Abstract
Salmonellae often have the ability to express two different flagellar antigen specificities (phase 1 and phase 2). At the cell level, only one flagellar phase is expressed at a time. Two genes, fliC, encoding phase-1 flagellin, and fljB, encoding phase-2 flagellin, are alternatively expressed. Flagellin genes from 264 serovars of Salmonella enterica were amplified by two phase-specific PCR systems. Amplification products were subjected to restriction fragment length polymorphism (RFLP) analysis by using endonucleases HhaI and HphI. RFLP with HhaI and HphI yielded 64 and 42 different restriction profiles, respectively, among 329 flagellin genes coding for 26 antigens. The phase-1 gene showed 46 patterns with HhaI and 30 patterns with HphI. The phase-2 gene showed 23 patterns with HhaI and 17 patterns with HphI. When the data from both enzymes were combined, 116 patterns were obtained: 74 for fliC, 47 for fljB, and 5 shared by both genes. Of these combined patterns, 80% were specifically associated with one flagellar antigen and 20% were associated with more than one antigen. Each flagellar antigen was divided into 2 to 18 different combined patterns. In the sample of strains used, determination of the phase-1 and phase-2 flagellin gene RFLP, added to the knowledge of the O antigen, allowed identification of all diphasic serovars. Overall, the diversity uncovered by flagellin gene RFLP did not precisely match that evidenced by flagellar agglutination.
In developed countries, salmonellosis is a major economic problem for the food industry, as well as a public health hazard for the consumer. In developing countries, the death toll from salmonellosis (typhoid and diarrhea in children) is very high. Individualization of strains of the pathogen is essential to the study of the association between clinical cases and possible sources of infection.
The genus Salmonella is composed of two species, “Salmonella enterica” (quotes indicate pending nomenclatural status) and S. bongori (11, 17). The primary basis for the typing of “S. enterica” is a serotyping scheme (the White-Kauffmann-Le Minor [WKL] scheme) in which 2,375 serovars have been recognized on the basis of the antigenic properties of the cell wall lipopolysaccharide (O antigen), the phase-1 flagellar protein (H1), and the phase-2 flagellar protein (H2) (15, 16).
The flagellar protein or flagellin constitutes the subunit of the helical filament that forms the flagellar organelle. Salmonella flagellin consists of extremely conserved terminal regions and a variable central region (7, 23). This central region of the molecule carries the antigenic specificity (14). For the phase-1 flagellin, 63 antigens have been distinguished. For the phase-2 flagellin, 37 antigens have been described. Some of these antigens are defined by a single factor (antigen i, d, or r); others are defined by several subfactors (e.g., antigens l,v; l,w; g,m; and e,n,x) (15).
The antigenic specificities of phase-1 and phase-2 flagellins are encoded by flagellin genes fliC and fljB, respectively. These flagellar genes are found at two different locations on the chromosome. At one location is the gene fliC. At another location is an operon containing the genes hin, encoding the Hin recombinase; fljB, encoding phase-2 flagellin; and fljA, encoding a repressor for fliC. The Hin recombinase catalyzes the reversible inversion of a 993-bp segment of the chromosome containing a promoter. In one orientation, the promoter directs transcription of the fljB and fljA genes. Phase-2 flagellin and the repressor are produced (thus repressing fliC). In the other orientation, repression of the fliC gene is relieved and phase-1 flagellin is expressed (6, 25).
Salmonella isolates expressing two antigenically distinct types of flagellin are biphasic. Monophasic Salmonella strains expressing only one type of flagellar antigen include many clinically and epidemiologically important salmonellae, e.g., serovar Typhi, the agent of typhoid fever, and serovar Enteritidis, a major foodborne pathogen associated with poultry and eggs (19). One serovar, Gallinarum, is always nonflagellated (12). Occasionally, nonmotile (nonflagellated) variants of normally motile serovars are isolated from specimens. These isolates cannot be identified with a known serovar by serotyping.
Molecular techniques such as restriction fragment length polymorphism (RFLP) could reflect the flagellar antigenic diversity of salmonellae at the genetic level (9). The purposes of this study were to determine whether (i) 26 flagellar antigens (carried by 237 serovars) could be differentiated by flagellin gene RFLP, (ii) genes coding for phase-1 and phase-2 antigens with the same designation could have identical RFLP patterns, (iii) flagellar antigens could be subdivided into RFLP patterns, and (iv) serovars could be identified by using flagellin gene RFLP. The results obtained showed these purposes to have been partially achieved.
MATERIALS AND METHODS
Collection of strains.
The 237 reference strains from different serovars of S. enterica subsp. enterica (subsp. I) and salamae (subsp. II) used in this study were from the World Health Organization Collaborating Center for Reference and Research on Salmonella (from M. Y. Popoff, Institut Pasteur, Paris, France). We also studied 27 strains received at the Centre National de Reference des Salmonella et Shigella or the Centre National de Reference pour le Typage Moleculaire Enterique (both centers are located in the Unité des Entérobactéries, Institut Pasteur). These included 7 isolates of serovar Typhi representing different ribotypes; 11 isolates of serovar Typhimurium, corresponding to six phage types (12 atypical, 29, 113, 114 atypical, 120, and 153); 1 isolate of serovar Bovismorbificans; and 7 nonmotile isolates. Biochemical confirmation and serotyping were done by conventional methods.
Serovars were selected to include (i) most frequently encountered phase-1 flagellar antigens i, r, and the g series (f, g, m, p, q, s, t, u, and z51); (ii) most frequently encountered phase-2 flagellar antigens 1,2 and 1,5; (iii) flagellar antigen d, which is associated with medically important serovars; and (iv) flagellar antigens l,v and l,w, which are found in both phase 1 and phase 2. Since most strains were diphasic, a number of flagellar antigens other than the selected ones were de facto included in the study. Strains of S. enterica subsp. II were included in the study because they share the selected antigens with S. enterica subsp. I. Flagellar antigens are listed in Table 1.
TABLE 1.
