Abstract
The NAM, ATAF1/2, and CUC2 (NAC) domain transcription factor VND‐INTERACTING2 (VNI2) negatively regulates xylem vessel formation by interacting with another NAC domain transcription factor, VASCULAR‐RELATED NAC‐DOMAIN7 (VND7), a master regulator of xylem vessel formation. Here, we screened interacting proteins with VNI2 using yeast two‐hybrid assay and isolated two NAC domain transcription factors, Arabidopsis thaliana ACTIVATION FACTOR 2 (ATAF2) and NAC DOMAIN CONTAINING PROTEIN 102 (ANAC102). A transient gene expression assay showed that ATAF2 upregulates the expression of genes involved in leaf senescence, and VNI2 effectively inhibits the transcriptional activation activity of ATAF2. vni2 mutants accelerate leaf senescence, whereas ataf2 mutants delay leaf senescence. In addition, the accelerated leaf senescence phenotype of the vni2 mutant is recovered by simultaneous mutation of ATAF2. Our findings strongly suggest that VNI2 interacts with and inhibits ATAF2, resulting in negatively regulating leaf senescence.
Keywords: Arabidopsis thaliana , leaf senescence, NAC domain protein, protein–protein interaction, transcription factor
1. INTRODUCTION
NAC domain transcription factors, one of the plant‐specific transcription factor families, have been known to be associated with a wide variety of functions in plants, such as cell proliferation, biotic and abiotic responses, flowering time, secondary cell wall biosynthesis, and phloem development (Furuta et al., 2014; Kim et al., 2006; Podzimska‐Sroka et al., 2015; Souer et al., 1996; Yamaguchi et al., 2008). It has been reported that the function of several NAC domain transcription factors is post‐translationally regulated by modifications such as phosphorylation (Hamasaki et al., 2019), ubiquitination (Miao et al., 2016), and S‐nitrosylation (Kawabe et al., 2018; Ohtani et al., 2018). In addition, NAC domain transcription factors form homo‐dimer and/or hetero‐dimer complexes with other NAC domain transcription factors (Jensen & Skriver, 2014; Mohanta et al., 2020; Olsen et al., 2005; Puranik et al., 2012). In peaches, the NAC transcription factor BLOOD (BL) can interact with NAC1 to amplify its regulatory effect on anthocyanin biosynthesis (Zhou et al., 2015).
An Arabidopsis NAC domain transcription factor, VASCULAR‐RELATED NAC‐DOMAIN PROTEIN7 (VND7), acts as a master regulator of xylem vessel element differentiation (Nakano et al., 2015; Yamaguchi & Demura, 2010). Transcriptome analysis demonstrated that VND7 directly regulates a broad range of genes involved in xylem vessel differentiation (Yamaguchi et al., 2011). VND7 forms homo‐dimers and hetero‐dimers with other VND proteins (Yamaguchi et al., 2008). In addition, other NAC domain transcription factors, VND‐INTERACTING1 (VNI1), VNI2, and ANAC103, were isolated as interacting proteins with VND7 (Yamaguchi et al., 2015; Yamaguchi, Ohtani, et al., 2010). Further analyses showed that VNI2 inhibits the transcriptional activation activities of VND7 by forming protein complexes, negatively regulating xylem vessel formation (Ailizati et al., 2022; Yamaguchi, Ohtani, et al., 2010).
Promoter analysis demonstrated that VNI2 expression was observed in various types of tissues and cells, such as phloem and guard cells as well as xylem vessel precursor cells (Yamaguchi, Ohtani, et al., 2010). In addition, VNI2 expression was induced in response to abiotic stress or abscisic acid (ABA) treatment (Yang et al., 2011). These expression profiles suggested that VNI2 plays roles in various biological processes (Yamaguchi, Ohtani, et al., 2010; Yang et al., 2011). Indeed, a vni2 mutant exhibited early leaf senescence and was susceptible to salt stress (Yang et al., 2011). It has also been reported that VNI2 interacts with a geminiviral replication initiator protein (Suyal et al., 2014). However, the molecular function of VNI2 remains to be fully elucidated.
Leaf senescence involves a series of intra‐cellular changes to breakdown chlorophyll and macromolecules in order to remobilize nutrients to reproductive, young, or even storage tissues. Some environmental changes such as drought, light, herbivores, or pathogens trigger leaf senescence (Sade et al., 2018). Regulation of leaf senescence involves a number of NAC domain transcription factors. Around 20 NAC domain transcription factors, including ORESARA1 (ORE1) (Kim et al., 2009), ORESARA1 SISTER1 (ORS1) (Balazadeh et al., 2011), AtNAP, ANAC046 (Oda‐Yamamizo et al., 2016), ATAF1 (Garapati et al., 2015), ANAC019 (Hickman et al., 2013), and ANAC072 (Li et al., 2016), are known to be positive regulators of leaf senescence. By contrast, JUNGBRUNNEN1 (JUB1) (Wu et al., 2012), ANAC017, ANAC082/VNI1, ANAC090 (Kim et al., 2018), and VNI2 (Yang et al., 2011) are reported as negative regulatory NAC domain transcription factors. These transcription factors consist of gene regulatory networks that control leaf senescence (Kim et al., 2014, 2018; Podzimska‐Sroka et al., 2015). Furthermore, it has been reported that these NAC domain transcription factors form various combinations of protein complexes during the leaf senescence process (Kim et al., 2018).
ATAF2 and ANAC102, which are close homologs of each other, belong to the stress‐responsive NAC subfamily SNAC‐A (Nakashima et al., 2012; Nuruzzaman et al., 2013). ANAC102 is involved in low‐oxygen stress responses (Christianson et al., 2009), high‐light stress responses (D'Alessandro et al., 2018), and brassinosteroid catabolism (Peng & Neff, 2021). It has also been reported that ANAC102 localizes in chloroplast nucleoids and regulates gene expression in chloroplasts (Xin et al., 2021). ATAF2 is involved in various processes such as biotic stress responses (Delessert et al., 2005), brassinosteroid catabolism (Peng et al., 2015; Peng & Neff, 2020, 2021), auxin biosynthesis (Huh et al., 2012), photomorphogenesis (Peng et al., 2015, 2020), circadian clock (Peng et al., 2015, 2020; Peng & Neff, 2020, 2021), and ethylene biosynthesis and responses (Peng et al., 2022). Recently, we found that ATAF2 upregulates several senescence regulators such as ORE1, ORS1, VNI2, and ANAC046 (Nagahage et al., 2018, 2020). In addition, ATAF2 overexpressing plants and ataf2 mutants exhibited accelerated and delayed leaf senescence, respectively (Nagahage et al., 2020). Thus, these results demonstrated that ATAF2 promotes leaf senescence.
Here, to understand the biological roles of VNI2 more deeply, we screened for proteins that interact with VNI2. Then, we isolated two NAC domain transcription factors, ATAF2 and ANAC102. VNI2 interacts with and inhibits the transcriptional activation activities of ATAF2, as in the case of VND7. Furthermore, ataf2 mutation suppressed the early leaf senescence phenotype observed in the vni2 mutant. These data strongly suggested that VNI2 regulates leaf senescence through inhibition of ATAF2.
2. RESULTS
2.1. NAC domain proteins, ATAF2 and ANAC102, were isolated as interacting proteins with VNI2
An Arabidopsis NAC domain transcription factor, VNI2, negatively regulates xylem vessel formation by interacting with and inhibiting another NAC domain transcription factor, VND7, a master regulator of xylem vessel element differentiation (Yamaguchi, Ohtani, et al., 2010). Because VNI2 expression was observed in various tissues and cells, and was regulated in response to endogenous and exogenous signals (Yamaguchi, Ohtani, et al., 2010; Yang et al., 2011), it is likely that VNI2 regulates some other biological processes besides xylem vessel formation. To further investigate the functions and biological roles of VNI2, we attempted to isolate proteins that interact with VNI2 using yeast two‐hybrid screening. Plant transcription factors often have transcription activities even in yeast cells (Yamaguchi et al., 2008). Thus, transcription factors lacking the transcriptional activation domain are usually used as bait for yeast two‐hybrid systems (Yamaguchi, Ohtani, et al., 2010). By contrast, budding yeast AH109 cells expressing full‐length VNI2 fused to the GAL4 DNA‐binding domain (GAL4‐BD‐VNI2) could not grow on selective medium lacking histidine with 1 mM 3‐AT (Figure 1a). Therefore, we used GAL4‐BD‐VNI2 as bait for screening. From at least 5 × 105 independent cells transformed with the cDNA library prepared from Arabidopsis primary roots (Yamaguchi, Ohtani, et al., 2010), we identified four positive clones containing cDNAs encoding two distinct NAC domain transcription factors, ATAF2 and ANAC102 (3 clones). ATAF2 cDNA included the 5′ and 3′ untranslated regions and full‐length coding sequence, whereas all obtained ANAC102 cDNAs partly lacked the N‐terminal sequence but included the whole NAC domain (Figure S1). We confirmed that VNI2 also interacts with the full‐length coding region of ATAF2 or ANAC102 without untranslated regions (Figure 1a). ATAF2 and ANAC102 are the closest homologs in the Arabidopsis genome (Figure 1b). ATAF2 is known to be involved in biotic stress responses, brassinosteroid catabolism, auxin biosynthesis, light‐mediated development pathway together with the circadian clock, and leaf senescence (Delessert et al., 2005; Nagahage et al., 2018, 2020; Peng et al., 2015, 2020, 2022; Peng & Neff, 2020, 2021; Wang et al., 2009; Wang & Culver, 2012), whereas ANAC102 is involved in low‐oxygen responses and gene expression in chloroplasts (Christianson et al., 2009; Klok et al., 2002).
FIGURE 1.
