Summary
Immunosuppressive myeloid cell populations have been documented in small cell lung cancer (SCLC) subtypes, playing a key role in remolding the tumor microenvironment (TME). However, the cancer-associated transcriptional features of monocytes and tumor-associated macrophages (TAMs) in SCLC remain poorly understood. Herein, we analyzed the molecular features and functions of monocyte/macrophage subsets aiming to inhibit monocyte recruitment and pro-tumor behavior of macrophages. We observe that NEUROD1-high SCLC subtype (SCLC-N) exhibits subtype-specific hypersialylation induced by the unique target c-Myc (MYC) of NEUROD1. The hypersialylation can alter macrophage phenotypes and pro-tumor behavior by regulating the expression of the immune-inhibiting lectin receptors on monocyte-derived macrophages (MDMs) in SCLC-N. Inhibiting the aberrant sialic acid metabolic pathways in SCLC can significantly enhance the phagocytosis of macrophages. This study provides a comprehensive overview of the cancer-specific immune signature of monocytes and macrophages and reveals tumor-associated biomarkers as potential therapeutic targets for SCLC.
Subject areas: Biochemistry, Immunology, Cell biology, Cancer
Graphical abstract

Highlights
-
•
SCLC TME is infiltrated by immunosuppressive monocytes/macrophages
-
•
Tumors alter circulating monocyte populations and promote monocyte differentiation
-
•
TAMs facilitate the tumor immune evasion via sialic acid-Siglec7/9 axis
-
•
MYC promotes SCLC hypersialylation by inducing the expression of sialyltransferases
Biochemistry; Immunology; Cell biology; Cancer
Introduction
SCLC is a malignant pulmonary tumor of neuroendocrine origin with rapid tumor growth, high metastasis, and dismal clinical outcomes. SCLC remains one of the most malignant tumors, with 5-year survival under 30% for limited-stage disease.1,2 In recent years, the introduction of immunotherapy to first-line chemotherapy has only benefited a minority of patients, mainly ascribed to the complex and heterogeneous tumor microenvironment in SCLC.3,4
There is an increasing consensus that SCLC can be classified into subtypes based on the differential RNA expression of transcription factors (ASCL1, NEUROD1, POU2F3, and YAP1),5,6 facilitating the in-depth exploration of the TME features of different SCLC subtypes. Neuroendocrine (NE)-high (SCLC-A and SCLC-N) subtypes exhibit less functional immune cell infiltration7 and greater accumulation of immunosuppressive cells, such as exhausted CD8+T cells and myeloid populations than NE-low subtypes (SCLC-P and SCLC-Y).8 Monocytes/macrophages represent the major subpopulations in the myeloid cells that play a key role in the TME.9,10 As we have reported before, targeting macrophages has become an issue of close attention for reversing the immunosuppressive status.11,12,13 It is well-established that monocytes/macrophages express more immune suppression-associated genes in NE-high SCLC subtypes. However, we still lack a complete understanding of molecular characteristics and distribution of monocytes/macrophages in SCLC subtypes.
Monocytes are highly plastic and heterogeneous in tumors. Classical monocytes (CD14+CD16−) and non-classical monocytes (CD14dimCD16+) exhibit obvious diversity in distribution and function in tumor tissues.14,15 It is widely thought that classical monocytes are recruited to tumor sites and differentiate into TAMs via distinct chemokine signaling pathways. Non-classical monocytes exhibit a greater enrichment in certain cancer cells than tumor-adjacent normal tissues, accelerating M2-type macrophage polarization.16 Identifying subset-specific biomarkers from the molecular signature of the pro-tumor behavior would provide a more effective strategy for blocking monocyte recruitment. However, the strategies for blocking the monocyte chemotaxis and differentiation are limited due to the lack of documented monocyte-specific markers and a complete understanding of the transcriptome characteristic of monocytes in SCLC subtypes. MDMs are polarized and accumulated in the TME, exhibiting tumor-promoting properties. In addition to the MDMs, a group of self-renewed TRMs reside in tumor lesions. A recent study revealed that TRMs but not MDMs play a critical role during early tumor progression in NSCLC.17 Other studies showed that MDMs might contribute more to tumor progression and metastasis in breast cancer and PDAC.9,18 However, the distribution of macrophages and how these macrophage lineages contribute to the NE-high SCLC TME remain unclear.
The aberrant glycosylation in tumor cells can trigger the expression of glycan-binding receptors known as lectins on immune cells.19 Glycans, such as sialic acid structures, can be recognized by inhibitory receptors expressed on the immune cells to promote tumor progression and immune evasion.20 Hypersialylation has been documented in breast, ovarian and lung cancers.21,22,23 It has been reported that knocking down the ST6GalNAC1 gene, a sialyltransferase in sialic acid metabolism, reduced MUC5AC sialylation as observed by low STn on the glycoprotein in lung cancer.23 However, sialylation in SCLC and the interaction between tumor cells and immune cells based on sialic acid modification in SCLC are poorly understood.
Herein, we revealed that the hypersialylation of SCLC-N can induce the increased expression of SIGLEC7 and SIGLEC9 on monocytes and macrophages. Inhibiting the aberrant sialic acid metabolic pathways24 in SCLC can significantly enhance the phagocytosis of macrophages. In addition, we identified the recruitment signature of monocytes in SCLC-N. Monocytes in SCLC-N are recruited and differentiate into TAMs mainly through the CXCL12/CXCR4 signaling pathway. These results lay the groundwork for future studies on valuable therapeutic targets for SCLC subtype-specific monocytes/macrophages and candidate diagnostic/prognostic biomarkers.
Results
Small cell lung cancer tumors alter the transcriptome of human monocytes
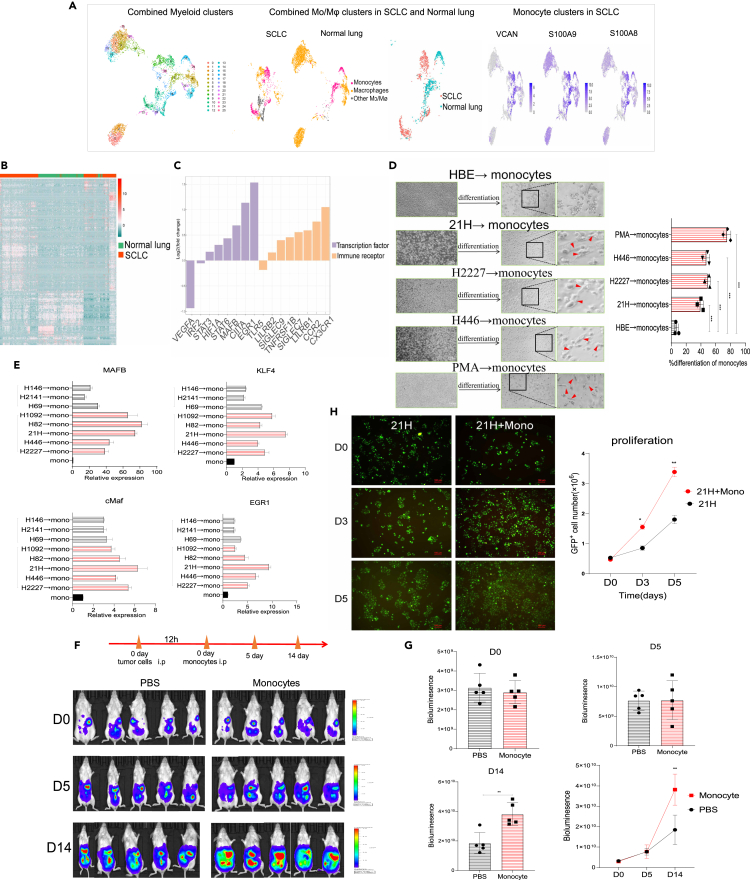
To better characterize the immunophenotype and function of monocytes in SCLC subtypes, we first analyzed the molecular features of myeloid subgroups from SCLC and tumor-adjacent normal lung samples using the available single-cell RNA sequencing (scRNA-seq) dataset published by Rudin et al.8 We screened and identified the monocytes that expressed canonical monocyte genes such as VCAN, S100A8 and S100A9 (Figure 1A). SCLC tumors exhibit greater monocyte enrichment. We defined monocytes in tumors as tumor-associated monocytes (TAMo). TAMo are highly enriched in the SCLC tumor tissues with differentially expressed genes (DEGs) compared with monocytes in the normal tissues (Figures 1B and 1C). Genes encoding transcription factors (TFs) involved in the monocyte differentiation into macrophages (MAFB and EGR1) were upregulated, and genes encoding immune regulatory molecules involved in immune-suppressive receptors (SIGLEC7, SIGLEC9 and LILRB1) were also increased in TAMo. GO and KEGG pathway analyses showed strong enrichment of innate immunity-related functions in TAMo (Figures S1A and S1B).
Figure 1.
SCLC tumors alter the transcriptome of human monocytes
(A) Analysis of macrophage and monocyte clusters from the myeloid cell population in the available scRNA-seq data of tumor samples and normal lung samples from 21 patients with SCLC (left and middle). The monocyte clusters defined by the expression of VCAN, S100A8 and S100A9 in the monocytes (right).
