Abstract
Mycoplasma gallisepticum (MG) is one of the important pathogens in poultry industry and has led to major economic losses. Understanding the epidemiology is crucial to improve the control and eradication program of MG. This study collected 1,250 chicken samples, including trachea and lung, from China in 2022 to investigate the epidemiology of MG. Among the collected samples, 938 samples were positive for MG infection, resulting in an average positive rate of 75.04%. Additionally, 570 samples were positive for both MG and Mycoplasma synoviae (MS) coinfection, with an average positive rate of 45.60%. A total of 183 MG infection positive samples in this study were selected for genotyping, and the multilocus sequence typing (MLST) method based on 7 housekeeping genes was used. As a result, 183 samples belonged to 11 sequence types (STs), with ST-78 being the most prevalent. After BURST analysis, all 183 sequences were divided into group 3. Besides, 119 reference sequences from database and 183 sequences of this study were selected to construct the phylogenetic tree using the neighbor-joining method. The results revealed that the sequences from China, total 196 sequences, were classified into 4 branches. The findings suggest that the MG strains in China exhibit diverse genotypes, which may be related to international trade and the use of live vaccines. Furthermore, we detected the drug susceptibility of 10 isolated strains randomly, which may be helpful to guide the clinical use of drugs to control MG infection.
Key words: Mycoplasma gallisepticum, epidemiology, MLST genotype, drug susceptibility
INTRODUCTION
Mycoplasma gallisepticum (MG) as one of the virulent avian Mycoplasma species affects chickens and turkeys worldwide and is listed and notifiable to the World Organization for Animal Health (OIE) (Malik et al., 2021). MG infection causes chronic respiratory disease in chickens and turkeys, characterized by nasal discharge, tracheal rales, coughing, and dyspnea, resulting major economic losses in terms of reduced weight gain, egg production and hatchability, downgrading carcass quality, and the infected birds become susceptible to other diseases (Sawicka et al., 2020; Yadav et al., 2021). MG transmits horizontally by direct or indirect contact and vertically through the egg (Kleven, 2008; Matucci et al., 2020).
According to molecular analysis of reported cases worldwide, a molecular epidemiological map of the MG infection has been established. The results show that MG infections occur in chicken coops in different regions and of varying scales. Specifically, the United States (Staley et al., 2018), Europe (Michiels et al., 2016; Felice et al., 2020), and Asia (Norouzian et al., 2019; Limsatanun et al., 2022) are high-incidence areas. In addition, there is also seasonal variation in incidence of this disease (Feberwee et al., 2022), with autumn and winter being the high-risk seasons.
With the development of molecular biology technology, DNA fingerprinting techniques were used to genotype of MG strain frequently (Charlton et al., 1999, Marois et al., 2001). However, these techniques are time-consuming, laborious, and poorly repeatable. To improve the reproducibility, reliability and applicability on clinical samples, and reduce labor intensity, sequence-based genotyping methods have been described, such as sequencing variable surface proteins (mgc2, pvpA, gapA and MGA_0319) or variable intergenic spacer region (IGSR) between 23S rRNA and 16S rRNA (Jiang et al., 2009; Sprygin et al., 2010). However, these methods showed insufficient discriminatory power to differentiate related strains (Delaney et al. 2012, Staley et al. 2018).
Among sequence-based genotyping methods, multilocus sequence typing (MLST) (Beko et al., 2019) and core genome multilocus sequence typing (cgMLST) (Ghanem et al., 2018) provide several advantages for genotyping bacterial species (Larsen et al., 2012; Kwong et al., 2016). Compared to MLST, whole genome sequencing is more expensive and time-consuming. MLST is based on the nucleotide sequences of internal fragments of housekeeping genes, in which mutations are assumed to be largely neutral (Jolley et al., 2018). Until now, 2 MLST typing method of MG were established based on 6 loci (atpG, dnaA, fusA, rpoB, ruvB, uvrA) (Beko et al., 2019) and 7 loci (ugpA, atpG, DUF3196, mraW, plsC, dppC, lgT) (Ghanem and El-Gazzar, 2019) respectively.
In this study, we collected 1,250 chicken samples, including trachea and lung, from 15 provinces in China in 2022 to detect MG infection. In order to investigate the dominant genotypes of MG, MLST genotype based on 7 housekeeping genes was carried out. This study will promote to understanding of the prevalence and evolutionary relationship of MG in China, and support the effective prevention and control of it.
MATERIALS AND METHODS
Sample Collection, Genomic DNA Extraction, and MG Isolation
The samples used in this study were collected from commercial broiler chicken farms in Jiangsu, Hubei, Shandong, Anhui, Guangxi, Guangdong, Yunnan, Henan, Hebei, Zhejiang, Hunan, Chongqing, Sichuan, Fujian, and Guizhou provinces of China in 2022, detail information showed in Table 1. Five chickens were collected from one flock. All tissues collected were trachea and lung of chickens that looks healthy. Tissues from one chicken were seemed as one sample, so 5 samples were collected from one flock. Tissues from each flock were collected under aseptic conditions, and placed into grinding tubes for nucleic acid extraction. After ground, tissue samples were centrifuged and the supernatant was added to the MagaBio Plus Genomic DNA Extraction Kit (Bioer Technology, Hangzhou, China) according to the manufacturer's instructions. For MG isolation, cut the trachea and lung tissue and add it to the modified mycoplasma medium, in a sterile environment (Bradbury and Howell, 1974), and shaken at 37°C for 3 to 4 h, then filtered into new sterile tubes using a 0.45 μm filter. The same volume of fresh medium was added and cultured at 37°C until the color changed from red to orange-yellow. The 200 μL of culture was plated on MG solid media, containing 10% porcine serum, 3.0 g/L glucose, 100 mg/L L-cysteine, and 100 mg/L NAD at 37°C for 3 to 7 days and constantly observed. Fried-egg-like single MG clones were selected and cultured in liquid medium until the color changes. Then, 200 μL of the culture was plated on MG solid media at 37°C again. Purified MG colonies were obtained after repeating these operations 3 times.
Table 1.