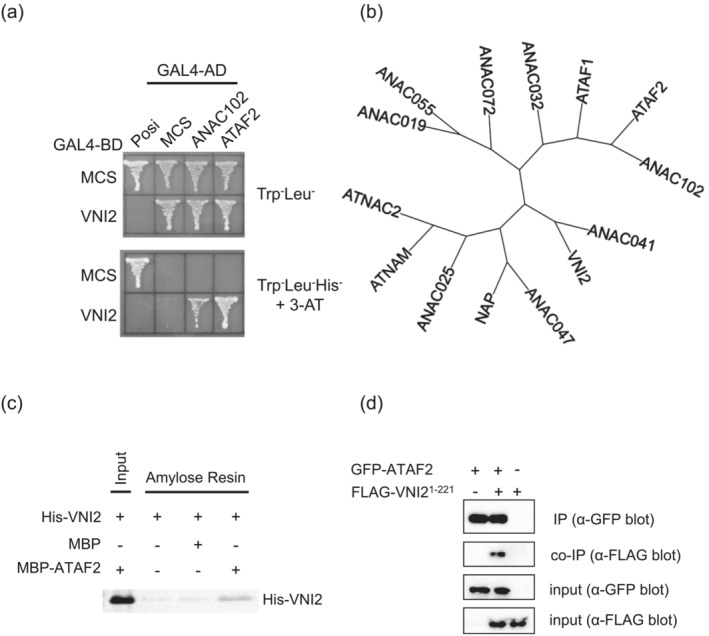
VNI2 interacts with ANAC102 and ATAF2. (a) Interaction of ANAC102 and ATAF2 together with VNI2 in yeast. The multi cloning site (MCS) and VNI2 fused to GAL4‐BD, and MCS,ANAC102, ATAF2 fused to GAL4‐AD were introduced into AH109 and grown on control (Trp‐Leu‐) and selective media (Trp‐Leu‐His‐). Plasmids containing MCS fused to GAL‐BD or GAL4‐AD were used as negative controls, and pBD‐wt and pAD‐wt were used as positive controls. (b) Phylogenetic tree of Arabidopsis NAC domain family involved in environmental stress. ANAC102 and ATAF2, isolated as interacting factors with VNI2, are closely related to each other. (c) In vitro binding of VNI2 to ATAF2. Proteins immobilized with the resin were subjected to immunoblot analysis. His‐VNI2 protein was detected with an anti‐His antibody. (d) In vivo binding of VNI21‐221 and ATAF2. YFP‐ATAF2 and FLAG‐VNI21‐221 were transiently co‐expressed in Nicotiana benthamiana leaves and subjected to co‐immunoprecipitation assay. Representative data from at least three replicates are shown.
To investigate the transcriptional activities of ATAF2 and ANAC102, a transient expression assay was carried out (Figure S2). The effector constructs containing VND7, VNI2, ATAF2, or ANAC102 fused to GAL4‐BD under the control of Cauliflower Mosaic Virus (CaMV) 35S promoter and reporter constructs containing firefly Luciferase (LUC) linked to GAL4‐binding sites were delivered to protoplasts obtained from Arabidopsis leaves. As previously reported, GAL4‐BD‐VND7 and GAL4‐BD‐ATAF2, but not GAL4‐BD‐VNI2, upregulated the LUC activity (Nagahage et al., 2018; Yamaguchi, Ohtani, et al., 2010). As with GAL4‐BD‐VNI2, GAL4‐BD‐ANAC102 did not upregulate the LUC activity (Figure S2).
2.2. ATAF2 interacts with VNI2 in vitro and in vivo
To understand which region of VNI2 is responsible for binding to ATAF2, yeast two‐hybrid screening was carried out using various lengths of VNI2 as bait (Figure S3). When GAL4‐BD fused VNI21‐170, containing the whole NAC domain, was co‐transformed with GAL4‐AD‐ATAF2, the yeast cells grew on the selective medium. By contrast, transformants expressing GAL4‐BD‐VNI21‐138 or GAL4‐BD‐VNI2147‐252, lacking parts of the NAC domain, together with GAL4‐AD‐ATAF2 were unable to grow on the selective medium (Figure S3). This result indicates that the whole NAC domain of VNI2 is required for binding to ATAF2.
To confirm the interaction observed in yeast cells, we conducted an in vitro pull‐down assay. ATAF2 fused to MBP‐ATAF2 and His‐VNI2 was prepared from Escherichia coli and incubated with amylose‐resin. Proteins bound to the amylose‐resin were subjected to immunoblot analysis with an anti‐His antibody. His‐VNI2 protein was effectively retained on the resin incubated with MBP‐ATAF2, whereas the resin immobilized with MBP hardly retained the His‐VNI2 (Figure 1c). These data indicated that ATAF2 binds to VNI2 in vitro.
To investigate the interaction in vivo, transient expression in Nicotiana benthamiana leaves was carried out. VNI2 has a PEST motif known to be the target of protein degradation. We previously demonstrated that VNI2 protein lacking the C‐terminal 30 amino acid region containing part of the PEST motif (VNI21‐221) is more stable than the full length VNI2 (Yamaguchi, Ohtani, et al., 2010). When protein extracts from leaves expressing GFP‐fused ATAF2 (GFP‐ATAF2) and FLAG‐tagged VNI21‐221 (FLAG‐VNI21‐221) were immuno‐precipitated with anti‐GFP antibody, FLAG‐VNI21‐221 was co‐precipitated (Figure 1d), indicating that VNI2 forms a protein complex with ATAF2 in plant cells.
2.3. VNI2 represses transcriptional activation activity of ATAF2
ATAF2 regulates the expression of genes associated with leaf senescence, including ORE1 and ORS1 (Nagahage et al., 2020). In addition, it has been reported that VNI2 inhibits the transcriptional activation activity of VND7 by forming protein complexes (Yamaguchi, Ohtani, et al., 2010). To investigate how VNI2 regulates ATAF2 function, we performed a transient reporter gene expression assay using protoplasts of Arabidopsis mesophyll cells. Effector constructs containing CaMV35S linked to VNI2 and ATAF2 genes and reporter constructs containing firefly LUC driven by the ORE1 or ORS1 promoter were delivered into the protoplasts (Figure 2a). When ATAF2 was used as the effector, the reporter gene expression was increased compared to the control, as previously shown (Nagahage et al., 2020). Whereas, when both ATAF2 and VNI2 were used as the effectors, the reporter gene expression was comparable with the control (Figure 2b). These data suggested that VNI2 inhibits ATAF2 function by forming a protein complex, as in the case of VND7 (Yamaguchi, Ohtani, et al., 2010). It is noteworthy that ATAF2 upregulates VNI2 expression (Nagahage et al., 2020). The transient assay showed that VNI2 also inhibited the reporter gene expression under control of its own promoter transactivated by ATAF2 (Figure 2b), suggesting the existence of negative feedback regulation for the VNI2 gene. It is also noteworthy that when VNI2 was used as the effector, expression level of the LUC under controls of VNI2 promoter was decreased compared to the control (Figure 2b). It might be possible that VNI2 interacts with and inhibits endogenous transcription factors.
FIGURE 2.
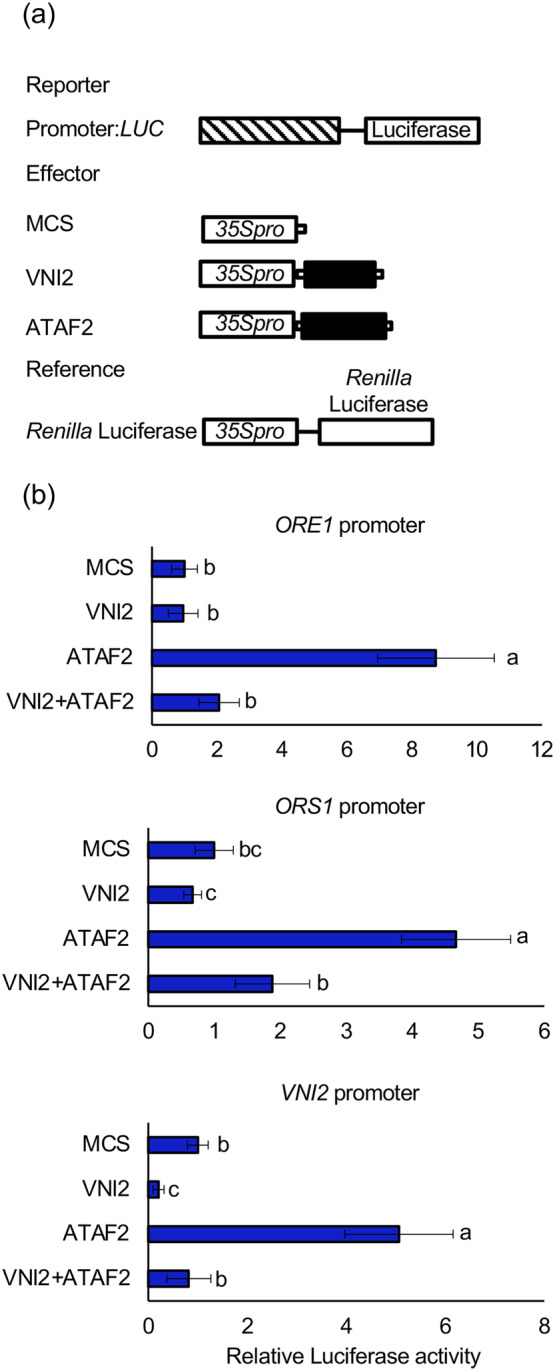
VNI2 inhibits the transcriptional activity of ATAF2. (a) Schematic diagram of the effector and reporter constructs. The reporter constructs contained the promoters of ORE1, ORS1, or VNI2 and a firefly LUCIFERASE reporter gene. The effector constructs contained either the multi cloning site (MCS) fragment or ATAF2 or full‐length VNI2 downstream from the CaMV35S promoter. (b) Relative luciferase activities after transfection with individual reporter constructs and the effector constructs to protoplasts. Firefly luciferase activity was normalized using the activity of Renilla luciferase. Values and error bars indicate means ± SD (n = 4). Different letters indicate significant differences at P < .05, as determined by one‐way analysis of variance (ANOVA) with Tukey's post test.