(B) Heatmap showing differentially expressed genes profiles in monocytes from SCLC and normal lung tissues.
(C) Relative expression of seclected DEG involved in the transcription factors and immune receptors in the TAMo compared with the monocytes in the normal tissues.
(D) Representative microscopic pictures showing the morphology of monocytes co-cultured with the SCLC cell lines. Positive control: monocytes treated with PMA. Bar = 100/200 μm.
(E) The expression of MAFB, KLF4, MAF and EGR1 in the monocytes induced by tumors (Student’s t test and the Kruskal–Wallis test were used to calculate the p values, SCLC-A versus SCLC-N, p < 0.05). NE-high SCLC cells (SCLC-A cell lines including H69, H146 and H2141, SCLC-N cell lines including 21H, H2227, H446, H82 and H1092).
(F) Bioluminescent imaging pictures showing tumor development in mice inoculated with tumors and monocytes.
(G) Quantification of tumor burden (total flux) by bioluminescent imaging.
(H) Living cell imaging system showing the tumor cell proliferation of CFSE dye labled 21H cells incubated with monocytes. Bar = 100 μm. Data are presented as the mean ± SEM of two or three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S1 and Table S1.
We further validated the differentiation of monocytes in vitro; a co-culture system was set up in which we cultured SCLC cells with monocytes derived from iPSCs (Figure S1C) and THP-1. First, we subclassified and selected the NE-high SCLC cell lines based on the RNA expression profiles of transcription factors in the CCLE database25 (Figure S1D). In the co-culture assay, significant alterations in cell morphology attracted our attention. The number of monocytes adhered to the plate with visible protrusions increased significantly in the tumor co-culture groups, compared with the monocytes co-cultured with normal human bronchial epithelial cells (HBE) (Figure 1D). In addition, monocytes significantly increased the mRNA levels of MAFB, KLF4, MAF and EGR1, which drive monocyte differentiation into macrophages,26 specifically in the monocytes incubated with SCLC-N cells (Figure 1E). Interestingly, we found high and relatively homogeneous expression of SIGLEC7 and SIGLEC9 encoding the immune-inhibiting lectin receptors in monocytes incubated with SCLC-N cells (Figure S1E). Moreover, monocytes showed high expression of M2-associated markers IL-10 and CD206 in SCLC-N cell lines (Figures S1F and S1G).
Next, we analyzed the effect of monocytes on the tumor development in vivo. In the mouse model, the luciferase-expressing tumors were infused via intraperitoneal injection. The tumor-bearing mice were treated 12h later with the intraperitoneal injection of iPSCs-derived monocytes. Monocyte-treated mice showed temporary inhibition of the tumor growth due to the strongest expansion of monocytes. However, tumor cell proliferation was significantly enhanced with the differentiation of monocytes to macrophages after 5days (Figures 1F and 1G). Then, we analyzed the phenotypes of monocytes infiltrated in the tumor tissues by IHC staining with anti-CD11b, anti-CD206, and anti-CD86 antibodies. The IHC showed that the macrophage marker CD11b and M2-type marker CD206 were expressed in the tumor tissues, suggesting that the tumors promote the differentiation of monocytes to M2-type macrophages (Figure S1H). In vitro, the proliferation of tumor cells co-cultured with monocytes was significantly enhanced compared to the tumor cells alone using the live cell imaging system (Figure 1H). Taken together, SCLC lesions exhibit greater monocyte enrichment, and the monocytes in SCLC-N are generally more inclined to differentiate into M2-type macrophages with pro-tumor function (Figure S1I).
Small cell lung cancer subtypes exhibit distinct monocyte subsets recruitment
Monocytes are highly plastic and heterogeneous, and have distinct functional phenotypes in response to environmental stimulation. To understand the molecular features and distribution of monocytes subpopulation, we analyzed the classical (CD14+CD16−) and non-classical (CD14dimCD16+) monocytes recruitment in SCLC subtypes. Non-classical monocytes have been reported to differentiate into M2-like macrophages, which secrete anti-inflammatory cytokines and contribute to tumor progression.16 In vitro, we first sorted monocytes co-cultured with SCLC cell lines, and analyzed the monocyte subsets stained with CD14 and CD16 by FASC. We found that the naive monocytes mainly expressed CD14, with few CD14dimCD16+ monocytes accounting for 13.8%. Upon the stimulation with the SCLC cells, the number of CD14dimCD16+monocytes was significantly higher than in the naive groups. SCLC-N cells induced a higher proportion of non-classical monocytes than other SCLC subtypes (SCLC-A and SCLC-Y) (Figure 2A). Next, we made use of an existing scRNA-seq dataset to map the monocytes subsets distribution in SCLC.8 We identified distinct monocyte subpopulations in patients with cancer versus normal tissues (Figure 2B). The distribution of monocytes in tumor-adjacent normal lung tissues was similar to other normal tissues, most of which were classical monocytes, with only a few non-classical monocytes. In SCLC lesions, the proportion of classical monocytes decreased, with increased recruitment of non-classical monocytes compared with the normal controls (Figure 2C). Next, we sought to assess the polarization phenotypes of classical and non-classical monocytes in the SCLC at the single-cell level. Classical monocytes in SCLC express both M1-type and M2-type markers, which are the mixed subgroup and cannot be categorized into binary states in SCLC. However, non-classical monocytes in SCLC were more likely to express M2-type markers, such as immune suppressive receptors CD163, SIGLEC7, SIGLEC10, TREM2, and CSF1R (Figures 2D and 2E), implying that subset-specific markers of monocytes are potential targets for SCLC progression.
Figure 2.
SCLC subtypes exhibit distinct monocyte subsets recruitment
(A) Monocytes subsets stained with CD14 and CD16 by FASC, co-cultured with SCLC-N, SCLC-A and SCLC-Y cell lines.
(B) Analysis of classical and non-classical clusters from the monocytes in the scRNA-seq data of tumor samples and normal lung samples from 21 patients with SCLC.
(C) The proportion of classical and non-classical monocytes in tumors and tumor-adjacent normal lung tissues.
(D and E) Dot plots show genes encoding M1 macrophage markers (D) and M2 macrophage markers in the monocytes (E). Data are presented as the mean ± SEM of two or three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Small cell lung cancer-N tends to recruit monocytes via CXCL12/CXCR4 signaling pathway
We next assessed the chemotaxis features of monocytes in NE-high SCLC cells. Analysis of the tumor tissues using the scRNA-seq data,8 we found that CXCR4 was one of the top upregulated genes encoding chemokine receptors in monocytes of SCLC-N compared to the SCLC-A (Figure 3A). CXCR4 expression in SCLC-N was significantly higher than in normal tissues (Figure 3B). Moreover, we found that CXCR4 was mainly expressed in the classical monocytes (Figure S2A). Subsequently, we assessed the expression of the ligands of chemotaxis receptors in SCLC-N and found that CXCL12 as the ligand of CXCR4 was highly expressed in SCLC-N tumors (Figure 3C). CCLE database also showed that the expression of CXCL12 was significantly higher in SCLC-N cells than in SCLC-A cell lines (Figure S2B). Validation by qPCR confirmed the significantly high expression of CXCL12 (Figure S2C) in SCLC-N cell lines, and elevated expression of CXCR4 in monocytes co-cultured with SCLC-N tumor cells or treated with CXCL12 (20 ng/mL, 50 ng/mL) as positive controls (Figure 3D).
Figure 3.
SCLC-N tends to recruit monocytes via CXCL12/CXCR4 signaling pathway
(A) Dot plots show genes encoding chemotaxis molecules in monocytes of NE-high SCLC tissues.
(B) Relative expression of CXCR4 in monocytes of SCLC and normal lung tissues.
(C) Dot plots show genes encoding chemotaxis molecules in SCLC-A and SCLC-N tumors.
(D) The mRNA levels of CXCR4 in the monocytes incubated with SCLC cell lines or treated with CXCL12.
(E) The number of td-tomato+monocytes infiltrating through the membrane into the tumors cells groups (21H and CXCL12 KD 21H) in the lower part. Bar = 100 μm.
(F) The mRNA levels of iNOS, GSK3β and IL-10 in the monocytes co-cultured with 21H.
(G) Western blots of molecules associated with signaling pathway (PKA/GSK3β) in the monocytes (mono: THP-1, imono: iPSC-derived monocytes) and CXCR4 KD monocytes co-cultured with SCLC-N cell lines or treated with CXCL12.
(H) Tumor cells activates the CXCR4/PKA/GSK3β signaling pathway in monocytes. Data are presented as the mean ± SEM of two or three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S2 and Table S1.
To verify the role of CXCL12/CXCR4 axis in monocyte recruitment in SCLC-N, we generated SCLC-N cell lines with CXCL12 knockdown and monocytes with CXCR4 knockdown by lentivirus transfection (Figures S2D and S2E). The CXCR4 expression was decreased in the monocytes co-cultured with CXCL12 KD tumor cells (Figure S2F). In a transwell system, the number of td-tomato+monocytes infiltrating through the membrane into the CXCL12 KD tumor cells in the lower chamber significantly decreased compared with the control groups using fluorescence microscopy at different times (Figure 3E). These results suggest that CXCL12/CXCR4 signaling pathway contributes to the recruitment of monocytes in SCLC-N.