Flagellar antigens represented in this study
| Flagellar antigen(s)a | No. of serovars | No. of strains
|
|
|---|---|---|---|
| Subsp. I | Subsp. II | ||
| Phase 1 | |||
| a | 1 | 1 | |
| b | 2 | 1 | 1 |
| c | 1 | 1 | |
| d | 22 | 24 | 1 |
| j | 1 | 5 | |
| e,h | 3 | 3 | |
| e,n,x | 1 | 1 | |
| g series | 49 | 36 | 13 |
| i | 25 | 36 | |
| r | 8 | 8 | |
| r,i | 8 | 9 | |
| l,v | 51 | 51 | |
| l,w | 17 | 17 | |
| z | 1 | 1 | |
| z6 | 1 | 1 | |
| z10 | 1 | 1 | |
| z35 | 1 | 1 | |
| z39 | 1 | 1 | |
| z42 | 1 | 1 | |
| Phase 2 | |||
| l,w | 70 | 70 | |
| 1,2 | 8 | 8 | |
| 1,5 | 17 | 17 | |
| 1,6 | 6 | 6 | |
| 1,7 | 7 | 7 | |
| 1,2,7 | 1 | 1 | |
| e,n,x | 6 | 6 | |
| e,n,x,z15 | 1 | 1 | |
| e,n,z15 | 10 | 10 | |
| z6 | 6 | 6 | |
| z35 | 2 | 2 | |
| Total: 26 | 237 | 245 | 19 |
Selected antigens are in boldface.
Preparation of DNA.
Isolates were grown in Trypto casein soy agar (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France). A single colony was grown in a shaking incubator for 18 h at 37°C in Trypto casein soy broth (Sanofi Diagnostics Pasteur). The culture was centrifuged at 10,000 rpm for 10 min. The pellet was suspended in 580 μl of lysis buffer (Tris-HCl at 0.05 M, EDTA at 0.05 M, NaCl at 0.1 M, pH 8) with 3 μl of a 20-mg/ml aqueous solution of pronase (Calbiochem, La Jolla, Calif.) and 32 μl of a 10% (wt/vol) sodium dodecyl sulfate solution and incubated for 1 h at 60°C to allow cell lysis. DNA was extracted with an AutoGen 540 automated DNA extraction system (AutoGen Instruments, Beverly, Mass.).
PCR amplification of fliC (phase 1).
For amplification of the phase-1 flagellin gene, the primers used were CAAGTCATTAATACMAACAGCC (FSa1; M = A or C) and TTAACGCAGTAAAGAGAGGAC (rFSa1). Primer FSa1 was selected on the basis of fliC gene conservation among sequences of Salmonella, Escherichia coli, and Shigella flagellin genes (GenBank and EMBL accession no. M23773, M84972, M84973, M23772, M23774, X04505, M11332, M84976, M84978, M84979, Z15064, Z15065, Z15066, Z15069, Z15086, Z15070, Z15071, Z15072, L21912, L07387, D18821, and D16819). Primer rFSa1 was selected to amplify only the Salmonella fliC gene. The target of primer FSa1 was located at positions 18 to 40, and the target of primer rFSa1 was located at positions 1530 to 1510 of the serovar Muenchen fliC gene (accession no. M23774). The expected size of the amplified fragment was about 1.5 kbp, except for the H1-j flagellin gene of variant serovar Typhi, which contained a deletion of 261 bp (5).
DNA amplification by PCR was performed in a reaction volume of 100 μl consisting of 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 1.5 mM MgCl2, 0.01% (wt/vol) gelatin; 2.5 U of Hi-Taq DNA polymerase (Bioprobe, Montreuil-sous-Bois, France); 200 μM each dATP, dTTP, dCTP, and dGTP; 50 pmol of each primer; and 1 μl of sample DNA. The reaction mixture was overlaid with 50 μl of mineral oil. Initial denaturation was carried out for 5 min at 94°C. Thirty-five cycles of amplification were performed in a PTC-100 thermal cycler (MJ Research, Watertown, Mass.). Each cycle consisted of three steps: denaturation for 1 min at 94°C, annealing for 1 min at 55°C, and extension for 1 min at 72°C. An additional step of extension for 5 min at 72°C was performed at the end of the amplification to complete extension of the primers. Amplification products were detected by electrophoresis in 0.8% (wt/vol) agarose gels in Tris-acetate buffer (0.04 M Tris-acetate, 0.002 M EDTA, pH 8.1), with the 1-kbp DNA Ladder (Gibco BRL, Gaithersburg, Md.) as a molecular size marker.
PCR amplification of fljB (phase 2).
The primers designed for amplification of the phase-2 flagellin gene were CAAGTAATCAACACTAACAGTC (FSa2) and TTAACGTAACAGAGACAGCAC (rFSa2). The target of primer FSa2 was located at positions 7 to 28, and the target of primer rFSa2 was located at positions 1506 to 1486 of the serovar Abortusequi fljB gene (accession no. D13690). This PCR will be referred to as fljB amplification.
Because the GenBank and EMBL international databases contained only one fljB gene sequence, primer selectivity was assessed by the following procedure. A first amplification of DNA from serovars Abortusequi, Bloomsbury, Rubislaw, Typhimurium, Goldcoast, Anatum, Brandenburg, and Verona was done with primers ST-HIN-L and SA-FLJA-R (2). These primers selectively amplified a part of the Salmonella flagellar operon consisting of the hin, fljB, and fljA genes. This PCR will be referred to as hin-fljB-fljA amplification. The reaction required extracted DNA from bacteria expressing the phase-2 flagellar antigen. Amplification involved predenaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 47°C for 1 min, and 72°C for 1 min, with a final step of elongation at 72°C for 5 min. The 2,976-bp expected fragment was extracted from a 0.8% (wt/vol) agarose gel by using the JETsorb kit (Bioprobe) prior to fljB amplification (with primers FSa2 and rFSa2) to eliminate genomic DNA from the PCR mixture. The specific PCR with primers FSa2 and rFSa2 was performed by 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min. For elongation of the PCR product, a final step of 72°C for 5 min was included. The amplified products were detected by electrophoresis as described above.
fljB amplification was also done directly on the same strain DNAs with the same PCR parameters. The identities of amplified fragments were determined by RFLP as described below. After this primer specificity control, fljB amplification was done directly on Salmonella genomic DNA.
RFLP analysis.
Endonucleases HhaI and HphI were chosen after study of restriction maps of eight sequences of fliC and fljB from the EMBL and GenBank databases (accession no. M23774, M84973, D13690, Z15068, M23772, X04505, M11332, and L21912). Maps were obtained with the Mapdraw program of the Lasergene software (DNAstar, Madison, Wis.). The enzyme HhaI recognized and cleaved the sequence GCGC, while HphI recognized the sequence TCACC or GGTGA and cleaved the sequence 8 or 9 bp further (10, 18). HhaI restriction sites were regularly distributed on the flagellin sequences, while HphI preferentially cleaved the genes in the hypervariable region.
In a microtube, 10-μl portions of PCR mixtures containing amplified flagellin genes were digested. Digestion was done for 2 h at 37°C for both restriction enzymes. RFLPs were determined by electrophoresis of the digested DNA in 1% (wt/vol) agarose (Bioprobe) plus 1% (wt/vol) Nusieve agarose (FMC Bioproducts, Rockland, Maine) gels for 5 h at 4.8 V/cm. The 1-kbp DNA Ladder (Gibco BRL) was used as a molecular size marker. The restricted fragments were stained with ethidium bromide.