2.4. Acceleration of age‐dependent leaf senescence in vni2 is suppressed by ataf2 mutation
To examine genetic interactions between ATAF2 and VNI2, we observed loss‐of‐function mutants. RT‐PCR analysis showed that transcripts of VNI2 or ATAF2 were hardly detected in vni2 and ataf2 mutants, respectively (Figure S4). In addition, expression of both VNI2 and ATAF2 expression was not found in vni2ataf2 double mutants (Figure S4). Under long‐day conditions, vni2 mutants exhibited an accelerated leaf senescence phenotype compared with the wild‐type, as shown in a previous report (Yang et al., 2011) (Figure 3a). On the other hand, ataf2 mutants did not show clear difference compared to the wild‐type. Interestingly, simultaneous mutations of VNI2 and ATAF2 attenuated the acceleration of leaf senescence as observed in vni2 (Figure 3a). Consistent with the overall phenotypes, chlorophyll content was lowest in the vni2 mutant, and that in vni2ataf2 double mutants was comparable with that of the wild‐type (Figure 3b). These data indicated that decrease of the chlorophyll content in vni2 mutants is dependent on the existence of ATAF2.
FIGURE 3.
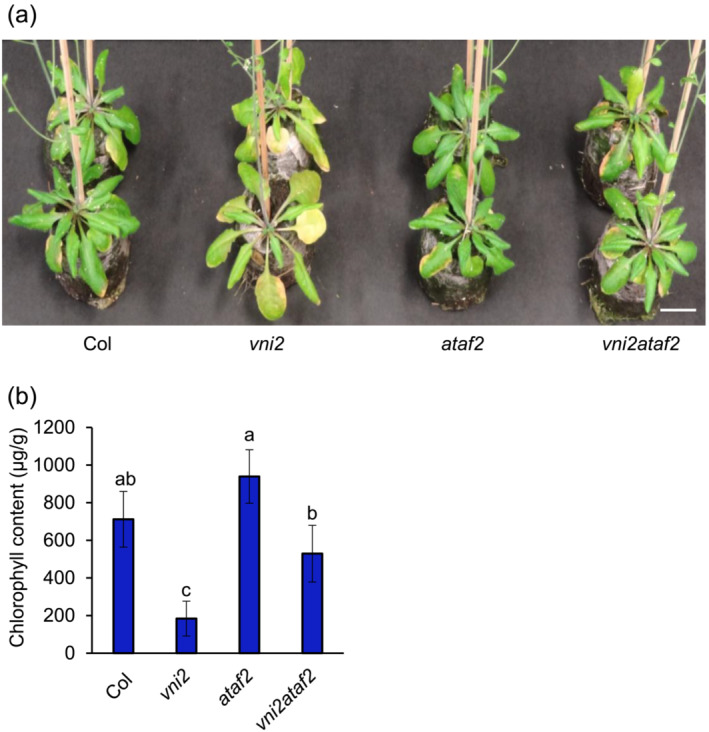
Effect of ATAF2 and VNI2 on age‐dependent leaf senescence. (a) 55‐day‐old wild‐type (Col), vni2, and ataf2, vni2ataf2 plants grown under long‐day conditions. Bar = 2.5 cm. (b) Chlorophyll content of fifth leaves from 55‐day‐old plants. Error bars indicate SD (n = 5).
To investigate the expression levels of genes involved in leaf senescence, quantitative RT‐PCR was performed by using the fifth leaves of different ages of plants. In the wild‐type, increased expression of VNI2 and ATAF2 was observed in 42‐day‐old and 49‐day‐old plants, respectively, compared with 30‐day‐old plants (Figure 4). In addition, although VNI2 expression was comparable between the wild‐type and ataf2 mutant, the expression level of ATAF2 in 30‐day‐old vni2 plants was higher than that in wild‐type plants (Figure 4). Similarly, expression levels of ORE1, ORS1, and ANAC046, which are NAC domain transcription factors upregulated by ATAF2 (Nagahage et al., 2020), were higher in 30‐day‐old vni2 mutants compared with the wild‐type, suggesting that VNI2 represses age‐dependent leaf senescence through inhibition of senescence‐associated genes regulated by ATAF2 (Figure 4). Interestingly, expression of ORE1 and ORS1 in 30‐day‐old vni2ataf2 mutants was higher or lower than those of wild‐type plants or vni2 mutants, respectively (Figure 4). Expression levels of SAG12, SAG13, and SAG29/SWEET15 genes, known as senescence marker genes (Balazadeh et al., 2010; Matallana‐Ramirez et al., 2013), were lower in vni2ataf2 mutants than those of vni2 mutants at 49 days old (Figure 4). These results suggested that ATAF2 at least partly contributes to upregulation of the gene expression observed in vni2 mutants. However, expression levels of ANAC046 and SAG29 in 42‐day‐old vni2 mutants were lower than those of vni2ataf2 mutants and/or wild‐type plants (Figure 4). It may be possible that ANAC046 and SAG29 genes are regulated by different transcription factors through the developmental stage.
FIGURE 4.
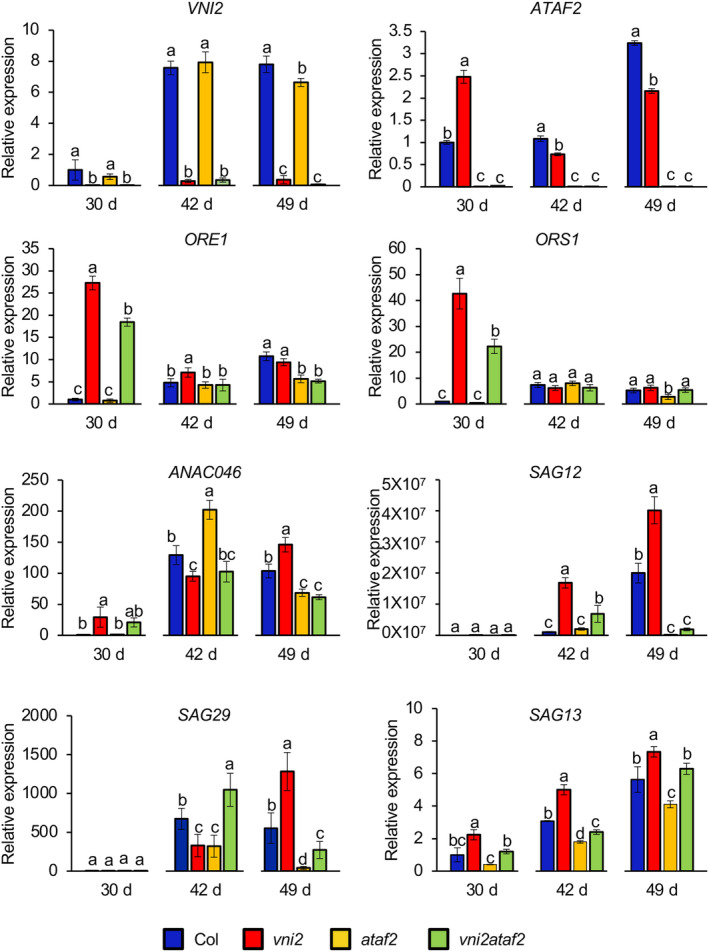
Expression analysis of the genes associated with leaf senescence. The fifth leaves of approximately 16 plants per each genotype were harvested at the indicated time points for total RNA extraction. Quantitative RT‐PCR analysis was performed, and four biological replicates were averaged. Values and error bars indicate means ± SD (n = 4). Different letters indicate significant differences at P < .05, as determined by one‐way analysis of variance (ANOVA) with Tukey's post test.
2.5. Acceleration of dark‐induced leaf senescence in vni2 is suppressed by ataf2 mutation
ATAF2 promotes not only age‐dependent leaf senescence but also dark‐induced leaf senescence (Nagahage et al., 2020). Thus, we investigated the relationship of VNI2 and ATAF2 during dark‐induced leaf senescence. The fifth leaves from 4‐week‐old wild‐type, vni2, ataf2, and vni2ataf2 mutants were incubated under continuous light or dark conditions for 5 days. No phenotypic changes were observed among the lines incubated under the light condition (Figure 5a). By contrast, under the dark condition, vni2 mutants showed a leaf yellowing phenotype, and the leaves of ataf2 mutants remained green. The leaves of vni2ataf2 mutants were similar to those of wild‐type plants (Figure 5a). Consistent with the appearance, the lowest and highest chlorophyll contents were observed in vni2 and ataf2 mutants, respectively, and the chlorophyll contents of wild‐type and vni2ataf2 double mutants were intermediate (Figure 5b).
FIGURE 5.
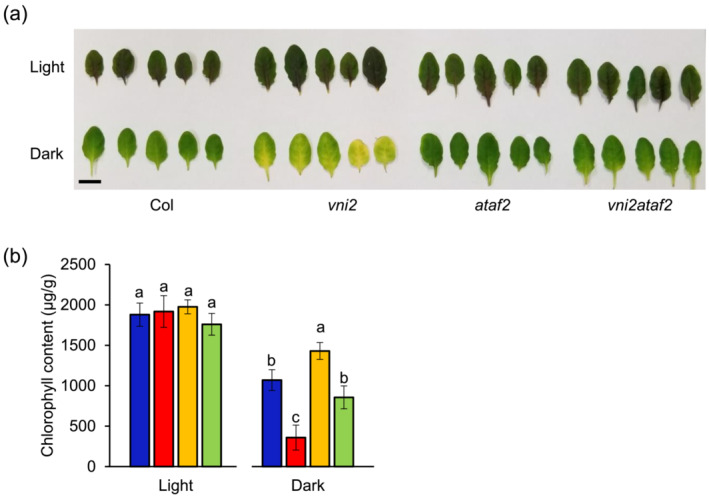
Dark‐induced leaf senescence in vni2, ataf2, and vni2ataf2. (a) The fifth leaves of 4‐week‐old plants incubated under continuous light or dark conditions for 5 days. Bar = 1 cm. (b) Chlorophyll content of the fifth leaves of 4‐week‐old plants incubated under continuous light or dark conditions for 5 days. Values and error bars indicate means ± SD (n = 5). Different letters indicate significant differences at P < .05, as determined by one‐way analysis of variance (ANOVA) with Tukey's post test.