The results of MG and MS infection among 1,250 samples collected in China in 2022.
| MG infection |
MG and MS coinfection |
||||
|---|---|---|---|---|---|
| Region | Sample numbers | Positive number | Positive rate | Positive number | Positive rate |
| Henan | 25 | 20 | 80% | 9 | 36% |
| Hebei | 50 | 46 | 92% | 7 | 14% |
| Shandong | 50 | 21 | 42% | 20 | 40% |
| Jiangsu | 345 | 237 | 68.70% | 149 | 43.19% |
| Anhui | 100 | 96 | 96% | 86 | 86% |
| Zhejiang | 15 | 10 | 66.67% | 9 | 60% |
| Hunan | 70 | 41 | 58.57% | 17 | 24.29% |
| Hunbei | 75 | 50 | 66.67% | 21 | 28% |
| Chongqing | 55 | 52 | 94.55% | 15 | 27.27% |
| Yunnan | 55 | 42 | 76.36% | 25 | 45.45% |
| Sichuan | 100 | 88 | 88% | 35 | 35% |
| Guizhou | 45 | 43 | 95.56% | 33 | 73.33% |
| Guangdong | 75 | 50 | 66.67% | 48 | 64% |
| Guangxi | 140 | 101 | 72.14% | 59 | 42.14% |
| Fujian | 50 | 41 | 82% | 37 | 74% |
| Total | 1250 | 938 | 75.04% | 570 | 45.60% |
RT-qPCR Detection
The genomic DNA extracted from tissue samples was used for MG-specific and MS-specific RT-qPCR detection respectively (Raviv and Kleven, 2009). Briefly, THUNDERBIRD probe qPCR Mix (TOYOBO, Shanghai, China) 10 μL, 300 nM of each forward primer, 300 nM of each reverse primer, 150 nM of each probe, 0.4 μL of 50 × ROX reference, 2 μL of template, ddH2O were added up to 20 μL. The qPCR amplification program consisted of 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The primer sequences used for detecting MG and Mycoplasma synoviae (MS) in this assay are as follows: MG forward primer: 5′-TTGGGTTTAGGGATTGGGATT-3′; MG reverse primer: 5′-CCAAGGGATTCAACCATCTT-3′; MG probe: 5′-FAM-TGATGATCCAAGAACGTGAAGAACACC-BHQ1-3′; MS forward primer: 5′-CTAAATACAATAGCCCAAGGCAA-3′; MS reverse primer: 5′-CCTCCTTTCTTACGGAGTACA-3′; MS probe: 5′-CY5-AGCGATACACAACCGCTTTTAGAAT-BHQ1-3′.
MLST Gene Amplification
In this study, MG positive samples with Ct values less than 28 were selected for genotyping. The 7 housekeeping genes used for MLST were ugpA, atpG, DUF3196, mraW, plsC, dppC and lgT, the primer sequences are available on the PubMLST website (https://pubmlst.org/static/organisms/mycoplasma-gallisepticum/primers.pdf) (Ghanem and El-Gazzar, 2019). The PCR reaction mixture contained 25 μL Premix Taq (LA Taq Version 2.0 Plus dye), 0.5 μL of each 10 μM primer and 2 μL of genomic DNA template in a total volume of 50 μL. The PCR amplification was performed with an initial denaturation step of 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 90 s. The PCR products were then subjected to Sanger sequencing (Sangon Biotech, Shanghai, China).
The DNA sequences of the 7 housekeeping genes were compared and assembled using Lasergene 7.1, and then submitted to the PubMLST database. For each new sequence, an allele ID was assigned. Each MG sample generated an allele profile consisting of 7 loci. Based on the obtained allele patterns, sequence types (STs) were determined and compared with information from 119 reference sequences in the PubMLST database. These reference sequences from the United States, Australia, the United Kingdom, Israel, Jordan, Japan, and China. Based on the STs, BURST analysis was applied (Feil et al., 2004) to group strains into clonal complexes with 4 or more matching alleles.
In Vitro Antimicrobial Sensitivity Test
The drug sensitivity of the isolated MG strains was tested using antibiotics including enrofloxacin (EN), doxycycline (DO), chlortetracycline (CH), tiamulin (TIA), valnemulin (VAL), tylosin (TY), tylvalosin (TYL), Timicosin (TIM), spectinomycin (SPE), and lincomycin (LIN). The concentration of antibiotic solution was diluted to 128 μg/mL. Then the solutions were sterilized with a 0.22-μm millipore filter membrane. The MIC (minimum inhibitory concentration) test was carried out in 96-well microdilution plates (Zhang et al., 2022). Briefly, the MG culture was diluted in mycoplasma broth medium to 104 ccu/mL. The 128 μg/mL diluted antibiotic solution was usually diluted 2-fold continuous gradient with 100 μL Mycoplasma broth medium. After dilution, the 100 μL diluted MG culture was inoculated into each well. MG culture and antibiotics were included in all tests as negative controls and antibiotic controls, respectively. Plates were incubated at 37°C. The lowest concentration of antibiotic to show a color change denoted MIC. The MIC was read when the phenol red indicator in the negative control had just turned orange-yellow.
Results
Epidemiological Information of Clinical Samples
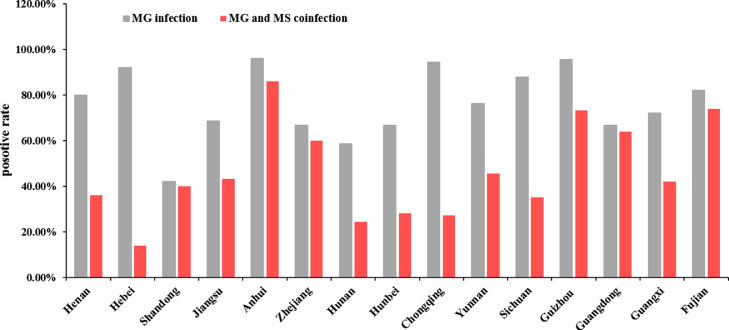
Total of 1,250 samples were collected from Henan, Hebei, Shandong, Jiangsu, Anhui, Zhejiang, Hunan, Hubei, Chongqing, Yunnan, Sichuan, Guizhou, Guangdong, Guangxi and Fujian provinces in China in 2022. Considering that MS and MG co-infection is common in poultry farms (Sid et al., 2015; Giram et al., 2022), we also detected for MS infection. After RT-qPCR detection, 938 samples were positive for MG infection with an average positivity rate of 75.04%, while 570 samples were positive for co-infections of MG and MS with an average positivity rate of 45.60% (Table 1). The positivity rates varied across different regions (Figure 1). The largest number of samples was 345 collected from Jiangsu province, among which 237 were positive for MG infection and 149 were positive for co-infection of MG and MS, with positivity rates of 68.7% and 43.19%, respectively. Among the 100 samples collected from Anhui province, the positivity rate of MG infection was the highest, reaching 96%, while the positivity rate of MG and MS co-infection was also the highest at 86%; in contrast, among the 50 samples collected from Shandong province, the positivity rate of MG infection was the lowest, about 42%. These data indicate that MG infection is very common and severe in China, especially when combined with MS infection, which should receive more attention. Among the 938 MG-positive samples, 183 samples were selected for sequencing and gene typing. The detail information about sequenced samples were listed in Table 2.