The RFLP patterns were scanned by using One-Scanner (Apple Computers, Cupertino, Calif.). Digitization and interpretation of RFLP profiles were done with the Taxotron package (Taxolab software; Institut Pasteur), including the programs RestrictoScan, RestrictoTyper, Adanson, and Dendrograf. Lanes and bands of the resulting TIFF images were detected with RestrictoScan. Fragment lengths were interpolated by using the Schaffer and Sederoff algorithm (20) implemented by RestrictoTyper. The program generated a normalized graph showing migration patterns. Fragments were considered identical if their sizes did not differ by more than 1% (percent tolerated error). The distance coefficient was calculated as the number of nonmatching fragments divided by the total number of bands in both patterns (complement of the Dice index [3]), and a distance matrix was built. The relationships between patterns were calculated by the average linkage (1), single linkage, and unweighted pair group using mathematical averages (UPGMA) (21) methods with the Adanson clustering program. Dendrograms were drawn by Dendrograf.
RESULTS
PCR amplification of fliC (phase 1).
A 1.5-kbp fragment was amplified from 259 strains and a 1.24-kbp fragment was amplified from five strains of variant serovar Typhi (H1:j), as expected. No other variation in fliC gene size was detectable on electrophoresis gels. Amplification occurred with strains from nonmotile serovar Gallinarum, as well as other nonmotile isolates.
PCR amplification of fljB (phase 2).
hin-fljB-fljA amplification generated 3-kbp amplification products from the eight serovars tested (Abortusequi, Bloomsbury, Rubislaw, Typhimurium, Goldcoast, Anatum, Brandenburg, and Verona). fljB amplification of total DNA and of hin-fljB-fljA amplification products yielded 1.5-kbp products. The fljB specificity of fljB PCR was demonstrated by HhaI and HphI restriction of fljB amplification products obtained from total DNA or from hin-fljB-fljA amplification products. For each strain tested, restriction profiles were identical in both cases (data not shown).
Thereafter, the specific fljB PCR was applied directly to Salmonella genomic DNA. For all of the diphasic serovars tested, the amplification product was invariably 1.5 kbp when amplification occurred. For the diphasic serovar Typhi strains tested (d:z66 and j:z66), fljB amplification failed to amplify a fragment. No fljB amplification was shown with the monophasic serovars Typhi, Paratyphi A, Enteritidis, Derby, Rissen, Agona, Borreze, Havana, Berta, Antarctica, Ona, Kingston, California, Congo, Giessen, Emek, Budapest, Dublin, Sylvania, Naestved, Essen, Gallinarum, Montevideo, Blegdam, Othmarshen, Rostock, Moscow, Senftenberg, Banana, Oranienburg, Hillingdon, Gateshead, Sangalkam, Ackwepe, and Keve.
RFLP analysis of flagellin genes.
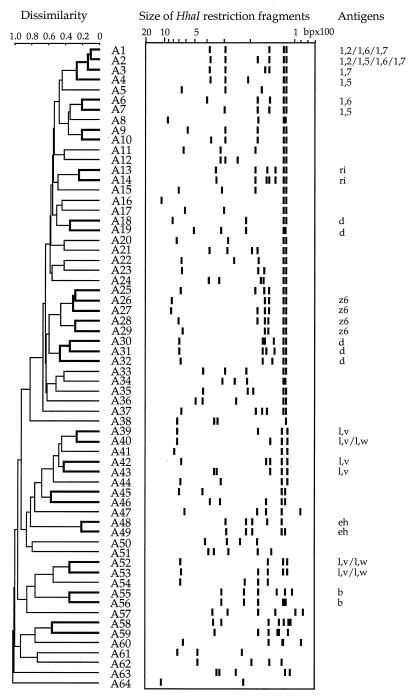
Restriction enzymes HhaI and HphI were used on PCR products from the fliC and fljB genes of each of the 264 Salmonella strains studied and yielded profiles consisting of two to seven fragments sized between 62 and 1,310 bp (Fig. 1 and 2).
FIG. 1.
Dissimilarity among restriction patterns obtained with HhaI. The dendrogram was generated by the UPGMA method. Clusters obtained with three clustering methods are indicated with thicker lines (robust clusters). Each branch of the tree faces each flagellin banding pattern. Antigenic specificities are indicated when typical of a cluster.
FIG. 2.
Dissimilarity among restriction patterns obtained with HphI. The dendrogram was generated by the UPGMA method. Clusters obtained with three clustering methods are indicated with thicker lines (robust clusters). Each branch of the tree faces each flagellin banding pattern. Antigenic specificities are indicated when typical of a cluster.
On repeated experiments, 42 strains tested with HhaI and 45 strains tested with HphI could be unambiguously assigned to the same pattern as that generated by the first experiment.
To ensure that no partial restriction occurred with the enzymes used, summation of fragment lengths for each profile was done. Surprisingly, the sum of the flagellin fragment lengths varied in accordance with the profiles studied and was often smaller than the size of the PCR product used for restriction. In studying profiles obtained from restriction maps of eight published sequences (accession no. M11332, M84973, Z15068, X03395-M23774, X03393-M23772, X04505, L21912, and D13690), we observed that only fragments larger than 60 bp were visible on the agarose gel. Product size, deduced from the sum of restriction fragment lengths, was reduced mostly with HhaI and slightly with HphI (Table 2). In these cases, part of the difference between the size of the PCR product and the sum of the restriction fragment lengths was due to fragments smaller than 60 bp. In comparing fragment lengths (greater than 60 bp) obtained from restriction maps of published sequences and those deduced from migration distances on electrophoresis gels for the same genes, two situations occurred (Table 2). In situation 1, five HhaI profiles and five HphI profiles gave the same number of fragments as the profiles deduced from restriction maps. Differences between the sums of fragment lengths from restriction maps and the experimental gel estimates ranged from 0.8 to 3.1% of the total size (experimental error). In situation 2, four HhaI profiles and three HphI profiles included fragments of equal sizes which appeared as single bands on agarose gels (comigrating fragments). Thus, for such profiles, the difference between sums of fragment lengths from restriction maps and experimental gel-based estimates ranged from 4.3 to 12.9% of the total size (comigration bias).
TABLE 2.