To examine the expression levels of genes involved in leaf senescence under dark conditions, quantitative RT‐PCR was performed. In the wild‐type, expression levels of all tested genes were increased after dark incubation (Figure 6). In addition, after dark incubation, expression levels of ORE1, ANAC046, SAG12, and SAG29 in vni2 mutants and ataf2 mutants were higher and lower, respectively, than those in the wild‐type (Figure 6). These data suggested that VNI2 represses the dark‐induced leaf senescence through inhibition of senescence‐associated genes regulated by ATAF2. Expression levels of ORS1, ANAC046, SAG12, and SAG13 in vni2 ataf2 mutants were lower than those in vni2 mutants after dark incubation, consistent with the chlorophyll contents (Figure 6). By contrast, expression levels of ORE1 and SAG29 genes in vni2ataf2 mutants were comparable with and higher than those of vni2 mutants, respectively, after dark incubation (Figure 6). It is possible that VNI2 inhibits the transcriptional activities of some other NAC domain transcription factors that highly contribute to upregulation of ORE1 and SAG29 gene expression during dark‐induced leaf senescence.
FIGURE 6.
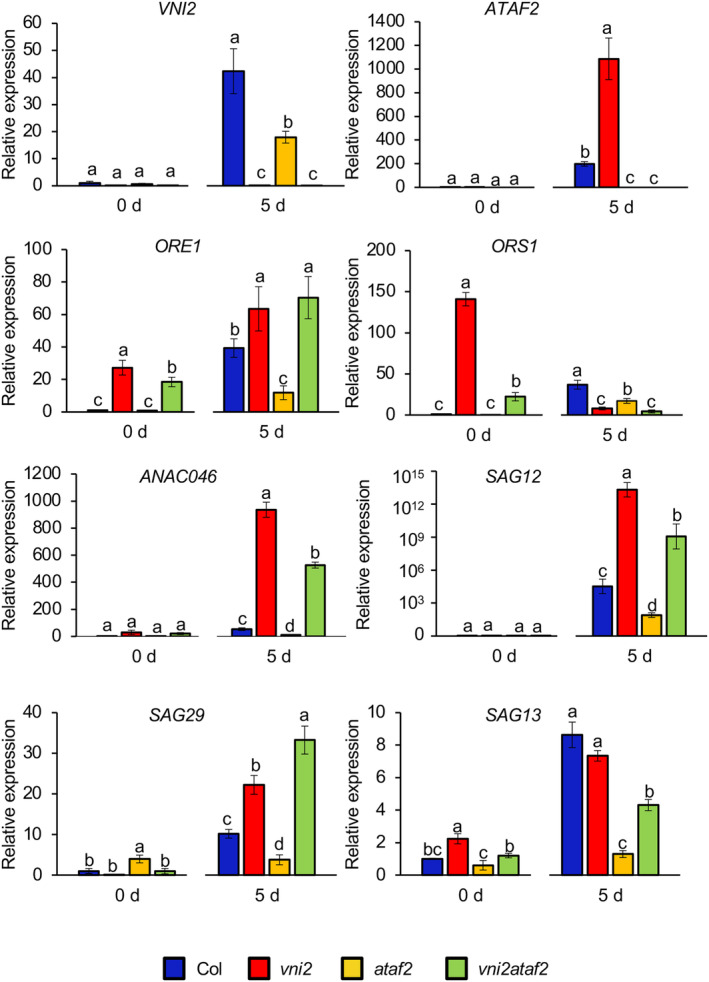
Expression of leaf senescence‐associated genes during dark‐induced senescence. The fifth leaves of approximately 16 plants per each genotype were harvested at the indicated time points for total RNA extraction. Quantitative RT‐PCR analysis was performed, and the biological replicates were averaged. Values and error bars indicate means ± SD (n = 4). Different letters indicate significant differences at P < .05, as determined by one‐way analysis of variance (ANOVA) with Tukey's post test.
3. DISCUSSION
3.1. VNI2 plays roles in leaf senescence and xylem vessel element differentiation by forming distinct NAC domain transcription factors
An NAC domain transcription factor, VNI2, was originally identified as an interacting factor with another NAC domain transcription factor, VND7, a key regulator of xylem vessel element differentiation (Yamaguchi, Ohtani, et al., 2010). VNI2 inhibits VND7 function, resulting in negative regulation of xylem vessel formation (Yamaguchi, Ohtani, et al., 2010). Here, we isolated other NAC domain transcription factors, ATAF2 and ANAC102, as interacting proteins with VNI2. ATAF2 and ANAC102 are known as stress regulatory NAC domain transcriptional factors and are closely related to each other. Yeast two‐hybrid analysis demonstrated that the whole NAC domain of VNI2 is necessary to interact with ATAF2 (Figure S3). Previously, it was reported that NAC subdomain V with the C‐terminal transcriptional activation domain of VNI2 was sufficient to interact with VND7 (Yamaguchi, Ohtani, et al., 2010), suggesting that VND7 and ATAF2 may bind to VNI2 in a different manner.
VND7 directly or indirectly regulates a broad range of genes involved in xylem vessel element differentiation (Yamaguchi et al., 2011; Zhong et al., 2010). Overexpression of VND7 induces transdifferentiation into xylem vessel elements resulting from upregulation of the downstream genes (Kubo et al., 2005; Yamaguchi, Goue, et al., 2010). VNI2 inhibits the transcriptional activation activities of VND7 by forming protein complexes (Yamaguchi, Ohtani, et al., 2010). Previously, we showed that ATAF2 upregulates genes associated with leaf senescence (Nagahage et al., 2020). Overexpression of ATAF2 promotes leaf senescence, whereas a knock‐out mutation of ATAF2 exhibits delay of leaf senescence (Nagahage et al., 2020). As in the case of VND7, the transient gene expression assay demonstrated that VNI2 inhibits the transcriptional activation activities of ATAF2 (Figure 2). It is known that some transcription factors have several distinct roles by forming complexes with different proteins, such as DELLA family and PHYTOCHROME‐INTERACTING FACTOR (PIF) proteins (de Lucas et al., 2008; Feng et al., 2008; Yoshida et al., 2014). Likewise, our data suggested that VNI2 plays roles in different biological processes by binding to and inhibiting distinct NAC domain transcription factors. Xylem vessel element differentiation accompanies programmed cell death as well as secondary cell wall formation (Nakano et al., 2015; Yamaguchi & Demura, 2010). Leaf senescence is characterized by a reversible phase followed by an irreversible phase. Up to the irreversible phase, senescence is temporally inhibited by genetic programs competing with promoting factors for plants to adapt to the new environmental condition. However, beyond that point, leaf senescence cannot be reversed and progresses to leaf death (Guiboileau et al., 2010). It is possible that VNI2 has a common role to prevent progression to the irreversible stages during xylem vessel formation and leaf senescence by inhibiting the promoting factors.
3.2. Genetic relationship between VNI2 and ATAF2
Overexpression of VNI2 delays leaf senescence (Yang et al., 2011), whereas that of ATAF2 promotes leaf senescence (Nagahage et al., 2020). Conversely, single mutation of VNI2 and ATAF2 promotes and delays leaf senescence phenotypes, respectively (Nagahage et al., 2020; Yang et al., 2011). In this study, observation of single and double mutants suggested that the acceleration of the leaf senescence phenotype of vni2 mutants is suppressed by ataf2 mutation. It is likely that ATAF2 is released from inhibition of VNI2 in the vni2 mutant background. However, vni2ataf2 double mutants did not completely recover the acceleration of leaf senescence (Figures 3, 4, 5, 6). ATAF2 belongs to the stress‐responsive NAC subfamily SNAC‐A, which includes 6 other members (Nakashima et al., 2012; Nuruzzaman et al., 2013). It has been reported that members of the SNAC‐A subfamily also promote leaf senescence in response to environmental stresses (Garapati et al., 2015; Li et al., 2016; Mahmood et al., 2016; Takasaki et al., 2015). The phenotype of the vni2ataf2 double mutant suggests a possibility that VNI2 also interacts with and inhibits some of the other SNAC‐A subfamily members. To prove the hypothesis, we should generate multiple mutants lacking VNI2 and/or members of the SNAC‐A subfamily and observe the leaf senescence phenotypes of these mutants.
3.3. Transcriptional network during leaf senescence
Recently, we have shown that VND7 negatively regulates VNI2 expression (Ailizati et al., 2021). By contrast, ATAF2 upregulates VNI2 gene expression (Nagahage et al., 2020). In this study, VNI2 inhibits its own expression that is activated by ATAF2 (Figure 2), indicating the negative feedback regulation for VNI2 expression during the leaf senescence process (Figure 7). The INDETERMINATE DOMAIN (IDD) protein family activates the expression of the gibberellin‐positive regulator SCARECROW‐LIKE 3 (SCL3), and SCL3 inhibits its own expression by forming protein complexes with IDD proteins (Yoshida et al., 2014). Likewise, some members of AUXIN RESPONSE FACTORs (ARFs) activate transcription of Auxin (Aux)/INDOLE 3‐ACETIC ACID (IAA) genes, while Aux/IAAs hetero‐dimerize with and inhibit ARFs (Freire‐Rios et al., 2020; Ito et al., 2016; Okushima et al., 2005). It is likely that the negative feedback regulations are important throughout plant development in response to various signals.
FIGURE 7.