Figure 1.
MG and MS infection rate in different regions in China.
Table 2.
Information of 183 sequenced samples.
| Number | IDs | Sample | Region | atpG | dppC | DUF3196 | lgT | mraW | plsC | ugpA | ST |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 296 | Guangxi/2022/HLG | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 2 | 297 | Guangxi/2022/LGF-1 | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 3 | 298 | Guangxi/2022/LGF-2 | Guangxi | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 4 | 299 | Guangxi/2022/FHC-1 | Guangxi | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 5 | 300 | Guangxi/2022/FHC-2 | Guangxi | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 6 | 301 | Guangxi/2022/T2-1 | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 7 | 302 | Guangxi/2022/T2-2 | Guangxi | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 8 | 303 | Guangxi/2022/H4-1 | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 9 | 304 | Guangxi/2022/H4-2 | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 10 | 305 | Guangxi/2022/H5-1 | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 11 | 306 | Guangxi/2022/H5-2 | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 12 | 307 | Guangxi/2022/LH-1 | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 13 | 308 | Guangxi/2022/LH-2 | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 14 | 309 | Guangxi/2022/CTX | Guangxi | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 15 | 310 | Guangxi/2022/MQL | Guangxi | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 16 | 311 | Guangxi/2022/WYP | Guangxi | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 17 | 312 | Guangxi/2022/WMD | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 18 | 313 | Guangxi/2022/LZH | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 19 | 314 | Guangxi/2022/HXX | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 20 | 315 | Guangxi/2022/ZXY | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 21 | 316 | Guangxi/2022/LS | Guangxi | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 22 | 317 | Guangxi/2022/DRS1 | Guangxi | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 23 | 318 | Guangxi/2022/TXL | Guangxi | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 24 | 319 | Guangxi/2022/WKX | Guangxi | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 25 | 320 | Guangxi/2022/OTS | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 26 | 321 | Guangxi/2022/DCM | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 27 | 322 | Guangxi/2022/LRC | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 28 | 323 | Guangxi/2022/YZL | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 29 | 324 | Guangxi/2022/LYZ | Guangxi | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 30 | 325 | Fujian/2022/ZW | Fujian | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 31 | 326 | Fujian/2022/XWF | Fujian | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 32 | 327 | Fujian/2022/ZXQ | Fujian | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 33 | 328 | Fujian/2022/GQY | Fujian | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 34 | 329 | Fujian/2022/XLN | Fujian | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 35 | 330 | Fujian/2022/ZXP | Fujian | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 36 | 331 | Fujian/2022/HYS2983 | Fujian | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 37 | 332 | Fujian/2022/HYS2986 | Fujian | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 38 | 333 | Hebei/2022/WZP | Hebei | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 39 | 334 | Hebei/2022/WST | Hebei | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 40 | 335 | Hebei/2022/LCX | Hebei | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 41 | 336 | Hebei/2022/XFQ | Hebei | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 42 | 337 | Hebei/2022/WHZ | Hebei | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 43 | 338 | Hebei/2022/TT | Hebei | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 44 | 339 | Shandong/2022/HQH | Shandong | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 45 | 340 | Zhejiang/2022/WAP | Zhejiang | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 46 | 341 | Zhejiang/2022/ZAQ | Zhejiang | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 47 | 342 | Anhui/2022/CJH | Anhui | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 48 | 343 | Anhui/2022/LFC | Anhui | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 49 | 344 | Anhui/2022/WSY | Anhui | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 50 | 345 | Anhui/2022/LJX | Anhui | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 51 | 346 | Anhui/2022/ZCH | Anhui | 3 | 13 | 29 | 43 | 21 | 28 | 1 | 76 |
| 52 | 347 | Anhui/2022/LP | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 53 | 348 | Anhui/2022/LB | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 54 | 349 | Anhui/2022/WLZ | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 55 | 350 | Anhui/2022/XWF | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 56 | 351 | Anhui/2022/ZYS | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 57 | 352 | Anhui/2022/GGL | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 58 | 353 | Anhui/2022/ZYF | Anhui | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 59 | 354 | Anhui/2022/CHL | Anhui | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 60 | 355 | Anhui/2022/FGS | Anhui | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 61 | 356 | Anhui/2022/WXG | Anhui | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 62 | 357 | Anhui/2022/ZHJ | Anhui | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 63 | 358 | Anhui/2022/ZZB | Anhui | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 64 | 359 | Anhui/2022/GHY | Anhui | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 65 | 360 | Hunan/2022/DMY | Hunan | 2 | 36 | 23 | 44 | 21 | 18 | 23 | 77 |
| 66 | 361 | Hunan/2022/CDC | Hunan | 2 | 36 | 23 | 44 | 21 | 18 | 23 | 77 |
| 67 | 362 | Hunan/2022/XSY | Hunan | 2 | 36 | 23 | 44 | 21 | 18 | 23 | 77 |
