitochondrial volume and increased sphericity (Figures 4I–4K). At the same time, the number of mitochondrial organelles surprisingly remained similar (Figure 4H, center) even though the mitochondrial content was increased (Figure 4I). This deviation in mitochondria number did not align with the expected changes for mitochondrial connectivity seen in WT ONC microglia (Figures 2K and 2L), indicating differences in UCP2KO mitochondrial network adaptations.
Figure 4.
Mitochondrial alterations occur in responsive UCP2KO IPL microglia after ONC
(A–J) Comparison of microglial and mitochondrial response in naive UCP2KO (tan) and 5 days after ONC UCP2KO (gold). (A–B) Representative IBA1-immunostained microglia (magenta) co-labeled with CD68 (cyan) from naive UCP2KO (A) or ONC UCP2KO microglia (B). Next, corresponding 3D-surfaces. Below: Zoom-in of region of interest (dashed line) from image and 3D-surface. Scale bar: 10 μm, zoom-in: 3 μm. (C) Percentage of total CD68 volume per microglia volume. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond points: outliers outside 1.5 IQRs. Wilcoxon rank-sum test: p < 0.0001. (D) CD68 vesicle localization. Scatterplot depicting CD68 vesicle volume vs. distance from the cell soma (0, origin) of the population of vesicles in naive UCP2KO (left) and ONC UCP2KO (right, gold) conditions. Point density, pseudo-colored. (E) 3D-filament tracings of microglia from naive UCP2KO (left) or ONC UCP2KO (right). Scale bar: 10 μm. Line plot for mean number of Sholl intersections per radial distant from the soma (μm) with 95% confidence interval band. Linear mixed effects model: p < 0.0001. (F–J) Mito-Dendra2 (green) expression in the IBA1-immunostained microglia from (A–B) for naive UCP2KO microglia (F) or ONC UCP2KO microglia (G). Next, corresponding 3D-surfaces. Below: Zoom-in of region of interest (dashed line) from image and 3D-surface. Scale bar: 10 μm, zoom-in: 3 μm. (H-J) Mitochondrial parameters. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. (H) Mitochondrial network connectivity determined by median mitochondrial volume (left, Wilcoxon rank-sum test: p < 0.0001), number of organelles within a single cell (center, Wilcoxon rank-sum test: p = 0.930881) and mean mitochondrial sphericity (right, Student’s t test: p < 0.0001). (I) Percentage of mitochondrial volume per microglial volume. Wilcoxon rank-sum test: p = 0.0003. (J) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in naive UCP2KO (left) or ONC UCP2KO (right) microglia. Point density, pseudo-colored. Percentage of total mitochondrial volume localized within the perinuclear region. Student’s t test: p < 0.0001. ∗∗∗p < 0.001, n.s. p > 0.05: not significant. See Table S4 for retina and cell numbers, statistical tests and corresponding data.
Loss of UCP2KO elicits a sexually dimorphic microglial response in ONC
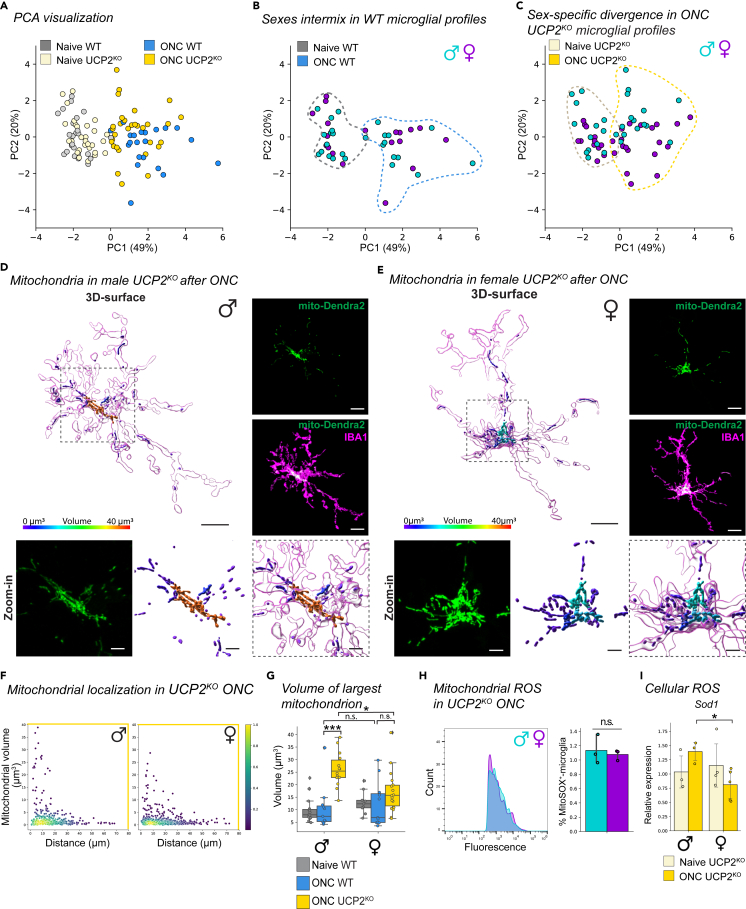
Next, we looked more closely at the relationship between conditions (naive, ONC) and genotypes (WT, UCP2KO) using principal component (PC) analysis. PC analysis allows unbiased visualization of six parameters for each analyzed microglia (Figure S1 and STAR Methods) and identification of patterns within the dataset. In naive conditions, WT and UCP2KO microglial profiles intermingled in the same PC space (Figure 5A), corroborating the results from Figure 3. Naive and ONC microglial profiles separated along the first PC for both WT and UCP2KO, aligning the with microglial-mitochondrial response in Figures 2 and 4. However, the ONC microglial profiles showed less prominent intermingling between genotypes, where UCP2KO showed greater spread along the second PC (Figure 5A). A recent study indicated that UCP2 loss in microglia results in anxiety phenotypes in male mice,52 therefore we annotated sex as an additional factor in the PC space. Whereas sexes were intermingled for WT profiles in both naive and ONC conditions (Figure 5B), UCP2KO microglial profiles diverged along the second PC only after ONC (Figure 5C), suggesting a sexually dimorphic response to ONC in UCP2KO microglia.
Figure 5.
Male UCP2KO microglia resort to mitochondrial hyperfusion for stress mitigation
(A–C) Principal component analysis (PCA) of microglia from WT and UCP2KO in naive and ONC conditions. The first two principle components (PC) are visualized. Each dot represents a single microglial profile defined by the following six parameters: microglial Sholl index, CD68 volume, median mitochondrial volume, number of mitochondria, % mitochondrial volume per microglia, % mitochondria perinuclear obtained from aforementioned genotype and condition (Figure S1). PC1 and PC2 describe 49% and 20% of the explained variance across the population, respectively. Each subpanel highlights the following comparisons: (A) Naive microglial profiles for WT (gray) and UCP2KO (tan) and ONC microglial profiles for WT (blue) and UCP2KO (gold). (B–C) Naive and ONC microglial profiles in male (turquoise) and female (purple) for WT (B) and for UCP2KO (C). Dashed line: Reference of distributions shown in (A).
(D and E) Representative 3D-surface renderings of IBA1 (magenta) and mito-Dendra2 (statistics-based volume spectral coloring) microglia from ONC-induced male UCP2KO (D) and female UCP2KO (E). Right: corresponding confocal images of IBA1-immonostained (magenta) microglia and mito-Dendra2 (green) expression. Below: Zoom-in of region of interest (dashed outline) of mito-Dendra2 (green) and corresponding 3D-surface with volume spectral coloring. Scale bar: 10 μm, zoom-in: 3 μm. Volume spectrum: 0 μm3 (blue) – 10 μm3 (red).
(F) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in ONC UCP2KO male (left) or female (right) microglia. Point density, pseudo-colored.
(G) Boxplot of largest mitochondrion per cell separated by sex. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. Naive WT: gray. ONC WT: blue. ONC UCP2KO: gold. Kruskal-Wallis test: p < 0.0001. Selected Conover’s post-hoc comparisons with Holm p-adjustment: ONC ♂WT vs. ♂UCP2KO, p < 0.0001; ONC ♂WT vs. ♀WT, p = 1.0; ONC ♂UCP2KO vs. ♀UCP2KO, p = 0.0387; ONC ♀WT vs. ♀UCP2KO, p = 0.1364.
(H) Frequency plot of MitoSOX fluorescence from FACSed mito-Dendra2+ retinal microglia for ONC UCP2KO male (turquoise) and female (purple). Corresponding bar plot of percentage of microglia that are mitoSOX+. Student’s t test: 0.6707. (I) Bar plot depicting relative Sod1 transcript expression from FACSed mito-Dendra2+ microglia in UCP2KO naive and ONC microglia. Student’s t test: p = 0.0377. ∗p < 0.05, n.s. p > 0.05: not significant. See Table S5 for retina and cell numbers, statistical tests, other post-hoc comparisons and corresponding data.
Male UCP2KO microglia induce mitochondrial hyperfusion to mitigate stress
To identify which of the six parameters differed between sexes, we first referenced the PC loadings where the highest contributing factor along the second PC was mitochondrial content (Figure S5A). When separating the volumetric analyses by sex, UCP2KO males showed significantly higher mitochondrial content per cell compared to female ONC UCP2KO conditions (Figure S5B). Additionally, male ONC UCP2KO exhibited a greater median mitochondrial volume and reduced mitochondrial number compared to female ONC UCP2KO microglia, aligning with a less fragmented network (Figure S5C). To resolve these sex-based differences, we revisited the UCP2KO microglial and mitochondrial images (Figures 5D and 5E, and Video S3) and separated the localization scatterplots. Here, we found that male ONC UCP2KO microglia had a higher frequency of mitochondria with large volumes close to the cell soma relative to female ONC UCP2KO (Figure 5F). When we compared the largest-volume mitochondrion per cell as a metric for highly connected or hyperfused mitochondria, we found that male UCP2KO had significantly greater volumes than female UCP2KO or WT after ONC (Figure 5G), indicating a sexually dimorphic hyperfused mitochondrial phenotype in UCP2KO microglia.