Comparison of fragment sizes from restriction maps and fragment sizes interpolated from migration distances on gels for the same sequences
| Enzyme | Map-, gel-based sizes (bp)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Dublin g,p antigen | Enteritidis g,m antigen | Rubislaw r antigen | Muenchen d antigen | Paratyphi A a antigen | Typhimurium i antigen | Typhi d antigen | Abortusequi e,n,x antigen | |
| HhaI | ||||||||
| 706, 681 | 706, 707 | 676, 672 | 709, 705 | 484, 484 | 667, 683 | 508, 520 | 399, 417 | |
| 431, 425 | 431, 441 | 306, 317 | 309, 315 | 195, 199 | 306, 309 | 309, 320 | 273, 289 | |
| 115, 122 | 115, 125 | 174, 184 | 174, 179 | 154, 151 | 174, 122 | 204, 218 | 174, 182 | |
| 104, 117 | 104, 119 | 115, 122 | 115, 122 | 146, —b | 104, 118 | 115, 122 | 115, 122 | |
| 104, 117 | 104, 118 | 113, 125 | 113, — | 113, — | ||||
| 104, 118 | 104, 117 | 104, 117 | ||||||
| Sums (SD [%]) | 1,356, 1,345 (0.8) | 1,356, 1,392 (2.5) | 1,375, 1,412 (2.6) | 1,411, 1,439 (1.9) | 1,196, 1,063 (12.5) | 1,251, 1,232 (1.5) | 1,353, 1,297 (4.3) | 1,178, 1,127 (4.5) |
| HphI | ||||||||
| 463, 465 | 463, 457 | 831, 806 | 756, 756 | 830, 817 | 706, 698 | 759, 771 | 488, 498 | |
| 365, 373 | 365, 368 | 195, 199 | 186, 194 | 194, 193 | 333, 332 | 333, 337 | 363, 379 | |
| 318, 324 | 318, 323 | 186, 188 | 186, — | 186, — | 306, 316 | 183, 196 | 192, 201 | |
| 224, 239 | 224, 240 | 147, 151 | 147, 154 | 147, 161 | 130, 144 | 130, 138 | 186, — | |
| 93, 110 | 93, 110 | 130, 135 | 130, 139 | 130, 146 | 107, 112 | 147, 156 | ||
| 107, 112 | 130, 138 | |||||||
| Sums (SD [%]) | 1,463, 1,511 (3.1) | 1,463, 1,498 (2.3) | 1,489, 1,479 (0.7) | 1,512, 1,355 (11.6) | 1,487, 1,317 (12.9) | 1,475, 1,490 (1) | 1,512, 1,554 (2.7) | 1,506, 1,372 (9.8) |
The values in boldface are significantly high.
—, band not seen.
Correspondence between patterns and antigens.
Restriction of 329 (195 phase-1 plus 134 phase-2) flagellin genes yielded 64 HhaI profiles and 42 HphI profiles (Table 3). Patterns were designated by a letter indicating the restriction enzyme used (A for HhaI or P for HphI) followed by a number (Fig. 1 and 2). Phase-1 genes showed 46 patterns with HhaI and 30 patterns with HphI. Phase-2 genes showed 23 patterns with HhaI and 17 patterns with HphI. Forty-one HhaI patterns and 25 HphI patterns were only associated with the fliC gene in this study. Eighteen HhaI patterns and 12 HphI patterns were only associated with the fljB gene. Five patterns obtained with HhaI (A15, A39, A40, A52, and A53) and five obtained with HphI (P1, P2, P8, P24, and P42) were associated with both the fliC and fljB genes.
TABLE 3.
Diversity of flagellin gene restriction patterns obtained with HhaI and HphI
| Flagellar antigen(s) | No. of patterns given by:
|
No. of combined patterns given by HhaI + HphI | No. of serovars | |
|---|---|---|---|---|
| HhaI | HphI | |||
| H:1 | ||||
| a | 1 | 1 | 1 | 1 |
| b | 2 | 2 | 2 | 2 |
| c | 1 | 1 | 1 | 1 |
| d | 8 | 6 | 12 | 22 |
| j | 1 | 2 | 2 | 1 |
| e,h | 2 | 1 | 2 | 3 |
| e,n,x | 1 | 1 | 1 | 1 |
| g series | 8 | 12 | 17 | 49 |
| i | 9 | 7 | 13 | 25 |
| r | 4 | 2 | 4 | 8 |
| r,i | 5 | 7 | 8 | 8 |
| l,v | 8 | 4 | 11 | 51 |
| l,w | 2 | 3 | 5 | 17 |
| z | 1 | 1 | 1 | 1 |
| z6 | 1 | 1 | 1 | 1 |
| z10 | 1 | 1 | 1 | 1 |
| z35 | 1 | 1 | 1 | 1 |
| z39 | 1 | 1 | 1 | 1 |
| z42 | 1 | 1 | 1 | 1 |
| Total | 46 | 30 | 74 | 195 |
| H:2 | ||||
| l,w | 4 | 10 | 18 | 70 |
| 1,2 | 2 | 3 | 3 | 8 |
| 1,5 | 8 | 4 | 10 | 17 |
| 1,6 | 3 | 3 | 4 | 6 |
| 1,7 | 4 | 3 | 5 | 7 |
| 1,2,7 | 1 | 1 | 1 | 1 |
| e,n,x | 1 | 3 | 3 | 6 |
| e,n,z15 | 3 | 4 | 5 | 10 |
| e,n,x,z15 | 1 | 1 | 1 | 1 |
| z6 | 5 | 4 | 5 | 6 |
| z35 | 1 | 1 | 1 | 2 |
| Total | 23 | 17 | 47 | 134 |
Fifty HhaI and 20 HphI patterns were each specifically associated with a single antigen. For example, the HhaI patterns found were A42 for the l,v flagellar antigen, A19 and A31 for d, A25 for i, A34 for j, and A54 for phase-2 l,w antigens; the HphI patterns found were P38 and P41 for i, P17 for j, P18 for g,z51, and P4 and P5 for phase-2 l,w antigens. The other patterns were each associated with more than one antigen. Figures 1 and 2 show results of cluster analyses of restriction patterns. Dendrograms produced clusters of similar restriction patterns. It is striking that some clusters corresponded to related flagellar antigens.
In combining the data obtained with both restriction enzymes, 116 combined patterns (74 combined patterns for fliC, 47 combined patterns for fljB, and 5 combined patterns shared by both genes) were identified among the 26 antigens examined (195 phase-1 and 134 phase-2 genes were tested). HhaI divided 21 HphI patterns into 92 combined patterns. HphI divided 16 HhaI patterns into 66 combined patterns. Two to 18 different combined patterns were observed for each of the following flagellar antigens: d; e,n,x; e,n,z15; i; r,i; l,v; l,w; z6; 1,2; 1,5; 1,6; and 1,7 (Tables 3 and 4). The restriction profiles of the fliC and fljB genes revealed a molecular diversity greater than the antigenic diversity.
