Abstract
In Belgium an increase in the incidence of meningococcal disease has been noted since the early 1990s. Four hundred twenty clinical strains isolated during the period from 1990 to 1995, along with a set of 30 European reference strains, and 20 Dutch isolates were examined by random-primer and repetitive-motif-based PCR. A subset was investigated by multilocus enzyme electrophoresis and pulsed-field gel electrophoresis. The data were compared with results obtained by serotyping (M. Van Looveren, F. Carion, P. Vandamme, and H. Goossens, Clin. Microbiol. Infect. 4:224–228, 1998). Both phenotypic and molecular epidemiological data suggest that the lineage III of Neisseria meningitidis, first encountered in The Netherlands in about 1980, has been introduced in Belgium. The epidemic clone, as defined by oligonucleotide D8635-primed PCR, encompasses mainly phenotypes B:4:P1.4 and B:nontypeable:P1.4, but strains with several other phenotypes were also encountered. Therefore, serotyping alone would underestimate the prevalence of the epidemic clone.
Infections due to Neisseria meningitidis are associated with high rates of morbidity and mortality, therefore posing an important health problem (22). Despite substantial advances in antimicrobial therapy and intensive care, the case fatality rate has fallen little over the past few decades. The highest prevalence of meningococcal disease was and still is in children (20, 31).
Meningococci are classified into serogroups on the basis of the immunological specificities of their capsular polysaccharides and are further subdivided into serotypes and subtypes on the basis of their outer membrane proteins (12). Of the dozen serogroups within N. meningitidis, three predominate, with strains of serogroups A, B, and C accounting for 90% of the cases of meningococcal disease worldwide (13). Of these, serogroup B meningococci had generally been associated only with sporadic cases and localized outbreaks. However, in the past two decades, disease caused by this serogroup has become a major health concern in Europe (21, 30). In Belgium, serogroup B meningococci currently represent more than 80% of the isolates (28).
Since the beginning of the 1990s, the number of meningococcal isolates received at the Belgian Meningococcal Reference Center has increased, rising from 77 in 1990 to 200 in 1995 (4). We have shown that this elevated number of meningococcal infections was primarily due to phenotype B:4:P1.4 and B:nontypeable (NT):P1.4 strains (28). Strains of phenotype B:4:P1.4 have been prevalent in the southern provinces of The Netherlands since the early 1980s (20) and were first noticed in Belgium near the Dutch border in the early 1990s. We have hypothesized that the phenotype B:4:P1.4 and B:NT:P1.4 strains represent a new epidemic clone that originated in The Netherlands and that spread south and west through Belgium between 1990 and 1995 (28).
To test this hypothesis, Belgian isolates were further characterized by several genotyping methods and compared with reference strains and with recent isolates from The Netherlands.
MATERIALS AND METHODS
Bacterial strains.
The following strains were examined: (i) 420 isolates of N. meningitidis received at the Belgian Meningococcal Reference Center between January 1990 and December 1995, (ii) a set of 30 reference strains (L93/2701 through L93/2730) provided to all European Meningococcal Reference Centers for quality assurance, and (iii) 20 clinical isolates of phenotype B:4:P1.4 isolated in The Netherlands between 1980 and 1990. The set of 30 reference strains included isolates from patients with meningitis and/or septicemia, as well as isolates from carriers. The strains were recovered in 16 European countries, and they represent 26 multilocus genotypes (5). All Dutch strains are representative of lineage III (electrophoretic type [ET] 24) described by Caugant et al. (8).
PCR-based typing.
Genomic DNA was extracted by the rapid procedure described by Pitcher et al. (19).
Seven arbitrary primers, comprising four short (10 nucleotides) primers (primers 1254, 1281, 1283, and 1290) (2, 32) and three long (≥17 nucleotides) primers (primers D14307, D11344, and D8635) (2), and two primers for amplification of repeat motifs, the enterobacterial repetitive intergenic consensus (ERIC) motifs 1R and 2 (29), were evaluated for their suitability in differentiating meningococci. DNA amplification was performed in a DNA thermal cycler (Perkin-Elmer GeneAmp PCR System 9600; Perkin-Elmer, Zaventem, Belgium). The 100-μl PCR mixtures consisted of 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 2.5 mM MgCl2, 0.1% Triton X-100, and 0.01% gelatin. Deoxyribonucleotide triphosphates were each used at a final concentration of 0.2 mM. Per reaction mixture, 0.6 U of Goldstar DNA polymerase (Eurogentec, Seraing, Belgium), 100 pmol (primers 1254, 1281, 1283, 1290, D14307, D11344, and D8635) or 50 pmol (primers ERIC-1R and ERIC-2) of primer, and 100 ng of extracted DNA were added. Primers 1254, 1281, 1283, and 1290 were used as described previously (32). PCR conditions for arbitrary primers D14307, D11344, and D8635 consisted of an initial four cycles of 94°C for 5 min, 40°C for 5 min, and 72°C for 5 min followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min and 1 cycle of 72°C for 10 min. The PCR conditions for ERIC primers consisted of an initial step of 95°C for 5 min, followed by 4 cycles of 94°C for 1 min, 26°C for 1 min, and 72°C for 2 min and 40 cycles of 94°C for 30 s, 40°C for 30 s, and 72°C for 1 min. After amplification, 25-μl aliquots of PCR products were electrophoresed (100 V, 3 h) in 1.5% pronarose D1 gels (Sphaero Q, Burgos, Spain) and 0.5× TBE (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA) running buffer containing 0.5 mg of ethidium bromide per ml. The patterns were visualized under UV light and were digitized with the Gel Doc 1000 documentation system (Bio-Rad Laboratories, Nazareth, Belgium). Conversion, normalization, and densitometric analysis of the patterns were done with GelCompar Software (version 4.0; Applied Maths, Kortrijk, Belgium). The similarity between all pairs of traces was expressed by the Pearson product-moment correlation coefficient, and clustering was performed by the unweighted pair group method with average linkage (24).