Previously, mitochondrial hyperfusion has been described as a transient phenotype in stress-induced environments that protects cellular and mitochondrial health.12,13 To investigate whether the observed hyperfused mitochondrial networks occurred in response to differences in ROS levels between sexes, we quantified MitoSOX+-microglia after ONC in UCP2KO for each sex. Males and females showed a comparable level of mitochondrial superoxide (Figure 5H), suggesting a similar production of intrinsic mitochondrial ROS. Interestingly, we detected opposing trends in Sod1 transcript expression in UCP2KO microglia after ONC, where males had significantly greater Sod1 expression compared to females after ONC (Figure 5I), indicating differences between sexes in the strategy for superoxide detoxification. This difference was more pronounced in WT microglia after ONC, where only males significantly increase Sod1, yet the production of mitochondrial superoxide remained similar in females (Figures S5D and S5E), suggesting sex-based differences in microglial ROS mitigation.
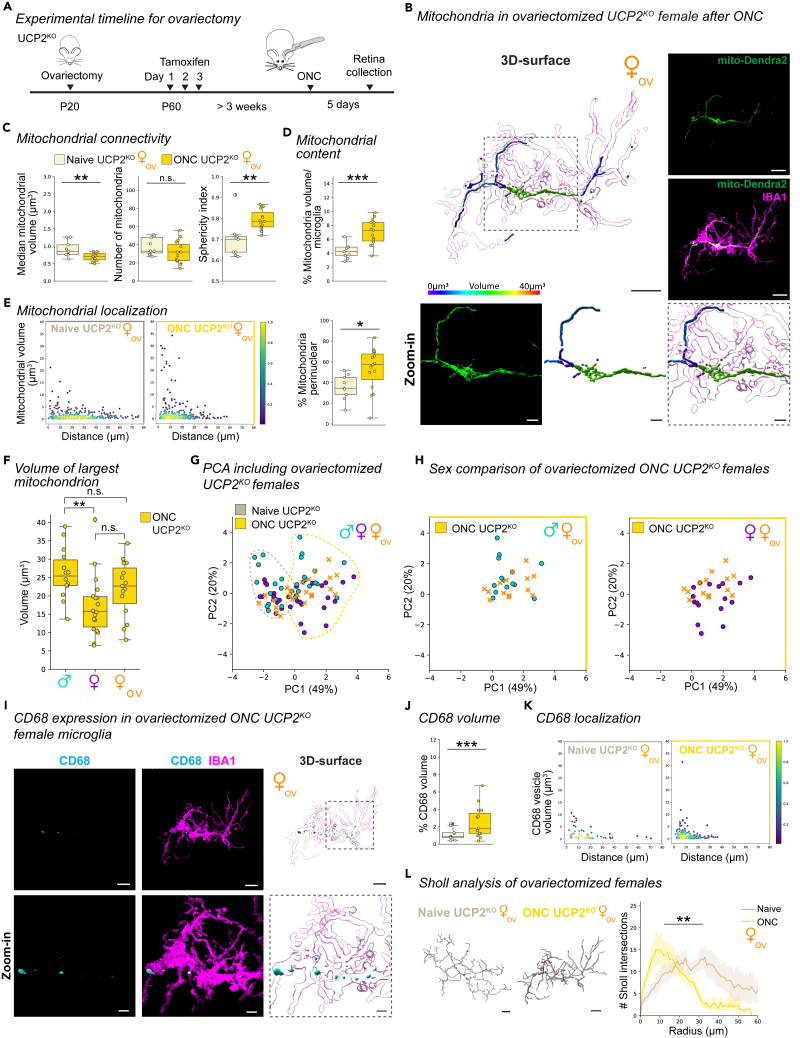
Ovariectomized female UCP2KO microglia evoke a male mitochondrial phenotype after optic nerve crush
Estrogens have been implicated to reduce oxidative stress and play a protective role in autoimmune and neurodegenerative conditions.54,55,56 Furthermore, both microglia and mitochondria express estrogen receptors.57,58 Since females did not implement stress-mitigation strategies via mitochondrial hyperfusion (Figures 5E–5G) or increased Sod1 transcription (Figures 5I and S5D), we evaluated whether circulating estrogens contribute to the sexually divergent mitochondrial phenotype in UCP2KO after ONC. Thus, we repeated the experiment with ovariectomized UCP2KO female mice (Figure 6A) and quantified the mitochondrial networks in naive and ONC (Figure 6B). The mitochondrial connectivity in ovariectomized UCP2KO female microglia exhibited decreased median mitochondrial volume, no change in mitochondria number, and significantly increased mitochondrial sphericity (Figure 6C). Furthermore, the mitochondrial content and their percentage in the perinuclear region increased after ONC (Figures 6D and 6E), together indicating mitochondrial adaptations to ONC in ovariectomized UCP2KO females. Remarkably, the mitochondrial distribution in ovariectomized ONC UCP2KO females reflected a similar increase in frequency of large-volume organelles close to the microglia soma (Figure 6E) as observed for UCP2KO males (Figure 5F). When we analyzed the largest-volume mitochondrion per cell, UCP2KO ovariectomized females had similarly large volumes as UCP2KO males (Figure 6F).
Figure 6.
Ovariectomized female UCP2KO microglia exhibit mitochondrial hyperfusion phenotype after ONC
(A) Experimental timeline. Ovariectomy performed at postnatal day 20 (P20) and three consecutive tamoxifen injections were administered daily for three days starting at P60. Optic nerve crush (ONC) was performed at least 3 weeks after tamoxifen injection and retinas collected five days after ONC.
(B) Representative 3D-surface renderings of IBA1 (magenta) and mito-Dendra2 (statistics-based volume spectral coloring) from ONC-induced ovariectomized female UCP2KO microglia. Right: corresponding confocal images of IBA1-immonostained (magenta) and mito-Dendra2 (green) expression. Below: Zoom-in of region of interest (dashed outline) of mito-Dendra2 (green) and corresponding 3D-surface with volume spectral coloring. Scale bar: 10 μm, zoom-in: 3 μm. Volume spectrum: 0 μm3 (blue) – 10 μm3 (red).
(C–G) Comparison of mitochondrial response in ovariectomized females. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. Naive UCP2KO: tan. ONC UCP2KO: gold.
(C) Mitochondrial network connectivity determined by median mitochondrial volume (left, Student’s t test: p = 0.0046), number of organelles within a single cell (center, Student’s t test: p = 0.1939) and mean mitochondrial sphericity (right, Wilcoxon rank-sum test: p = 0.0024).
(D) Percentage of mitochondrial volume per microglial volume. Student’s t test: p = 0.0004.
(E) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in microglia. Point density, pseudo-colored. Percentage of total mitochondrial volume localized within the perinuclear region. Student’s t test: p = 0.0320.
(F) Largest mitochondrion per cell separated by sex. One-way ANOVA: p = 0.0088. Selected Tukey’s post-hoc comparisons: ONC ♂UCP2KO vs. ONC ♀UCP2KO, p = 0.0065; ONC ♂UCP2KO vs. ONC ♀ovUCP2KO, p = 0.3297, ONC ♀UCP2KO vs. ONC ♀ovUCP2KO, p = 0.1938.
(G) Principal component analysis (PCA) of microglia from male, female and ovariectomized UCP2KO microglia in naive and ONC conditions. The first two principle components (PC) are visualized. Naive and ONC UCP2KO microglial profiles in male (turquoise), female (purple) and ovariectomized females (orange x-marker). Dashed outline: Reference of distributions shown in (Figure 5A).
(H) Comparison of ovariectomized ONC UCP2KO female (orange x-marker) microglial profiles with male (turquoise, left) or female (purple, right) ONC UCP2KO microglial profiles.
(I) Ovariectomized female ONC UCP2KO IBA1-immunostained microglia (magenta) from Figure 6B co-labeled with CD68 (cyan) and corresponding 3D-surfaces. Below: Zoom-in of region of interest (dashed line) from image and 3D-surface. Scale bar: 10 μm, zoom-in: 3 μm.
(J) Percentage of total CD68 volume per microglia volume in WT and UCP2KO ovariectomized females in naive and ONC conditions. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond points: outliers outside 1.5 IQRs. Wilcoxon rank-sum test: p < 0.0001.
(K) CD68 vesicle localization. Scatterplot depicting CD68 vesicle volume vs. distance from the cell soma (0, origin) of the population of vesicles from ovariectomized females in UCP2KO naive (left) and UCP2KO ONC (right) conditions. Point density, pseudo-colored.
(L) 3D-filament tracings of microglia from UCP2KO naive (left) or ONC (right). Scale bar: 10 μm. Line plot for mean number of Sholl intersections per radial distant from the soma (μm) with 95% confidence interval band. Linear mixed effects model: p = 0.0018. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. p > 0.05: not significant. See Table S6 for retina and cell numbers, statistical tests and corresponding data.
To substantiate that ovariectomized ONC UCP2KO females exhibit a mitochondrial phenotype similar to males, we represented all UCP2KO microglial profiles in the PC space. As anticipated, the ovariectomized UCP2KO females separated along the first PC into the microglial profiles for the respective naive or ONC condition (Figure 6G). In the ONC condition, ovariectomized UCP2KO females trended toward males along the second PC (Figure 6H). Aligning with the ONC-responsive mitochondrial phenotype, ovariectomized UCP2KO females showed increased CD68 volume with greater distribution and reduced branching complexity (Figures 6I–6L). Together, this data illustrates that circulating estrogens play a role in maintaining mitochondrial networks in female microglia, and are a contributing factor to the sexually divergent stress-mitigation strategies in UCP2KO microglia.