We next evaluated whether CXCL12/CXCR4 pathway contributes to the polarization states of monocytes in SCLC cells. As shown in Result 1, the monocytes in SCLC-N are generally inclined to differentiate into M2-type macrophages with pro-tumoral function. However, we found that the expression of iNOS and GSK3β associated with anti-tumour phenotype27,28 of macrophages was increased, and the expression of IL-10 decreased in the CXCR4 KD monocytes co-cultured with SCLC-N tumor cells (Figure 3F). Moreover, the expression of iNOS and GSK3β was increased in the monocytes response to the CXCL12 KD tumor cells (Figure S2G). We next examined the mechanism of CXCL12/CXCR4-induced polarization in the monocytes. Western blotting revealed that the catalytic subunit of cAMP-dependent Protein kinase A (PKA), PKAα/β/γ, was activated in monocytes upon tumor cells (21H) and CXCL12 (50 ng/mL) stimulation (Figure 3G). In addition, PKAα/β/γ expression was significantly reduced in CXCR4 KD monocytes. The expression of phospho-GSK3βSer9 was significantly decreased in the monocytes co-cultured with tumors, and was negatively regulated by PKA,29 suggesting that the activation of CXCR4/PKA impairs macrophage M1 polarization in SCLC (Figure 3H).
Monocyte-to-macrophages exhibit cancer-specific molecules in small cell lung cancer
To analyze the components and phenotypes of TAMs that populate human SCLC lesions, we screened and identified the macrophages in SCLC using the public data8 (Figure 4A). GO and KEGG analyses indicated that immune effector process including “phagocytosis” and “negative regulation of defense response” were both significantly enriched in TAMs (Figures S3A and S3B). TAMs exhibited DEGs compared with macrophages in the normal tissues (Figure S3C). The expression of immune receptors, transmembrane molecules, and soluble factors were altered in SCLC (Figure 4B), including increased expression of anti-inflammatory cytokines, immunosuppressive receptors (SIGLEC7, SIGLEC9, and LILRB1), and decreased expression of M1-type genes encoding pro-inflammatory cytokines. qPCR and flow cytometry confirmed high expression of SIGLEC7 and SIGLEC9 in macrophages, especially in the macrophages co-cultured with SCLC-N (Figures 4C and 4D). The scRNA-seq data showed that SIGLEC7 and SIGLEC9 were mainly expressed in monocyte and macrophage subgroups of all myeloid populations in SCLC (Figures 4E and 4F). To characterize the expression of these immunosuppressive molecules in macrophage lineages, we analyzed the immune signature of macrophage lineages. First, we identified tissue-resident macrophage clusters that highly expressed the cell-cycle genes STMN1, alveolar macrophage-related genes PPARG and scavenger receptors SIGLEC1, as well as expressing classical macrophage genes such as CD68, HLA-DRA, MRC1 and C1QA.17,30 Monocyte-derived macrophage clusters were defined by the classical macrophage genes and lacked the expression of TRM-related genes. We found that the macrophages in adjacent normal tissues were predominantly TRMs clusters and rarely MDMs, while in the tumor tissues, both TRMs and MDMs exhibited comparable enrichment levels (Figure 4G). Notably, most immunosuppression-related genes were expressed in MDMs but not in the TRMs, for example, SIGLEC7 and SIGLEC9 were upregulated in MDMs, while TREM1 and CD276 were highly expressed mainly in TRMs (Figures 4H and S3D). These results suggest that targeting lineages-specific immune checkpoints may be more effective for immunotherapy; for example, blocking SIGLEC7 and SIGLEC9 may be more useful for MDM subgroups (Figure 4I).
Figure 4.
Monocyte-to-macrophages exhibit cancer-specific molecules in SCLC
(A) Analysis of macrophage clusters from the myeloid cell population in the scRNA-seq data.
(B) Relative expression of seclected DEG associated in the TAMs compared with the macrophages in the normal tissues.
(C and D) The expression of SIGLEC7 and SIGLEC9 in macrophages incubated with tumor cells, qPCR (C) (Student’s t test and the Kruskal–Wallis test were used to calculate the p values, SCLC-A versus SCLC-N, p < 0.001), and flow cytometry (D).
(E and F) Expression of SIGLECs in the different immune cells (E) and myeloid cells (F).
(G) Characterization and proportion of TRMs and MDMs in the SCLC in the scRNA-seq data.
(H) Dot plots show relative frequency and expressing cells of genes encoding M2-type markers in TRMs and MDMs.
(I) Schematic representation of TRMs and MDMs in tumors. Data are presented as the mean ± SEM of two or three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S3 and Table S1.
Small cell lung cancer exhibits hypersialylation as the contributor to immune-inhibiting lectin receptors of tumor-associated macrophages
Given that SCLC can significantly induce the expression of the lectin receptors SIGLEC7 and SIGLEC9 in TAMs, we further analyzed the signature of the sialylation, which serve as the ligands for letin receptors in SCLC (Figure 5A). Sialic acids present as a terminal sugar on the periphery of oligosaccharides on the cell surface through different glycosidic linkages (namely α2,3, α2,6 and α2,8). CMP-sialic acid metabolic pathways include a series of enzymes that catalyze the biosynthesis, activation, and conversion of sialic acids to glycoconjugates, as well as the removal and degradation of sialic acid (Figure 5B).
Figure 5.
SCLC exhibits hypersialylation as the contributor to immune-inhibiting lectin receptors of TAMs
(A) Schematic representation of sialic acid-Siglec axis in tumors.
(B) The metabolic pathway of sialic acids.
(C) Evaluation of α2,3 and α2,6 sialic acid expression in the lysates from SCLC cells by ELISA (Student’s t test and the Kruskal–Wallis test were used to calculate the p values, SCLC-A versus SCLC-N, p < 0.05).
(D) The relative expression of ST3GAL5, ST6GALNAC3/5 in SCLC cells (Student’s t test and the Kruskal–Wallis test were used to calculate the p values, SCLC-A versus SCLC-N, p < 0.05).
(E) The mean normalized expression of α2,3 and α2,6 sialyltransferases in SCLC-A and SCLC-N subtypes in the scRNA-seq data.
(F) Analysis of the altered α2,3 and α2,6 sialic acid expression in the lysates from ST3GAL5 KD cells and ST6GALNAC5 KD cells respectively.
(G and H) The relative expression of SIGLEC7 and SIGLEC9 in the macrophages co-cultured with ST3GAL5 KD cells, ST6GALNAC5 KD cells and control groups.
(I) Flow cytometry showing phagocytosis of ST3GAL5 KD or ST6GALNAC5 KD cells by macrophages. Dye-labeled (red) mac co-incubated with GFP+ tumor cells and collected for FACS analysis. Double-positive cells represent the Mac cells that engulfed cancer cells. Data are presented as the mean ± SEM of two or three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S4 and Table S1.
To examine the level of α2,3 and α2,6 sialylation in NE-high SCLC subtypes, we first quantified the expression of biotinylated plant lectins derived from Maackia amurensis lectin II (MALII) and Sambucus nigra (SNA), which represented the expression of α2,3 and α2,6 sialylation, respectively.18 The results showed that MALII and SNA were highly expressed in the lysates of SCLC cells (Figure 5C). Moreover, MALII and SNA could be detected in the medium of SCLC cells, indicating that sialic acid-containing glycans could be secreted by the tumor cells (Figure S4A). Remarkably, we found that the level of α2,3 sialylation was higher in SCLC-N tumor cells than in SCLC-A cells. These results indicated that α2,3 sialytion may be the main sialic acid pathway that induces high expression of SIGLEC7 and SIGLEC9 on macrophages in the SCLC-N.
To further characterize the sialic acid metabolism in SCLC subtypes, we analyzed the expression of the enzymes involved in the sialic acid metabolic pathways. The expression of GNE associated with sialic acid synthesis and CMAS involved in the activation of CMP-Neu5Ac was comparable in SCLC subtypes (Figure S4B). CMP-Neu5Ac is transferred to the glycoconjugates by a family of lineage-specific sialyltransferases after activation. Notably, we found that the expression of sialyltransferases ST3GAL2/5 for α2,3 sialic acid and ST6GALNAC3/5 for α2,6 sialic acid were significantly higher in SCLC-N than in the other subtypes (Figure S4C). Validation by qPCR confirmed the significantly higher expression of ST3GAL5 and ST6GALNAC3/5 mRNA observed in the CCLE database (Figure 5D). In the scRNA-seq data, we also confirmed that ST3GAL5 and ST6GALNAC5 were significantly elevated in SCLC-N subtypes, and other sialyltransferases expression were not significantly different between SCLC-N and SCLC-A (Figure 5E). For the degradation of sialoglycoconjugates, sialic acids on glycoconjugates can be released by sialidases (neuraminidases, NEU1-4). CCLE data showed no significant difference in NEU1-4 expression in different SCLC subtypes (Figure S4D).