| 68 | 363 | Hunan/2022/LHJ | Hunan | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 69 | 364 | Hunan/2022/LSB | Hunan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 70 | 365 | Hunan/2022/ZGB | Hunan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 71 | 366 | Hunan/2022/LZL | Hunan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 72 | 367 | Hunan/2022/PXN | Hunan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 73 | 368 | Hunan/2022/MMZ | Hunan | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 74 | 369 | Jiangsu/2022/TWX | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 75 | 370 | Jiangsu/2022/TYH | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 76 | 371 | Jiangsu/2022/SJS | Jiangsu | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 77 | 372 | Jiangsu/2022/ZJG | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 78 | 373 | Jiangsu/2022/SWX | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 79 | 374 | Jiangsu/2022/ZZB | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 80 | 375 | Jiangsu/2022/LP | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 81 | 376 | Jiangsu/2022/HJZ | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 82 | 377 | Jiangsu/2022/XFY | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 83 | 378 | Jiangsu/2022/LFJ | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 84 | 379 | Jiangsu/2022/ZQX | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 85 | 380 | Jiangsu/2022/YHC | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 86 | 381 | Jiangsu/2022/ZXXQ-1 | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 87 | 382 | Jiangsu/2022/ZXXQ-2 | Jiangsu | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 88 | 383 | Jiangsu/2022/ZXXQ-3 | Jiangsu | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 89 | 384 | Jiangsu/2022/ZXXQ-4 | Jiangsu | 3 | 13 | 29 | 43 | 21 | 28 | 1 | 76 |
| 90 | 385 | Jiangsu/2022/ZGC | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 91 | 386 | Jiangsu/2022/LXJ | Jiangsu | 3 | 13 | 4 | 20 | 19 | 18 | 18 | 83* |
| 92 | 387 | Jiangsu/2022/XQG | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 93 | 388 | Jiangsu/2022/XHJ | Jiangsu | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 94 | 389 | Jiangsu/2022/JXG | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 95 | 390 | Jiangsu/2022/SXZ | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 96 | 391 | Jiangsu/2022/MGM | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 97 | 392 | Jiangsu/2022/JH | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 98 | 393 | Jiangsu/2022/CJT | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 99 | 394 | Jiangsu/2022/DCM | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 100 | 395 | Jiangsu/2022/LL | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 101 | 396 | Jiangsu/2022/LSJ | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 102 | 397 | Jiangsu/2022/ZZM | Jiangsu | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 103 | 398 | Jiangsu/2022/XXS | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 104 | 399 | Jiangsu/2022/CL | Jiangsu | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 105 | 400 | Jiangsu/2022/XKF | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 106 | 401 | Jiangsu/2022/ZTC | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 107 | 402 | Jiangsu/2022/HZZ | Jiangsu | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 108 | 403 | Jiangsu/2022/HXM | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 109 | 404 | Jiangsu/2022/XHZ | Jiangsu | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 110 | 405 | Jiangsu/2022/GKQ | Jiangsu | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 111 | 406 | Jiangsu/2022/WLB | Jiangsu | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 112 | 407 | Jiangsu/2022/LZH | Jiangsu | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 113 | 408 | Sichuan/2022/YG | Sichuan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 114 | 409 | Sichuan/2022/LYB | Sichuan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 115 | 410 | Sichuan/2022/WKW | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 116 | 411 | Sichuan/2022/CTW | Sichuan | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 117 | 412 | Sichuan/2022/HY | Sichuan | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 118 | 413 | Sichuan/2022/WGYU | Sichuan | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 119 | 414 | Sichuan/2022/YXH | Sichuan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 120 | 415 | Sichuan/2022/LYM | Sichuan | 3 | 13 | 29 | 43 | 21 | 28 | 1 | 76 |
| 121 | 416 | Sichuan/2022/WGYN | Sichuan | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 122 | 417 | Sichuan/2022/WXG | Sichuan | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 123 | 418 | Sichuan/2022/ZXY | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 124 | 419 | Sichuan/2022/LJ | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 125 | 420 | Sichuan/2022/ZYH | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 126 | 421 | Sichuan/2022/HYH | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 127 | 422 | Sichuan/2022/LYL | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 128 | 423 | Sichuan/2022/ZGQ | Sichuan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 129 | 424 | Yunnan/2022/OLN | Yunnan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 130 | 425 | Yunnan/2022/HZX | Yunnan | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 131 | 426 | Yunnan/2022/DHQ | Yunnan | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 132 | 427 | Yunnan/2022/WHC | Yunnan | 3 | 13 | 29 | 43 | 21 | 28 | 1 | 76 |
| 133 | 428 | Yunnan/2022/PJD | Yunnan | 3 | 13 | 29 | 43 | 21 | 28 | 1 | 76 |
| 134 | 429 | Yunnan/2022/JWD | Yunnan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 135 | 430 | Yunnan/2022/BQY | Yunnan | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 136 | 431 | Yunnan/2022/GZY | Yunnan | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 137 | 432 | Guangdong/2022/Q3 | Guangdong | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 138 | 433 | Guangdong/2022/3-3 | Guangdong | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 139 | 434 | Guangdong/2022/CG6-1 | Guangdong | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 140 | 435 | Guangdong/2022/QYF | Guangdong | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 141 | 436 | Guangdong/2022/YL | Guangdong | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 142 | 437 | Guangdong/2022/SWX | Guangdong | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 143 | 438 | Guangdong/2022/OXZ | Guangdong | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 144 | 439 | Guangdong/2022/HRQ | Guangdong | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 145 | 440 | Guangdong/2022/CG5-5 | Guangdong | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 146 | 441 | Guangdong/2022/LRD5 | Guangdong | 3 | 13 | 23 | 44 | 21 | 21 | 1 | 85* |
| 147 | 442 | Guangdong/2022/LAH | Guangdong | 3 | 13 | 23 | 44 | 21 | 21 | 1 | 85* |
| 148 | 443 | Guangdong/2022/XML | Guangdong | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 149 | 444 | Guangdong/2022/XYX | Guangdong | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 81* |