Discussion
Our results provide a new perspective of mitochondrial networks in microglia in vivo, and demonstrate their adaptation to an injury environment and increased cellular stress. Importantly, we established sex-based differences in microglial stress mitigation strategies, which we uncovered at the mitochondrial network level upon UCP2 knockout.
Extensive in vitro studies have shown that cellular energy demands of immune cells critically depend on balanced mitochondrial networks equilibrated by mitochondrial fission and fusion.16 Insight into these dynamics in microglia in vivo are mostly unknown due to technical limitations. Tissue-wide mitochondrial immunostaining requires advanced image segmentation techniques to decipher which mitochondria are within or outside the microglia.23 This necessitates high resolution images and experience in how to determine cell edges to avoid user-biases.59 In the retina, we performed high resolution imaging of TOMM20 immunostaining and were challenged to perform image segmentation of microglial mitochondria for both the retina (Figure 1D), and in 2D-mixed primary glial culture (Figure 1E). In contrast, our mouse model allowed clear image segmentation of microglial mitochondria (Figures 1C, 1F, and 1G). Alternative techniques such as electron microscopy can be used to assess cross-sectional mitochondrial areas,34,53 however resolving the entire mitochondrial network of an intact microglia would require time- and labor-intensive serial sectioning followed by image segmentation.60 Our mouse model visualizes the mitochondrial network of an entire microglia in vivo and allowed us to resolve elongated, tubular organelles in naive condition, which was absent in vitro (Figures 1F and 1G).21,24 Our results in ONC-responsive microglia in vivo corroborated a study reporting increased mitochondrial mass in responsive microglia isolated from brain tissue,23 indicating that results in vivo are achievable without microglial isolation and importantly, maintain the spatial and morphological information of the cell. To this end, we identified regional differences in mitochondrial network adaptations between microglia localized in the IPL and OPL after ONC, which are not attainable in cell-dissociation or in vitro studies (Figure 2). Together, this emphasizes that our mouse model provides a valuable strategy for assessing the mitochondrial network of microglia in a region-specific manner without cell-isolation.
Mitochondrial networks are most commonly assessed using connectivity parameters, including organelle volume, number, and sphericity.61 We found that these three parameters reliably identified fragmented networks in ONC-responsive IPL microglia (Figure 2), which was also the case when two of the three parameters were altered, highlighting the importance of using more than one metric to evaluate mitochondrial connectivity in the 3D environment. As an additional parameter, we also assessed the subcellular localization of each mitochondrion within individual microglia. Previous studies have shown that responsive microglia enhance their transcriptional activity,62 a cellular feature facilitated by perinuclear localization of mitochondria.10 We consistently detected an increased fraction of the mitochondria in the perinuclear region across genotypes and sexes after ONC (Figures 2, 4, 6, and S3). Interestingly, this localization was one of the only two mitochondrial parameters that significantly changed in OPL microglia (Figure 2M). The second parameter was decreased median mitochondrial volume (Figure 2K), which is indicative of mitochondrial fragmentation. Thus, both parameters suggest initial mitochondrial adaptations in microglia after ONC in the OPL niche more distant from the injury site. The combination of subcellular localization and the mitochondrial content per cell were critical to identify the hyperfused mitochondrial network (Figure 5). Hyperfused mitochondrial networks have been described in vitro as either a stress-induced pro-survival mechanism in conditions of nutrient deprivation or as a result of non-functional fission machinery.12,13,63,64,65,66 We found that this phenotype also occurs in microglia in vivo, which we identified with significant increases in large-volume mitochondria (Figures 5F–5H). This metric was most informative for identifying hyperfusion in microglia since the unique, 3D morphology of microglia makes it difficult to use a subjective classification method without quantification to identify hyperfused networks, which has been the common methodology for defining mitochondrial hyperfusion in in vitro studies.12,13,63
Our approach to evaluate the effects of increased cellular stress on mitochondrial networks in vivo was to endogenously manipulate stress levels through selective-knockout of UCP2, a negative regulator of ROS. UCP2KO microglia showed increased intrinsic stress and CD68 expression in the naive environment (Figures 3B, 3C, and 3I–3K), aligning with previous literature.34,52 Although these stress levels were increased, we did not detect any significant mitochondrial network alterations in the naive environment (Figure 3). It is possible that mitochondrial adaptations occurred earlier after knockout. However, our model limited us to a three week timepoint after UCP2-depletion when circulating Cx3Cr1+-monocytes have repopulated.67 This ensures that UCP2-knockout and mitochondrial-labeling is restricted to the resident microglia and thus confirms that our mitochondrial analysis is exclusive to this population in the event of monocyte infiltration after ONC injury. Nevertheless, we did detect mitochondrial fragmentation and a microglial response in the UCP2KO ONC environment (Figures 4I and 4J–4L), which was surprising since previous studies reported that UCP2 loss prevented these changes in the hypothalamus by assessing cross-sectional area and number of mitochondria in electron microscopy preparations.34,53 On the other hand, Kim et al.34 reported the prevention of mitochondrial fragmentation only in male mice, while females were largely unaffected in diet-induced obesity paradigms and their mitochondrial phenotypes were not reported. Thus, our results align with these previous reports and refine that UCP2 loss affects mitochondrial networks in a sex-specific manner, such that UCP2KO induces hyperfusion in male microglia after ONC, while female UCP2KO microglia still exhibit mitochondrial fragmentation.
Since male and female microglia responded differently to increased stress via UCP2KO (Figures 5 and S5), we suspect that estrogens may explain the difference in the female response. Circulating estrogens provide a protective role for neurons under oxidative stress or in injury conditions55,68 and enhance antioxidant gene expression in female mitochondria.69,70 Indeed, in absence of circulating estrogens, both male and ovariectomized female microglia rely on the same mechanism of mitochondrial hyperfusion to mitigate excess stress from UCP2KO 12,13. On the other hand, estrogen may not be the only mechanism. In the absence of circulating estrogens in vitro, female but not male neurons were shown to utilize lipids as a pro-survival fuel source,71 and in a separate study, it was shown that female microglia support protection from diet-induced obesity by an estrogen-independent increase in CX3CR1 signaling.72 This suggests that sex differences in microglial stress responses may be driven by a combination of circulating estrogens and dimorphisms in transcriptional programs.57,58,73,74
In conclusion, our study highlights the substantial importance of sex-mediated effects in microglia which are reflected in the mitochondrial network, and provides a foundation for mitochondrial network analysis of microglia in vivo.
Limitations of the study
Our mouse model allows microglia-specific labeling of the mitochondria, however we recognize the limitation requiring tamoxifen administration to both control and experimental cohorts for induced expression of the mitochondrial tag. Long-term tamoxifen treatment can result in human retinopathy75 or provide protection in retinal degenerative mouse models,76 however we did not detect any effects in our model with three consecutive days of tamoxifen administration combined with a 3 weeks waiting period before analysis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-IBA1, 1:500 | GeneTex | Cat#: GTX100042; RRID: AB_1240434 |
| Goat anti-IBA1, IHC: 1:250, WB: 1μg/ml | Abcam | Cat#: ab 5076; RRID: AB_2224402 |
| Rat anti-CD68, 1:500 | Bio-Rad | Cat#: MCA1957; RRID: AB_322219 |
| Guinea pig anti-RBPMS, 1:200 | PhosphoSolutions | Cat#: 1832-RBPMS; RRID: AB_2492226 |
| Rabbit anti-RBPMS, 1:200 | Abcam | Cat#: ab194213; RRID: AB_2920590 |
| Rabbit anti- Cleaved caspase 3, 1:400 | Cell Signaling Technology | Cat#: 9661S; RRID: AB_2341188 |
| UCP2, 1:1000 | Pohl Lab, purified at Pineda | Cat#: NT-Tier2 |
| SDHA, 1:5000 | Abcam | Cat#: ab14715; RRID: AB_301433 |
| β-actin, 1:5000 | Sigma | Cat#: A5441; RRID: AB_476744 |
| Goat anti-rabbit IgG (H+L) Alexa Fluor 568, 1:2000 | ThermoFisher | Cat#: A11036; RRID: AB_10563566 |
| Goat anti- rat IgG (H+L) Alexa Fluor 647, 1:2000 | ThermoFisher | Cat#: A21247; RRID: AB_141778 |