Multilocus enzyme electrophoresis (MLEE).
Methods of protein extract preparation, starch gel electrophoresis, and selective enzyme staining were similar to those described by Selander et al. (23). The 14 enzymes assayed were malic enzyme, glucose-6-phosphate dehydrogenase, peptidase, isocitrate dehydrogenase, aconitase, NADP-linked glutamate dehydrogenase, NAD-linked glutamate dehydrogenase, alcohol dehydrogenase, fumarase, alkaline phosphatase, two indophenol oxidases, adenylate kinase, and an unknown dehydrogenase. Each isolate was characterized by its combination of alleles at 14 enzyme loci, and distinctive multilocus genotypes were designated ETs. The ET numbers corresponded to those assigned previously (8, 21).
PFGE.
Strains were grown overnight at 37°C in 5% CO2 on Columbia agar (GIBCO, Life Technologies, Paisley, Scotland) supplemented with 5% defibrinated horse blood. One loopful of each strain was washed three times in EET buffer [100 mM EDTA, 10 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 10 mM Tris-HCl (pH 8.0)]. The pelleted cells were resuspended in lysis buffer (6 mM Tris-HCl [pH 7.6], 1 M NaCl, 100 mM EDTA [pH 8.0], 0.5% Brij 58, 0.2% deoxycholate, 0.5% N-lauroylsarcosine), adjusted to a density of 4 × 109 CFU/ml, mixed with an equal volume of 1.6% (wt/vol) low-melt preparative-grade agarose (Bio-Rad Laboratories, Nazareth, Belgium) in lysis buffer, and dispensed into plastic molds (Plexi-Labo, Drongen, Belgium). The solidified plugs were incubated overnight at 37°C in 0.5 ml of lysis buffer containing 2.88 mg of lysozyme (Sigma, Bornem, Belgium) per ml. The inserts were then transferred to 1 ml of EET buffer–3.3 mg pronase E (Sigma) per ml–1.6% (wt/vol) sodium dodecyl sulfate, and the mixture was incubated overnight at 37°C. Subsequently, the agarose plugs were washed four times in EET buffer and twice in T10E0.1 buffer (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]). Finally, they were equilibrated for 1 h in 250 μl of the appropriate restriction buffer. For SpeI (Eurogentec, Seraing, Belgium), restriction was carried out overnight at 37°C in 250 μl of fresh restriction buffer containing 30 U of restriction enzyme. For SfiI (Sigma), restriction occurred at 50°C in 150 μl of restriction buffer containing 30 U of enzyme. The digestion reaction was stopped by the addition of 0.5 ml of 0.5 M EDTA (pH 8.0), and the plugs were stored at 4°C. The chromosomal restriction fragments were separated by pulsed-field gel electrophoresis (PFGE) in a CHEF MAPPER system (Bio-Rad Laboratories) by sealing small pieces of the plugs into slots of a 1% (wt/vol) Pulsed-Field Certified Agarose (Bio-Rad Laboratories) gel in 0.5× TBE buffer. The electrophoresis was performed in 0.5× TBE buffer, equilibrated at 14°C, for 24 h at a constant voltage of 6 V/cm. Separation of the genomic DNA digested with SpeI was achieved with pulse times that ramped alinearly (ramping factor, −1.243) from 3 to 30 s. For SfiI pulse times were ramped linearly from 5 to 35 s.
The SmaI-digested genome of Staphylococcus aureus NCTC 8325 (27) was used as a molecular size standard.
The gels were stained with ethidium bromide, visualized, digitized, and analyzed as described above for PCR. The similarities of the PFGE banding patterns were estimated with the Dice coefficient (11), and clustering was performed by the unweighted pair group method with average linkage.
RESULTS
PCR-based typing.
To identify primers generating informative arrays of PCR products, we tested DNA from five randomly selected clinical isolates of N. meningitidis with primers that had shown to be useful in other typing studies (2, 29, 32). First, four decamers that had been used before for the typing of meningococci (32) were tested. The results that we obtained were unsatisfactory in our hands: primers 1254 and 1290 were unable to generate a banding pattern, whereas primers 1281 and 1283 resulted in a few amplified bands for each of the five strains tested. Three long (≥17-nucleotide) arbitrary primers used to distinguish clinical isolates of Helicobacter pylori (2) were investigated as well. Primer D14307 generated only one band, but primers D8635 and D11344 displayed 10 to 15 well-separated bands. Finally, we evaluated the repetitive primers ERIC-1R and ERIC-2 (29). Both generated informative arrays of PCR products, with up to 10 easily distinguishable bands.