| 150 | 445 | Guangdong/2022/ZXK | Guangdong | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 151 | 446 | Guangdong/2022/ZX | Guangdong | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 152 | 447 | Guizhou/2022/LM | Guizhou | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 153 | 448 | Guizhou/2022/WPS | Guizhou | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 154 | 449 | Guizhou/2022/DDY | Guizhou | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 155 | 450 | Guizhou/2022/ZWX | Guizhou | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 156 | 451 | Guizhou/2022/ZWH | Guizhou | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 157 | 452 | Guizhou/2022/WSJ | Guizhou | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 158 | 453 | Guizhou/2022/ZQH | Guizhou | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 159 | 454 | Guizhou/2022/QCS | Guizhou | 3 | 13 | 29 | 43 | 21 | 28 | 1 | 76 |
| 160 | 455 | Guizhou/2022/YY | Guizhou | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 161 | 456 | Henan/2022/ZKK | Henan | 3 | 13 | 23 | 44 | 21 | 21 | 1 | 85* |
| 162 | 457 | Henan/2022/ZZZ | Henan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 163 | 458 | Henan/2022/WYZ | Henan | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 164 | 459 | Henan/2022/GAX | Henan | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 165 | 460 | Hubei/2022/MYM | Hubei | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 166 | 461 | Hubei/2022/HXP | Hubei | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 167 | 462 | Hubei/2022/WL | Hubei | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
| 168 | 463 | Hubei/2022/LYQ | Hubei | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 169 | 464 | Hubei/2022/QJH | Hubei | 3 | 13 | 23 | 44 | 21 | 21 | 1 | 85* |
| 170 | 465 | Hubei/2022/PAY | Hubei | 3 | 13 | 29 | 34 | 21 | 28 | 1 | 73 |
| 171 | 466 | Hubei/2022/GWB | Hubei | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 172 | 467 | Hubei/2022/LBX | Hubei | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 173 | 468 | Hubei/2022/WF | Hubei | 3 | 13 | 23 | 44 | 21 | 21 | 1 | 85* |
| 174 | 469 | Hubei/2022/WSC | Hubei | 3 | 13 | 23 | 44 | 21 | 9 | 1 | 78 |
| 175 | 470 | Chogqing/2022/ZDQ | Chogqing | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 176 | 471 | Chogqing/2022/LZF | Chogqing | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 177 | 472 | Chogqing/2022/ZZQ | Chogqing | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 178 | 473 | Chogqing/2022/DRC | Chogqing | 17 | 13 | 20 | 20 | 19 | 18 | 18 | 36 |
| 179 | 474 | Chogqing/2022/LSW | Chogqing | 2 | 36 | 23 | 42 | 21 | 18 | 1 | 75 |
| 180 | 475 | Chogqing/2022/ZGQ | Chogqing | 3 | 13 | 4 | 44 | 28 | 9 | 1 | 78 |
| 181 | 476 | Chogqing/2022/WYX | Chogqing | 3 | 36 | 20 | 21 | 21 | 28 | 10 | 84* |
| 182 | 477 | Chogqing/2022/HDG | Chogqing | 3 | 13 | 23 | 44 | 21 | 21 | 1 | 85* |
| 183 | 478 | Chogqing/2022/LHW | Chogqing | 2 | 36 | 23 | 34 | 21 | 18 | 23 | 82* |
Represents new STs.
Multilocus Sequence Typing Genotyping
After submitting the sample sequences to the PubMLST database to comparison, it was found that 130 sample sequences belonged to ST-36, ST-73, ST-75, ST-76, ST-77, and ST-78, while 53 sequences did not match any ST in the database, indicating that these 53 sequences belonged to new STs. After re-annotation by the database administrators, these 53 sequences were divided into ST-81, ST-82, ST-83, ST-84, and ST-85. Among all these genotypes, ST-78 had the highest proportion of 27.32% (50/183), followed by ST-36 with 18.03% (33/183), and then ST-81 with 16.39% (30/183). The detailed information was listed in Table 3.
Table 3.
The number of STs for 183 samples in our study.
| ST | Number | Percentage |
|---|---|---|
| ST-36 | 33 | 18.03% |
| ST-73 | 21 | 11.48% |
| ST-75 | 17 | 9.29% |
| ST-76 | 6 | 3.28% |
| ST-77 | 3 | 1.64% |
| ST-78 | 50 | 27.32% |
| ST-81* | 30 | 16.39% |
| ST-82* | 8 | 4.37% |
| ST-83* | 1 | 0.55% |
| ST-84* | 8 | 4.37% |
| ST-85* | 6 | 3.28% |
Represent new STs.
Allelic Variations of Genotypes
As shown in Table 4, there were 3 allelic variants of the atpG gene, among which atpG-3 was the most common, accounting for 66.67% (122/183). Two allelic variants of the dppC gene, among which dppC-13 was the most common, accounting for 80.33% (147/183). Four allelic variants of the DUF3196 gene, among which DUF3196-23 was the most common, accounting for 35.52% (65/183). Six allelic variants of the lgT gene, among which lgT-44 was the most common, accounting for 48.63% (89/183). Three allelic variants of the mraW gene, among which mraW-21 was the most common, accounting for 54.64% (100/183). Four allelic variants of the plsC gene, among which plsC-9 was the most common, accounting for 43.72% (80/183). Four allelic variants of the ugpA gene, among which ugpA-1 was the most common, accounting for 71.04% (130/183). No new allelic variant sequences were found in our samples.
Table 4.
The alleles of 7 loci and the distribution of these alleles.
| Gene | Allele | Frequency | Percentage | Number of alleles |
|---|---|---|---|---|
| atpG | 2 | 28 | 15.30% | 3 |
| 3 | 122 | 66.67% | ||
| 17 | 33 | 18.03% | ||
| dppC | 13 | 147 | 80.33% | 2 |
| 36 | 36 | 19.67% | ||
| DUF3196 | 4 | 50 | 27.32% | 4 |
| 20 | 41 | 22.40% | ||
| 23 | 65 | 35.52% | ||
| 29 | 27 | 14.75% | ||
| lgT | 20 | 34 | 18.58% | 6 |
| 21 | 8 | 4.37% | ||
| 34 | 29 | 15.85% | ||
| 42 | 17 | 9.29% | ||
| 43 | 6 | 3.28% | ||
| 44 | 89 | 48.63% | ||
| mraW | 19 | 34 | 18.58% | 3 |
| 21 | 100 | 54.64% | ||
| 28 | 49 | 26.78% | ||
| plsC | 9 | 80 | 43.72% | 4 |
| 18 | 62 | 33.88% | ||
| 21 | 6 | 3.28% | ||
| 28 | 35 | 19.13% | ||
| ugpA | 1 | 130 | 71.04% | 4 |
| 10 | 8 | 4.37% | ||
| 18 | 34 | 18.58% | ||
| 23 | 11 | 6.01% |
BURST Analysis
BURST analysis was used for genetic clustering analysis by setting allelic profiles matching at 4 or more loci to any other member of the group. A total of 302 sequences were analyzed, including 183 sequences from this study and 119 reference sequences from domestic and foreign sources obtained through the database. These 302 sequences belonged to 78 STs and were divided into 6 groups and one singleton group (Supplemental Table 1). The number of STs in different groups varied greatly (Supplemental Table 2). Group 3 was the largest group, containing 202 sequences and 21 STs, accounting for 66.89% (202/302) of the total sequences. Group 1 was the second-largest group, containing 39 sequences and 17 STs, accounting for 12.91% (39/302) of the total sequences. The singleton group contained 17 sequences and 15 STs, accounting for 5.63% (17/302) of the total sequences. As shown in Supplemental Table 2, all 183 sequences from this study belonged to Group 3.