| Donkey Anti-Guinea Pig IgG (H+L) Alexa Fluor 647, 1:1000 | Jackson ImmunoResearch | Cat#: 706-605-148; RRID: AB_2340476 |
| Donkey anti-rabbit IgG (H+L) Alexa Fluor 568, 1:2000 | ThermoFisher | Cat#: A10042; RRID: AB_2534017 |
| Donkey anti-goat IgG (H+L), 1:2000 | ThermoFisher | Cat#: A11057; RRID: AB_2534104 |
| Donkey anti-goat Alexa Fluor 568 IgG (H+L) Alexa Fluor 488 , 1:2000 | ThermoFisher | Cat#: A11055; RRID: AB_2534102 |
| Donkey anti-rabbit IgG (H+L) Alexa Fluor 488, 1:2000 | ThermoFisher | Cat#: A21206; RRID: AB_2535792 |
| Donkey anti-Rat IgG (H+L) Antibody DyLight 650, 1:2000 | ThermoFisher | Cat#: SA5-10029; RRID: AB_2556609 |
| Donkey anti-Rat IgG (H+L) Alexa Fluor 488, 1:2000 | ThermoFisher | Cat#: A21208; RRID: AB_2535794 |
| Donkey anti-rabbit IgG (H+L) Alexa Fluor 647, 1:2000 | ThermoFisher | Cat#: A31573; RRID: AB_2536183 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat#: T5648-5G |
| Proparacaine hydrochloride 0.5% | Ursapharm Arzneimittel GmbH | Cat#: 02029723 |
| Meloxacam | Boehringer Ingelheim | Cat#: KPOEH3R |
| Metamizol | Sanofi Avenis | Cat#Ay005 |
| 4-hydroxy tamoxifen | Sigma-Aldrich | Cat#: SML1666-1ML |
| Critical commercial assays | ||
| NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module | New England BioLabs | Cat#: E6421L |
| Luna® Universal qPCR Master Mix | New England BioLabs | Cat#: M3003L |
| Experimental models: organisms/strains | ||
| Mouse: B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J | The Jackson Laboratory | JAX: #020940 |
| Mouse: B6;129S-Gt(ROSA)26Sortm1(CAG-COX8A/Dendra2)Dcc/J | The Jackson Laboratory | JAX: #018385 |
| Mouse: B6;129S-Ucp2tm2.1Lowl/J | The Jackson Laboratory | JAX: #022394 |
| Oligonucleotides | ||
| Oligonucleotides for RT-qPCR | See Table S11 | N/A |
| Software and algorithms | ||
| Imaris v.9.1-9.3 | http://www.bitplane.com/imaris/imaris | RRID:SCR_007370 |
| FlowJo ™ v10.8 | BD Life Sciences | RRID: SCR_008520 |
| SVI Huygens Professional v21.10 | https://svi.nl/HomePage | RRID: SCR_014237 |
| R (version 4.1.0) | https://www.r-project.org/ | RRID:SCR_001905 |
| ggplot2 (version 3.0.0) | Wickham, 2016 | RRID:SCR_014601 |
| lme4 (version 1.1-17) | Bates et al.,93 | RRID:SCR_015654 |
| Python (Version 3.7) | https://www.python.org/ | RRID: SCR_008394 |
| MatPlotLib (Version 3.4.1) | https://matplotlib.org | RRID: SCR_008624 |
| statsmodels (Version 0.13.2) | http://www.statsmodels.org/ | RRID:SCR_016074 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sandra Siegert (ssiegert@ist.ac.at).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Animals
Animal housing and procedures were approved by the “Bundesministerium für Wissenschaft, Forschung und Wirtschaft (bmwfw) Tierversuchsgesetz 2012, BGBI. I Nr. 114/2012 (TVG 2012) under the number GZ: 2021-0.607.460. Mice were housed in a 12 hour light-dark cycle in individually ventilated cages in a controlled environment (room temperature 22 ± 1 °C; relative humidity 55 ± 10 %) with access to standard diet (rat/mouse maintenance diet (V1534-300) or mouse breeding diet (V1124-300), ssniff Spezialitäten GmbH) and autoclaved water provided ad libitum in the ISTA Preclinical facility. Founder mouse strains were obtained from Jackson Laboratories and backcrossed to C57Bl6/J (#000664) background for at least 10 generations: Cx3cr1CreERT2 (#020940), PhAMfl/fl (#018385,35), Ucp2fl/fl (#022394,77). Throughout the manuscript WT refers to Cx3cr1CreERT2/+/PhAMfl/fl, while UCP2KO refers to Cx3cr1CreERT2/+/PhAMfl/fl/Ucp2fl/fl. In addition to our confirmation of the mito-Dendra2 localization at the mitochondria (Figures 1D and 1E), the original study verifies this localization in the mouse model.35 For UCP2 protein knockout primary microglia culture experiments (Figures S4G–S4I) mice did not contain the Dendra2 mitochondria label, and instead were Cx3cr1CreERT2/+ and Cx3cr1CreERT2/+/UCP2fl/fl. Experiments used male and female adult mice between 2-5 months of age.
Primary glial culture
Mixed glia culture were prepared from the published protocol by Bronstein et al.78 Cortices from 3-5 murine pups aged P0-P2 of mixed sex were dissected under a sterile ventilation hood in ice-cold Hank’s buffered saline (ThermoFisher, #14025-050) and digested in 0.05% (v/v) Trypsin + 1X EDTA (ThermoFisher, #25300-054) for 15 minutes at 37°C. The trypsinization was stopped with the addition of serum-containing medium (DMEM, 10% (v/v) heat-inactivated FBS, 1% (v/v) Penicillin/Streptomycin, 1% (v/v) Non-essential amino acids (Sigma-Aldrich, M7145-100ML)). Cells were pelleted at 500xg for 5 min, washed one time with serum-containing medium, then passed through a 40μm cell strainer (Szabo-Scandic, #352340). For Western blotting, the resulting cell suspension was plated onto T75 cell culture flasks (TPP, #Z707503) and maintained at 37°C and 5% CO2 in a humidified incubator. The culture medium was replaced at day 3 and day 10.
Method details
Tamoxifen administration for Cre-induced recombination
Tamoxifen (Sigma Aldrich, T5648-5G) was dissolved in corn oil (Sigma Aldrich, C8267-500ML) at a concentration of 20 mg/mL and sonicated for 40 minutes. Freshly prepared tamoxifen was administered via intraperitoneal injection daily (150mg/kg) for three days. The animals did not show any abnormal behavior or discomfort after gene knockout. All experimental groups (WT and UCP2KO, naïve and ONC) were tamoxifen-injected for two reasons; first, expression of the Dendra2 mitochondria label is only possible after Cre recombinase excision of the stop cassette and second, Cx3Cr1CreERT2 reporter lines have been shown to include some ‘leakiness’, i.e. tamoxifen-independent Cre recombination.79 Cx3Cr1CreERT2 mice also express CreERT2 in monocyte populations, therefore to ensure no interference from UCP2KO circulating monocytes, ONC experiments were performed at least three weeks after tamoxifen induction when monocyte populations have turned over.67 This also provided sufficient time for adaptation of microglia to the UCP2 loss conditions and clearance of all tamoxifen metabolites.80
Optic nerve crush (ONC) procedure
Mice were anesthetized in an induction chamber with 5% (v/v) isoflurane (Zoetis) supplied with oxygen at a flow rate of 0.6 L/min. After lack of a foot pinch reflex, mice were maintained at 2.5% (v/v) isoflurane applied through a nose cone while on a heating pad to maintain body temperature at 37°C. Proparacaine hydrochloride 0.5% ophthalmic eye drops (Ursapharm Arzneimittel GmbH) were applied to numb the eyes, and subcutaneous injection of 5mg/kg Metacam alleviated pain (Meloxacam, Boehringer Ingelheim). The lateral canthus was de-vascularized by clamping with a hemostat (Fine Science Tools) for 10 seconds. Using a Leica dissection microscope, a lateral canthotomy allowed visualization of the posterior pole. While firmly holding the conjunctiva with a jeweler forceps, the conjunctiva was cut perpendicular to the posterior pole. The surrounding muscle was carefully dissected as to not puncture the vascular plexus. The optic nerve was pinched 1mm from the posterior pole for 4 seconds using a curved N7 self-closing forceps (Dumont). Triple antibiotic ointment was applied to the eye directly after the surgery to prevent infection.
Ovariectomy
Cx3cr1CreERT2/+/PhAMfl/fl or Cx3cr1CreERT2/+/PhAMfl/fl/Ucp2fl/fl prepubescent postnatal day 20 female mice were anesthetized using 5% (v/v) isofluorane supplemented with oxygen at a flow rate of 0.6L/min in an induction chamber.81 Mice were maintained in 2% (v/v) isoflurane with the same flow rate after absence of a foot pinch reflex. Above the lumbar spine, the skin was exposed by shaving the fur using an electric razor, then sterilized with 70% (v/v) ethanol. A 1cm midline incision on the lower back allowed gentle dissection of the subcutaneous tissue to expose the muscular fascia and ovarian fat pad. A small incision into the peritoneal cavity for entry exposed the fallopian tube, which was used as a guide to identify the ovary. The ovary was removed via cauterization of the oviduct and blood vessels to prevent bleeding. The muscular fascia was sutured after the fallopian tube was replaced into the peritoneal cavity. After repeating the procedure to remove the contralateral ovary, the midline incision of the skin was sutured. Mice received subcutaneous injection of Metamizol (Sanofi Avenis, 200 mg/kg) during the surgical procedure and meloxicam (Boehringer-Ingelheim, 5mg/kg) to alleviate pain.
Primary microglia isolation
Two T75 flasks containing mixed primary glia cultures prepared in tandem were treated with 1μM 4-hydroxy tamoxifen (Sigma-Aldrich, SML1666-1ML) or vehicle at day 10. Three days later, microglia were isolated using mild trypsinization (0.8mM CaCl2 in 1xTrypsin-EDTA, ThermoFisher #25300-054) is added to the flask after a washing step with pre-warmed 1X PBS to remove inhibiting serum, then incubate at 37°C for 10-20 minutes, or until the non-microglial cells have just lifted from the flask forming a floating layer. The floating cells were collected with the supernatant, spun down at 10,000xg, washed once with 1X PBS, pelleted and snap-frozen. The microglia remaining on the flask were overlaid with 5ml 1X PBS, removed using a cell scraper, pelleted, and snap-frozen.