TABLE 4.
Combined patterns and corresponding serovars
| Pattern | Antigen(s)
|
Serovar(s) | |
|---|---|---|---|
| Phase 1 | Phase 2 | ||
| A1P1 | 1,2 | Saint Paul | |
| 1,7 | Schwarzengrund, Banco | ||
| A1P24 | 1,2 | Typhimurium, Heidelberg | |
| 1,6 | Quentin | ||
| 1,7 | Strasbourg, Dieuppeul | ||
| A2P1 | 1,5 | Choleraesuis | |
| A2P3 | 1,6 | Agama, Togo, Westeinde | |
| 1,7 | Lika | ||
| A2P7 | 1,2 | Newport, Muenchen, Stanley, Gabon, Overchurch | |
| A2P12 | e,n,z15 | Tsevie | |
| A2P20 | 1,6 | Anatum | |
| A2P24 | 1,5 | Fulda | |
| A3P1 | 1,7 | Indiana | |
| A4P24 | 1,5 | Mono | |
| A5P13 | r | Brive | |
| A5P22 | i | Douala, Bandia | |
| A6P20 | 1,6 | Dumfries | |
| A7P24 | 1,5 | Ontario | |
| A8P2 | z35 | Coromandel | |
| A8P13 | r | Massakory | |
| i | Veneziana | ||
| A9P3 | 1,5 | Colorado | |
| A10P12 | e,n,x | Veneziana, Rubislaw | |
| e,n,z15 | Olten, Toulon | ||
| A10P15 | e,n,x | Farsta, Brooklyn, Abortusequi | |
| e,n,z15 | Duisburg, Avonmouth, Surat, Brandenburg | ||
| e,n,x,z15 | Euston | ||
| A10P19 | e,n,x | Hadar | |
| e,n,z15 | Moussoro, Lomnava | ||
| A11P21 | e,n,x | 9,12:e,n,x:−a | |
| A12P11 | 1,5 | Lagos | |
| A13P24 | r,i | Euston | |
| A14P23 | r,i | Fareham | |
| A15P1 | 1,5 | Alamo, Maricopa, Bovismorbificans, Victoria | |
| 1,2,7 | Eingedi | ||
| A15P12 | z6 | Verona | |
| A15P13 | r,i | Bovismorbificans | |
| A15P16 | d | Muenchen, Stanley, Duisburg, Manhattan, Caen | |
| i | Moussoro, Dieuppeul | ||
| r,i | Drogana | ||
| A15P24 | 1,5 | Isangi, Manhattan, Gligji, Zigong, Pasing | |
| A15P40 | d | Schwarzengrund | |
| r,i | Dumfries | ||
| A15P41 | i | Lagos, Tumodie | |
| A16P28 | f,g,t | 1,4,12:f,g,t:z6,z42a | |
| A16P30 | f,g,t | Eingedi | |
| A16P31 | g,s,t | Gateshead | |
| A16P33 | f,g | Derby, Rissen, Havana | |
| f,g,s | Agona | ||
| f,g,t | Berta | ||
| g,s,t | Ona, Kingston | ||
| g,t | Bloomsbury, Budapest, Senftenberg | ||
| A17P23 | i | Potto | |
| A18P1 | d | Strasbourg, Olten, Plymouth | |
| A18P25 | d | Quentin | |
| A18P40 | d | Ontario | |
| A19P40 | d | Typhi | |
| A20P22 | i | Hoboken | |
| A21P1 | 1,7 | Truro | |
| A22P39 | z35 | Pasing | |
| A23P17 | d | 6,7:d:z42a | |
| A24P35 | g,m,s,t | Congo | |
| A25P16 | i | Soerenga, Verona | |
| A26P1 | z6 | Potto | |
| A27P42 | z6 | Kentucky | |
| A28P1 | z6 | Tumodi | |
| A29P24 | z6 | Assine, Plymouth | |
| A30P16 | d | Tilburg | |
| A31P16 | d | Livingstone, Mons, Putten, Cullingworth, Dembe | |
| A31P40 | d | Birmingham | |
| A32P16 | d | Niamey | |
| A33P24 | 1,5 | Bovismorbificans | |
| A34P17 | j | Typhi | |
| A34P22 | j | Typhi | |
| A35P1 | 1,5 | Bloomsbury | |
| A36P27 | m,t | Sangalkam | |
| A36P34 | g,m,t | California | |
| m,t | Banana, Oranienburg | ||
| A37P35 | d | Isangi | |
| A38P41 | i | Agama, Farsta, Tsevie | |
| A39P24 | l,v | Coromandel | |
| z35 | Kotte | ||
| A40P2 | l,v | India, Toronto | |
| A40P24 | l,w | Brikama, Soerenga | |
| A41P2 | l,v | Sinchew | |
| A42P2 | l,v | Nchanga, Horsham, Geraldton | |
| A43P1 | l,v | Burgas | |
| A44P1 | i | Avonmouth | |
| A44P8 | l,v | Sinstorf | |
| A44P13 | i | Lika, Kedougou | |
| r | Bovismorbificans, Heidelberg, Goldcoast, Grampian, Bochum | ||
| f,g,s | Borreze | ||
| A44P22 | i | Kentucky, Gloucester, Norton, Magherafelt, Pisa, Kaneshie | |
| r,i | Zuilen | ||
| A44P23 | r,i | Africana | |
| A44P38 | i | Typhimurium | |
| A45P26 | g,m | Essen, Enteritidis, Gallinarum, Othmarshen | |
| g,p | Dublin, Sylvania | ||
| g,p,u | Rostock | ||
| g,m,p,s | Montevideo | ||
| g,q | Moscow | ||
| g,m,q | Blegdam | ||
| g,m,s | 30:g,m,s:e,n,xa, Giessen, Emek | ||
| g,p,s | Naestved | ||
| A45P28 | g,m,s,t | 1,13,23:g,m,s,t:1,5a | |
| A45P29 | g,z63 | Antarctica | |
| A45P32 | g,m | Hillingdon | |
| g,m,s | Macclesfield | ||
| A45P33 | f,g,m,t | 6,8:f,g,m,t:−a | |
| g,m,s | 40:g,m,s:e,n,xa | ||
| g,m,s,t | 9,12:g,s,t:e,n,xa; 1,9,12:g,m,s,t:e,n,xa | ||
| g,s,t | 28:g,s,t:e,n,xa | ||
| A45P34 | g,m,t | 4,12:g,m,t:z39a | |
| A46P35 | z10 | Hadar | |
| A47P24 | c | Choleraesuis | |
| A48P24 | e,h | Saint Paul | |
| A49P24 | e,h | Newport, Anatum | |
| A50P14 | e,n,z15 | Drogana | |
| A51P1 | r,i | Banco | |
| A52P1 | l,v | Koessen, Fyris, Azteca, Give, Gdansk, Zaiman, Goettingen, Svedvi, Fann | |
| l,w | Victoria, Assine | ||
| l,w | Sindelfingen, Meleagridis, Tilburg, Calabar, Ordonez, Worthington, Minna, Vegesack, Yoruba, Caen, Fareham, Brive, Shannon | ||
| A52P2 | l,v | Kimuenza, Loanda, Amherstiana, Manchester, Holcomb, Edmonton, Mendoza, Kapemba, London, Brandenburg, Bredeney, Ruzizi, Ngor, Parkroyal, Maracaibo, Bullbay, Lovelace, Borbeck, Nanga, Boecker | |