To identify the role of these sialyltransferases in catalyzing the transfer of sialic acids to glycoconjugates on tumor cell membrane, we knocked down these sialyltransferases using shRNA. ELISA showed that the expression of α2,3 and α2,6-linked sialic acids decreased obviously in ST3GAL5 and ST6GALNAC5 KD groups respectively (Figure 5F). The expression of α2,3 and α2,6 sialic acid was not significantly decreased in the ST3GAL2 and ST6GALNAC3 KD groups respectively (Figure S4E). These results suggest that ST3GAL5 and ST6GALNAC5 may be the main contributors to the transfer of sialic acid in SCLC-N cells. Subsequently, we analyzed the effects of ST3GAL5 and ST6GALNAC5 on the expression of SIGLEC7 and SIGLEC9 in the macrophages. We found that the expression of SIGLEC7 and SIGLEC9 significantly decreased in macrophages co-cultured with ST3GAL5 and ST6GALNAC5 KD groups using qPCR (Figures 5G and 5H). Next, we analyzed the effects of ST3GAL5 and ST6GALNAC5 on macrophage phagocytosis activity. Flow cytometry showed that macrophages co-cultured with the ST3GAL5 and ST6GALNAC5 KD SCLC-N cells experienced significantly enhanced phagocytosis (Figure 5I). These results suggest that both ST3GAL5 for α2,3 and ST6GALNAC5 for α2,6 sialic acid participate in the immunomodulation in SCLC-N through inducing SIGLEC7 and SIGLEC9 expression in TAMs.
Involvement of MYC in the induction of ST3GAL5 and ST6GALNAC5 to promote hypersialylation in small cell lung cancer-N
To explore the mechanism of the subtype-specific hypersialylation in SCLC cells, we searched for potential transcription factors involved in their transcriptional regulation. We first examined the role of the transcription factor NEUROD1 in regulating the expression of ST3GAL5 and ST6GALNAC5 by using shRNA to knock down NEUROD1 (NEU KD) in SCLC-N cell lines (Figure S5A). The expression of ST3GAL5 and ST6GALNAC5 was significantly reduced in NEU KD tumor cells by qPCR (Figure 6A). In addition, we found that the expression of SIGLEC7 and SIGLEC9 was significantly reduced in macrophages co-cultured with NEU KD SCLC cells (Figure 6B). These results suggest that NEUROD1 may be involved in the expression of ST3GAL5 and ST6GALNAC5 and regulates the sialic acid metabolic process.
Figure 6.
Involvement of MYC in the induction of ST3GAL5 and ST6GALNAC5 to promote hypersialylation in SCLC-N
(A) The relative expression of ST3GAL5 and ST6GALNAC5 in the NEU KD and control groups.
(B) The expression of SIGLEC7 and SIGLEC9 in the macrophages induced by NEU KD 21H cells and control groups.
(C) The relative expression of MYC in the NEU KD and control groups.
(D and E) The relative expression of ST3GAL5 and ST6GALNAC5 in the MYC KD (D), and MYC OV and control groups (E).
(F) ChIP analysis of MYC enrichment at the promoters of ST3GAL5 and ST6GALNAC5 in 21H cells.
(G and H) Co-IP and reverse co-IP evaluating the interaction between MYC and ST3GAL5 (G) and ST6GALNAC5 (H) in 21H cells.
(I) Representative microscopic pictures showing the morphology of monocytes co-cultured with the MYC KD 21H cells. Bar = 100 μm.
(J–L) The expression of SIGLEC7 (J), SIGLEC9 (K) and CD206 (L) in the MYC KD and control groups by qPCR and flow cytometry. Data are presented as the mean ± SEM of two or three independent experiments. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S5 and Table S1.
In a previous study, we reported that NEUROD1 could regulate the expression of the oncogene MYC.31 Moreover, ChIP-seq showed that the unique target MYC of NEUROD1 could distinguish ASCL1-bound regions.32 The expression of MYC was significantly decreased in NEU KD SCLC cells (Figure 6C). In this context, we hypothesize that MYC may be the direct TF that promotes sialic acid expression by the transcriptional induction of ST3GAL5 and ST6GALNAC5 in SCLC-N tumors. Next, we established stable cell lines with low expression and overexpression of MYC respectively (Figure S5B), then detected the effects of MYC knockdown (MYC KD) and overexpression (MYC OV) on the expression of α2,3 and α2,6-linked sialic acids. We found that MYC KD significantly inhibited ST3GAL5 and ST6GALNAC5 expression, while MYC OV increased the expression of ST3GAL5 and ST6GALNAC5 by qPCR (Figures 6D and 6E). Consequently, ELISA showed that α2,3 and α2,6 sialic acid expression was decreased in the MYC KD SCLC-N cell lines, but increased in the MYC OV groups (Figures S5C–S5E). ChIP assays showed enhanced recruitment of MYC to the promoter of ST3GAL5 and ST6GALNAC5 (Figures 6F, S5F, and S5G). Co-immunoprecipitation (co-IP) and reverse co-IP indicated that MYC interacts with ST3GAL5 and ST6GALNAC5 in 21H cells (Figures 6G and 6H). Overall, these results reveal a pivotal role of MYC in promoting α2,3 and α2,6 sialic acid accumulation through the transcriptional regulation of ST3GAL5 and ST6GALNAC5 in SCLC-N. To further explore the role of MYC in the altered phenotype and immune suppression of macrophages in SCLC-N, a co-culture system was set up in which MYC KD SCLC cell lines were co-cultured with macrophages. Monocytes co-cultured with wild-type SCLC-N tumor cells were more likely to induce monocyte differentiation with visible protrusions, while most of the monocytes co-cultured with MYC KD tumor cells remained in a monocyte-like suspension state (Figure 6I). We found that the expression of SIGLEC7 and SIGLEC9 on the surface of macrophages was significantly reduced in MYC KD cells (Figures 6J and 6K). In addition, the M2-type marker CD206 was significantly decreased in the MYC KD SCLC cell lines (Figure 6L). These results demonstrate that MYC can alter macrophage phenotype and pro-tumor behavior by regulating the sialylation level of SCLC-N.
Discussion
In recent years, many studies have revealed distinct RNA gene expression profiles of SCLC, highlighting the heterogenicity of the tumor microenvironment. NE-high and NE-low SCLC subtypes were associated with “immune desert” and “immune oasis” phenotypes,33,34,35,36 respectively. In NE-high SCLC, the TME was predominantly infiltrated with exhausted immune cells and less active immune cells.37 Notably, the SCLC-N subtype showed stronger immunosuppression with more Treg aggregation, fewer CD8+T infiltration and immunosuppressive monocyte/macrophage subpopulation. In addition to the T cell populations, the myeloid compartment was the main contributor to the immunosuppressive TME in SCLC.8 However, the characterization of the myeloid groups in SCLC subtypes has been largely understudied.
Herein, we analyzed the transcriptomes and composition of myeloid subgroups in SCLC TME using public data. The transcriptome profiles of monocytes in SCLC were significantly distinct from the monocyte population in normal tissues. Tumors alter the RNA gene expression profiles and distribution of monocytes in SCLC. Our results showed that monocytes in SCLC express high levels of immunosuppressive factors and chemokines, such as SIGLEC7/9, LILRB1, CSF1R and CXCR4. In the co-culture experiments, we substantiated that SCLC cell lines could significantly induce monocyte differentiation to IL-10-high and CD206-high macrophage subtypes. Analysis of monocyte subsets revealed that a mixture of classical and non-classical monocytes with an immunosuppressive phenotype infiltrated in SCLC TME. Non-classical monocytes were recruited to tumors and were more likely to differentiate into M2 macrophages. In SCLC TME, certain immunosuppressive molecules are more highly expressed in non-classical monocytes, such as SIGLEC7, CD163, TREM2 and CSF1R. Analysis of the monocyte chemotaxis signals in the NE-high SCLC showed that SCLC-N cells tend to recruit monocytes via CXCL12/CXCR4 pathway. SCLC-N promotes monocytes differentiation to IL-10-high and iNOS-low macrophages through the CXCR4/PKA/GSK3β pathway, which impairs M1-type polarization.
Analysis of the macrophage subpopulations in SCLC showed that tumor-associated TRMs and MDMs coexisted in the SCLC TME. Our analysis of the RNA gene expression profiles of TRMs and MDMs revealed that immunosuppressive molecules were mostly expressed on MDMs but not on TRMs. For example, the immunosuppressive molecule CD36, which supports Tregs survival and function,38 and CD93 which correlates with immune cell infiltration and affects immunotherapy,39 were both upregulated on MDMs. In the TRMs of SCLC, CD276 involved in the tumor-promotion T cell subset40 and suppressive monocyte enrichment was upregulated. Our analysis showed that immune-inhibiting lectin receptors, especially SIGLEC7 and SIGLEC9, were highly expressed in the MDMs in SCLC-N subtypes. Taken together, targeting macrophage lineages-specific biomarkers may be more efficient for the re-education and depletion of TAMs.