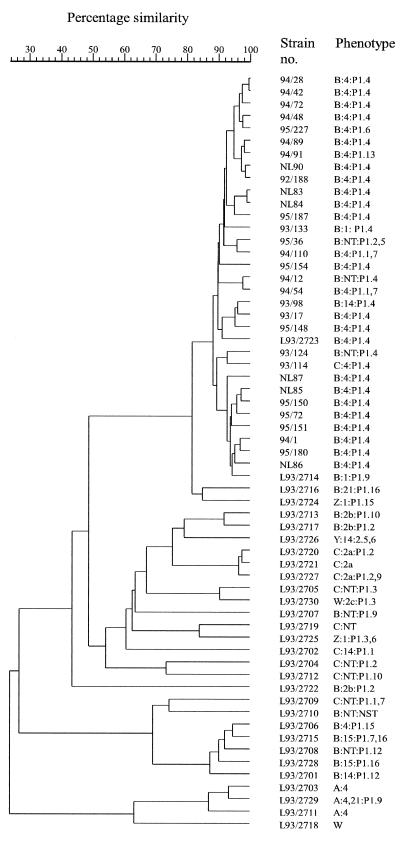
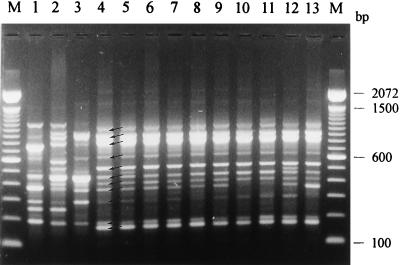
Four of these primers (primers D8635, D11344, ERIC-1R, and ERIC-2) were chosen to examine a set of 105 N. meningitidis strains comprising the 30 reference strains and a selection of 25 strains, isolated in the province of Antwerp, which predominantly had phenotypes B:4:P1.4 (27 strains) and B:NT:P1.4 (7 strains). Oligonucleotide D8635-primed PCR yielded the most discriminative pattern. In addition, it was the only primer able to unequivocally differentiate all but two reference strains (see Fig. 2). Isolates with the B:4:P1.4 or B:NT:P1.4 phenotype, including the Dutch B:4:P1.4 reference strain L93/2723, had very similar DNA amplification patterns, with the only differences primarily being differences in band intensity (cf. Fig. 1). This pattern, which we refer to as “pattern 2723,” was characterized by 11 bands ranging in size from 150 to 1,150 bp (bands marked by arrows in Fig. 1, lane 4). Pattern 2723 was clearly different from the amplification patterns of the remaining reference strains, and it was encountered in all B:4:P1.4 and B:NT:P1.4 strains. In spite of occasional minor reproducibility problems in the high-molecular-mass region, numerical analysis of the DNA amplification patterns clearly grouped all strains belonging to pattern 2723 in a single cluster (Fig. 2).
FIG. 2.
Dendrogram derived from the unweighted pair group average linkage of correlation coefficients between the PCR profiles obtained with arbitrary primer D8635 for 61 N. meningitidis strains comprising the 30 reference strains (strains L93/2701 through L93/2730), 25 Belgian pattern 2723 strains, and 6 Dutch B:4:P1.4 strains (strains NL83, NL84, NL85, NL86, NL87, and NL90). The correlation coefficient is represented as a percent similarity for convenience. NT, nonserotypeable.
FIG. 1.
PCR analysis with primer D8635 of a selection of reference and representative pattern 2723 N. meningitidis strains (phenotypes are given in parentheses). Lanes 1 to 4, reference strains L93/2703 (A:4), L93/2707 (B:NT:P1.9), L93/2715 (B:15:P1.7,16), and L93/2723 (B:4:P1.4), respectively; lanes 5 to 13, pattern 2723 strains 95/72 (B:4:P1.4), 95/154 (B:4:P1.4), 92/188 (B:4:P1.4), 94/89 (B:4:P1.4), 93/114 (C:4:P1.4), 93/77 (B:4:P1.13), 94/110 (B:4:P1.1,7), 95/227 (B:4:P1.6), and 93/133 (B:1:P1.4), respectively; lane M, 100-bp DNA ladder. The arrows indicate the bands characteristic of pattern 2723.
The arbitrary primer D11344 and the ERIC primers were not used for further typing studies, since they unsatisfactorily discriminated between the reference strains. In addition, their amplification patterns consisted of only about 8 bands, whereas primer D8635 generated up to 18 bands. They revealed few relationships between individual strains but could detect slight differences between strains with the B:4:P1.4 or B:NT:P1.4 phenotype and as such are noteworthy.
Consequently, primer D8635 was chosen to examine all of 420 strains isolated in Belgium between 1990 and 1995. Identification of pattern 2723 was performed by means of numerical analysis and visual comparison with the pattern of strain L93/2723, which was run on each gel. Of the 157 strains with the B:4:P1.4 or B:NT:P1.4 phenotype, all but 1 were characterized as having pattern 2723. In addition, 55 strains with other phenotypes were also characterized as having the same overall pattern (Table 1), although occasionally the banding pattern differed from the typical profile (Fig. 1, lane 4) by the absence of one of the bands (Fig. 1, lanes 10, 11, and 13), by the presence of an additional band (Fig. 1, lanes 5 and 12), or by a clear difference in the density of one of the bands (Fig. 1, lane 13). Most of the non-B:4:P1.4 or non-B:NT:P1.4 strains characterized as having pattern 2723 (21 strains [i.e., 40%]) had the B:4:P1.13 phenotype. In addition, 7 strains with the B:1:P1.4 phenotype, 6 strains with the B:4:P1.1,7 phenotype, and 5 strains that were neither serotypeable nor subtypeable also had pattern 2723. The remainder of the phenotypes were represented by only single isolates (Table 1). Remarkably, one serogroup C strain revealed pattern 2723 (Fig. 1, lane 9), and the 20 clinical isolates with the B:4:P1.4 phenotype, isolated in The Netherlands between 1980 and 1990, all had pattern 2723.
TABLE 1.