| l,w | Welikade, Los Angeles | ||
| l,w | Gabon, Fulda, Gligji, Zigong, Westeinde, Brooklyn, Lomnava, Africana, Norton, Hillsborough, Magherafelt, Dabou, Bamboye, Anderlecht, Livingstone, Uhlenhorst | ||
| A52P3 | l,w | Clerkenwell | |
| A52P4 | l,w | Gloucester, Krefeld | |
| A52P5 | l,w | Zuilen | |
| A52P6 | l,w | Cullingworth | |
| A52P8 | l,w | Cyprus | |
| A52P9 | l,w | Alexanderpolder | |
| A52P10 | r | Surat | |
| A52P24 | l,v | Irumu, Bonn, Clackamas, Concord | |
| l,w | Mono, Togo, Colorado, Toulon, Overchurch | ||
| l,w | Bruck, Wyldegreen, Goldcoast, Abidjan | ||
| A53P1 | l,w | Ackwepe | |
| l,w | Hallflod, Ohio, Bukuru, Langensalza, Broughton, Niamey, Douala, Fairfield, Moroto, Dembe, Massakory, Niarembe | ||
| A53P2 | l,v | Mkamba, Kortrijk, Shanghai, Salford | |
| l,w | Kewe, Ramsey | ||
| l,w | Mannheim | ||
| A53P3 | l,w | Kedougou | |
| A53P4 | l,w | Wien, Mura, Bochum, Nantes, Birmingham, Hoboken, Alkmaar, Carno, Kaneshie | |
| A53P5 | l,w | Grampian | |
| A53P7 | l,w | Kuru, Epicartes, Newrochelle, Putten, Vridi, Demerara, Bandia | |
| A53P24 | l,v | Potsdam, Pakistan, Litchfield | |
| A54P1 | l,w | Huettwillen, Mons, Preston | |
| A54P7 | l,w | Coleypark | |
| A55P36 | b | Kotte | |
| A56P42 | b | 6,7:b:z39a | |
| A57P24 | z6 | 6,7:z6:1,7a | |
| A58P13 | i | Truro | |
| A58P24 | z39 | 6,7:z39:1,5,7a | |
| A59P24 | z42 | 6,7:z42:1,7a | |
| A60P10 | z | Indiana | |
| A61P33 | g,t | 52:g,t:−a; 3,10:g,t:−a | |
| A62P24 | a | Paratyphi A | |
| A63P18 | g,z51 | Alamo, Maricopa | |
| A64P33 | g,z62 | 1,9,12:g,z62:−a; 4,12:g,z62:−a | |
Serovar of subsp. II.
When 14 antigens from 19 serovars of subsp. II were tested, 13 different combined patterns were obtained (Table 4). Of these combined patterns, 12 were specifically associated with subsp. II. The exception was a serovar of subsp. II (O30; g,m,s; e,n,x) which shared fliC pattern A45P26 with serovars Giessen and Emek of subsp. I (Table 4).
Of the 116 combined patterns, 93 (80%) were each specifically associated with a single antigen and 23 (20%) each corresponded to several antigens (Table 4). For 71 combined patterns associated with one antigen (among the antigens tested), one enzyme provided specificity. In the 22 other combined patterns associated with only one antigen, specificity was given by the combination. The combination increased the number of specific RFLPs (combined patterns) associated specifically with one antigen but did not differentiate all of the antigens. Of the 13, 4, 8, and 12 combined patterns obtained for the i; r; r,i; and d antigens, 5 corresponded to several antigens (Table 4). Pattern A15P40 corresponded to both the d and r,i antigens. Pattern A15P16 corresponded to the d; i; and r,i antigens. Pattern A44P22 corresponded to both the i and r,i antigens. Patterns A8P13 and A44P13 corresponded to both the i and r antigens.
When antigens shared antigenic subfactors, RFLP did not differentiate among them. Phase-2 antigens 1,2; 1,5; 1,6; 1,7; and 1,2,7 yielded four combined patterns (A1P1, A1P24, A2P3, and A15P1) without correspondence between antigenic factors and patterns. Antigens e,n,x and e,n,z15 shared three combined patterns (A10P12, A10P15, and A10P19). The sensitivity of the RFLP method for recognition of the g series of antigens was as high as 97.9%. Only one strain tested (serovar Borreze) shared pattern A44P13 with other antigens (r and i). However, the discrimination power of the RFLP method within the g series was very low. For 19 different antigens, only 17 combined patterns were obtained. Each of the combined patterns A16P33, A36P34, A45P26, A45P32, and A45P33 corresponded to two to eight antigens of the g series. For l,v and l,w antigens, 22 of 23 combined patterns corresponded to both antigens. Pattern A39P24 was shared by the l,v (serovar Coromandel) and z35 antigens. Of the 23 combined patterns obtained, only 6 seemed to be specifically associated with the phase-1 l,v antigen flagellin gene (A40P2, A41P2, A42P2, A43P1, A44P8, and A53P24) and 13 were specifically associated with the phase-2 l,w antigen flagellin gene (A40P24, A52P3, A52P4, A52P5, A52P6, A52P8, A52P9, A53P3, A53P4, A53P5, A53P7, A54P1, and A54P7). Some genes encoding l,v or l,w antigens shared the same comined patterns (A52P1, A52P2, A52P24, and A53P2). All of the combined patterns of genes encoding phase-1 l,w antigen (A52P1, A52P2, A52P24, A53P1, and A53P2) were found for phase-2 l,w antigen. RFLP analysis showed that phase-1 and phase-2 genes sharing l,w antigens were not differentiated by the endonucleases used.
Correspondence between patterns and serovars.