Western blotting
Total cellular protein isolation, production of recombinant UCP2 in E. coli, and western blot (WB) was performed as described previously.46,82 For WB analyses 10 μg of total protein were loaded on the SDS-gel, 1 ng of recombinant UCP2 was used as positive control for anti-UCP2 antibody detection. To ensure blotting homogeneity to nitrocellulose membranes, Ponceau S staining was performed. To relate UCP2 expression to loading control, the membranes were stripped, re-blocked and incubated with respective antibodies. The affinity purified antibody against the N-terminal sequence of mouse UCP2 (VGFKATDVPPTATVKF) was designed and evaluated in the Pohl laboratory.46 Peptide synthesis, rabbit immunization and affinity purification were performed by PINEDA Antibody-service GmbH (Berlin, Germany). All other antibody source and concentrations are provided in the key resources table. Recombinant murine UCP2 used as positive control was generated in the Pohl laboratory.83 Briefly, E. coli Rosetta DE3 strain (Merck) were transformed with murine UCP2 cDNA expression plasmids and protein was isolated after induction, high-pressure homogenization, and centrifugation. 1 mg of isolated inclusion bodies were solubilized in a TE/G buffer (2% N-lauroylsarcosine, 1.3% Triton X-114, 0.3% n-octylpolyoxyethylene, 1 mM DTT, and GTP at pH 7.5), with gradual addition of 50 mg lipid mixture (DOPC:DOPE:CL; 45:45:10 mol%). The mixture was dialyzed in buffer (50 mM Na2SO4, 10 mM MES, 10 mM Tris, and 0.6 mM EGTA at pH 7.34) after a concentration step. Bio-Beads SM-2 (Bio-Rad) application removed non-ionic detergents and hydroxyapatite columns removed unfolded and aggregated proteins from the dialysate. Micro BCA Protein Assay Kit (Thermo Fisher Scientific) was used for determining protein concentration.
Retina sample preparation for fluorescence-activated cell sorting (FACS)
Adult animals expressing the mito-Dendra2 tag were anesthetized with isoflurane and decapitated. Retinas were immediately explanted and dissected in 1X PBS on ice. Each retina was transferred to a 1.5ml Eppendorf low-adhesion tube filled with the 800μl digestion buffer (1:8:1 Cysteine/EDTA solution (2.5mM Cysteine, 0.5mM EDTA (ethylenediaminetetraacetic acid) in HBSS, 10mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) in HBSS, and 10mg/ml Papain, Roche #10108014001), incubated at 37°C for 10 minutes, then centrifuged for 2.5 min at 240xg. Samples were transferred on ice, the supernatant was discarded and 800μl 1mM EDTA in HBSS + 2% (v/v) FBS added. After two washes with 1mM EDTA in HBSS + 2% (v/v) FBS, the digested tissue was triturated 10-15 times with a pulled glass pipette, then filtered through a 70 μm strainer. For the measurement of mitochondrial reactive oxygen species, 5 μM MitoSox (ThermoFisher, #M36008) in HBSS was added to the digested tissue and pipetted to break apart the tissue prior to a 10 minute incubation at 37°C, then the sample was centrifuged for 3 minutes at 304xg at 4°C. Samples were carefully washed two times with 1mM EDTA in HBSS + 2% (v/v) FBS, triturated 10-15 times with a pulled glass pipette, then filtered through a 70 μm strainer.
Fluorescent activated cell sorting (FACS)
FACS was performed using a SONY SH800SFP equipped with a 100 μm nozzle and sorting speed of approximately 8,000 events per second in normal purity mode. Microglia were gated from the population by setting the forward and side scatter and by the fluorescence of the mito-Dendra2 tag. From each retina, 100 microglia were sorted and immediately collected in one well of a 96-well plate (Eppendorf) filled with 5 μl cold lysis buffer from the NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module (New England Bio Labs, #E6421L). After the sorting, the plate was shortly spun down to ensure all cells were collected at the bottom of the well and was immediately processed for cDNA synthesis. For MitoSOX based flow cytometry approximately 500,000 events were acquired for each experimental condition. All collected data were gated based first on forward and side scatter, and then on the fluorescence of the mito-Dendra2 tag and the MitoSOX. Data were analyzed using FlowJo ™ v10.8 Software (BD Life Sciences).
cDNA synthesis
Sorted microglia were processed with the NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module (New England Bio Labs, #E6421L) according to the manufacturer’s protocol.
Reverse transcription quantitative real-time PCR and gene expression analysis
Primers (Table S11) were designed with the free PrimerQuest Tool from Integrated DNA Technologies (https://eu.idtdna.com/PrimerQuest/Home/Index). To ensure the target sequence of each primer, primers were blasted (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Their self-complementarity and folding probability were investigated using the UNAFold Web server (http://www.unafold.org/). Primer efficiencies were validated from the slope of four to five serial 1:4 dilutions of cDNA template according to Equation 1. Primers with efficiencies between 90-110% were used.
| (Equation 1) |
For gene expression analysis, RT-qPCR (Luna® Universal qPCR Master Mix; New England BioLabs; M3003L) was performed in 384-well plates (Bio-Rad; HSR4805) on a Roche Lightcycler 480 according to the manufacturer’s manual. Total reaction volume was 10μl containing 1μl of 1:10 diluted cDNA as template and a final concentration of 0.25μM for each primer. Cycle conditions were 60 seconds at 95°C for initial denaturation, followed by 40 cycles of denaturation (15 seconds; 95°C) and annealing/extension (30 seconds; 60°C). Each run was completed with a melting curve analysis to confirm amplification of only one amplicon. Each PCR reaction was run in triplicate from which a mean Cq value was calculated and used for further analysis.
Analysis of RT-qPCR results
Fold change differences between each condition and WTnaive were calculated according to delta-delta Ct method.84 dCq values were obtained by normalizing mean Cq values to the reference housekeeping gene (GAPDH) measured within the same experiment (Equation 2). ddCq values were then calculated by normalizing dCq values to the respective control condition (WTnaive) within each experimental repetition (Equation 3). Fold changes were obtained by transforming ddCq values from log2-scale to linear scale (Equation 4). These fold changes were used for data visualization. Exclusion criteria were based on IBA1 and GFAP fold change expression to determine purity of cell isolation, which would indicate substantial contamination from astrocytes and Müller glia.
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
Retina dissection and fixation
Animals were briefly anesthetized with isoflurane (Zoetis) until breathing slowed, then cervical dislocation was performed. Retinas were immediately dissected from the enucleated eyes in 1X phosphate buffered saline (PBS) and fixed in 4% (w/v) paraformaldehyde for 30 minutes. After 3x PBS washes, retinas were put in 30% (w/v) sucrose in 1X PBS overnight at 4°C for either -80°C freezer storage or immunostaining.
Immunostaining
Staining was carried out in 24-well tissue culture plates (TPP, #92024) always protected from light to preserve the endogenous mito-Dendra2 fluorophore. Retinas saturated in sucrose (30% sucrose (w/v) in 1X PBS) underwent three freeze/thaw cycles using dry ice, then were washed 3x with 1X PBS. Next, a blocking step using blocking solution (1% (w/v) bovine serum albumin (Sigma A9418), 5% (v/v) Triton X-100 (Sigma T8787), 0.01% (v/v) sodium azide (VWR 786-299), and 10% (v/v) goat (Millipore S26) or donkey (Millipore S30) serum) was carried out on an orbital shaker for 1 hour at room temperature. Primary antibodies (key resources table) were diluted in antibody solution (1% (w/v) bovine serum albumin, 5% (v/v) triton X-100, 0.01% (v/v) sodium azide, 3% (v/v) goat or donkey serum) and incubated for three nights at 4°C on an orbital shaker. Following 3x PBS washes for 30 mins each, secondary solutions were prepared in the antibody solution and incubated at room temperature for 2 hours. After three washes, Hoechst 334422 (1:5,000, Thermo Fisher Scientific, H3570) for 10 mins, and three more 1X PBS washes. Retinas were mounted using an antifade solution (10% (v/v) mowiol (Sigma, 81381), 26% (v/v) glycerol (Sigma, G7757), 0.2M tris buffer pH 8, 2.5% (w/v) Dabco (Sigma, D27802)) on glass microscope slides with parafilm spacers to prevent tissue distortion and overlaid with coverslips (VWR, #631-0147).
Confocal microscopy
Single cell images for deconvolution were acquired on Zeiss LSM800 microscope using a Plan-Apochromat 40X objective NA 1.4 (#420762-9900) at 1.5x digital zoom to acquire a pixel size of 0.05 μm in X and Y resolution and 0.150 μm in Z. Images were acquired from the peripheral region of the retina, in two opposing quadrants, and inner plexiform layer microglia were selected by setting the z-stack between the ganglion cell and inner nuclear layer Hoechst staining. Microglia in the ganglion cell layer were excluded due to their low population in naïve environment, and known migration from the optic nerve head to the nerve fiber layer after ONC.85 After IPL selection, the IBA1 channel was visualized and the nearest IBA1+ cell was selected for imaging, thereby reducing bias in cell selection and blinded to the mitochondrial architecture within the cell. Tile scans (2x2) were necessary to acquire the area of single IPL microglia in naïve condition, resulting in images of dimension 202 x 202 μm. For each animal, 23 images for from two opposing quadrants were acquired. Images were stitched using Zen 2.3 or 3.5 desktop version.
Tiled (2x2) images for overview images were acquired using a Plan-Apochromat 40X objective NA 1.4 (#420762-9900) with pixel size of 0.156 μm in X and Y and 0.21 μm in Z plane for a region measuring 303 x303 μm. Tiled images (2x2) for microglia and retinal ganglion cell counting were acquired using a Plan-Apochromat 20X objective NA 0.8 (#420650-9901) with pixel resolution 0.312 x 0.312 x 0.5 μm to obtain a region measuring 605 x 605 μm in four peripheral quadrants of each retinal wholemount.
Image processing
SVI Huygens Professional v21.10 (https://svi.nl/HomePage) was used for batch deconvolution processing of stitched confocal images. Microscope parameters for batch settings were as follows: 50 nm sampling intervals for X and Y, 150 nm in Z. The refractive index and embedding medium were set to 1.515 and 1.49, respectively. Objective quality was listed as ‘good.’ For channel settings, the back projected pinhole was set to 250 nm, and the excitation fill factor was 2. Excitation (ex) and emission (em) settings for the four channels were as follows: 405/420 nm, 488/520 nm, 561/602 nm, 633/650 nm. Deconvolution parameters were the default settings for the ‘confocal low template’ for deconvolution using the classic MLE (maximum likelihood estimation) algorithm with maximum 30 iterations. Images were output in the Imaris file format. All confocal images presented in figures were masked in Imaris based on the IBA1-immunostained microglia of interest to remove microglia and mitochondria labeling not belonging to the analyzed cell. Supplementary videos were animated using Imaris.