Recent studies have described the roles of sialylation in tumor growth and metastasis; the aberrant sialylation on tumor cell surfaces promotes tumor proliferation and metastasis and facilitates the immune escape of tumors.21,41 Analysis of sialylated structures revealed that both α2,3 and α2,6 sialylation are expressed in the NE-high SCLC, with the SCLC-N cells exhibiting higher α2,3 sialylation. Our results reveal that α2,3 and α2,6 linked sialylated structures are the major contributor to stimulate the high expression of SIGLEC7 and SIGLEC9 on macrophages in the SCLC-N. Analysis of the enzymes involved in the sialic acid metabolic pathway in SCLC showed significant differences in the expression of sialyltransferases ST3GAL5 and ST6GALNAC5 between NE-high SCLC subtypes, both of which were highly expressed in SCLC-N. We identified the expression of ST3GAL5 and ST6GALNAC5 as main contributors to the synthesis of sialylated structures sensed by the myeloid cell receptors SIGLEC7 and SIGLEC9, facilitating the immune evasion in SCLC.
Multiple studies have implicated an increase in MYC in chemotherapy-resistant mouse and human tumors and have correlated high MYC with immune evasion, more aggressive and shorter patient survival.42,43,44 In our previous study, we have reported that NEUROD1 could bind to the oncogene MYC in SCLC. SCLC-N, as the MYC-high subtype, exhibited hypersialylation regulated by MYC. Analysis of sialylation alteration showed downregulated expression of ST3GAL5 and ST6GALNAC5 and decreased sialylation in the MYC KD SCLC cells. The ChIP assay identified increased recruitment of MYC in the promoters of ST3GAL5 and ST6GALNAC5, and the Co-IP assay revealed the interactions. Overall, these data demonstrate that MYC contributes to the α2,3 and α2,6 hypersialylation by the transcriptional induction of ST3GAL5 and ST6GALNAC5.
In summary, we analyzed the molecular signature and lineage-specific distribution of monocytes and macrophages in SCLC. We identified a mechanism of immune evasion where SCLC cells induce immune suppressive networks in the TAMs via the sialic acid-Siglec axis. In addition, targeting tumor subtype-specific chemotaxis signaling pathways may be more effective in preventing the infiltration of monocytes in SCLC. Such data support the concept of monocytes and macrophages in the controlling immune suppression,45 and identification of the subtype-specific gene expression provides useful targeting strategies, including inhibiting monocyte infiltration, improving the phagocytosis of macrophages and activating the tumor immune microenvironment (Figure 7).
Figure 7.
Schematic representation of the immunotherapy strategies targeting the monocytes and macrophages in SCLC
Limitations of the study
In this study, we analyzed the immunosuppressive profile of macrophages in SCLC, demonstrating that the hypersialylation of tumors can alter macrophage phenotypes and pro-tumor behavior by regulating the expression of the immune-inhibiting lectin receptors on MDMs. However, the cancer-associated sialic acid-binding glycoproteins or glycolipids on the tumors cell surface are unclear, we still unknow which sialylated proteins can be recognized by the lectin receptors. In addition, the mechanism of immune suppression of SIGLEC7 and SIGLEC9 in macrophages has not been deeply investigated here. Another limitation is lack of a humanized immune system mouse model for validation, and we further will establish a mouse model of SCLC with human immune reconstitution to analyze the characteristics of myeloid clusters and examine whether mouse model can provide the results consistent with those in vitro.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-human CD14 Antibody | Biolegend | clone HCD14; Cat# 325603; RRID: AB_830676 |
| FITC anti-human CD11b Antibody | Biolegend | clone ICRF44; Cat# 301330; RRID: AB_2561703 |
| APC anti-human CD206 (MMR) Antibody | Biolegend | clone 15-2; Cat# 321109; RRID: AB_571884 |
| Anti-CD11b (IHC) | Abcam | Cat# ab133357; RRID: AB_2650514 |
| Anti-CD206 (IHC) | Abcam | Cat# ab64693; RRID: AB_1523910 |
| Anti-CD86 (IHC) | Cell Signaling Technology Inc | Cat# 91882; RRID: AB_2797422 |
| APC anti-human CD328 (Siglec-7) Antibody | Biolegend | Clone 6-434; Cat# 339205; RRID: AB_2565238 |
| PE anti-human Siglec9 Antibody | R&D SYSTEMS | Clone 191240; Cat# FAB1139P-025; RRID: AB_2187294 |
| APC anti-human CD14 Antibody | Biolegend | Clone M5E2; Cat# 301808; RRID: AB_314190 |
| FITC anti-human CD16 Antibody | Biolegend | Clone 3G8; Cat# 302005; RRID: AB_314205 |
| c-Myc (WB/IP) | Cell Signaling Technology Inc | Cat# 18583; RRID: AB_2895543 |
| ST3GAL5 (WB) | Proteintech | Cat# 14614-1-AP; RRID: AB_2194414 |
| ST3GAL5 (co-IP) | Santa Cruz Biotechnology | Cat# sc-365329; RRID: AB_10846084 |
| ST6GALNAC5 (WB/co-IP) | Thermo Scientific | Cat# PA5-112276; RRID: AB_2867012 |
| Anti-PKA alpha/beta/gamma (WB) | Abcam | Cat# ab75991; RRID: AB_1524202 |
| Anti-PKA R2 (WB) | Abcam | Cat# ab32390; RRID: AB_779040 |
| Anti-GSK3 beta (WB) | Abcam | Cat# ab32391; RRID: AB_2115066 |
| Anti-GSK3 beta+alpha (WB) | Abcam | Cat# ab68476; RRID: AB_10013745 |
| Anti-GSK3 beta (pS9) (WB) | Abcam | Cat# ab75814; RRID: AB_1310289 |
| Bacterial and virus strains | ||
| Subcloning Efficiency™ DH5α | Invitrogen | Cat# 18265017 |
| psPAX2 packaging plasmid | addgene | Cat# 12260 |
| pMD2.G packaging plasmid | addgene | Cat# 12259 |
| GV248 lentiviral vector | Shanghai Genechem Co.,Ltd | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Human CXCL12 | PeproTech, Inc | Cat# 300-28A |
| Recombinant Human BMP-4 | PeproTech, Inc | Cat# 120-05 |
| Recombinant Human bFGF | PeproTech, Inc | Cat# 100-18B |
| Recombinant Human VEGF | PeproTech, Inc | Cat# 100-20 |
| Recombinant Human SCF | PeproTech, Inc | Cat# 300-07 |
| Recombinant Human IGF-1 | PeproTech, Inc | Cat# 100-11 |
| Recombinant Human IL-3 | PeproTech, Inc | Cat# 200-03 |
| Recombinant Human M-CSF | PeproTech, Inc | Cat# 300-25 |
| Recombinant Human GM-CSF | PeproTech, Inc | Cat# 300-23 |
| Lipofectamine 2000 | ThermoFisher | Cat# 11668019 |
| PMA | Sigma-Aldrich | Cat# 16561-29-8 |
| Puromycin | Sigma-Aldrich | Cat# P8833 |
| Polybrene | Sigma-Aldrich | Cat# TR-1003-G |
| TRIzol | Takara | Cat# 9109 |
| HiScript II Q RT SuperMix for qPCR | Vazyme | Cat# R222-01 |
| Rock inhibitor Y27632 | STEMCELL Technologies | Cat# 72304 |
| Matrigel Matrix | Corning | Cat# 354277 |
| Critical commercial assays | ||
| SimpleChIP Plus Enzymatic Chromatin IP Kit | Cell Signaling Technology Inc | Cat# 9005 |
| Cell-Tracker Three-color Trial Kit | Maokangbio,Co., Ltd | Cat# MX4111 |
| Human SNA ELISA Kit | Enzyme-linked Biotechnology Co., Ltd | Cat# YJ956321 |
| Human MALII ELISA Kit | Enzyme-linked Biotechnology Co., Ltd | Cat# YJ915220 |
| Human CXCL12 Kit | Elabscience, Co., Ltd | Cat# E-EL-H0052c |
| SYBR® green qPCR kit | Roche | Cat# 0488735200 |
| CellTracker Green CMFDA | ThermoFisher | Cat# C2925 |
| STEMdiff™ APEL™2 Medium | STEMCELL Technologies | Cat# 05270 |
| StemPro™-34 SFM medium | Gibco | Cat#10639011 |
| TrypLE | Gibco | Cat#12604-021 |
| mTeSR medium | STEMCELL Technologies | Cat#85851 |
| Deposited data | ||
| Single-cell RNA sequencing (scRNA-seq) dataset | Chan, J.M. et al.8 | https://data.humantumoratlas.org |
| Transcriptome data of SCLC cell lines | Tlemsani, C. et al.25 | GSE36139 |
| Experimental models: Cell lines | ||
| H446 | ATCC | Cat# HTB-171 |
| H2227 | ATCC | Cat# CRL-5934 |
| H82 | ATCC | Cat# HTB-175 |
| H1092 | ATCC | Cat# CRL-5855 |
| H69 | ATCC | Cat# HTB-119 |
| H196 | ATCC | Cat# CRL-5823 |
| H1339 | ATCC | Cat# CRL-5959 |
| DMS-114 | ATCC | Cat# CRL-2066 |
| H146 | ATCC | Cat# HTB-173 |
| H2141 | ATCC | Cat# CRL-5927 |
| 293T | ATCC | Cat# CRL-3216 |
| THP-1 | ATCC | Cat# TIB-202 |
| Experimental models: Organisms/strains | ||
| NOD-PrkdcscidIl2rgem1/Smoc (M-NSG) mice | Shanghai Model Organisms Center, Inc. | N/A |
| Oligonucleotides | ||
| Primers for qPCR, Refer Table S1 |
Tsingke Biotechnology | N/A |
| Primers for ChIP-pcr, Refer Table S1 |
Tsingke Biotechnology | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.2.1 software | GraphPad Prism Software Inc. | https://www.graphpad.com/ |
| Flowjo v10.8.1 software | Becton, Dickinson & Company | https://www.flowjo.com/ |
| Adobe Illustrator 2023 software | Adobe Systems Inc. | https://www.adobe.com/cn/ |
| Seurat v4.2.0 | New York Genome Center, Satija Lab | https://satijalab.org/seurat/ |
| R package | R CRAN | http://www.r-project.org/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ying Cheng (chengying@csco.org.cn).