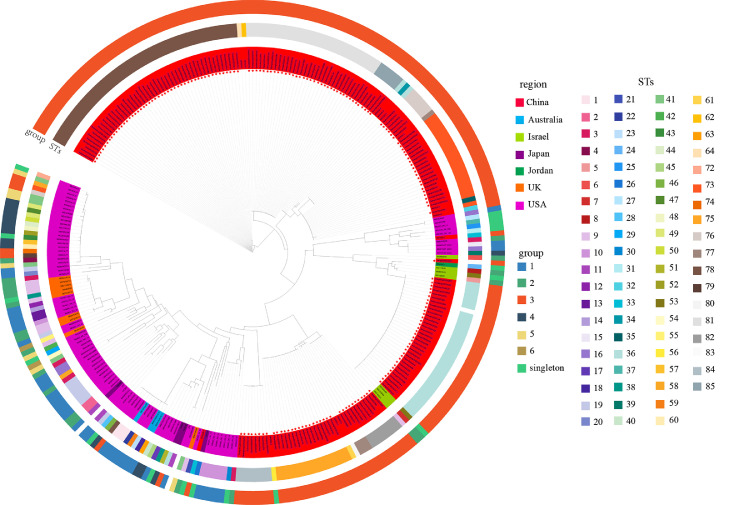
Furthermore, a phylogenetic tree was generated using the concatenated nucleotide sequences of 7 housekeeping genes, and neighbor-joining method was used for clustering analysis of the 302 sequences, among which 196 sequences were originated from China (Figure 2), the 183 sequences from our study were marked with red stars. From this phylogenetic tree, it can be seen that 196 sequences from China were more similar to each other but divided into 4 clades, 39 sequences from China were relatively close to most foreign samples from the United States, the United Kingdom, Japan, and Australia, and 33 sequences from China, 7 from Israel, and 1 from Jordan clustered together and were related to F strain.
Figure 2.
MLST neighbor joining tree of 302 MG samples. The rings (starting from the innermost) contain information on isolate identifiers, the red stars mean the samples collected in this study, the different background colors of isolates represent different groups. The outer rings from inside to outside are ST type and group.
Minimal Inhibitory Concentration
Ten isolated MG strains were randomly selected for drug susceptibility determination. The results are shown in Table 5. All isolates had low MIC values for pleuromutilin, the MICs for tiamulin ranging from 0.015625 to 0.0625 μg/mL and for valnemulin ranging from 0.015625 to 0.03125 μg/mL. The MG isolates displayed variance in MICs for tetracyclines, MIC values for doxycycline ranging from 0.0625 to 1μg/mL, while for chlortetracycline ranging from 8 to 32 μg/mL. Besides, The MG isolates also displayed variant MIC values for macrolides, MICs for tylosin and tylvalosin was similar, 9/10 MG strains for tylosin and 8/10 srtains for tylvalosin ranged from 1 to 4 μg/mL, while the lowest MICs for tilmicosin was 8 μg/mL. The MICs for enrofloxacin ranging from 1 to 4 μg/mL, for spectinomycin ranging from 0.25 to 4 μg/mL. High MIC value was detected in all MG strains for lincomycin ranging from 16 to 128 μg/mL.
Table 5.
MICs of MG strains.
| Strains | MICs (μg/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EN | DO | CH | TIA | VAL | TY | TYL | TIM | SPE | LIN | |
| AH-LJX | 4 | 1 | 32 | 0.015625 | 0.015625 | 1 | 0.25 | 8 | 2 | 128 |
| CQ-LHW | 4 | 0.25 | 16 | 0.03125 | 0.015625 | 2 | 0.5 | 32 | 4 | 32 |
| SC-ZGQ | 4 | 0.125 | 8 | 0.03125 | 0.015625 | 4 | 4 | 128 | 4 | 32 |
| JS-TYH | 2 | 0.25 | 16 | 0.03125 | 0.0625 | 2 | 2 | 32 | 2 | 16 |
| GD-Q3 | 4 | 0.0625 | 16 | 0.03125 | 0.015625 | 1 | 1 | 8 | 0.5 | 16 |
| GD-3-3 | 4 | 0.5 | 32 | 0.03125 | 0.015625 | 32 | 1 | 128 | 2 | 128 |
| GZ-LM | 4 | 1 | 16 | 0.03125 | 0.03125 | 4 | 2 | 32 | 2 | 32 |
| HB-TT | 1 | 0.0625 | 16 | 0.015625 | 0.015625 | 2 | 1 | 32 | 0.25 | 16 |
| GX-DRS | 2 | 0.25 | 16 | 0.0625 | 0.015625 | 4 | 2 | 16 | 2 | 16 |
| FJ-XLN | 4 | 0.125 | 16 | 0.03125 | 0.015625 | 4 | 1 | 32 | 2 | 16 |
Abbreviations: CH, chlortetracycline; DO, doxycycline; EN, enrofloxacin; LIN, lincomycin; SPE, spectinomycin; TIA, tiamulin; TIM, Timicosin; TY, tylosin; TYL, tylvalosin; VAL, valnemulin.
DISCUSSION
MG and MS are the most important Mycoplasma species in global poultry industry, causing significant economic losses (Felice et al., 2020). This study collected 1,250 samples from 15 provinces in China in 2022 to detect the infection rate of MG and MS. Among the 1,250 samples, 938 were positive for MG, with an average positivity rate of 75.04%, and 570 were positive for both MG and MS, with an average positivity rate of 45.60% (Table 1). The situation of MG and MS infection in China is not optimistic and should be given more attention.