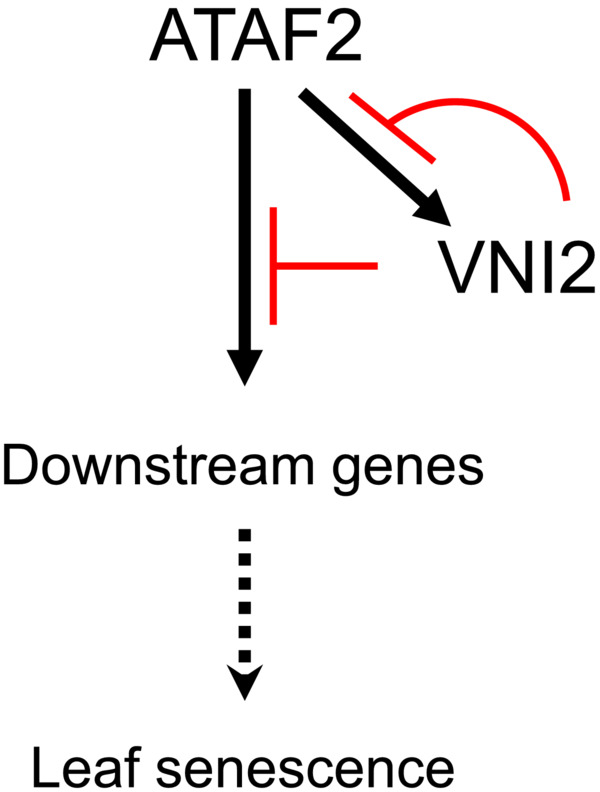
Schematic model of the relationship between ATAF2 and VNI2 during leaf senescence.
Leaf senescence is a well‐programmed process that includes macronutrient catabolism, detoxification, and transportation to young vegetative or reproductive parts and is essential for nutrient relocation and efficient plant growth. Thus, the senescence rate is an important aspect of the overall productivity of plants (Long et al., 2006). ATAF2 upregulates genes encoding not only promoting factors but also repressing factors of leaf senescence (Nagahage et al., 2020). It is possible that ATAF2 initiates leaf senescence by induction of promoting factors and maintains sufficient time for the nutrient recycling process by induction of VNI2. In addition, as described above, leaf senescence is an irreversible process. It is also possible that VNI2 induced by ATAF2 temporarily prolongs the reversible stages of leaf senescence. In order to confirm whether developmental or environmental conditions are sufficient to progress to the irreversible stage.
It has been reported that NAC domain transcription factors regulating leaf senescence form homodimer and/or heterodimer complexes (Kim et al., 2018). Taken together with these reports, our data also support the hypothesis that transcriptional regulations and various combinations of protein complexes among the NAC domain transcription factors play important roles in fine‐tuning the progression of leaf senescence.
In this study, we demonstrated that VNI2 interacts with and inhibits ATAF2, which is known to be involved in leaf senescence. We have shown that VNI2 regulates xylem vessel formation by forming the protein complex with VND7 (Yamaguchi, Ohtani, et al., 2010). Taken together with these reports, our data proposed a hypothesis that VNI2 plays roles in different biological events by forming protein complexes with distinct NAC domain transcription factors. As described above, regulation of leaf senescence involves a number of NAC domain transcription factors (Kim et al., 2014, 2018; Podzimska‐Sroka et al., 2015). Thus, it is necessary to investigate the relationship between VNI2 and these NAC domain transcription factors to understand the biological roles of VNI2 during leaf senescence more deeply. Furthermore, VNI2 is expressed in various types of cells and stress conditions (Yamaguchi, Ohtani, et al., 2010; Yang et al., 2011). VNI2 may play roles in biological processes other than xylem vessel element differentiation and leaf senescence by forming protein complexes. A cDNA library composed only of cDNAs of Arabidopsis transcription factors has been established (Mitsuda et al., 2010). It is useful to identify novel interacting proteins with VNI2 to unveil other biological roles of VNI2.
4. EXPERIMENTAL PROCEDURES
4.1. Vector constructions
The promoter fragments and cDNAs were subcloned into the pENTR/D‐TOPO vector (Thermo Fisher Scientific) to generate entry clones. The resultant clones were integrated into GATEWAY destination vectors for yeast two‐hybrid assay (pBD‐GAL4‐GWRFC and pAD‐GAL4‐GWRFC; Yamaguchi et al., 2008), for expression of maltose‐binding protein (MBP) fusion proteins in E. coli (pMAL‐GWRFC; Yamaguchi, Ohtani, et al., 2010), for transient expression analysis (pAG35, pA35BDG, and pAGL; Endo et al., 2015; Yamaguchi et al., 2015), and for co‐immunoprecipitation (pGWB406 and pGWB412; Nakagawa et al., 2007). The GATEWAY destination vectors containing the nucleotide sequence of the multicloning site (MCS) fragments were used as controls (Yamaguchi et al., 2008). The nucleotide sequences of the MCS fragment and primers used in this study are shown in Table S1.
4.2. Plant materials
The Columbia ecotype of Arabidopsis thaliana (Col) was used for all the experiments. The ATAF2 T‐DNA insertion mutant SALK_136355 and the VNI2 T‐DNA insertion mutant SALK_143793 were obtained from the Arabidopsis Biological Resource Center. Plants were grown under long‐day conditions (16/8 h light/dark; 100 μmol m−2 s−1 light intensity) at 22°C. The vni2ataf2 double mutant was generated by crossing VNI2 and ATAF2 T‐DNA lines.
4.3. Yeast two‐hybrid screening
The S.c. EasyCompTM Transformation Kit (Invitrogen) was used according to the protocol (https://www.lifetechnologies.com/order/catalog/product/K505001), and pBD‐GAL4‐GWRFC and/or pAD‐GAL4‐GWRFC plasmids were introduced into Saccharomyces cerevisiae strain AH109 by using S.c. solution III. Transformants were incubated at 30°C on MVD medium without tryptophan and leucine for 3 days. pBD‐wt and pAD‐wt harboring fragment C of lambda cI repressor (amino acid region 132–236) were used as positive controls (Agilent Technologies). Yeast successfully grown as above was transferred to MVD medium lacking tryptophan, leucine, and histidine and incubated at 30°C for 3 days. pBD‐GAL4‐MCS and pAD‐GAL4‐MCS were used as negative controls; 3AT (1 mM) was added to minimize the background false positive effect.
4.4. Phylogenetic tree analysis
An unrooted phylogenetic tree was constructed using the neighbor‐joining (NJ) method, and the bootstrap test was carried out with 1000 iterations for Arabidopsis NAC domain proteins.
4.5. Dual luciferase transient expression assay
Effector constructs, reporter constructs, and reference constructs were generated as described in Nagahage et al. (2020). Leaves of 3‐ to 4‐week‐old Arabidopsis plants without bolting were peeled using the Tape‐Arabidopsis Sandwich method (Sakamoto et al., 2016). The peeled leaves were subjected to an enzymatic solution (1% [w/v] Cellulase “Onozuka” R10 [Yakult Pharmaceutical Industry], .25% (w/v) macerozyme R10 [Yakult Pharmaceutical Industry], 400‐mM mannitol, 20‐mM MES [pH 5.7], 20‐mM KCl, 10‐mM CaCl2, and 5‐mM 2‐mercaptoethanol) for 1 h at 22°C with shaking at 60 rpm and then filtrated with 70‐μm nylon membran (Cellstraner 70 μm, BD Falcon). The protoplast cells were collected by centrifugation at 100 × g for 5 min and rinsed twice with W5 buffer (150‐mM NaCl, 125‐mM CaCl2, 5‐mM KCl, and 2‐mM MES [pH 5.7]) and incubated on ice for 10 min. The protoplast cells were resuspended with MMg buffer (400‐mM mannitol, 15‐mM MgCl2, and 4‐mM MES [pH 5.7]). Ten microliter plasmid solution was mixed with 35‐μl suspension of the protoplast cells and 45‐μl PEG solution (40% [w/v] PEG 4000 [Sigma‐Aldrich], 200‐mM mannitol, and 100‐mM CaCl2) and incubated for 10 min at room temperature. The protoplast cells were washed three times with W5 buffer and incubated for 16–20 h at 22°C under dark condition. Luciferase activity was measured with the Dual‐Luciferase Reporter Assay System (Promega, http://www.promega.com) using a Mithras LB940 Multimode Microplate Reader (Berthold, http://berthold.com) according to the manufactures' protocols.
4.6. In vitro pull‐down assay
Poly‐His tagged VNI2 (His‐VNI2) was prepared according to a previous report (Yamaguchi, Ohtani, et al., 2010). ATAF2 cDNA was integrated into the pMAL‐GWRFC. MBP‐ATAF2 was expressed in E. coli strain BL21 trxB (DE3; Cosmo Bio) in the presence of isopropyl ß‐D‐thiogalactoside (IPTG) and purified with amylose resin (New England Biolabs). His‐VNI2 and/or MBP‐ATAF2 proteins were incubated with the amylose resin for 90 min at 4°C. The proteins immobilized with the resin were subjected to immunoblot analysis. His‐VNI2 protein was detected with an anti‐His antibody (Santa Cruz Biotechnology) and an anti‐rabbit IgG antibody (Amersham Biosciences).
4.7. In vivo co‐immunoprecipitation assay
Transient protein co‐expression in N. benthamiana and co‐immunoprecipitation assay were performed as described previously with some modifications (Fujiwara et al., 2014; Kim et al., 2007). Briefly, protein extracts were incubated with an anti‐GFP antibody (A‐11120, Thermo Fisher Scientific) and Dynabeads Protein A (Thermo Fisher Scientific) at 4°C for 1 h with gentle agitation. The proteins retained on the beads were separated by SDS‐PAGE. The blots were probed with the anti‐GFP antibody (ab6556, Abcam, 1:2000) and an anti‐DYKDDDDK tag antibody (019‐22394, Fujifilm, 1:20,000) and detected with ECL Prime (Cytiba Amersham).
4.8. Analysis of leaf senescence
The experiment was conducted as described previously in Nagahage et al. (2020). For dark‐induced leaf senescence analysis, the fifth rosette leaves of 4‐week‐old plants were excised and placed on moistened filter paper. The Petri dishes were sealed with surgical tape, wrapped with aluminum foil or not, and incubated at 22°C under continuous light conditions. Chlorophyll was extracted using dimethyl formamide at 4°C overnight. The chlorophyll extract was subjected to absorbance measurements via spectroscopy at wavelengths of 645 and 663 nm (Porra et al., 1989).