The discrimination power of flagellin gene RFLP analysis alone was insufficient to distinguish all of the serovars tested. Among the 237 serovars studied, 112 combined patterns were each assigned to 1 serovar (Table 4). Each combined pattern was given by 1 to 22 serovars. For 51 serovars, combined patterns of the flagellin phase-1 gene were serovar specific. For 28 serovars, specificity involved only combined patterns from the fljB gene. For 33 serovars, combined patterns obtained from both genes were required. The specificity of the fliC gene combined pattern was given by HhaI for 27 serovars, by HphI for 11 serovars, and by both enzymes for 13 serovars. The specificity of the fljB gene combined pattern was given by HhaI for 12 serovars, by HphI for 7 serovars, and by both enzymes for 9 serovars. For r,i antigens, the discrimination of RFLP was so high that each serovar studied had its own specific combined pattern.
Specific patterns characterized some important serovars. Regular serovar Typhi (H1:d) showed specific pattern A19 with HhaI. Variant serovar Typhi (H1:j) showed specific patterns A34 with HhaI and P17 with HphI. This result was tested on nine strains, seven of which showed different ribotypes. Strains from serovar Typhi harbored three types of specific combined patterns: A19P40 (regular Typhi), A34P17, and A34P22 (variant Typhi).
Serovar Typhimurium showed specific pattern P38 (H1:i) with HphI. This result was confirmed for 11 strains, 6 of which had different phage types. Strains from serovar Typhimurium consistently showed combined pattern A44P38-A1P24 (phase-1 and phase-2 genes).
Some other epidemiologically important serovars were also differentiated (phase-1/phase-2 genes): Hadar (A46P35-A10P19), Heidelberg (A44P13-A1P24), Indiana (A60P10-A3P1), Newport (A49P24-A2P7), Choleraesuis (A47P24-A2P1), SaintPaul (A48P24-A1P1), Goldcoast (A44P13-A52P24), Paratyphi A (A62P24/−), and Bovismorbificans H1: r,i (A15P13-A33P24).
Thus, among 170 strains studied in both phases, 134 strains (112 serovars) could be identified at the serovar level with the flagellin gene combined patterns. Lack of pattern specificity was observed for 36 serovars. For 25 serovars, the phase-2 gene was missing (monophasic strains) and for 11 diphasic serovars, phase-1 and phase-2 gene combined patterns were insufficient for serotype identification. The knowledge of the O antigen and both flagellin gene RFLPs contributed to the identification of these 11 diphasic serovars (in the sample of serovars studied). For example, serovars Brooklyn and Brandenburg shared the A52P2-A10P15 flagellin gene combined pattern but had the O16 and O4 antigens, respectively.
Nonmotile isolates.
A strain of serovar Gallinarum, a serovar failing to express flagella (lack of flagellar antigens), was assigned to fliC combined pattern A45P26. Other nonmotile isolates were studied. Two isolates (O9:H−:Vi−) had combined pattern A45P26, which was observed for the flagellar antigen g series. These two isolates originated from poultry.
Four of the other isolates tested matched the A19P40 combined pattern that was observed for the H1:d antigen of serovar Typhi. Three of these isolates were O9,12:H−:Vi, and one was rough. No amplification of fljB was obtained.
One isolate (O4,5:H−) had a unique phase-1 flagellin gene combined pattern not described in this report.
DISCUSSION
This work shows that exploring the genetic diversity of Salmonella flagellin genes by RFLP is far more complicated than originally thought (9).
Subsp. II was the only subspecies other than subsp. I included in this study. This is because subsp. II strains share both O and H antigens with subsp. I strains. With a single exception, patterns of subsp. II flagellin genes were different from that of subsp. I. RFLP patterns suggest that flagellin genes from both subsp. I and II form separate evolutionary groups and are in support of the subspecies concept.
Phase-1 and phase-2 flagellin genes can be amplified separately. The phase-2 specificity of the fljB PCR system was verified by amplification of the hin-fljB-fljA region. It is remarkable that assignment of flagellin antigens to either phase was generally correct. There is still a question about serovars Kotte and Coromandel. Coromandel fliC and Kotte fljB have the same RFLP pattern (A39P24). However, the Coromandel phase-1 antigen is l,v whereas the Kotte phase-2 antigen is z35. Since the Coromandel phase-2 antigen is z35 (the Kotte phase-1 antigen is b), there is a possibility that the antigen phase assignment is wrong for Coromandel. More strains with the z35 and b antigens need to be studied to strengthen this hypothesis, and gene sequencing should be done.
Five patterns associated with antigen l,w were given by fliC and fljB amplified from different strains. In these cases, in spite of the lack of RFLP gene differentiation, some sequence difference must occur since the primers for amplification were different.
No fljB gene could be amplified from known monophasic serovars. This is in agreement with previous studies which showed that serovars Enteritidis, Typhi (monophasic), Berta, Agona, and Montevideo do not possess the fljA, hin, or fljB gene (2). It is striking that we were unable to amplify fljB from the (rarely occurring) serotype Typhi strains expressing second-phase z66. In contrast with the lack of a phase-2 gene in monophasic serotypes, the fliC gene could always be amplified from nonmotile isolates.
The high diversity of restriction profiles was attributed to variability within an internal region of the flagellin genes, whereas regions at the 5′ and 3′ ends are more conserved (6). In most cases, diversity highlighted by flagellin gene RFLP exceeded the diversity showed by antigens. This finding on the genetic variation of flagellin genes agrees with recently reported observations within populations of related strains of E. coli or Pseudomonas aeruginosa (4, 24). However, the flagellin gene sequence information was reduced by comigrating fragments. In addition, the choice of an endonuclease with restriction sites preferentially located in the variable region (HphI) did not yield better discrimination. Endonuclease HphI showed fewer profiles than HhaI and fewer antigen- or serovar-specific patterns than HhaI. The use of a restriction enzyme which targets a highly variable region seems to increase the probability of generating falsely identical fragments, i.e., unrelated fragments with similar sizes in different patterns.
The correlation between flagellar antigens and flagellin RFLP patterns is difficult to assess. It should be noted that building the WKL scheme has involved many historical and arbitrary decisions. The choice of strains for immunization and absorption was determining. Since antigenic factors could often be split further into subfactors, decisions had to be made about when to stop splitting. The WKL scheme is only a simplified summary of antigenic relationships among Salmonella serovars. When two antigens have the same designation, it indicates which antisera are likely to react, not that these antigens are identical. For example, serovars with H1:d may have different subfactors, such as d,d1 (Typhi), d,d3 (Stanley), and d,d3,d4 (Muenchen) (8). Flagellins of the g series have even more complex structures, such as g,o,m,z1,z2 for Enteritidis (summarized as g,m), g,o,m,q,z1 for Blegdam (summarized as g,m,q), or g,o,q,z3 for Moscow (summarized as g,q) (8). Phase-2 flagellins 1,5 and 1,7 are also very complex (8). On the other hand, i and r antigens have known antigenic relationships. Thus, it seems that RFLP patterns often split flagellar types in a different way than serotyping.