MG has become one of the most important pathogens threatening the global poultry industry, causing respiratory and reproductive disorders. Efficient monitoring and epidemiological investigations are crucial. This study used MLST method developed by Ghanem and El-Gazzar (2019) to genotype 183 MG samples. MLST is a validated molecular typing method that has been successfully applied to many bacterial species, including Mycoplasma spp., such as Mycoplasma bovis (Menghwar et al., 2017), Mycoplasma iowae (Ghanem and El-Gazzar, 2016), M. gallisepticum (Ghanem and El-Gazzar, 2019), Mycoplasma hyopneumoniae (Zhang et al., 2021), Mycoplasma agalactiae (McAuliffe et al., 2011), and Mycoplasma hyorhinis (Foldi et al., 2020). We found 5 new STs relative to the database. Among the 183 samples collected in 2022, ST-78 had the highest proportion, followed by ST-36 and ST-81. This study selected 7 housekeeping genes atpG, dppC, DUF3196, lgT, mraW, plsC, and ugpA to classify these 183 samples. All 7 genes had at least 2 alleles, with the highest genetic variability detected in the lgT gene, which had 6 alleles. No new allele sequences were found. By analyzing STs and alleles, we found that the genotype of MG is very diverse in China.
The MLST database (PubMLST) can be accessed for free on the internet and contains reference data provided by other researchers for molecular epidemiology and molecular evolutionary analysis. After BURST analysis, 302 sequences belonging to 78 STs were identified, divided into 6 groups and a singleton group (Supplemental Table 1), of which 196 Chinese sequences belonged to Group 3. Phylogenetic tree analysis revealed that some sequences from China were closely related to foreign sequences, and some sequences had high homology with vaccine strain F. We speculate that the diversity of Chinese MG genotypes is related to international trade and the use of live vaccines.
By measuring the drug resistance of different strains, it is possible to effectively guide the use of medication for treatment. After test, all strains are sensitive to pleuromutilin; there are differences in resistance to fluoroquinolones, tetracyclines, and macrolides drugs. All strains have developed resistance to tiamulin, valnemulin, and doxycycline. This results here are consistent with other research conclusions. However, the representativeness of the 10 strains is relatively poor, and more drug sensitivity tests are needed.
Taken together, this study detected the prevalence of MG infection in 1,250 samples collected from China in 2022, and genotyped 183 MG samples using MLST method based on 7 housekeeping genes, and analyzed the genetic evolutionary relationship between these 183 samples and 119 reference sequences, the genotype of MG in China is diverse. All MG isolated strains are sensitive to pleuromutilin. Through the analysis of epidemiological and molecular epidemiological data continuously, as well as the understanding of the pathogen transmission routes, the use of vaccines and antibiotics, will help to improve the control and eradication plan of MG in China.
ACKNOWLEDGMENTS
This work was supported by the Key Research and Development Program of Guangdong Province (2019B1515210008).
Author Contributions: WXN, ZQ and CF conceived and designed the experiments, carried out the experiments, analyzed the data, and wrote the manuscript. ZQ and LHZ participated in performing experiments. ZQY participated in revising the manuscript. WDA and YZQ collected the tissue samples. All the authors have read and approved the final version of the manuscript.
DISCLOSURES
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102930.
Appendix. Supplementary materials
REFERENCES
- Beko K., Kreizinger Z., Sulyok K.M., Kovacs A.B., Grozner D., Catania S., Bradbury J., Lysnyansky I., Olaogun O.M., Czanik B., Ellakany H., Gyuranecz M. Genotyping Mycoplasma gallisepticum by multilocus sequence typing. Vet. Microbiol. 2019;231:191–196. doi: 10.1016/j.vetmic.2019.03.016. [DOI] [PubMed] [Google Scholar]
- Bradbury J.M., Howell L.J. Delayed growth and atypical colony formation of Mycoplasma synoviae. Res. Vet. Sci. 1974;17:408–410. [PubMed] [Google Scholar]
- Charlton B.R., Bickford A.A., Walker R.L., Yamamoto R. Complementary randomly amplified polymorphic DNA (RAPD) analysis patterns and primer sets to differentiate Mycoplasma gallisepticum strains. J. Vet. Diagn. Invest. 1999;11:158–161. doi: 10.1177/104063879901100209. [DOI] [PubMed] [Google Scholar]
- Delaney N.F., Balenger S., Bonneaud C., Marx C.J., Hill G.E., Ferguson-Noel N., Tsai P., Rodrigo A., Edwards S.V. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feberwee A., de Wit S., Dijkman R. Clinical expression, epidemiology, and monitoring of Mycoplasma gallisepticum and Mycoplasma synoviae: an update. Avian Pathol. 2022;51:2–18. doi: 10.1080/03079457.2021.1944605. [DOI] [PubMed] [Google Scholar]
- Feil E.J., Li B.C., Aanensen D.M., Hanage W.P., Spratt B.G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice V., Lupini C., Mescolini G., Silveira F., Guerrini A., Catelli E., Di Francesco A. Molecular detection and characterization of Mycoplasma gallisepticum and Mycoplasma synoviae strains in backyard poultry in Italy. Poult. Sci. 2020;99:719–724. doi: 10.1016/j.psj.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldi D., Beko K., Felde O., Kreizinger Z., Kovacs A.B., Toth F., Banyai K., Kiss K., Biksi I., Gyuranecz M. Genotyping Mycoplasma hyorhinis by multi-locus sequence typing and multiple-locus variable-number tandem-repeat analysis. Vet. Microbiol. 2020;249 doi: 10.1016/j.vetmic.2020.108836. [DOI] [PubMed] [Google Scholar]