Distribution of pattern 2723, as determined by D8635-primed PCR, among 420 N. meningitidis strains isolated in Belgium between 1990 and 1995, 20 Dutch isolates, and 30 reference strains
| Strain and phenotypea | No. of strains with the following D8635-primed PCR DNA amplification pattern:
|
|||||
|---|---|---|---|---|---|---|
| Pattern 2723
|
Other patterns | |||||
| Pattern 2723b | Absence of one band | Presence of an additional band | Difference in density of one band | Absence of one band and difference in density of one band | ||
| Belgian isolates | ||||||
| B:4:P1.4 | 112 | 14 | 6 | 9 | 0 | 0 |
| B:NT:P1.4 | 12 | 2 | 0 | 1 | 0 | 1 |
| B:4:P1.13 | 15 | 3 | 0 | 3 | 0 | 1 |
| B:NT:NST | 4 | 1 | 0 | 0 | 0 | 13 |
| B:1:P1.4 | 4 | 1 | 0 | 1 | 1 | 0 |
| B:NT:P1.2,5 | 1 | 0 | 0 | 0 | 0 | 6 |
| B:4:P1.1,7 | 2 | 2 | 0 | 2 | 0 | 1 |
| B:NT:P1.1,7 | 0 | 0 | 0 | 1 | 0 | 5 |
| B:14:P1.4 | 2 | 0 | 0 | 0 | 0 | 0 |
| B:14:P1.15 | 0 | 0 | 0 | 1 | 0 | 1 |
| B:4:P1.6 | 0 | 0 | 1 | 0 | 0 | 1 |
| B:4:P1.7 | 2 | 0 | 0 | 0 | 0 | 1 |
| B:NT:P1.7 | 2 | 1 | 0 | 1 | 0 | 3 |
| B:4:NST | 2 | 0 | 0 | 0 | 0 | 1 |
| C:4:P1.4 | 1 | 0 | 0 | 0 | 0 | 0 |
| NG:NT:NST | 0 | 1 | 0 | 0 | 0 | 0 |
| Others | 0 | 0 | 0 | 0 | 0 | 175 |
| Total | 159 | 25 | 7 | 19 | 1 | 209 |
| Dutch isolates (B:4:P1.4) | 16 | 2 | 1 | 1 | 0 | 0 |
| Reference strains | 2 | 0 | 0 | 0 | 0 | 28 |
NG, nongroupable; NT, nonserotypeable; NST, nonsubtypeable.
Pattern 2723 is described in the text.
Strains representative of the various phenotypes characterized by pattern 2723 (among which six were Dutch B:4:P1.4 strains) were included in a numerical comparison of DNA amplification patterns, as illustrated in Fig. 2. All pattern 2723 strains formed a single cluster above the 85% similarity level. The reproducibility of the method was tested with primer D8635. Multiple DNA extracts of 10 randomly chosen strains always generated identical patterns when they were included in the same PCR assay. In general, the intrarun reproducibility was excellent. However, the reproducibility between different amplification assays was not always satisfactory because, occasionally, minor DNA fragments, especially in the high-molecular-mass region, were not reproducibly amplified. Identification of pattern 2723 was therefore always confirmed by visual inspection of the DNA banding patterns (a strain with pattern 2723 was run on each gel). When examined by numerical analysis, reproducibility within and between gels was at least 90%.
MLEE.
A subset of 61 strains was selected for determination of their ETs. This subset included 52 pattern 2723 strains and 9 strains that had a different amplification pattern. The latter 9 strains belonged to phenotypes that can also have representatives among the pattern 2723 strains.
The 52 pattern 2723 strains included a random stratified sample of B:4:P1.4 (n = 26), B:NT:P1.4 (n = 6), B:4:P1.13 (n = 4), B:NT:nonsubtypeable (NST) (n = 1), B:1:P1.4 (n = 2), B:NT:P1.2,5 (n = 1), B:4:P1.1,7 (n = 4), B:NT:P1.1,7 (n = 1), B:14:P1.4 (n = 1), B:14:P1.15 (n = 1), B:4:P1.6 (n = 1), B:4:P1.7 (n = 1), B:NT:P1.7 (n = 2), and C:4:P1.4 (n = 1) isolates. The other strains included one B:NT:P1.4, one B:4:P1.13, one B:NT:P1.13, two B:NT:NST, one B:4:P1.1,7, one B:14:P1.15, and two B:NT:P1.6 strains.
Among the 61 strains analyzed by MLEE, 10 very closely related clones that differed from each other at two or fewer alleles were identified as belonging to lineage III (Table 2) (8). Of these, ETs 24, 24.3, and 25 were previously encountered in The Netherlands (8, 21). Thirty-five strains were ET 24, and among these 35 strains 2 were non-pattern 2723 strains (strains 95/159 and 93/31): 1 isolate was ET 24.3, and 1 isolate was ET 25. The remaining seven ETs of lineage III had not been identified in The Netherlands.
TABLE 2.