4.9. Gene expression analysis
For each genotype, the fifth leaves of about 16 plants were collected and averaged across four biological replicates. Total RNA was isolated by the RNeasy Plant Mini Kit (Qiagen) and treated with DNase I (Qiagen). The synthesis of cDNA was performed by using SuperScript II reverse transcriptase (Invitrogen). Quantitative RT‐PCR was performed by using Power SYBR Green PCR Master Mix and the 7300 Real‐Time PCR system (Applied Biosystems). The primers used for the expression analyses are listed in Table S1.
AUTHOR CONTRIBUTIONS
Isura S.P. Nagahage and Masatoshi Yamaguchi conceived the original research plans. Isura S.P. Nagahage performed most of the experiments. Kohei Matsuda, Misato Ohtani, Ko Kato, Taku Demura, and Masatoshi Yamaguchi screened the interacting proteins with VNI2. Isura S.P. Nagahage conducted transient expression assays. Isura S.P. Nagahage and Takuya Yamada contributed to the pull‐down assay. Kyoko Miyashita and Sumire Fujiwara performed co‐immunoprecipitation assays. Chanaka Mannapperuma performed phylogenetic tree analysis. Isura S.P. Nagahage, Kyoko Miyashita, Sumire Fujiwara, Chanaka Mannapperuma, Takuya Yamada, Shingo Sakamoto, Toshiki Ishikawa, Minoru Nagano, Misato Ohtani, Ko Kato, Hirofumi Uchimiya, Nobutaka Mitsuda, Maki Kakawi‐Yamada, Taku Demura, and Masatoshi Yamaguchi interpreted the results. Isura S.P. Nagahage and Masatoshi Yamaguchi wrote the manuscript with input from all authors.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no known competing interests that could have appeared to influence this work.
Supporting information
Data S1. Peer Review.
Figure S1. Schematic diagram of NAC domain transcription factors. The gray boxes indicate subdomains I to V of the NAC domains. The black bar corresponds to the shortest encoded region of ANAC102 isolated by screening.
Figure S2. ATAF2 has transcriptional activation activity. (a) Schematic diagram of the constructs used in the dual luciferase transient assay. The reporter construct contained the firefly luciferase reporter gene under the control of five repeats of the upstream activation sequence of GAL4 (5 X GAL4 UAS) fused to a minimal CaMV35S promoter (min pro). The effector constructs contained GAL4‐BD bound to an empty multiple cloning site (GAL4‐BD‐MCS) or to coding sequences corresponding to full length VND7, VNI2, ATAF2, and ANAC102 driven by the CaMV35S promoter (35Spro). (b) Results of the transient transfection assay. Firefly luciferase activity was normalized to Renilla luciferase activity. Error bars indicate SD (n = 4). Different letters indicate significant differences at P < .05, as determined by one‐way ANOVA with Tukey's post‐test.
Figure S3. The whole NAC domain of VNI2 is necessary for interaction with ATAF2. (a) Schematic diagram of full length and truncated VNI2 used for the yeast two‐hybrid assay. (b) Result of yeast two‐hybrid assay. Full length ATAF2 fused to GAL4‐BD, and full length or truncated VNI2 fused to GAL4‐AD were introduced into AH109 yeast cells. The transformed cells were grown on control (Trp−Leu−) and selective media (Trp−Leu−His− with .1 mM 3‐AT). Plasmids containing MCS fused to GAL‐BD or GAL4‐AD were used as negative controls, and pBD‐wt and pAD‐wt were used as positive controls.
Figure S4. VNI2 and ATAF2 T‐DNA insertion lines. (a) Schematic diagram of the T‐DNA insertion sites of vni2 and ataf2. Grey, black, and white boxes indicate untranslated regions, coding regions, and introns, respectively. Arrows indicate the locations of the primers used for RT‐PCR. (b) RT‐PCR analysis results. Analysis was performed using the seedlings of 7‐day‐old plants.
Table S1. Oligonucleotides used in this study.
ACKNOWLEDGMENTS
We thank Hiromi Ogawa, Utako Kanda, and Kaori Tashiro (Saitama University, Japan) for their excellent technical assistance. We also thank Prof. Tsuyoshi Nakagawa (Shimane University, Japan) for providing the pGWB406 and pGWB412 vectors. This work was supported by JST, PRESTO to M.Y. (JPMJPR11BB) and Grants‐in‐Aid from the Ministry of Education, Science, Sports, and Culture of Japan to M.Y. (25840098, 15K07094, and 18K06277).
Nagahage, I. S. P. , Matsuda, K. , Miyashita, K. , Fujiwara, S. , Mannapperuma, C. , Yamada, T. , Sakamoto, S. , Ishikawa, T. , Nagano, M. , Ohtani, M. , Kato, K. , Uchimiya, H. , Mitsuda, N. , Kawai‐Yamada, M. , Demura, T. , & Yamaguchi, M. (2023). NAC domain transcription factors VNI2 and ATAF2 form protein complexes and regulate leaf senescence. Plant Direct, 7(9), e529. 10.1002/pld3.529
Contributor Information
Isura Sumeda Priyadarshana Nagahage, Email: isura.nagahage@umu.se.
Masatoshi Yamaguchi, Email: yamagu@mail.saitama-u.ac.jp.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- Ailizati, A. , Nagahage, I. S. P. , Miyagi, A. , Ishikawa, T. , Kawai‐Yamada, M. , Demura, T. , & Yamaguchi, M. (2021). An Arabidopsis NAC domain transcriptional activator VND7 negatively regulates VNI2 expression. Plant Biotechnology (Tokyo), 38, 415–420. 10.5511/plantbiotechnology.21.1013a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailizati, A. , Nagahage, I. S. P. , Miyagi, A. , Ishikawa, T. , Kawai‐Yamada, M. , Demura, T. , & Yamaguchi, M. (2022). VND‐INTERACTING2 effectively inhibits transcriptional activities of VASCULAR‐RELATED NAC‐DOMAIN7 through a conserved sequence. Plant Biotechnology (Tokyo), 39, 22.0122a. 10.5511/plantbiotechnology.22.0122a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh, S. , Kwasniewski, M. , Caldana, C. , Mehrnia, M. , Zanor, M. I. , Xue, G. P. , & Mueller‐Roeber, B. (2011). ORS1, an H(2)O(2)‐responsive NAC transcription factor, controls senescence in Arabidopsis thaliana . Molecular Plant, 4, 346–360. 10.1093/mp/ssq080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh, S. , Siddiqui, H. , Allu, A. D. , Matallana‐Ramirez, L. P. , Caldana, C. , Mehrnia, M. , Zanor, M. I. , Kohler, B. , & Mueller‐Roeber, B. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt‐promoted senescence. The Plant Journal, 62, 250–264. 10.1111/j.1365-313X.2010.04151.x [DOI] [PubMed] [Google Scholar]
- Christianson, J. A. , Wilson, I. W. , Llewellyn, D. J. , & Dennis, E. S. (2009). The low‐oxygen‐induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low‐oxygen treatment. Plant Physiology, 149, 1724–1738. 10.1104/pp.108.131912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro, S. , Ksas, B. , & Havaux, M. (2018). Decoding beta‐cyclocitral‐mediated retrograde signaling reveals the role of a detoxification response in plant tolerance to photooxidative stress. Plant Cell, 30, 2495–2511. 10.1105/tpc.18.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas, M. , Daviere, J. M. , Rodriguez‐Falcon, M. , Pontin, M. , Iglesias‐Pedraz, J. M. , Lorrain, S. , Fankhauser, C. , Blazquez, M. A. , Titarenko, E. , & Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature, 451, 480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Delessert, C. , Kazan, K. , Wilson, I. W. , van der Straeten, D. , Manners, J. , Dennis, E. S. , & Dolferus, R. (2005). The transcription factor ATAF2 represses the expression of pathogenesis‐related genes in Arabidopsis. The Plant Journal, 43, 745–757. 10.1111/j.1365-313X.2005.02488.x [DOI] [PubMed] [Google Scholar]
- Endo, H. , Yamaguchi, M. , Tamura, T. , Nakano, Y. , Nishikubo, N. , Yoneda, A. , Kato, K. , Kubo, M. , Katayama, Y. , Ohtani, M. , & Demura, T. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR‐RELATED NAC‐DOMAIN7, a master switch of xylem vessel differentiation. Plant & Cell Physiology, 56, 242–254. 10.1093/pcp/pcu134 [DOI] [PubMed] [Google Scholar]
- Feng, S. , Martinez, C. , Gusmaroli, G. , Wang, Y. , Zhou, J. , Wang, F. , Chen, L. , Yu, L. , Iglesias‐Pedraz, J. M. , Kircher, S. , Schafer, E. , Fu, X. , Fan, L. M. , & Deng, X. W. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature, 451, 475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire‐Rios, A. , Tanaka, K. , Crespo, I. , van der Wijk, E. , Sizentsova, Y. , Levitsky, V. , Lindhoud, S. , Fontana, M. , Hohlbein, J. , Boer, D. R. , Mironova, V. , & Weijers, D. (2020). Architecture of DNA elements mediating ARF transcription factor binding and auxin‐responsive gene expression in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 117, 24557–24566. 10.1073/pnas.2009554117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, S. , Sakamoto, S. , Kigoshi, K. , Suzuki, K. , & Ohme‐Takagi, M. (2014). VP16 fusion induces the multiple‐knockout phenotype of redundant transcriptional repressors partly by Med25‐independent mechanisms in Arabidopsis. FEBS Letters, 588, 3665–3672. 10.1016/j.febslet.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Furuta, K. M. , Yadav, S. R. , Lehesranta, S. , Belevich, I. , Miyashima, S. , Heo, J. O. , Vaten, A. , Lindgren, O. , de Rybel, B. , van Isterdael, G. , Somervuo, P. , Lichtenberger, R. , Rocha, R. , Thitamadee, S. , Tahtiharju, S. , Auvinen, P. , Beeckman, T. , Jokitalo, E. , & Helariutta, Y. (2014). Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science, 345, 933–937. 10.1126/science.1253736 [DOI] [PubMed] [Google Scholar]