Materials availability
All reagents were purchased commercially. This study did not generate new unique reagents.
Experimental model and study participant details
Cell cells
ALL SCLC cell lines, 21H, H2227, H446, H82, H1092, H69, H146, H2141, H196, H1339, SBC-5 and DMS-114 were originally purchased from ATCC and subsequently maintained in our laboratory. All cell lines were authenticated through the STR characterization method and frequently tested for Mycoplasma using PCR. All the cell lines except for H446 were cultured in RPMI 1640 (Gibco), and supplemented with 10% v/v Fetal Calf Serum (Gibco), 2 mM L-Glutamine (Gibco) and 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco). The H446 cell line was cultured in high-glucose DMEM (Gibco) with 10% v/v serum. THP-1 cell lines were cultured in RPMI1640 with 10% v/v serum. Human iPSCs were cultured in mTeSR medium (STEMCELL Technologies) with Matrigel Matrix plate. 293T were cultured in high-glucose DMEM containing 10% FBS, 100 μM nonessential amino acids (GENOM) and 100 U/ml penicillin, 100 μg/ml streptomycin.
Monocyte and macrophage differentiation from iPSCs
Undifferentiated iPSCs were treated with TrypLE (Gibco, #12604-021) for 1 min to digest them into single cells. The cells were collected and transferred to low-attachment plates (Corning, #3471) to allow the formation of embryoid bodies (EBs) in mTeSR medium (STEMCELL Technologies, #85851) supplemented with Rock inhibitor Y27632 (STEMCELL Technologies, #72304). Formation of EBs was facilitated by an overnight incubation at 37°C in 5% CO2 with shaking at 50 rpm. Briefly, mesodermal progenitor cells generated from EBs were cultured in APEL II medium (05270, STEMCELL Technologies) with BMP-4 (10 ng/ml) and bFGF (5 ng/ml) on day 1. Next, hematopoietic specification was completed in the addition of BMP-4 (10 ng/ml), bFGF (5 ng/ml), VEGF (50 ng/ml) and SCF (100 ng/mL) in APEL II medium on day 2 to day 7. Then, addition of bFGF (10 ng/ml), VEGF (50 ng/ml), SCF (50 ng/mL), IGF-1 (10 ng/mL), IL-3 (25 ng/mL), M-CSF (50 ng/mL) and GM-CSF (50 ng/mL) in APEL II medium promotes the differentiation of myeloid lineage on day 8 to day 9. On day 10, culture EBs onto 6-well plate pre-coated with Matrigel Matrix (354277, Corning) in StemPro™-34 SFM medium (10639011, Gibco) with bFGF (5 ng/ml), VEGF (50 ng/ml), SCF (50 ng/mL), IGF-1 (10 ng/mL), IL-3 (25 ng/mL), M-CSF (50 ng/mL) and GM-CSF (50 ng/mL) for 10 days. On day 21 and 22, collect the cells to new empty Matrigel plates in myeloid maturation medium (MM medium) containing bFGF (5 ng/ml), VEGF (50 ng/ml), SCF (50 ng/mL), IGF-1 (10 ng/mL), IL-3 (25 ng/mL), M-CSF (100 ng/mL) and GM-CSF (100 ng/mL). From day 22 to day 27, cells were cultured with MM medium removal of IL-3. Well-differentiated iPSC-macrophages were cultured in RPMI1640 medium with 10% FBS containing M-CSF (100 ng/mL) and GM-CSF (100 ng/mL) from day 28.12
Macrophage differentiation from THP-1
THP-1 (5∼8x105 cells/ml in 6-well plates) cells were differentiated into macrophages using 5ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) for 48hr. Differentiation of PMA-treated cells was enhanced after the initial 2d stimulus by removing the PMA-containing media and then incubating the cells in fresh RPMI 1640 with 10% FBS and 1% L-glutamine.
Animals
In the present study, male NOD-PrkdcscidIl2rgem1/Smoc (M-NSG) mice were used. Animals (4-6 weeks old, 19±1.5 g) were obtained from Shanghai Model Organisms Center, Inc and kept in a room under pathogen-free conditions with a 12/12 h light-dark cycle, controlled temperature (22 ± 2°C) and humidity (50 ± 5%). All animals were housed 3-4 per cage in Plexiglas cages with free access to food and tap water. All procedures were approved by the Institutional Animal Care and Use Committee of Zhejiang Academy of Medical Sciences.
Method details
Detection for sialoglycans and cytokines
Lysates and culture supernatant were harvested from tumor cells for ELISA-based assay. Tumor cells (1x106 cells/mL in 6-well plates) were washed with ice cold PBS twice and incubated with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.6, 0.5% Nonidet P-40) with 1 mM protease inhibitors. The concentration of protein was tested by the bicinchoninic acid (BCA) assay (Beyotime). We also tested the concentration of sialic acid secreted into the culture supernatant; we performed an ELISA in FBS-free culture supernatant from tumor cells. 1x106 cells were cultured for 24 hr with RPMI 1640 supplemented with 2 mM L-Glutamine and 1000 U/mL Penicillin-Streptomycin. Supernatants were obtained and observed at -20°C. Protein concentration was determined by BCA assay. The levels of α2,3 and α2,6 sialylation in tumors were quantified by MALII and SNA ELISA kits (MLbio, YJ956321 and YJ915220) following the manufacturer’s instructions. The culture supernatant of tumor cells was harvested to quantify the secretion of cytokine IL-10 (Elabscience, E-EL-H6154), CXCL12 (Elabscience, E-EL-H0052c) and CSF-1 (Elabscience, E-EL-H0097c) by ELISA kits following the manufacturer’s instructions.
RNA extraction, cDNA and real-time PCR
Tumor cells and FACS-sorted monocytes or macrophages were lysed, and total RNA was extracted from cells using RNA-easy Isolation Reagent (Vazyme, R701) following the manufacturer’s instructions. Typically 1 μg RNA was reverse transcribed to cDNA with HiScript II Q RT Super Mix (Vazyme, R223). Target gene expression was normalized to the expression of the housekeeping gene GAPDH and analyzed with the SYBR-Green qPCR Master mix. Relative gene expression was calculated using the standard 2-ΔΔCT method. The primers used for qPCR were listed in Table S1.
Stable cell line construction
All shRNA experiments were carried out by stable lentiviral transduction to establish stable cells. shRNAs targeting sequences were cloned into the GV248 vectors (Shanghai GeneChem Co., Ltd.) using restriction enzyme cloning and ligation. Lentiviruses were generated by transfecting HEK293T cells in 6-well plates transfected with 0.5 μg pMD2.G, 1.5 μg psPAX2 and the lentiviral backbone vector containing the constructs of the target using Lipofectamine 2000 (Invitrogen). The lentivirus producted by HEK293T cells was collected per 12 hours for 3 days, filtered with a 0.22 μm filter (Millipore), and added with 30% PEG8000 overnight at 4°C. The lentivirus concentrate was centrifuged at 10000 rpm for 30 min at 4°C. The collected virus was added to the tumor cells, iPSC, monocytes or macrophages. For tumor cells, 5x105 cells were transduced with lentivirus -NEUROD1, c-Myc, ST3GAL5, ST6GALNAC3, ST6GALNAC5, CXCL12, and CSF1. The cells were treated with puromycin (1μg/mL) when more than half of the cells expressed fluorescent proteins to select successfully transduced cells. For iPSCs, the concentrated virus was added to iPSCs and cells were treated with puromycin (0.25 μg/ml) 48 hours post- infection for 3 days to select successfully transduced clones. We then conducted macrophage differentiation to produce monocytes and macrophages. The suspended THP-1 cells (1∼2x106) were transduced with lentivirus, treating with polybrene (0.5 μg/ml) by centrifugation at 800×g for 30 min, and subsequently transduced THP-1 cells were cultured. Then, the stably transduced cells were analyzed and selected with flow cytometry using fluorescent proteins.