Allele profiles and serogroups:serotypes for 61 Belgian N. meningitidis strains
| Cluster | ETa | Strainb | Allele at the indicated enzyme locusa,c:
|
Serogroup: serotype | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAE | G6P | PEP | IDH | ACO | GD1 | GD2 | ADH | FUM | ALK | IP1 | IP2 | ADK | UDH | ||||
| Lineage III | 24 | 92/148, 92/155, 92/188, 93/1, 93/18, 93/81, 93/86, 93/146, 94/1, 94/4, 94/28, 94/42, 94/48, 94/72, 94/89, 95/17, 95/72, 95/150, 95/151, 95/154, 95/180, 95/219 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.4 |
| 24 | 92/154 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.4 | |
| 24 | 93/77, 93/130, 94/91, 95/159,d 95/171 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.13 | |
| 24 | 93/31d | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.13 | |
| 24 | 95/227 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.6 | |
| 24 | 94/22 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.7 | |
| 24 | 93/40, 94/11 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.7 | |
| 24 | 94/97 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:NST | |
| 24 | 93/114 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | C:4:P1.4 | |
| 24.3 | 93/124 | 1 | 3 | 5 | 12 | 2 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.4 | |
| 24.5 | 92/139 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 1 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.4 | |
| 24.6 | 93/17 | 5 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 5 | 2 | 3 | 2 | 3 | B:4:P1.4 | |
| 24.7 | 93/132 | 5 | 3 | 5 | 12 | 0 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.4 | |
| 24.8 | 94/80 | 5 | 3 | 5 | 13 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:1:P1.4 | |
| 24.9 | 94/110, 95/165, 95/176 | 5 | 1 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.1,7 | |
| 24.9 | 95/80 | 5 | 1 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.1,7 | |
| 24.9 | 95/36 | 5 | 1 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.2,5 | |
| 24.10 | 95/148, 95/187 | 5 | 3 | 5 | 9 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:4:P1.4 | |
| 24.11 | 93/29, 94/12, 95/184 | 1 | 3 | 5 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.4 | |
| 93/98 | B:14:P1.4 | ||||||||||||||||
| 25 | 93/133 | 5 | 3 | 5 | 12 | 4 | 2 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:1:P1.4 | |
| Other lineages | A | 94/54 | 1 | 3 | 4 | 5 | 0 | 1 | 3 | 2 | 1 | 8 | 2 | 3 | 2 | 3 | B:4:P1.1,7 |
| B | 93/92d | 3 | 1 | 4 | 5 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.6 | |
| C | 94/37d | 3 | 5 | 4 | 9 | 0 | 1 | 3 | 0 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.4 | |
| D | 95/106d | 3 | 1 | 9 | 9 | 4 | 1 | 3 | 3 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:NST | |
| E | 95/129d | 3 | 3 | 4 | 12 | 0 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:14:P1.15 | |
| F | 94/57d | 3 | 3 | 4 | 12 | 4 | 1 | 3 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:NST | |
| G | 95/194 | 3 | 3 | 4 | 12 | 4 | 1 | 3 | 2 | 1 | 8 | 2 | 3 | 2 | 3 | B:14:P1.15 | |
| H | 95/136d | 3 | 4 | 2 | 7 | 4 | 1 | 1 | 2 | 1 | 3 | 2 | 3 | 2 | 3 | B:NT:P1.6 | |
| I | 95/93d | 4 | 3 | 1 | 7 | 4 | 1 | 3 | 0 | 1 | 3 | 2 | 5 | 2 | 3 | B:4:P1.1,7 | |
ET numbers for lineage III strains are as described by Scholten et al. (21), with new ETs having new consecutive decimal numbers.
The first two numbers indicate the year of isolation.
Enzyme abbreviations: MAE, malic enzyme; G6P, glucose-6-phosphate dehydrogenase; PEP, peptidase; IDH, isocitrate dehydrogenase; ACO, aconitase; GD1, NADP-linked glutamate dehydrogenase; GD2, NAD-linked glutamate dehydrogenase; ADH, alcohol dehydrogenase; FUM, fumarase; ALK, alkaline phosphatase; IP1 and IP2, two indophenol oxidases; ADK, adenylate kinase; UDH, an unknown dehydrogenase.
Non-pattern 2723 strains.
Two strains (strains 94/54 and 95/194) classified as belonging to pattern 2723 by the PCR method did not belong to lineage III. Strain 94/54 had an ET that was not earlier encountered, and strain 95/194 was a representative of lineage IV, described by Caugant et al. (8). Reference strain L93/2714, which had pattern 2723, also did not belong to lineage III (5).
Remarkably, the single non-pattern 2723 B:NT:P1.4 strain (strain 94/37) did not belong to lineage III (Table 2), confirming the results of the PCR analysis.
PFGE-based typing.
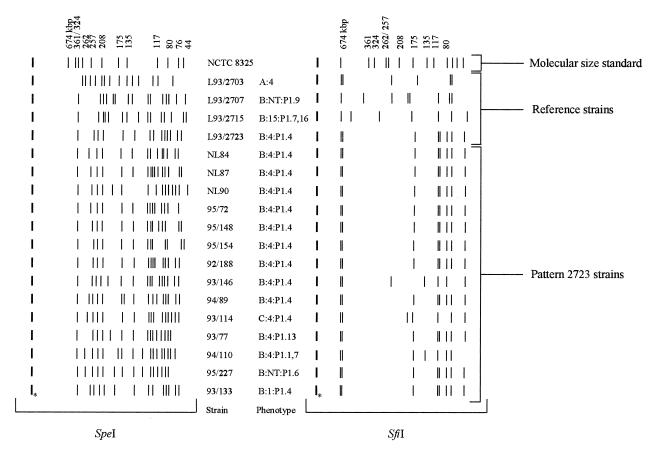
The SpeI macrorestriction fragment distribution of the same set of 61 strains investigated by MLEE was studied. In addition, we included the 30 reference strains and 6 Dutch B:4:P1.4 isolates.
In general, SpeI produced 14 to 20 distinct genomic DNA fragments ranging in size from 6 to 340 kbp (Fig. 3). Visual comparison of the banding patterns of the reference strains and field isolates revealed some epidemiologically related clusters of indistinguishable (no band differences), closely related (two to three band differences), or possibly related (four to six band differences) strains as defined by Tenover et al. (27). However, the majority of the strains was classified as not related. Most pattern 2723 strains (including reference strain L93/2723), as determined by D8635-primed PCR, were characterized by several common bands in their PFGE patterns (Fig. 3). The remaining 29 reference strains had clearly different macrorestriction restriction profiles and were classified as being unrelated, as determined by Tenover et al. (27).
FIG. 3.
PFGE of SpeI- and SfiI-cleaved genomic DNAs of reference strains and representative pattern 2723 strains (including three Dutch pattern 2723 strains [strains NL84, NL87, and NL90]). The SmaI-digested genome of Staphylococcus aureus NCTC 8325 (lane M) was used as molecular size standard. ∗, places where plugs were inserted.