Quantification and statistical analysis
Filament tracing
Microglia morphology was traced in 3-dimensions using the Filament wizard in IMARIS. The starting point was detected with diameter 10 μm and 0.6 μm was used to set seeding points and the disconnected segment filtering smoothness. Final filament traces were manually edited to remove incorrect segments from the semi-automatic Filament tracing.
Mitochondria and CD68 surfaces in single microglia
Surface rendering analysis was performed using IMARIS v9.1-9.3 (Bitplane, Imaris). Individual cells were cropped from the original tiled image when necessary to reduce processing time. Then, 3-dimensional surfaces were created for microglia, mitochondria and CD68 vesicles using the Surface wizard with surface details settings of 0.5 μm, 0.2 μm, and 0.4 μm, respectively. Each surface was manually edited to resolve any inconsistencies in surface creation. Mitochondrial surfaces were carefully edited to verify connectivity throughout the entirety of the network, often requiring manual splitting of surfaces which were joined by the thresholding within the wizard.
Mitochondrial organelle and CD68 vesicle localization in single microglia
Completed Surfaces for either mitochondria or CD68 were ‘split’ using the Xtension Split Surfaces available via Imaris bitplane (https://imaris.oxinst.com/open/). This generates a Split Surface group where each individual organelle/vesicle becomes a new uniquely named surface nested within the Split Surface group. A spot 5 μm in diameter was manually added to identify the nucleus. Then, the Spots and Surfaces distance XTension calculates the distance between the nucleus spot and the center point of each individual surface in the Split surface group. The Xtension was modified from the original version to allow for up to 100 distance calculations and to output a csv file.
Data compilation and metric calculations
Python version 3.7 with packages pandas,86 matplotlib87 and seaborn88 for data compilation and visualization. From Imaris, mitochondria, microglia, and CD68 surfaces, microglia filament tracings, and mitochondrial and CD68 localization Statistics files for each cell were exported to comma separated values file format with a unique identifier for ease in compiling cell-specific data for downstream analysis. Metric calculations for each surface type are detailed below:
Filament tracing
The ‘Number of Filament Sholl intersections’ from the Statistics file was used to plot the number of intersections per increasing radii of 1 μm.89 Plotted Sholl curves were truncated at 60 μm.
Median mitochondrial volume and number
From the Statistics file, the volume of each mitochondrial Surface within a cell was used to determine the median mitochondrial volume per cell. Here, the total number of organelles per cell was also be extracted.
Sphericity
From the Statistics file, the sphericity metric of each mitochondrial Surface within a cell was used to determean the mean sphericity per cell.
Percentage of CD68 volume
From the Statistics file, the volume from each CD68 Surface within a cell was summed. Additionally, the sum volume of the microglia Surface was extracted from the microglia Statistics file. The percentage of CD68 volume was calculated from total CD68 volume per total microglia volume for each cell.
Percentage of mitochondrial volume
From the Statistics file, the volume from each mitochondrial Surface within a cell was summed. Additionally, the sum volume of the microglia Surface was extracted from the microglia Statistics file. The percentage of mitochondrial volume was calculated from total mitochondria volume per total microglia volume for each cell.
Mitochondrial perinuclear localization
The Statistics file for localization data includes the distance of each organelle from the cell soma (μm) and a unique Surface ID that is derived from the original mitochondrial Surface. From the Statistics file of mitochondrial volume, the Surface ID can be used to compile distance and volume for each unique organelle within the cell.
The percentage of mitochondrial volume in the perinuclear region was determined by calculating the sum of organelle volumes within a 10 μm radius of the cell soma, and dividing by the total mitochondria volume in that cell. The microglia soma ranges from 5-10 μm in diameter due to its non-uniform shape, therefore a 10 μm radius from the center of the soma Spot in IMARIS includes ∼2.5-5 μm of soma, allowing an estimation of mitochondrial localized in the perinuclear region to be at maximum 5-7 μm from the nuclear membrane.
We corroborated this perinuclear boundary distance by principal component analysis of the mitochondrial distances. Mitochondrial distances were binned in increments of 10 μm, and the sum volume in each bin was normalized to total mitochondria volume within a cell. PC loadings of the normalized values confirmed that the two bins contributing the most to principal component loadings for distances between 0-10 μm, and 71-80 μm.
Identifying Sholl radii for PCA analysis
The Sholl curve (see STAR Methods: filament tracing) is a high-dimensional, vector representation of the microglial morphology. To incorporate filament tracing into the PCA without overpowering the other scalar parameters, we first determined which Sholl radius contributed most to the differences in Sholl intersection curves. Here, Sholl intersections with 10 μm radial distances were normalized for each cell. We observed the greatest PC loadings in the 11-20 μm bin. We repeated this analysis with 5 μm radial distances and confirmed the greatest PC loadings in 11-15 μm bin. As the PC loading was greater for the 10 μm radial increments, we implemented the normalized ratio of Sholl intersections at 11-20 μm for each cell and refer to this as the Sholl index.
Principal Component Analysis (PCA)
PCA analysis was performed using scikit-learn in Python.90 Mitochondrial sphericity was omitted from analysis since it is derived from mitochondrial volume. PCA was then implemented on the remaining six metrics; median mitochondrial volume was included as a median value, while number of mitochondria, percentage of mitochondrial volume per microglia and percentage of CD68 volume per microglia volume were included as mean values for each cell. Mitochondria and CD68 localization values were included as percentage values. Data from all experimental groups were included for the PCA, where Figure 5 shows only male and female within all conditions and genotypes and Figure 6 also shows ovariectomized groups.
Cell counting
RBPMS+-cells or cleaved-caspase3+/RBPMS+-cells were counted using the Spots function of Imaris v9.3, with additional manual editing. Each of these metrics was quantified from four peripheral quadrants of each analyzed retina and represented as a median value.
Statistics
Statistical tests were performed using the Python packages statsmodels91 and Pingouin92 as indicated in the figure legends. Groups for comparison were first tested for normal distribution and equal variance using the Shapiro test and Levene test, respectively. No data were excluded from analysis. Where applicable, post-hoc tests were performed using either Tukey’s multiple comparison of means or Conover’s test in Python packages statsmodels ‘multicomp’ or Pingouin. Significance levels are indicated using the following notation (n.s. p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). For Sholl analysis comparisons, linear regression models from lme4 package93 were performed in R. The default contrast for unordered variables was changed to ‘contr.sum’, allowing type III ANOVA comparison. Statistical analysis and accompanying data for all main and supplemental figure panels are found in the Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10.
Acknowledgments
We thank the Scientific Service Units (SSU) of ISTA through resources provided by the Imaging and Optics Facility (IOF), the Lab Support Facility (LSF), and the Pre-Clinical Facility (PCF) team, specifically Sonja Haslinger and Michael Schunn for excellent mouse colony management and support. This research was supported by the FWF Sonderforschungsbereich F83 (to E.E.P). We thank Bálint Nagy, Ryan John A. Cubero, Marco Benevento and all members of the Siegert group for constant feedback on the project and article.
Author contributions
M.M. and S.S. conceived and developed experimental design collaboratively. M.M. wrote the initial draft with subsequent editorial input from S.S. M.M. performed experiments, created and implemented python data import for 3-dimensional analysis, performed statistical analysis, and created figures. M.M., F.S. and E.E.P. conceived and verified UCP2 protein knockout. G.C. performed FACS-based RT-qPCR experiments, and data analysis. F.S.U. and M.M. performed FACS-based MitoSOX experiments and data analysis. A.V. performed ovariectomy surgeries. All authors provided comment and discussion on the article and approved the final version.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more authors of this paper self-identifies as a gender minority in their field of research.
Published: August 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107780.
Supplemental information
Data and code availability
-
•
The data reported in this paper is available from the lead contact upon request.
-
•
The paper does not report original code.