- Garapati, P. , Xue, G. P. , Munne‐Bosch, S. , & Balazadeh, S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiology, 168, 1122–1139. 10.1104/pp.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau, A. , Sormani, R. , Meyer, C. , & Masclaux‐Daubresse, C. (2010). Senescence and death of plant organs: Nutrient recycling and developmental regulation. Comptes Rendus Biologies, 333, 382–391. 10.1016/j.crvi.2010.01.016 [DOI] [PubMed] [Google Scholar]
- Hamasaki, H. , Kurihara, Y. , Kuromori, T. , Kusano, H. , Nagata, N. , Yamamoto, Y. Y. , Shimada, H. , & Matsui, M. (2019). SnRK1 kinase and the NAC transcription factor SOG1 are components of a novel signaling pathway mediating the low energy response triggered by ATP depletion. Frontiers in Plant Science, 10, 503. 10.3389/fpls.2019.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, R. , Hill, C. , Penfold, C. A. , Breeze, E. , Bowden, L. , Moore, J. D. , Zhang, P. , Jackson, A. , Cooke, E. , Bewicke‐Copley, F. , Mead, A. , Beynon, J. , Wild, D. L. , Denby, K. J. , Ott, S. , & Buchanan‐Wollaston, V. (2013). A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. The Plant Journal, 75, 26–39. 10.1111/tpj.12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, S. U. , Lee, S. B. , Kim, H. H. , & Paek, K. H. (2012). ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Molecules and Cells, 34, 305–313. 10.1007/s10059-012-0122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, J. , Fukaki, H. , Onoda, M. , Li, L. , Li, C. , Tasaka, M. , & Furutani, M. (2016). Auxin‐dependent compositional change in mediator in ARF7‐ and ARF19‐mediated transcription. Proceedings of the National Academy of Sciences of the United States of America, 113, 6562–6567. 10.1073/pnas.1600739113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, M. K. , & Skriver, K. (2014). NAC transcription factor gene regulatory and protein‐protein interaction networks in plant stress responses and senescence. IUBMB Life, 66, 156–166. 10.1002/iub.1256 [DOI] [PubMed] [Google Scholar]
- Kawabe, H. , Ohtani, M. , Kurata, T. , Sakamoto, T. , & Demura, T. (2018). Protein S‐Nitrosylation regulates xylem vessel cell differentiation in Arabidopsis. Plant & Cell Physiology, 59, 17–29. 10.1093/pcp/pcx151 [DOI] [PubMed] [Google Scholar]
- Kim, H. J. , Hong, S. H. , Kim, Y. W. , Lee, I. H. , Jun, J. H. , Phee, B. K. , Rupak, T. , Jeong, H. , Lee, Y. , Hong, B. S. , Nam, H. G. , Woo, H. R. , & Lim, P. O. (2014). Gene regulatory cascade of senescence‐associated NAC transcription factors activated by ETHYLENE‐INSENSITIVE2‐mediated leaf senescence signalling in Arabidopsis. Journal of Experimental Botany, 65, 4023–4036. 10.1093/jxb/eru112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. J. , Park, J. H. , Kim, J. , Kim, J. J. , Hong, S. , Kim, J. , Kim, J. H. , Woo, H. R. , Hyeon, C. , Lim, P. O. , Nam, H. G. , & Hwang, D. (2018). Time‐evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 115, E4930–E4939. 10.1073/pnas.1721523115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. H. , Woo, H. R. , Kim, J. , Lim, P. O. , Lee, I. C. , Choi, S. H. , Hwang, D. , & Nam, H. G. (2009). Trifurcate feed‐forward regulation of age‐dependent cell death involving miR164 in Arabidopsis. Science, 323, 1053–1057. 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Kim, W. Y. , Fujiwara, S. , Suh, S. S. , Kim, J. , Kim, Y. , Han, L. , David, J. , Putterill, J. , Nam, H. G. , & Zomers, D. E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature, 449, 356–360. 10.1038/nature06132 [DOI] [PubMed] [Google Scholar]
- Kim, Y. S. , Kim, S. G. , Park, J. E. , Park, H. Y. , Lim, M. H. , Chua, N. H. , & Park, C. M. (2006). A membrane‐bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell, 18, 3132–3144. 10.1105/tpc.106.043018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok, E. J. , Wilson, I. W. , Wilson, D. , Chapman, S. C. , Ewing, R. M. , Somerville, S. C. , Peacock, W. J. , Dolferus, R. , & Dennis, E. S. (2002). Expression profile analysis of the low‐oxygen response in Arabidopsis root cultures. Plant Cell, 14, 2481–2494. 10.1105/tpc.004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, M. , Udagawa, M. , Nishikubo, N. , Horiguchi, G. , Yamaguchi, M. , Ito, J. , Mimura, T. , Fukuda, H. , & Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes & Development, 19, 1855–1860. 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Gao, J. , Yao, L. , Ren, G. , Zhu, X. , Gao, S. , Qiu, K. , Zhou, X. , & Kuai, B. (2016). The role of ANAC072 in the regulation of chlorophyll degradation during age‐ and dark‐induced leaf senescence. Plant Cell Reports, 35, 1729–1741. 10.1007/s00299-016-1991-1 [DOI] [PubMed] [Google Scholar]
- Long, S. P. , Zhu, X. G. , Naidu, S. L. , & Ort, D. R. (2006). Can improvement in photosynthesis increase crop yields? Plant, Cell & Environment, 29, 315–330. 10.1111/j.1365-3040.2005.01493.x [DOI] [PubMed] [Google Scholar]
- Mahmood, K. , El‐Kereamy, A. , Kim, S. H. , Nambara, E. , & Rothstein, S. J. (2016). ANAC032 positively regulates age‐dependent and stress‐induced senescence in Arabidopsis thaliana . Plant & Cell Physiology, 57, 2029–2046. 10.1093/pcp/pcw120 [DOI] [PubMed] [Google Scholar]
- Matallana‐Ramirez, L. P. , Rauf, M. , Farage‐Barhom, S. , Dortay, H. , Xue, G. P. , Droge‐Laser, W. , Lers, A. , Balazadeh, S. , & Mueller‐Roeber, B. (2013). NAC transcription factor ORE1 and senescence‐induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Molecular Plant, 6, 1438–1452. 10.1093/mp/sst012 [DOI] [PubMed] [Google Scholar]
- Miao, M. , Niu, X. L. , Kud, J. , Du, X. R. , Avila, J. , Devarenne, T. P. , Kuhl, J. C. , Liu, Y. S. , & Xiao, F. M. (2016). The ubiquitin ligase SEVEN IN ABSENTIA (SINA) ubiquitinates a defense‐related NAC transcription factor and is involved in defense signaling. The New Phytologist, 211, 138–148. 10.1111/nph.13890 [DOI] [PubMed] [Google Scholar]
- Mitsuda, N. , Ikeda, M. , Takada, S. , Takiguchi, Y. , Kondou, Y. , Yoshizumi, T. , Fujita, M. , Shinozaki, K. , Matsui, M. , & Ohme‐Takagi, M. (2010). Efficient yeast one−/two‐hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana . Plant and Cell Physiology, 51, 2145–2151. 10.1093/pcp/pcq161 [DOI] [PubMed] [Google Scholar]
- Mohanta, T. K. , Yadav, D. , Khan, A. , Hashem, A. , Tabassum, B. , Khan, A. L. , Abd Allah, E. F. , & Al‐Harrasi, A. (2020). Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE, 15, e0231425. 10.1371/journal.pone.0231425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahage, I. S. P. , Sakamoto, S. , Nagano, M. , Ishikawa, T. , Kawai‐Yamada, M. , Mitsuda, N. , & Yamaguchi, M. (2018). An NAC domain transcription factor ATAF2 acts as transcriptional activator or repressor dependent on promoter context. Plant Biotechnol‐Nar, 35, 285–289. 10.5511/plantbiotechnology.18.0507a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahage, I. S. P. , Sakamoto, S. , Nagano, M. , Ishikawa, T. , Mitsuda, N. , Kawai‐Yamada, M. , & Yamaguchi, M. (2020). An Arabidopsis NAC domain transcription factor, ATAF2, promotes age‐dependent and dark‐induced leaf senescence. Physiologia Plantarum, 170, 299–308. 10.1111/ppl.13156 [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Kurose, T. , Hino, T. , Tanaka, K. , Kawamukai, M. , Miwa, Y. , Toyooka, K. , Matsuoka, K. , Jinbo, Y. , & Kimura, T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering, 104, 34–41. 10.1263/jbb.104.34 [DOI] [PubMed] [Google Scholar]
- Nakano, Y. , Yamaguchi, M. , Endo, H. , Rejab, N. A. , & Ohtani, M. (2015). NAC‐MYB‐based transcriptional regulation of secondary cell wall biosynthesis in land plants. Frontiers in Plant Science, 6, 288. 10.3389/fpls.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, K. , Takasaki, H. , Mizoi, J. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta, 1819, 97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman, M. , Sharoni, A. M. , & Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers in Microbiology, 4, 248. 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda‐Yamamizo, C. , Mitsuda, N. , Sakamoto, S. , Ogawa, D. , Ohme‐Takagi, M. , & Ohmiya, A. (2016). The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Scientific Reports‐Uk, 6, 23609. 10.1038/srep23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani, M. , Kawabe, H. , & Demura, T. (2018). Evidence that thiol‐based redox state is critical for xylem vessel cell differentiation. Plant Signaling & Behavior, 13, e1428512. 10.1080/15592324.2018.1428512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y. , Overvoorde, P. J. , Arima, K. , Alonso, J. M. , Chan, A. , Chang, C. , Ecker, J. R. , Hughes, B. , Lui, A. , Nguyen, D. , Onodera, C. , Quach, H. , Smith, A. , Yu, G. , & Theologis, A. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell, 17, 444–463. 10.1105/tpc.104.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A. N. , Ernst, H. A. , Leggio, L. L. , & Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends in Plant Science, 10, 79–87. 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Peng, H. , & Neff, M. M. (2020). CIRCADIAN CLOCK ASSOCIATED 1 and ATAF2 differentially suppress cytochrome P450‐mediated brassinosteroid inactivation. Journal of Experimental Botany, 71, 970–985. 10.1093/jxb/erz468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, H. , & Neff, M. M. (2021). Two ATAF transcription factors ANAC102 and ATAF1 contribute to the suppression of cytochrome P450‐mediated brassinosteroid catabolism in Arabidopsis. Physiologia Plantarum, 172, 1493–1505. 10.1111/ppl.13339 [DOI] [PubMed] [Google Scholar]