The restriction enzyme SfiI produced seven to eight fragments ranging in size from 30 to 700 kbp (Fig. 3). All pattern 2723 strains showed very similar banding patterns. Among the 52 Belgian pattern 2723 strains tested, 35 (67%) had the same banding pattern as the Dutch reference strain, 9 were closely related, 4 were possibly related, and 4 were unrelated to strain L93/2723 (27). The six Dutch strains investigated all had the same banding pattern as the B:4:P1.4 reference strain.
All except three of the reference strains had easily distinguishable banding patterns. The fingerprints of the two reference strains L93/2716 (B:21:P1.16) and L93/2730 (W:2c:P1.3) were identical and closely related to that of the Dutch reference strain, respectively.
For the nine non-pattern 2723 strains tested, six were unrelated to the Dutch reference strain (including strain 94/37, again confirming the results of the PCR analysis). Strains 93/31 and 95/159, which did not have pattern 2723 by PCR but which were of ET 24 of lineage III on the basis of MLEE, were closely and possibly related to the Dutch reference strain, respectively. Finally, strain 94/57 was possibly related to the Dutch reference strain.
The reproducibility of the PFGE patterns was examined by analysis of the same bacterial strain on several occasions, and identical patterns were observed upon repeat analysis. A reproducibility level of 100% was obtained when the similarity of the PFGE banding patterns was calculated with the Dice coefficient.
DISCUSSION
Much effort has been devoted to the typing of meningococcal strains in order to trace outbreaks and epidemics, compare isolates from patients with disease and isolates from carriers, identify virulent strains, and assess the genetic relatedness among isolates of this species (1, 6, 9). Classical typing schemes for meningococci involve serological analyses. Inherent to this phenotyping system are problems which include the inability to subtype all isolates with the available reagents, poor expression or masking of surface antigens, and phenotypic variations of the organisms that affect typeability (6, 7). MLEE is used as a standard method for meningococcal typing (6) and allows investigators to study changes in meningococcal populations occurring in connection with outbreaks or epidemics (23).
At present, relatedness among isolates is often assessed by DNA-based typing techniques. Random-primer- and repetitive-motif-based PCR analysis is fast and appears to be increasingly useful for the differentiation of strains within species (2, 16, 29, 32). PFGE of genomic macrorestriction fragments is highly discriminatory and is the reference method for several bacterial species (10, 14, 15, 18). It was proven to be useful for establishing genetic relationships among serogroup A, B, and C meningococci (3, 25, 33, 34).
In the present study, we primarily used arbitrarily primed PCR to characterize a collection of 420 strains isolated between 1990 and 1995 in Belgium. All of these isolates were serogrouped, serotyped, and serosubtyped at the Meningococcal Reference Center, Department of Bacteriology, Scientific Institute for Public Health Louis-Pasteur, Brussels, Belgium, the details of which are presented elsewhere (28). We demonstrated that the increase in the number of meningococcal infections in Belgium was primarily due to strains with the B:4:P1.4 and B:NT:P1.4 phenotypes and hypothesized that this “hyperendemic wave” (17) of N. meningitidis infection was due to a bacterial clone which originated in The Netherlands, entered Belgium in the early 1990s, and spread through Belgium in the following years (28).
Comparative analysis of the 420 clinical meningococcal isolates with primer D8635 revealed highly similar DNA amplification patterns for 211 strains, among which were included all 141 strains with the B:4:P1.4 phenotype and 15 of 16 strains with the B:NT:P1.4 phenotype (one isolate did not have pattern 2723) (Table 1). In addition, 55 strains of 14 other phenotypes had pattern 2723; of the latter, 10 phenotypes also had representatives that did not have pattern 2723. One serogroup C strain also had pattern 2723. This was later confirmed in the MLEE and PFGE analyses. Also, a single B:NT:P1.4 strain did not have pattern 2723, and this was later confirmed by the other analyses. Close genetic relationships between strains of serogroup B and C were already demonstrated in 1986 (6), and recently, Swartley and coworkers (26) have shown that two meningococcal isolates of the same clone can express different capsular serogroups just by exchanging the synD gene encoding the polysialyltransferase. This genotype-based approach indicated that all strains with the B:4:P1.4 phenotype and all but one of the strains with the B:NT:P1.4 phenotype belong to a single epidemic lineage. However, it also demonstrated that the epidemic lineage is not confined to strains with these phenotypes, because 26% of the strains with pattern 2723 had other phenotypes.
This is in agreement with the findings of Scholten and coworkers (21), who stated that clones that are newly introduced in a certain geographic area will be homogeneous with regard to surface characteristics, but clones that have been prevalent n a population for a long period might have undergone changes in their surface characteristics due to mutation and/or recombination of genes encoding surface markers. In that case, the clones become increasingly heterogeneous with regard to phenotype. Caugant et al. (8) reported on a new lineage of N. meningitidis (lineage III) that appeared in The Netherlands in about 1980. At its appearance, the lineage was very homogeneous with regard to both ET (ET 24) and phenotype (B:4:P1.4). After 1984, other clones appeared in the lineage (clones of ET 24.1, ET 24.2, ET 24.3, ET 24.4, and ET 25), and the various clones acquired other serotypes (serotypes 14 and 15) and subtypes (subtypes P1.2, P1.7, and P1.12), indicating the frequent exchange of genetic material between clones (21). Until 1984, strains with the B:4:P1.4 phenotype were predominantly recovered in the southwestern part of The Netherlands. Afterward, the phenotype dispersed throughout the country, but the incidence remained most pronounced in the southern provinces. In Belgium, an increase in the incidence of meningococcal disease was first noticed in the province of Antwerp, near the border with The Netherlands (28), and the first strain with the B:4:P1.4 phenotype was noticed in 1990. Thus, there was a time period of 10 years between the introduction of B:4:P1.4 strains in The Netherlands and the first detection of strains with that phenotype in Belgium. It therefore seems logical that the epidemic lineage entering Belgium in the 1990s is not a single clone but a cluster of closely related clones. This was substantiated by the findings of the present study.