-
•
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
References
- 1.Giacomello M., Pyakurel A., Glytsou C., Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020;21:204–224. doi: 10.1038/S41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 2.Collier J.J., Oláhová M., McWilliams T.G., Taylor R.W. Mitochondrial signalling and homeostasis: from cell biology to neurological disease. Trends Neurosci. 2023;46:137–152. doi: 10.1016/J.TINS.2022.12.001. [DOI] [PubMed] [Google Scholar]
- 3.West A.P., Shadel G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017;176:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugiura A., McLelland G.-L., Fon E.A., McBride H.M. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/EMBJ.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornmann B., Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J. Cell Sci. 2010;123:1389–1393. doi: 10.1242/JCS.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youle R.J., van der Bliek A.M. Mitochondrial Fission, Fusion, and Stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisner V., Picard M., Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 2018;20:755–765. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palikaras K., Lionaki E., Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Naturen. 2015;5217553:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 9.Kasahara A., Cipolat S., Chen Y., Dorn G.W., Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and notch signaling. Science. 2013;342:734–737. doi: 10.1126/SCIENCE.1241359. [DOI] [PubMed] [Google Scholar]
- 10.Al-Mehdi A.B., Pastukh V.M., Swiger B.M., Reed D.J., Patel M.R., Bardwell G.C., Pastukh V.V., Alexeyev M.F., Gillespie M.N. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park M.K., Ashby M.C., Erdemli G., Petersen O.H., Tepikin A.V. Perinuclear, perigranular and sub-plasmalemmal mitochondria have distinct functions in the regulation of cellular calcium transport. EMBO J. 2001;20:1863–1874. doi: 10.1093/EMBOJ/20.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/EMBOJ.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes L.C., Di Benedetto G., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambold A.S., Kostelecky B., Elia N., Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 2011;108:10190–10195. doi: 10.1073/PNAS.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonian N.A., Coyle J.T. Oxidative stress in neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 1996;36:83–106. doi: 10.1146/ANNUREV.PA.36.040196.000503. [DOI] [PubMed] [Google Scholar]
- 16.Rambold A.S., Pearce E.L. Mitochondrial Dynamics at the Interface of Immune Cell Metabolism and Function. Trends Immunol. 2018;39:6–18. doi: 10.1016/j.it.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Caputa G., Castoldi A., Pearce E.J. Metabolic adaptations of tissue-resident immune cells. Nat. Immunol. 2019;20:793–801. doi: 10.1038/s41590-019-0407-0. [DOI] [PubMed] [Google Scholar]
- 18.Salter M.W., Stevens B. Microglia emerge as central players in brain disease. Nat. Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 19.Hanisch U.-K., Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 20.Baik S.H., Kang S., Lee W., Choi H., Chung S., Kim J.I., Mook-Jung I. A Breakdown in Metabolic Reprogramming Causes Microglia Dysfunction in Alzheimer’s Disease. Cell Metabol. 2019;30:493–507.e6. doi: 10.1016/J.CMET.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Nair S., Sobotka K.S., Joshi P., Gressens P., Fleiss B., Thornton C., Mallard C., Hagberg H. Lipopolysaccharide-induced alteration of mitochondrial morphology induces a metabolic shift in microglia modulating the inflammatory response in vitro and in vivo. Glia. 2019;67:1047–1061. doi: 10.1002/glia.23587. [DOI] [PubMed] [Google Scholar]
- 22.Bernier L.P., York E.M., Kamyabi A., Choi H.B., Weilinger N.L., MacVicar B.A. Microglial metabolic flexibility supports immune surveillance of the brain parenchyma. Nat. Commun. 2020;11:1559–1617. doi: 10.1038/s41467-020-15267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erny D., Dokalis N., Mezö C., Castoldi A., Mossad O., Staszewski O., Frosch M., Villa M., Fuchs V., Mayer A., et al. Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metabol. 2021;33:2260–2276.e7. doi: 10.1016/j.cmet.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Park J., Choi H., Min J.-S., Park S.-J., Kim J.-H., Park H.-J., Kim B., Chae J.-I., Yim M., Lee D.-S. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J. Neurochem. 2013;127:221–232. doi: 10.1111/jnc.12361. [DOI] [PubMed] [Google Scholar]
- 25.Scheiblich H., Dansokho C., Mercan D., Schmidt S.V., Bousset L., Wischhof L., Eikens F., Odainic A., Spitzer J., Griep A., et al. Microglia jointly degrade fibrillar alpha-synuclein cargo by distribution through tunneling nanotubes. Cell. 2021;184:5089–5106.e21. doi: 10.1016/J.CELL.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gosselin D., Skola D., Coufal N.G., Holtman I.R., Schlachetzki J.C.M., Sajti E., Jaeger B.N., O’Connor C., Fitzpatrick C., Pasillas M.P., et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh S.E., Walker A.J., Kamath T., Dissing-Olesen L., Hammond T.R., de Soysa T.Y., Young A.M.H., Murphy S., Abdulraouf A., Nadaf N., et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 2022;25:306–316. doi: 10.1038/s41593-022-01022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakina O., Conrad T., Mitic N., Oliveira Vidal R., Obrusnik T., Sai S., Nolte C., Semtner M., Kettenmann H., Berlin U. In situ Patch-seq analysis of microglia reveals a lack of stress genes as found in FACS-isolated microglia. bioRxiv. 2023 doi: 10.1101/2023.03.22.533782. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colombo G., Cubero R.J.A., Kanari L., Venturino A., Schulz R., Scolamiero M., Agerberg J., Mathys H., Tsai L.H., Chachólski W., et al. A tool for mapping microglial morphology, morphOMICs, reveals brain-region and sex-dependent phenotypes. Nat. Neurosci. 2022;25:1379–1393. doi: 10.1038/s41593-022-01167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Schlamp C.L., Nickells R.W. Experimental induction of retinal ganglion cell death in adult mice. Invest. Ophthalmol. Vis. Sci. 1999;40:1004–1008. [PubMed] [Google Scholar]
- 31.Lehmann U., Heuss N.D., McPherson S.W., Roehrich H., Gregerson D.S. Dendritic cells are early responders to retinal injury. Neurobiol. Dis. 2010;40:177–184. doi: 10.1016/J.NBD.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodeutsch N., Siebert H., Dermon C., Thanos S. Unilateral injury to the adult rat optic nerve causes multiple cellular responses in the contralateral site. J. Neurobiol. 1999;38:116–128. doi: 10.1002/(sici)1097-4695(199901)38:1<116::aid-neu9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 33.Ježek P., Holendová B., Garlid K.D., Jabůrek M. Mitochondrial Uncoupling Proteins: Subtle Regulators of Cellular Redox Signaling. Antioxidants Redox Signal. 2018;29:667–714. doi: 10.1089/ars.2017.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J.D., Yoon N.A., Jin S., Diano S. Microglial UCP2 Mediates Inflammation and Obesity Induced by High-Fat Feeding. Cell Metabol. 2019;30:952–962.e5. doi: 10.1016/J.CMET.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham A.H., McCaffery J.M., Chan D.C. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yona S., Kim K.-W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neurohr J.M., Paulson E.T., Kinsey S.T. A higher mitochondrial content is associated with greater oxidative damage, oxidative defenses, protein synthesis and ATP turnover in resting skeletal muscle. J. Exp. Biol. 2021;224:jeb242462. doi: 10.1242/JEB.242462/272326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damoiseaux J.G., Döpp E.A., Calame W., Chao D., MacPherson G.G., Dijkstra C.D. Rat macrophage lysosomal membrane antigen recognized by monoclonal antibody ED1. Immunology. 1994;83:140–147. [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer J., Sminia T., Wouterlood F.G., Dijkstra C.D. Phagocytic activity of macrophages and microglial cells during the course of acute and chronic relapsing experimental autoimmune encephalomyelitis. J. Neurosci. Res. 1994;38:365–375. doi: 10.1002/jnr.490380402. [DOI] [PubMed] [Google Scholar]
- 40.Levkovitch-Verbin H., Harris-Cerruti C., Groner Y., Wheeler L.A., Schwartz M., Yoles E. RGC death in mice after optic nerve crush injury: Oxidative stress and neuroprotection. Invest. Ophthalmol. Vis. Sci. 2000;41:4169–4174. [PubMed] [Google Scholar]
- 41.Wohl S.G., Schmeer C.W., Witte O.W., Isenmann S. Proliferative response of microglia and macrophages in the adult mouse eye after optic nerve lesion. Invest. Ophthalmol. Vis. Sci. 2010;51:2686–2696. doi: 10.1167/iovs.09-4537. [DOI] [PubMed] [Google Scholar]
- 42.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y.R., Imrichova H., Wang H., Chao T., Xiao Z., Gao M., Rincon-Restrepo M., Franco F., Genolet R., Cheng W.C., et al. Disturbed mitochondrial dynamics in CD8+ TILs reinforce T cell exhaustion. Nat. Immunol. 2020;21:1540–1551. doi: 10.1038/s41590-020-0793-3. [DOI] [PubMed] [Google Scholar]
- 44.Siegert S., Cabuy E., Scherf B.G., Kohler H., Panda S., Le Y.Z., Fehling H.J., Gaidatzis D., Stadler M.B., Roska B. Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci. 2012;15:487–495.S1-S2. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- 45.Vozza A., Parisi G., De Leonardis F., Lasorsa F.M., Castegna A., Amorese D., Marmo R., Calcagnile V.M., Palmieri L., Ricquier D., et al. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc. Natl. Acad. Sci. USA. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rupprecht A., Bräuer A.U., Smorodchenko A., Goyn J., Hilse K.E., Shabalina I.G., Infante-Duarte C., Pohl E.E. Quantification of Uncoupling Protein 2 Reveals Its Main Expression in Immune Cells and Selective Up-Regulation during T-Cell Proliferation. PLoS One. 2012;7:e41406. doi: 10.1371/journal.pone.0041406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brand M.D., Esteves T.C. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metabol. 2005;2:85–93. doi: 10.1016/J.CMET.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Bechmann I., Diano S., Warden C.H., Bartfai T., Nitsch R., Horvath T.L. Brain mitochondrial uncoupling protein 2 (UCP2): a protective stress signal in neuronal injury. Biochem. Pharmacol. 2002;64:363–367. doi: 10.1016/S0006-2952(02)01166-8. [DOI] [PubMed] [Google Scholar]