In several cases, RFLP was unable to discriminate among fliC genes encoding antigens with different designations such as d, i, or r,i antigens. This may be due to an unfortunate choice of endonucleases, although in these cases, sequences are not available for comparison. In other cases, lack of discrimination was due to excessive genetic similarity. The discrimination among antigens g,m (as in serovar Enteritidis) and g,p (as in serovar Dublin), which is essential in epidemiology, could not be achieved by RFLP analysis, since the corresponding genes differ by only six nucleotides (13). Genes encoding flagellin antigens 1,2; 1,5; and 1,6 are more than 96.2% related (22).
Although serotyping is the “gold standard” of Salmonella typing, all absorbed sera that are necessary for complete serotyping are not commercially available. This limits complete serotyping to National Reference Centers. A number of sera have to be prepared by Reference Centers. Since this preparation and serotyping itself (with phase inversion) are expensive and labor intensive, alternative methods which could be applied by clinical laboratories need to be sought. The dream that flagellar antigens could be deduced from flagellin gene RFLP has not been realized by this work. However, some important flagellin antigens, and even some serovars, can be deduced from RFLP patterns. When this is confirmed with more strains and combined with some PCR approach to O-antigen typing, then some major Salmonella serotypes could be identified by PCR and restriction, leaving serotyping for less common serotypes.
ACKNOWLEDGMENTS
Thanks are due to Michel Y. Popoff of the Collaborating Center for Reference and Research on Salmonella, World Health Organization, for reference strains and to Philippe J. M. Bouvet and F. Grimont for helpful discussions.
REFERENCES
- 1.Barthelemy J P, Guenoche A. Les arbres et les représentations de proximités. Paris, France: Masson; 1988. [Google Scholar]
- 2.Burnens A P, Stanley J, Sechter I, Nicolet J. Evolutionary origin of a monophasic Salmonella serovar, 9,12:l,v:−, revealed by IS200 profiles and restriction fragment polymorphisms of the fljB gene. J Clin Microbiol. 1996;34:1641–1645. doi: 10.1128/jcm.34.7.1641-1645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dice L R. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 4.Fields P I, Blom K, Hugues H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel G, Newton S M C, Schoolnik G K, Stocker B A D. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 1989;8:3149–3152. doi: 10.1002/j.1460-2075.1989.tb08468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iino T. Genetics of structure and function of bacterial flagella. Annu Rev Genet. 1977;11:161–182. doi: 10.1146/annurev.ge.11.120177.001113. [DOI] [PubMed] [Google Scholar]
- 7.Joys T M. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J Biol Chem. 1985;260:15758–15761. [PubMed] [Google Scholar]
- 8.Kauffmann F. The bacteriology of Enterobacteriaceae. Copenhagen, Denmark: Munksgaard; 1966. [Google Scholar]
- 9.Kilger G, Grimont P A D. Differentiation of Salmonella phase 1 flagellar antigen types by restriction of the amplified fliC gene. J Clin Microbiol. 1993;31:1108–1110. doi: 10.1128/jcm.31.5.1108-1110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleid D, Humayun Z, Jeffrey A, Ptashne M. Novel properties of a restriction endonuclease isolated from Haemophilus parahaemolyticus. Proc Natl Acad Sci USA. 1976;73:293–297. doi: 10.1073/pnas.73.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Minor L, Popoff M Y. Designation of Salmonella enterica sp. nov., nom. rev., as the type and only species of the genus Salmonella. Int J Syst Bacteriol. 1987;37:465–468. [Google Scholar]
- 12.Li J, Smith NH, Nelson K, Crichton P B, Old D C, Whittam T S, Selander R K. Evolutionary origin and radiation of the avian-adapted non-motile salmonellae. J Med Microbiol. 1993;38:129–139. doi: 10.1099/00222615-38-2-129. [DOI] [PubMed] [Google Scholar]
- 13.Masten B J, Joys T M. Molecular analysis of the Salmonella g… flagellar antigen complex. J Bacteriol. 1993;175:5359–5365. doi: 10.1128/jb.175.17.5359-5365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parish C R, Wistar R, Ada G L. Cleavage of bacterial flagellin with cyanogen bromide: antigenic properties of the protein fragments. Biochem J. 1969;113:501–506. doi: 10.1042/bj1130501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popoff M Y, LeMinor L. Antigenic formulas of the Salmonella serovars, 6th revision. WHO Collaborating Centre for Reference and Research on Salmonella. Paris, France: Institut Pasteur; 1992. [Google Scholar]
- 16.Popoff M Y, Bockemühl J, McWhorter-Murlin A. Supplement 1993 (no. 37) to the Kauffmann-White scheme. Res Microbiol. 1994;145:711–716. doi: 10.1016/0923-2508(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 17.Reeves M W, Evins G M, Heiba A A, Plikaytis B D, Farmer J J., III Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts R J, Myers P A, Morrison A, Murray K. A specific endonuclease from Haemophilus haemolyticus. J Mol Biol. 1976;103:199–208. doi: 10.1016/0022-2836(76)90060-7. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigue D C, Tauxe R V, Rowe B. International increase in S. enteritidis: a new pandemic. Epidemiol Infect. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffer H E, Sederoff R R. Improved estimation of DNA fragment lengths from agarose gels. Anal Biochem. 1981;115:113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]
- 21.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1973. [Google Scholar]
- 22.Vanegas R A, Joys T M. Molecular analyses of the phase-2 antigen complex 1,2,.. of Salmonella spp. J Bacteriol. 1995;177:3863–3864. doi: 10.1128/jb.177.13.3863-3864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L N, Joys T M. Covalent structure of three phase-1 flagellar filament proteins of Salmonella. J Mol Biol. 1985;186:791–803. doi: 10.1016/0022-2836(85)90397-3. [DOI] [PubMed] [Google Scholar]
- 24.Wistangley C, Coulson M A, Wepner B, Alun J, Morgan W, Hart C A. Flagellin gene and protein variation amongst clinical isolates of Pseudomonas aeruginosa. Microbiology. 1996;142:2145–2151. doi: 10.1099/13500872-142-8-2145. [DOI] [PubMed] [Google Scholar]
- 25.Zieg J, Silverman M, Hilmem M, Simon M. Recombination switch for gene expression. Science. 1977;196:170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]