The PCR patterns of the Belgian epidemic strains correspond to that of Dutch reference strain L93/2723 and to the patterns of the 20 clinical isolates from The Netherlands, indicating that the same epidemic lineage is present in both countries and suggesting that the Belgian hyperendemic wave originated in The Netherlands. Investigation of 52 pattern 2723 strains by MLEE showed that all but two pattern 2723 strains indeed belonged to lineage III, and 10 closely related clones were identified: the majority of the investigated pattern 2723 strains belonged to ETs 24, 24.3, and 25, three ETs previously encountered in The Netherlands. However, 15 strains representing 7 different variants were not found previously in The Netherlands. Surprisingly, two of the pattern 2723 strains (strains 94/54 and 95/194) did not belong to lineage III. When PFGE was applied to these strains, their SpeI macrorestriction profiles had no bands in common with those of the other pattern 2723 strains. However, when the restriction enzyme SfiI was used, strain 95/194 had the same pattern as the Dutch reference strain and strain 94/54 remained unrelated to the pattern 2723 strains. Alternatively, among the nine non-pattern 2723 strains tested by MLEE, two strains (strains 93/31 and 95/159) were found to belong to lineage III. The SpeI macrorestriction profiles of these two strains shared several bands with the pattern 2723 strains, and PFGE with the restriction enzyme SfiI demonstrated that they were both related to the Dutch reference strain, which is in agreement with the results obtained by MLEE.
In the PFGE analysis, a total of 97 strains, comprising the 30 reference strains, 6 Dutch isolates, and 61 clinical isolates, were investigated. Analysis of these strains with the restriction enzyme SpeI revealed a high degree of pattern diversity among the reference strains and among the strains with pattern 2723. All strains with pattern 2723, as delineated by D8635-primed PCR, shared several bands (Fig. 3). However, without prior knowledge of relationships revealed by randomly amplified polymorphic DNA analysis or MLEE, allocation of isolates to an epidemic type defined by PFGE is hardly possible. In addition, pairwise comparison of the PFGE patterns of isolates presumed to be part of the hyperendemic wave by using the criteria defined by Tenover et al. (27) would classify the majority of these isolates as unrelated. The data obtained with the restriction enzyme SfiI were remarkably different. Of the 52 Belgian pattern 2723 strains investigated, 48 were related to the Dutch reference strain when the criteria of Tenover et al. (27) were applied. The results for the remaining four strains were also inconsistent with those of PCR typing, and a total of seven discrepancies for PCR typing and five discrepancies for MLEE typing were noticed. The restriction enzyme SfiI is less discriminative, and for the meningococci, SfiI gives a fingerprint characteristic for the subgroups and complexes of ETs, as defined by MLEE (17).
Conclusions.
When using primer D8635, a similar DNA amplification pattern is observed for all strains with the B:4:P1.4 phenotype and all except one of the B:NT:P1.4 strains. However, strains with several other serotypes and subtypes and even one serogroup C strain also had the same pattern. Application of MLEE and PFGE with the restriction enzyme SfiI confirmed the data obtained by PCR analysis. PFGE with the restriction enzyme SpeI, however, classified the majority of the strains as being nonrelated, which is contradictory to the results obtained by all other typing methods used in the investigation. This is due to the fact that SpeI is far too sensitive (discriminative) for population studies of meningococci. The differences between the different typing methods presumably reflect differences in the nature of the data used by each technique. Discrepancies could be caused by differences between the chromosomal locations of the genes encoding the proteins used in MLEE, PFGE restriction sites, or primer-binding sites in the PCR assays or by changes in chromosome structure which did not affect the phenotypic characters used in MLEE or serological studies. The absence of a full agreement between any of the various typing methods used in the investigation highlights the need to carefully evaluate the suitability of the typing method(s) used for a particular organism and suggests that conclusions should be based on evidence provided by multiple typing methods.
Finally, our study demonstrates that the current increase in the incidence of meningococcal disease in Belgium is not restricted to one phenotype. Therefore, serotyping alone would underestimate the prevalence of the epidemic clone. Comparison of Dutch and Belgian isolates suggests a southward migration of a genetically well-delineated lineage of strains causing disease in The Netherlands since the early 1980s and in Belgium since the early 1990s.
ACKNOWLEDGMENTS
This study was supported in part by a grant from Glaxo Belgium to M.V.L.
P.V. is indebted to the Fund for Scientific Research-Flanders (Belgium) for a position as a postdoctoral research fellow.
REFERENCES
- 1.Achtman M. Clonal spread of serogroup A meningococci: a paradigm for the analysis of microevolution in bacteria. Mol Microbiol. 1994;11:15–22. doi: 10.1111/j.1365-2958.1994.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz N, Bukano N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bygraves J A, Maiden M C J. Analysis of the clonal relationships between strains of Neisseria meningitidis by pulsed-field gel electrophoresis. J Gen Microbiol. 1992;138:523–531. doi: 10.1099/00221287-138-3-523. [DOI] [PubMed] [Google Scholar]
- 4.Carion, F. Unpublished data.
- 5.Caugant, D. A. Unpublished data.
