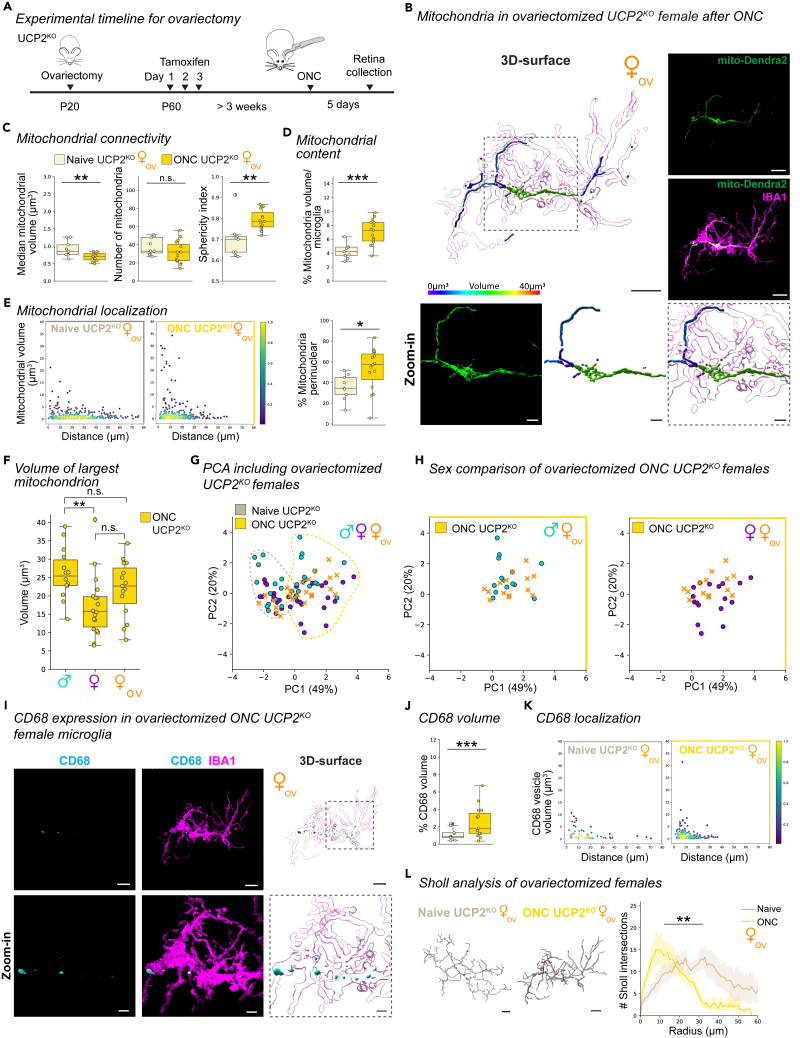
Figure 6.
Ovariectomized female UCP2KO microglia exhibit mitochondrial hyperfusion phenotype after ONC
(A) Experimental timeline. Ovariectomy performed at postnatal day 20 (P20) and three consecutive tamoxifen injections were administered daily for three days starting at P60. Optic nerve crush (ONC) was performed at least 3 weeks after tamoxifen injection and retinas collected five days after ONC.
(B) Representative 3D-surface renderings of IBA1 (magenta) and mito-Dendra2 (statistics-based volume spectral coloring) from ONC-induced ovariectomized female UCP2KO microglia. Right: corresponding confocal images of IBA1-immonostained (magenta) and mito-Dendra2 (green) expression. Below: Zoom-in of region of interest (dashed outline) of mito-Dendra2 (green) and corresponding 3D-surface with volume spectral coloring. Scale bar: 10 μm, zoom-in: 3 μm. Volume spectrum: 0 μm3 (blue) – 10 μm3 (red).
(C–G) Comparison of mitochondrial response in ovariectomized females. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond: outliers outside of 1.5 IQRs. Naive UCP2KO: tan. ONC UCP2KO: gold.
(C) Mitochondrial network connectivity determined by median mitochondrial volume (left, Student’s t test: p = 0.0046), number of organelles within a single cell (center, Student’s t test: p = 0.1939) and mean mitochondrial sphericity (right, Wilcoxon rank-sum test: p = 0.0024).
(D) Percentage of mitochondrial volume per microglial volume. Student’s t test: p = 0.0004.
(E) Mitochondrial localization. Scatterplot depicting mitochondria volume vs. distance from the cell soma (0, origin) of the population of mitochondria in microglia. Point density, pseudo-colored. Percentage of total mitochondrial volume localized within the perinuclear region. Student’s t test: p = 0.0320.
(F) Largest mitochondrion per cell separated by sex. One-way ANOVA: p = 0.0088. Selected Tukey’s post-hoc comparisons: ONC ♂UCP2KO vs. ONC ♀UCP2KO, p = 0.0065; ONC ♂UCP2KO vs. ONC ♀ovUCP2KO, p = 0.3297, ONC ♀UCP2KO vs. ONC ♀ovUCP2KO, p = 0.1938.
(G) Principal component analysis (PCA) of microglia from male, female and ovariectomized UCP2KO microglia in naive and ONC conditions. The first two principle components (PC) are visualized. Naive and ONC UCP2KO microglial profiles in male (turquoise), female (purple) and ovariectomized females (orange x-marker). Dashed outline: Reference of distributions shown in (Figure 5A).
(H) Comparison of ovariectomized ONC UCP2KO female (orange x-marker) microglial profiles with male (turquoise, left) or female (purple, right) ONC UCP2KO microglial profiles.
(I) Ovariectomized female ONC UCP2KO IBA1-immunostained microglia (magenta) from Figure 6B co-labeled with CD68 (cyan) and corresponding 3D-surfaces. Below: Zoom-in of region of interest (dashed line) from image and 3D-surface. Scale bar: 10 μm, zoom-in: 3 μm.
(J) Percentage of total CD68 volume per microglia volume in WT and UCP2KO ovariectomized females in naive and ONC conditions. Boxplot minimum and maximum: InterQuartile Range (IQR) around median (center line). Whiskers: 1.5 IQRs. Black diamond points: outliers outside 1.5 IQRs. Wilcoxon rank-sum test: p < 0.0001.
(K) CD68 vesicle localization. Scatterplot depicting CD68 vesicle volume vs. distance from the cell soma (0, origin) of the population of vesicles from ovariectomized females in UCP2KO naive (left) and UCP2KO ONC (right) conditions. Point density, pseudo-colored.
(L) 3D-filament tracings of microglia from UCP2KO naive (left) or ONC (right). Scale bar: 10 μm. Line plot for mean number of Sholl intersections per radial distant from the soma (μm) with 95% confidence interval band. Linear mixed effects model: p = 0.0018. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. p > 0.05: not significant. See Table S6 for retina and cell numbers, statistical tests and corresponding data.