Transwell assay
A co-culture system was established to observe the monocyte chemotaxis using 12-well cell culture inserts (LABSELECT, 14221) with a 3μm polycarbonate membrane. A total of 3x105 tumor cells were plated in the lower compartment with RPMI-1640 containing 10% FBS. Then, 1x105 td-tomato+monocytes were plated in the upper compartment and incubated at 37°C for 24 h. After incubation, we recorded the number of monocytes expressing red fluorescence in the mixed cells of the lower chamber.
Flow cytometry - Sorting and analysis
Monocytes or macrophages co-cultured with tumor cells were sorted by FACS (BD FASC Canto™), using CD14-FITC (Biolegend, 325603), CD11b-PE antibodies (Biolegend, 982606) or pre-labeled fluorescent dyes. At the end of each isolation a sorting purity check was performed. The sorted cells were cultured with the corresponding medium.
For the cytofluorimetric analysis assay, the sorted monocytes or macrophages were harvested and re-suspended in PBS with 1% BSA. Fc receptors blocker was used to reduce the non-specific immunofluorescent staining by incubating samples for 20min on ice. 1x106 cells were stained in a final volume of 100 μL using the following antibodies at 1:100 dilutions: CD11b-FITC (Biolegend, 301330), CD206-APC (Biolegend, 321109), SIGLEC7-APC (Biolegend,339205), SIGLEC9-PE (R&D, FAB1139P-025), CD14-APC (Biolegend, 301808), CD16-FITC (Biolegend, 302005).
For the in vitro phagocytosis assay by flow cytometry, we constructed a co-culture system in which tumor cells were co-cultured with macrophages in a 1:3-5 ratio. After 24 hours, the cells were harvested and analyzed by flow cytometry.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed with SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, #9005) according to the manufacturer’s instructions. Briefly, tumor cells (2x107) were cross-linked with 1% formaldehyde. After cell lysis, the isolated nuclei were sonicated to obtain chromatin fragments. Chromatin samples were precipitated with Anti-c-Myc antibody (1:100, #18583, CST) or normal Rabbit IgG control. Chromatin-antibody complexes were captured using magnetic protein A/G beads. Purified DNAs were subjected to qPCR. The primers used for ChIP-PCR were listed in Table S1.
Co-immunoprecipitation
For immunoblot analysis, cells were lysed with RIPA buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% (v/v) NP-40, 50 mmol/L EDTA, 0.1% SDS] supplemented with 1 mM PMSF. Protein concentrations were determined using an enhanced BCA Protein Assay Kit with BSA as a standard. For co-immunoprecipitation, lysates were then incubated with Anti-c-Myc antibodies for overnight at 4°C. Protein A/G–agarose was added and incubated for another 1 h at 4°C. After incubation, beads were washed three times with IP buffer and analyzed by Western blot assay. Western blots were performed according to standard protocols.
Immunohistochemistry
Briefly, tissues from the tumor areas were fixed with 4% paraformaldehyde at RT; dehydrated in 70%, 80%, 90%, and 100% ethanol (v/v); cleaned in xylene; and embedded in paraffin. Tissue sections (5 μm) following the antigen restoration were incubated with 3% hydrogen peroxide (H2O2) solution at RT for 10 minutes, immunostaining with CD11b (1:100, Abcam, ab133357), CD206 (1:500, Cell Signaling technology, 24595S) or CD86 (1:50, Cell Signaling technology, 91882S) at 4°C overnight. Development of section was performed with 3,3'-Diaminobenzidine (DAB, Abcam) and using haematoxylin as counterstain. Data were analyzed using the image acquisition and analysis system.
Public single-cell RNA-seq analysis
A available scRNA-seq dataset of human SCLC previously published by Rudin et al. was downloaded on the Human Tumor Atlas Network (HTAN, SYN23591073) data portal at (https://data.humantumoratlas.org) as pre-processed raw data. The obtained single-cell gene expression matrix was analyzed by Seurat v4.2.0 (https://satijalab.org/seurat/). The gene expression was normalized using LogNormalize method of the “NormalizeData” function of the Seurat software (scale.factor = 10,000). Highly variable genes were determined with the FindVariableFeatures function (selection.method=“vst,” nfeatures=2000). RunPCA was applied to identify significant principal components (PCs), and the P-value distribution was visualized using the JackStraw and ScoreJackStraw functions. We used top 30 PCs for downstream analysis. Clustering analysis was performed with FindClusters function (resolution=1.5) from the R package Seruat. The clusters were projected into a two-dimensional plot for visualization using the “RunTSNE” and “RunUMAP” functions. DEGs of each cluster were screened using the “FindAllMarkers” function with logfc.threshold = 0.5 in Seurat. The cell types were identified based on the expression of classic markers. The gene expression was visualized with dot plot, violin plot, feature plot, and heatmap.
In vivo experiment
In brief, 4×105 of luciferase-expressing tumor cells were resuspended in 0.1 mL of PBS, and subcutaneously injected into M-NSG mice via IP to establish the tumor model. Then, mice were treated 12 hours later with IP injection of PBS or iPSCs-derived monocytes. Tumor burden was measured by bioluminescence (total flux) using a IVIS Lumina series III small animal imaging system.
Quantification and statistical analysis
Statistical analysis of the differences between two groups was calculated by two-tailed Student’s t test, and three or more groups comparisons were analysed using one-way or two-way ANOVA when data follow a normal distribution. A Kruskal–Wallis test was used to determine statistically significant differences in non-normally distributed data. All experiments were done at least three times (independent biological replicates). All the data are presented as the mean ± standard error of the mean (SEM). The significance thresholds were defined as statistically significant (∗ p < 0.05), highly significant (∗∗ p < 0.01), or extremely significant (∗∗∗ p < 0.001). ns, not significant. Statistical analysis was performed using GraphPad Prism 8.2.1.
Acknowledgments
This work is financially supported by the Jilin Natural Science Foundation projects of China (Grant No. YDZJ202201ZYTS146, YDZJ202202CXJD009, 20210303002SF, 20210204031YY), Health Commission of Jilin Province (Grant No. 2021JC094), Guangdong Association of Clinical Trials (GACT) / Chinese Thoracic Oncology Group (CTONG) (Grant No. CTONG-YC20220112) and Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (Grant No. 2017B030314120), and Merit-based funding for postdoctoral researchers of Jilin Province (Grant No. 2023-337, TL). We thank Dr. Hua Yu (westlake genomics and bioinformatics lab, Westlake university) for the suggestions of bioinformatics analysis.
Author contributions
L.T. and Y.C. designed and supervised the study. L.T., H.L., P.Z., Y.L. (Yan Liu), Y.L. (Yuanhua Lu), R.Z., and Y.J. performed the experiments and analyzed the data. L.T. and T.T. performed the animal experiments and provided key reagents.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107771.
Supplemental information
Document S1. Figures S1–S5
All primer sequences for differentially expressed genes in SCLC cells and monocytes/macrophages used for qRT-PCR and primers explicitly designed to target the ST3GAL5 or ST6GALNAC5 promoter region in SCLC cells for ChIP-pcr.