- 6.Caugant D A, Frøholm L O, Bøvre K, Holten E, Frasch C E, Mocca L F, Zollinger W D, Selander R K. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc Natl Acad Sci USA. 1986;83:4927–4931. doi: 10.1073/pnas.83.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caugant D A, Mocca L F, Frasch C E, Frøholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caugant D A, Bol P, Høiby E A, Zanen H C, Frøholm L O. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958–1986. J Infect Dis. 1990;162:867–874. doi: 10.1093/infdis/162.4.867. [DOI] [PubMed] [Google Scholar]
- 9.Crowe B A, Wall R A, Kusecek B, Neumann B, Olyhoek T, Abdillahi H, Hassan-King M, Greenwood B M, Poolman J T, Achtman M. Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in The Gambia, West Africa. J Infect Dis. 1989;159:686–700. doi: 10.1093/infdis/159.4.686. [DOI] [PubMed] [Google Scholar]
- 10.De Moissac Y R, Ronald S L, Peppler M S. Use of pulsed-field gel electrophoresis for epidemiological study of Bordetella pertussis in a whooping cough outbreak. J Clin Microbiol. 1994;32:398–402. doi: 10.1128/jcm.32.2.398-402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dice L R. Measure of the amounts of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 12.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 13.Frasch, C. E. 1989. Vaccines for prevention of meningococcal disease. Clin. Microbiol. Rev. 2(Suppl.):S134–S138. [DOI] [PMC free article] [PubMed]
- 14.Gordillo M E, Singh K V, Baker C J, Murray B E. Typing of group B streptococci: comparison of pulsed-field gel electrophoresis and conventional electrophoresis. J Clin Microbiol. 1993;31:1430–1434. doi: 10.1128/jcm.31.6.1430-1434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harsono K D, Kaspar C W, Luchansky J B. Comparison and genomic sizing of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1993;59:3141–3144. doi: 10.1128/aem.59.9.3141-3144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski J R, Weinstock G M. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiden M C J, Feavers I M. Population genetics and global epidemiology of the human pathogen Neisseria meningitidis. In: Baumberg S, Young J P W, Saunders S R, Wellington E M H, editors. Population genetics of bacteria. Cambridge, Great Britain: Cambridge University Press; 1995. pp. 269–293. [Google Scholar]
- 18.Miranda A G, Singh K V, Murray B E. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J Clin Microbiol. 1991;29:2752–2757. doi: 10.1128/jcm.29.12.2752-2757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 20.Scholten R J P M, Bijlmer H A, Poolman J T, Kuipers B, Caugant D A, Van Alphen L, Dankert J C, Valkenburg H. Meningococcal disease in The Netherlands, 1958–1990: a steady increase in the incidence since 1982 partially caused by new serotypes and subtypes of Neisseria meningitidis. Clin Infect Dis. 1993;16:237–246. doi: 10.1093/clind/16.2.237. [DOI] [PubMed] [Google Scholar]
- 21.Scholten R J P M, Poolman J T, Valkenburg H A, Bijlmer H A, Dankert J, Caugant D A. Phenotypic and genotypic changes in a new clone complex of Neisseria meningitidis in The Netherlands, 1958–1990. J Infect Dis. 1994;169:673–676. doi: 10.1093/infdis/169.3.673. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz, B., P. S. Moore, and C. V. Broome. 1989. Global epidemiology of meningococcal disease. Clin. Microbiol. Rev. 2(Suppl.):S118–S124. [DOI] [PMC free article] [PubMed]
- 23.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman & Co.; 1973. [Google Scholar]
- 25.Strathdee C A, Tyler S D, Ryan J A, Johnson W M, Ashton F E. Genomic fingerprinting of Neisseria meningitidis associated with group C meningococcal disease in Canada. J Clin Microbiol. 1993;31:2506–2508. doi: 10.1128/jcm.31.9.2506-2508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swartley J S, Marfin A A, Edupuganti S, Liu L-J, Cieslak P, Perkins B, Wenger J D, Stephens D S. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Looveren M, Carion F, Vandamme P, Goossens H. Surveillance of meningococcal infections in Belgium. Clin Microbiol Infect. 1998;4:224–228. doi: 10.1111/j.1469-0691.1998.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 29.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wedege E, Kolberg J, Delvig A, Høiby E A, Holten E, Rosenqvist E, Caugant D A. Emergence of a new virulent clone within the electrophoretic type 5 complex of serogroup B meningococci in Norway. Clin Diagn Lab Immunol. 1995;2:314–321. doi: 10.1128/cdli.2.3.314-321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whalen C M, Hockin J C, Ryan A, Ashton F. The changing epidemiology of invasive meningococcal disease in Canada, 1985 through 1992. Emergence of a virulent clone of Neisseria meningitidis. JAMA. 1995;273:390–394. [PubMed] [Google Scholar]
- 32.Woods J P, Kersulyte D, Tolan R W, Jr, Berg C M, Berg D E. Use of arbitrarily primed polymerase chain reaction analysis to type disease and carrier strains of Neisseria meningitidis isolated during a university outbreak. J Infect Dis. 1994;169:1384–1389. doi: 10.1093/infdis/169.6.1384. [DOI] [PubMed] [Google Scholar]
- 33.Yakubu D E, Abadi F J R, Pennington T H. Molecular epidemiology of recent United Kingdom isolates of Neisseria meningitidis serogroup C. Epidemiol Infect. 1994;113:53–65. doi: 10.1017/s0950268800051463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakubu D E, Pennington T H. Epidemiological evaluation of Neisseria meningitidis serogroup B by pulsed-field gel electrophoresis. FEMS Immunol Med Microbiol. 1995;10:185–190. doi: 10.1111/j.1574-695X.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]