- Peng, H. , Phung, J. , Stowe, E. C. , Dhingra, A. , & Neff, M. M. (2022). The NAC transcription factor ATAF2 promotes ethylene biosynthesis and response in Arabidopsis thaliana seedlings. FEBS Letters, 596, 1586–1599. 10.1002/1873-3468.14317 [DOI] [PubMed] [Google Scholar]
- Peng, H. , Phung, J. , Zhai, Y. , & Neff, M. M. (2020). Self‐transcriptional repression of the Arabidopsis NAC transcription factor ATAF2 and its genetic interaction with phytochrome A in modulating seedling photomorphogenesis. Planta, 252, 48. 10.1007/s00425-020-03456-5 [DOI] [PubMed] [Google Scholar]
- Peng, H. , Zhao, J. , & Neff, M. M. (2015). ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development, 142, 4129–4138. 10.1242/dev.124347 [DOI] [PubMed] [Google Scholar]
- Podzimska‐Sroka, D. , O'Shea, C. , Gregersen, P. L. , & Skriver, K. (2015). NAC transcription factors in senescence: From molecular structure to function in crops. Plants (Basel), 4, 412–448. 10.3390/plants4030412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R. J. , Thompson, W. A. , & Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous‐equations for assaying chlorophyll‐a and chlorophyll‐B extracted with 4 different solvents—Verification of the concentration of chlorophyll standards by atomic‐absorption spectroscopy. Biochimica et Biophysica Acta, 975, 384–394. 10.1016/S0005-2728(89)80347-0 [DOI] [Google Scholar]
- Puranik, S. , Sahu, P. P. , Srivastava, P. S. , & Prasad, M. (2012). NAC proteins: Regulation and role in stress tolerance. Trends in Plant Science, 17, 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Sade, N. , del Mar Rubio‐Wilhelmi, M. , Umnajkitikorn, K. , & Blumwald, E. (2018). Stress‐induced senescence and plant tolerance to abiotic stress. Journal of Experimental Botany, 69, 845–853. 10.1093/jxb/erx235 [DOI] [PubMed] [Google Scholar]
- Sakamoto, S. , Takata, N. , Oshima, Y. , Yoshida, K. , Taniguchi, T. , & Mitsuda, N. (2016). Wood reinforcement of poplar by rice NAC transcription factor. Scientific Reports, 6, 19925. 10.1038/srep19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer, E. , van Houwelingen, A. , Kloos, D. , Mol, J. , & Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell, 85, 159–170. 10.1016/S0092-8674(00)81093-4 [DOI] [PubMed] [Google Scholar]
- Suyal, G. , Rana, V. S. , Mukherjee, S. K. , Wajid, S. , & Choudhury, N. R. (2014). Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) rep protein. Virus Genes, 48, 486–493. 10.1007/s11262-013-1028-6 [DOI] [PubMed] [Google Scholar]
- Takasaki, H. , Maruyama, K. , Takahashi, F. , Fujita, M. , Yoshida, T. , Nakashima, K. , Myouga, F. , Toyooka, K. , Yamaguchi‐Shinozaki, K. , & Shinozaki, K. (2015). SNAC‐as, stress‐responsive NAC transcription factors, mediate ABA‐inducible leaf senescence. The Plant Journal, 84, 1114–1123. 10.1111/tpj.13067 [DOI] [PubMed] [Google Scholar]
- Wang, X. , & Culver, J. N. (2012). DNA binding specificity of ATAF2, a NAC domain transcription factor targeted for degradation by tobacco mosaic virus. BMC Plant Biology, 12, 157. 10.1186/1471-2229-12-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Goregaoker, S. P. , & Culver, J. N. (2009). Interaction of the tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. Journal of Virology, 83, 9720–9730. 10.1128/JVI.00941-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, A. , Allu, A. D. , Garapati, P. , Siddiqui, H. , Dortay, H. , Zanor, M. I. , Asensi‐Fabado, M. A. , Munne‐Bosch, S. , Antonio, C. , Tohge, T. , Fernie, A. R. , Kaufmann, K. , Xue, G. P. , Mueller‐Roeber, B. , & Balazadeh, S. (2012). JUNGBRUNNEN1, a reactive oxygen species‐responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell, 24, 482–506. 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, K. , Pan, T. , Gao, S. , & Yan, S. (2021). A transcription factor regulates gene expression in chloroplasts. International Journal of Molecular Sciences, 22, 6769. 10.3390/ijms22136769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M. , & Demura, T. (2010). Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnology, 27, 237–242. 10.5511/plantbiotechnology.27.237 [DOI] [Google Scholar]
- Yamaguchi, M. , Goue, N. , Igarashi, H. , Ohtani, M. , Nakano, Y. , Mortimer, J. C. , Nishikubo, N. , Kubo, M. , Katayama, Y. , Kakegawa, K. , Dupree, P. , & Demura, T. (2010). VASCULAR‐RELATED NAC‐DOMAIN6 and VASCULAR‐RELATED NAC‐DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiology, 153, 906–914. 10.1104/pp.110.154013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M. , Kubo, M. , Fukuda, H. , & Demura, T. (2008). VASCULAR‐RELATED NAC‐DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. The Plant Journal, 55, 652–664. 10.1111/j.1365-313X.2008.03533.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M. , Mitsuda, N. , Ohtani, M. , Ohme‐Takagi, M. , Kato, K. , & Demura, T. (2011). VASCULAR‐RELATED NAC‐DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. The Plant Journal, 66, 579–590. 10.1111/j.1365-313X.2011.04514.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M. , Nagahage, I. S. P. , Misato, O. , Ishikawa, T. , Uchimiya, H. , Kawai‐Yamada, M. , & Demura, T. (2015). Arabidopsis NAC domain proteins VND‐INTERACTINGa and ANAC103 interact with multiple NAC domain proteins. Plant Biotechnology, 32, 119–123. 10.5511/plantbiotechnology.15.0208a [DOI] [Google Scholar]
- Yamaguchi, M. , Ohtani, M. , Mitsuda, N. , Kubo, M. , Ohme‐Takagi, M. , Fukuda, H. , & Demura, T. (2010). VND‐INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell, 22, 1249–1263. 10.1105/tpc.108.064048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. D. , Seo, P. J. , Yoon, H. K. , & Park, C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell, 23, 2155–2168. 10.1105/tpc.111.084913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H. , Hirano, K. , Sato, T. , Mitsuda, N. , Nomoto, M. , Maeo, K. , Koketsu, E. , Mitani, R. , Kawamura, M. , Ishiguro, S. , Tada, Y. , Ohme‐Takagi, M. , Matsuoka, M. , & Ueguchi‐Tanaka, M. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proceedings of the National Academy of Sciences of the United States of America, 111, 7861–7866. 10.1073/pnas.1321669111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. , & Ye, Z. H. (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Molecular Plant, 3, 1087–1103. 10.1093/mp/ssq062 [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Lin‐Wang, K. , Wang, H. , Gu, C. , Dare, A. P. , Espley, R. V. , He, H. , Allan, A. C. , & Han, Y. (2015). Molecular genetics of blood‐fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. The Plant Journal, 82, 105–121. 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Peer Review.
Figure S1. Schematic diagram of NAC domain transcription factors. The gray boxes indicate subdomains I to V of the NAC domains. The black bar corresponds to the shortest encoded region of ANAC102 isolated by screening.
Figure S2. ATAF2 has transcriptional activation activity. (a) Schematic diagram of the constructs used in the dual luciferase transient assay. The reporter construct contained the firefly luciferase reporter gene under the control of five repeats of the upstream activation sequence of GAL4 (5 X GAL4 UAS) fused to a minimal CaMV35S promoter (min pro). The effector constructs contained GAL4‐BD bound to an empty multiple cloning site (GAL4‐BD‐MCS) or to coding sequences corresponding to full length VND7, VNI2, ATAF2, and ANAC102 driven by the CaMV35S promoter (35Spro). (b) Results of the transient transfection assay. Firefly luciferase activity was normalized to Renilla luciferase activity. Error bars indicate SD (n = 4). Different letters indicate significant differences at P < .05, as determined by one‐way ANOVA with Tukey's post‐test.
Figure S3. The whole NAC domain of VNI2 is necessary for interaction with ATAF2. (a) Schematic diagram of full length and truncated VNI2 used for the yeast two‐hybrid assay. (b) Result of yeast two‐hybrid assay. Full length ATAF2 fused to GAL4‐BD, and full length or truncated VNI2 fused to GAL4‐AD were introduced into AH109 yeast cells. The transformed cells were grown on control (Trp−Leu−) and selective media (Trp−Leu−His− with .1 mM 3‐AT). Plasmids containing MCS fused to GAL‐BD or GAL4‐AD were used as negative controls, and pBD‐wt and pAD‐wt were used as positive controls.
Figure S4. VNI2 and ATAF2 T‐DNA insertion lines. (a) Schematic diagram of the T‐DNA insertion sites of vni2 and ataf2. Grey, black, and white boxes indicate untranslated regions, coding regions, and introns, respectively. Arrows indicate the locations of the primers used for RT‐PCR. (b) RT‐PCR analysis results. Analysis was performed using the seedlings of 7‐day‐old plants.
Table S1. Oligonucleotides used in this study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