Data and code availability
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. Transcriptomic data was downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) with the following accession numbers: GSE36139, GSE36133 and GSE36138. The single cell RNA-Seq data was downloaded from publicly available data on the Human Tumor Atlas Network (HTAN) data portal at https://data.humantumoratlas.org/.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Byers L.A., Rudin C.M. Small cell lung cancer: where do we go from here? Cancer. 2015;121:664–672. doi: 10.1002/cncr.29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horn L., Mansfield A.S., Szczęsna A., Havel L., Krzakowski M., Hochmair M.J., Huemer F., Losonczy G., Johnson M.L., Nishio M., et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018;379:2220–2229. doi: 10.1056/NEJMoa1809064. [DOI] [PubMed] [Google Scholar]
- 4.Rudin C.M., Brambilla E., Faivre-Finn C., Sage J. Small-cell lung cancer. Nat. Rev. Dis. Prim. 2021;7:3. doi: 10.1038/s41572-020-00235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudin C.M., Poirier J.T., Byers L.A., Dive C., Dowlati A., George J., Heymach J.V., Johnson J.E., Lehman J.M., MacPherson D., et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer. 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gay C.M., Stewart C.A., Park E.M., Diao L., Groves S.M., Heeke S., Nabet B.Y., Fujimoto J., Solis L.M., Lu W., et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M., Chen R., Jin Y., Li J., Hu X., Zhang J., Fujimoto J., Hubert S.M., Gay C.M., Zhu B., et al. Cold and heterogeneous T cell repertoire is associated with copy number aberrations and loss of immune genes in small-cell lung cancer. Nat. Commun. 2021;12:6655. doi: 10.1038/s41467-021-26821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.M., Quintanal-Villalonga Á., Gao V.R., Xie Y., Allaj V., Chaudhary O., Masilionis I., Egger J., Chow A., Walle T., et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021;39:1479–1496.e18. doi: 10.1016/j.ccell.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassetta L., Fragkogianni S., Sims A.H., Swierczak A., Forrester L.M., Zhang H., Soong D.Y.H., Cotechini T., Anur P., Lin E.Y., et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell. 2019;35:588–602.e10. doi: 10.1016/j.ccell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson N.R., Minutolo N.G., Gill S., Klichinsky M. Macrophage-Based Approaches for Cancer Immunotherapy. Cancer Res. 2021;81:1201–1208. doi: 10.1158/0008-5472.CAN-20-2990. [DOI] [PubMed] [Google Scholar]
- 11.Tian L., Lei A., Tan T., Zhu M., Zhang L., Mou H., Zhang J. Macrophage-Based Combination Therapies as a New Strategy for Cancer Immunotherapy. Kidney Dis. 2022;8:26–43. doi: 10.1159/000518664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Tian L., Dai X., Yu H., Wang J., Lei A., Zhu M., Xu J., Zhao W., Zhu Y., et al. Pluripotent stem cell-derived CAR-macrophage cells with antigen-dependent anti-cancer cell functions. J. Hematol. Oncol. 2020;13:153. doi: 10.1186/s13045-020-00983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeNardo D.G., Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019;19:369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapellos T.S., Bonaguro L., Gemünd I., Reusch N., Saglam A., Hinkley E.R., Schultze J.L. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front. Immunol. 2019;10:2035. doi: 10.3389/fimmu.2019.02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ożańska A., Szymczak D., Rybka J. Pattern of human monocyte subpopulations in health and disease. Scand. J. Immunol. 2020;92:e12883. doi: 10.1111/sji.12883. [DOI] [PubMed] [Google Scholar]
- 16.Auffray C., Fogg D., Garfa M., Elain G., Join-Lambert O., Kayal S., Sarnacki S., Cumano A., Lauvau G., Geissmann F. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 17.Casanova-Acebes M., Dalla E., Leader A.M., LeBerichel J., Nikolic J., Morales B.M., Brown M., Chang C., Troncoso L., Chen S.T., et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature. 2021;595:578–584. doi: 10.1038/s41586-021-03651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez E., Boelaars K., Brown K., Eveline Li R.J., Kruijssen L., Bruijns S.C.M., van Ee T., Schetters S.T.T., Crommentuijn M.H.W., van der Horst J.C., et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 2021;12:1270. doi: 10.1038/s41467-021-21550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Läubli H., Borsig L. Altered Cell Adhesion and Glycosylation Promote Cancer Immune Suppression and Metastasis. Front. Immunol. 2019;10:2120. doi: 10.3389/fimmu.2019.02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mereiter S., Balmaña M., Campos D., Gomes J., Reis C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell. 2019;36:6–16. doi: 10.1016/j.ccell.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Dobie C., Skropeta D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer. 2021;124:76–90. doi: 10.1038/s41416-020-01126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi M., Altea-Manzano P., Demicco M., Doglioni G., Bornes L., Fukano M., Vandekeere A., Cuadros A.M., Fernández-García J., Riera-Domingo C., et al. PHGDH heterogeneity potentiates cancer cell dissemination and metastasis. Nature. 2022;605:747–753. doi: 10.1038/s41586-022-04758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakshmanan I., Chaudhary S., Vengoji R., Seshacharyulu P., Rachagani S., Carmicheal J., Jahan R., Atri P., Chirravuri-Venkata R., Gupta R., et al. ST6GalNAc-I promotes lung cancer metastasis by altering MUC5AC sialylation. Mol. Oncol. 2021;15:1866–1881. doi: 10.1002/1878-0261.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner M.E. The enzymes of sialic acid biosynthesis. Bioorg. Chem. 2005;33:216–228. doi: 10.1016/j.bioorg.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Tlemsani C., Pongor L., Elloumi F., Girard L., Huffman K.E., Roper N., Varma S., Luna A., Rajapakse V.N., Sebastian R., et al. SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep. 2020;33:108296. doi: 10.1016/j.celrep.2020.108296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao S., Xia W., Yamaguchi H., Wei Y., Chen M.K., Hsu J.M., Hsu J.L., Yu W.H., Du Y., Lee H.H., et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin. Cancer Res. 2017;23:3711–3720. doi: 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W.L., Chiang F.T., Kao J.T.W., Chiou S.H., Lin H.L. GSK3 modulation in acute lung injury, myocarditis and polycystic kidney disease-related aneurysm. Biochim. Biophys. Acta. Mol. Cell Res. 2020;1867:118798. doi: 10.1016/j.bbamcr.2020.118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajishengallis G., Lambris J.D. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao P., Sun X., Li H., Liu Y., Cui Y., Tian L., Cheng Y. c-Myc Targets HDAC3 to Suppress NKG2DL Expression and Innate Immune Response in N-Type SCLC through Histone Deacetylation. Cancers. 2022;14:457. doi: 10.3390/cancers14030457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borromeo M.D., Savage T.K., Kollipara R.K., He M., Augustyn A., Osborne J.K., Girard L., Minna J.D., Gazdar A.F., Cobb M.H., Johnson J.E. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remon J., Aldea M., Besse B., Planchard D., Reck M., Giaccone G., Soria J.C. Small cell lung cancer: a slightly less orphan disease after immunotherapy. Ann. Oncol. 2021;32:698–709. doi: 10.1016/j.annonc.2021.02.025. [DOI] [PubMed] [Google Scholar]
- 34.Carvajal-Hausdorf D., Altan M., Velcheti V., Gettinger S.N., Herbst R.S., Rimm D.L., Schalper K.A. Expression and clinical significance of PD-L1, B7-H3, B7-H4 and TILs in human small cell lung Cancer (SCLC) J. Immunother. Cancer. 2019;7:65. doi: 10.1186/s40425-019-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonanno L., Pavan A., Dieci M.V., Di Liso E., Schiavon M., Comacchio G., Attili I., Pasello G., Calabrese F., Rea F., et al. The role of immune microenvironment in small-cell lung cancer: Distribution of PD-L1 expression and prognostic role of FOXP3-positive tumour infiltrating lymphocytes. Eur. J. Cancer. 2018;101:191–200. doi: 10.1016/j.ejca.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Li Z., Dong B., Sun W., Yang X., Liu R., Zhou L., Huang X., Jia L., Lin D. Prognostic significance of PD-L1 expression and CD8+ T cell infiltration in pulmonary neuroendocrine tumors. Diagn. Pathol. 2018;13:30. doi: 10.1186/s13000-018-0712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dora D., Rivard C., Yu H., Bunn P., Suda K., Ren S., Lueke Pickard S., Laszlo V., Harko T., Megyesfalvi Z., et al. Neuroendocrine subtypes of small cell lung cancer differ in terms of immune microenvironment and checkpoint molecule distribution. Mol. Oncol. 2020;14:1947–1965. doi: 10.1002/1878-0261.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Franco F., Tsui Y.C., Xie X., Trefny M.P., Zappasodi R., Mohmood S.R., Fernández-García J., Tsai C.H., Schulze I., et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020;21:298–308. doi: 10.1038/s41590-019-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y., Chen W., Torphy R.J., Yao S., Zhu G., Lin R., Lugano R., Miller E.N., Fujiwara Y., Bian L., et al. Blockade of the CD93 pathway normalizes tumor vasculature to facilitate drug delivery and immunotherapy. Sci. Transl. Med. 2021;13:eabc8922. doi: 10.1126/scitranslmed.abc8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C., Li Y., Jia L., Kim J.K., Li J., Deng P., Zhang W., Krebsbach P.H., Wang C.Y. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell. 2021;28:1597–1613.e7. doi: 10.1016/j.stem.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F., Ding J. Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein Cell. 2019;10:550–565. doi: 10.1007/s13238-018-0597-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel A.S., Yoo S., Kong R., Sato T., Sinha A., Karam S., Bao L., Fridrikh M., Emoto K., Nudelman G., et al. Prototypical oncogene family Myc defines unappreciated distinct lineage states of small cell lung cancer. Sci. Adv. 2021;7:eabc2578. doi: 10.1126/sciadv.abc2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhanasekaran R., Deutzmann A., Mahauad-Fernandez W.D., Hansen A.S., Gouw A.M., Felsher D.W. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022;19:23–36. doi: 10.1038/s41571-021-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ireland A.S., Micinski A.M., Kastner D.W., Guo B., Wait S.J., Spainhower K.B., Conley C.C., Chen O.S., Guthrie M.R., Soltero D., et al. MYC Drives Temporal Evolution of Small Cell Lung Cancer Subtypes by Reprogramming Neuroendocrine Fate. Cancer Cell. 2020;38:60–78.e12. doi: 10.1016/j.ccell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S5
All primer sequences for differentially expressed genes in SCLC cells and monocytes/macrophages used for qRT-PCR and primers explicitly designed to target the ST3GAL5 or ST6GALNAC5 promoter region in SCLC cells for ChIP-pcr.
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. Transcriptomic data was downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) with the following accession numbers: GSE36139, GSE36133 and GSE36138. The single cell RNA-Seq data was downloaded from publicly available data on the Human Tumor Atlas Network (HTAN) data portal at https://data.humantumoratlas.org/.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.