- 49.O’Koren E.G., Yu C., Klingeborn M., Wong A.Y.W., Prigge C.L., Mathew R., Kalnitsky J., Msallam R.A., Silvin A., Kay J.N., et al. Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity. 2019;50:723–737.e7. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoang T., Wang J., Boyd P., Wang F., Santiago C., Jiang L., Yoo S., Lahne M., Todd L.J., Jia M., et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science. 2020;370:eabb8598. doi: 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kizaki T., Suzuki K., Hitomi Y., Taniguchi N., Saitoh D., Watanabe K., Onoé K., Day N.K., Good R.A., Ohno H. Uncoupling protein 2 plays an important role in nitric oxide production of lipopolysaccharide-stimulated macrophages. Proc. Natl. Acad. Sci. USA. 2002;99:9392–9397. doi: 10.1073/PNAS.142206299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yasumoto Y., Stoiljkovic M., Kim J.D., Sestan-Pesa M., Gao X.-B., Diano S., Horvath T.L. Ucp2-dependent microglia-neuronal coupling controls ventral hippocampal circuit function and anxiety-like behavior. Mol. Psychiatr. 2021;26:2740–2752. doi: 10.1038/s41380-021-01105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toda C., Kim J.D., Impellizzeri D., Cuzzocrea S., Liu Z.-W., Diano S. UCP2 Regulates Mitochondrial Fission and Ventromedial Nucleus Control of Glucose Responsiveness. Cell. 2016;164:872–883. doi: 10.1016/j.cell.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Razmara A., Duckles S.P., Krause D.N., Procaccio V. Estrogen suppresses brain mitochondrial oxidative stress in female and male rats. Brain Res. 2007;1176:71–81. doi: 10.1016/J.BRAINRES.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakazawa T., Takahashi H., Shimura M. Estrogen has a neuroprotective effect on axotomized RGCs through ERK signal transduction pathway. Brain Res. 2006;1093:141–149. doi: 10.1016/J.BRAINRES.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 56.Di Florio D.N., Sin J., Coronado M.J., Atwal P.S., Fairweather D. Sex differences in inflammation, redox biology, mitochondria and autoimmunity. Redox Biol. 2020;31:101482. doi: 10.1016/J.REDOX.2020.101482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang S.-H., Liu R., Perez E.J., Wen Y., Stevens S.M., Valencia T., Brun-Zinkernagel A.-M., Prokai L., Will Y., Dykens J., et al. Mitochondrial localization of estrogen receptor beta. Proc. Natl. Acad. Sci. USA. 2004;101:4130–4135. doi: 10.1073/pnas.0306948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker A.E., Brautigam V.M., Watters J.J. Estrogen Modulates Microglial Inflammatory Mediator Production via Interactions with Estrogen Receptor β. Endocrinology. 2004;145:5021–5032. doi: 10.1210/EN.2004-0619. [DOI] [PubMed] [Google Scholar]
- 59.Costa L. da F., Cesar R.M. second edition second. Taylor & Francis Group, LLC; 2009. Shape Classification and Analysis: Theory and Practice. [Google Scholar]
- 60.Bolasco G., Weinhard L., Boissonnet T., Neujahr R., Gross C.T. Three-dimensional nanostructure of an intact microglia cell. Front. Neuroanat. 2018;12:105. doi: 10.3389/FNANA.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaudhry A., Shi R., Luciani D.S. A pipeline for multidimensional confocal analysis of mitochondrial morphology, function, and dynamics in pancreatic β-cells. Am. J. Physiol. Endocrinol. Metab. 2020;318:E87–E101. doi: 10.1152/AJPENDO.00457.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 2019;50:253–271.e6. doi: 10.1016/J.IMMUNI.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA. 2009;106:11960–11965. doi: 10.1073/PNAS.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abdullah M.O., Zeng R.X., Margerum C.L., Papadopoli D., Monnin C., Punter K.B., Chu C., Al-Rofaidi M., Al-Tannak N.F., Berardi D., et al. Mitochondrial hyperfusion via metabolic sensing of regulatory amino acids. Cell Rep. 2022;40:111198. doi: 10.1016/J.CELREP.2022.111198. [DOI] [PubMed] [Google Scholar]
- 65.Qian W., Choi S., Gibson G.A., Watkins S.C., Bakkenist C.J., Van Houten B. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J. Cell Sci. 2012;125:5745–5757. doi: 10.1242/JCS.109769/-/DC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. The Role of Dynamin-Related Protein 1, a Mediator of Mitochondrial Fission, in Apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/S1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 67.Goldmann T., Wieghofer P., Müller P.F., Wolf Y., Varol D., Yona S., Brendecke S.M., Kierdorf K., Staszewski O., Datta M., et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Q.G., Raz L., Wang R., Han D., De Sevilla L., Yang F., Vadlamudi R.K., Brann D.W. Estrogen Attenuates Ischemic Oxidative Damage via an Estrogen Receptor α-Mediated Inhibition of NADPH Oxidase Activation. J. Neurosci. 2009;29:13823–13836. doi: 10.1523/JNEUROSCI.3574-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borrás C., Sastre J., García-Sala D., Lloret A., Pallardó F.V., Viña J. Mitochondria from females exhibit higher antioxidant gene expression and lower oxidative damage than males. Free Radic. Biol. Med. 2003;34:546–552. doi: 10.1016/S0891-5849(02)01356-4. [DOI] [PubMed] [Google Scholar]
- 70.Pinto R.E., Bartley W. The nature of the sex-linked differences in glutathione peroxidase activity and aerobic oxidation of glutathione in male and female rat liver. Biochem. J. 1969;115:449–456. doi: 10.1042/BJ1150449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Du L., Hickey R.W., Bayir H., Watkins S.C., Tyurin V.A., Guo F., Kochanek P.M., Jenkins L.W., Ren J., Gibson G., et al. Starving Neurons Show Sex Difference in Autophagy. J. Biol. Chem. 2009;284:2383–2396. doi: 10.1074/JBC.M804396200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dorfman M.D., Krull J.E., Douglass J.D., Fasnacht R., Lara-Lince F., Meek T.H., Shi X., Damian V., Nguyen H.T., Matsen M.E., et al. Sex differences in microglial CX3CR1 signalling determine obesity susceptibility in mice. Nat. Commun. 2017;8:14556. doi: 10.1038/NCOMMS14556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Villa A., Gelosa P., Castiglioni L., Cimino M., Rizzi N., Pepe G., Lolli F., Marcello E., Sironi L., Vegeto E., Maggi A. Sex-Specific Features of Microglia from Adult Mice. Cell Rep. 2018;23:3501–3511. doi: 10.1016/j.celrep.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thion M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., Blecher R., Ulas T., Squarzoni P., Hoeffel G., et al. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 2018;172:500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim H.A., Lee S., Eah K.S., Yoon Y.H. Prevalence and Risk Factors of Tamoxifen Retinopathy. Ophthalmology. 2020;127:555–557. doi: 10.1016/j.ophtha.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 76.Wang X., Zhao L., Zhang Y., Ma W., Gonzalez S.R., Fan J., Kretschmer F., Badea T.C., Qian H.H., Wong W.T. Tamoxifen Provides Structural and Functional Rescue in Murine Models of Photoreceptor Degeneration. J. Neurosci. 2017;37:3294–3310. doi: 10.1523/JNEUROSCI.2717-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong D., Vong L., Parton L.E., Ye C., Tong Q., Hu X., Choi B., Brüning J.C., Lowell B.B. Glucose Stimulation of Hypothalamic MCH Neurons Involves KATP Channels, Is Modulated by UCP2, and Regulates Peripheral Glucose Homeostasis. Cell Metabol. 2010;12:545–552. doi: 10.1016/J.CMET.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bronstein R., Torres L., Nissen J.C., Tsirka S.E. Culturing microglia from the neonatal and adult central nervous system. J. Vis. Exp. 2013 doi: 10.103791/50647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Hove H., Antunes A.R.P., De Vlaminck K., Scheyltjens I., Van Ginderachter J.A., Movahedi K. Identifying the variables that drive tamoxifen-independent CreERT2 recombination: Implications for microglial fate mapping and gene deletions. Eur. J. Immunol. 2020;50:459–463. doi: 10.1002/EJI.201948162. [DOI] [PubMed] [Google Scholar]
- 80.Jahn H.M., Kasakow C.V., Helfer A., Michely J., Verkhratsky A., Maurer H.H., Scheller A., Kirchhoff F. Refined protocols of tamoxifen injection for inducible DNA recombination in mouse astroglia. Sci. Rep. 2018;8:5913–6011. doi: 10.1038/s41598-018-24085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoffmann H.M. Determination of Reproductive Competence by Confirming Pubertal Onset and Performing a Fertility Assay in Mice and Rats. J. Vis. Exp. 2018:58352. doi: 10.3791/58352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rupprecht A., Sittner D., Smorodchenko A., Hilse K.E., Goyn J., Moldzio R., Seiler A.E.M., Bräuer A.U., Pohl E.E. Uncoupling Protein 2 and 4 Expression Pattern during Stem Cell Differentiation Provides New Insight into Their Putative Function. PLoS One. 2014;9:e88474. doi: 10.1371/JOURNAL.PONE.0088474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Žuna K., Jovanović O., Khailova L.S., Škulj S., Brkljača Z., Kreiter J., Kotova E.A., Vazdar M., Antonenko Y.N., Pohl E.E. Mitochondrial uncoupling proteins (UCP1-UCP3) and adenine nucleotide translocase (ANT1) enhance the protonophoric action of 2,4-dinitrophenol in mitochondria and planar bilayer membranes. Biomolecules. 2021;11:1178. doi: 10.3390/BIOM11081178/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 85.Heuss N.D., Pierson M.J., Roehrich H., McPherson S.W., Gram A.L., Li L., Gregerson D.S. Optic nerve as a source of activated retinal microglia post-injury. Acta Neuropathol. Commun. 2018;6:66. doi: 10.1186/s40478-018-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mckinney W. 2010. Data Structures for Statistical Computing in Python. [Google Scholar]
- 87.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 88.Waskom M. seaborn: statistical data visualization. J. Open Source Softw. 2021;6:3021. doi: 10.21105/JOSS.03021. [DOI] [Google Scholar]
- 89.Sholl D.A. Vol. 87. 1953. (Dendritic organization in the neurons of the visual and motor cortices of the cat). [PMC free article] [PubMed] [Google Scholar]
- 90.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 91.Seabold S., Perktold J. Statsmodels: Econometric and Statistical Modeling with Python. Proc. 9th Python Sci. Conf. 2010 [Google Scholar]
- 92.Vallat R. Pingouin: statistics in Python. J. Open Source Softw. 2018;3:1026. doi: 10.21105/JOSS.01026. [DOI] [Google Scholar]
- 93.Bates D.W., Mächler M., Bolker B.M., Walker S.C. Fitting Linear Mixed-Effects Models Using lme4. BMJ Qual. Saf. 2015;24:1–3. doi: 10.18637/JSS.V067.I01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data reported in this paper is available from the lead contact upon request.
-
•
The paper does not report original code.
-
•
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.