- Ghanem M., El-Gazzar M. Development of multilocus sequence typing (MLST) assay for Mycoplasma iowae. Vet. Microbiol. 2016;195:2–8. doi: 10.1016/j.vetmic.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Ghanem M., El-Gazzar M. Development of a multilocus sequence typing assay for Mycoplasma gallisepticum. Avian Dis. 2019;63(4):693–702. doi: 10.1637/aviandiseases-D-19-00072. [DOI] [PubMed] [Google Scholar]
- Ghanem M., Wang L., Zhang Y., Edwards S., Lu A., Ley D., El-Gazzar M. Core genome multilocus sequence typing: A standardized approach for molecular typing of mycoplasma gallisepticum. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.01145-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giram P., Bhutada P., Prajapati C., Koratkar S.S., Patil S., Hooda D., Rale V., Tongaonkar S.S. Percent positivity and phylogenetic analysis of Mycoplasma gallisepticum and Mycoplasma synoviae in commercial poultry from the different states of India. Vet. World. 2022;15:1843–1851. doi: 10.14202/vetworld.2022.1843-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.X., Chen J.R., Yan H.L., Li X.N., Chen Z.L., Zeng Z.L. Molecular variability of DR-1 and DR-2 within the pvpA gene in Mycoplasma gallisepticum isolates. Avian Dis. 2009;53:124–128. doi: 10.1637/8338-042908-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleven S.H. Control of avian mycoplasma infections in commercial poultry. Avian Dis. 2008;52:367–374. doi: 10.1637/8323-041808-Review.1. [DOI] [PubMed] [Google Scholar]
- Kwong J.C., Mercoulia K., Tomita T., Easton M., Li H.Y., Bulach D.M., Stinear T.P., Seemann T., Howden B.P. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 2016;54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Ponten T., Ussery D.W., Aarestrup F.M., Lund O. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsatanun A., Pakpinyo S., Limpavithayakul K., Prasertsee T. Targeted sequencing analysis of Mycoplasma gallisepticum isolates in chicken layer and breeder flocks in Thailand. Sci. Rep. 2022;12:9900. doi: 10.1038/s41598-022-14066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.S., Arun Prince Milton A., Ghatak S., Ghosh S. Pages 171–181 in Role of Birds in Transmitting Zoonotic Pathogens. Springer Singapore; Singapore: 2021. Avian mycoplasmosis. [Google Scholar]
- Marois C., Dufour-Gesbert F., Kempf I. Molecular differentiation of Mycoplasma gallisepticum and Mycoplasma imitans strains by pulsed-field gel electrophoresis and random amplified polymorphic DNA. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2001;48:695–703. doi: 10.1046/j.1439-0450.2001.00496.x. [DOI] [PubMed] [Google Scholar]
- Matucci A., Stefani E., Gastaldelli M., Rossi I., De Grandi G., Gyuranecz M., Catania S. Molecular differentiation of Mycoplasma gallisepticum outbreaks: a last decade study on Italian farms using GTS and MLST. Vaccines (Basel) 2020;8:665. doi: 10.3390/vaccines8040665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe L., Gosney F., Hlusek M., de Garnica M.L., Spergser J., Kargl M., Rosengarten R., Ayling R.D., Nicholas R.A.J., Ellis R.J. Multilocus sequence typing of Mycoplasma agalactiae. J. Med. Microbiol. 2011;60(Pt 6):803–811. doi: 10.1099/jmm.0.028159-0. [DOI] [PubMed] [Google Scholar]
- Menghwar H., He C., Zhang H., Zhao G., Zhu X., Khan F.A., Faisal M., Rasheed M.A., Zubair M., Memon A.M., Ridley A., Robertson I.D., Chen Y., Guo A. Genotype distribution of Chinese Mycoplasma bovis isolates and their evolutionary relationship to strains from other countries. Microb. Pathog. 2017;111:108–117. doi: 10.1016/j.micpath.2017.08.029. [DOI] [PubMed] [Google Scholar]
- Michiels T., Welby S., Vanrobaeys M., Quinet C., Rouffaer L., Lens L., Martel A., Butaye P. Prevalence of Mycoplasma gallisepticum and Mycoplasma synoviae in commercial poultry, racing pigeons and wild birds in Belgium. Avian Pathol. 2016;45:244–252. doi: 10.1080/03079457.2016.1145354. [DOI] [PubMed] [Google Scholar]
- Norouzian H., Farjanikish G., Hosseini H. Molecular characterisation of Mycoplasma gallisepticum isolates from Iran in the period 2012-2017. Acta Vet. Hung. 2019;67:347–359. doi: 10.1556/004.2019.036. [DOI] [PubMed] [Google Scholar]
- Raviv Z., Kleven S.H. The development of diagnostic real-time TaqMan PCRs for the four pathogenic avian mycoplasmas. Avian Dis. 2009;53:103–107. doi: 10.1637/8469-091508-Reg.1. [DOI] [PubMed] [Google Scholar]
- Sawicka A., Durkalec M., Tomczyk G., Kursa O. Occurrence of Mycoplasma gallisepticum in wild birds: a systematic review and meta-analysis. PLoS One. 2020;15 doi: 10.1371/journal.pone.0231545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid H., Benachour K., Rautenschlein S. Co-infection with multiple respiratory pathogens contributes to increased mortality rates in Algerian poultry flocks. Avian Dis. 2015;59:440–446. doi: 10.1637/11063-031615-Case.1. [DOI] [PubMed] [Google Scholar]
- Sprygin A.V., Andreychuk D.B., Elatkin N.P., Zinyakov N.G., Kolosov S.N., Mudrak N.S., Irza V.N., Drygin V.V., Borisov A.V., Perevozchikova N.A. Genetic diversity of Mycoplasma gallisepticum field isolates using partial sequencing of the pvpA gene fragment in Russia. Avian Dis. 2010;54:899–904. doi: 10.1637/8989-070909-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Staley M., Bonneaud C., McGraw K.J., Vleck C.M., Hill G.E. Detection of Mycoplasma gallisepticum in house finches (Haemorhous mexicanus) from Arizona. Avian Dis. 2018;62:14–17. doi: 10.1637/11610-021317-Reg.1. [DOI] [PubMed] [Google Scholar]
- Yadav J.P., Tomar P., Singh Y., Khurana S.K. Insights on Mycoplasma gallisepticum and Mycoplasma synoviae infection in poultry: a systematic review. Anim. Biotechnol. 2021;33:1–10. doi: 10.1080/10495398.2021.1908316. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang Y., Gao L., Wang Y., Wei R. Genotype diversity of Mycoplasma hyopneumoniae in Chinese swine herds based on multilocus sequence typing. BMC Vet. Res. 2021;17:347. doi: 10.1186/s12917-021-03059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Guo M., Xie D., Chen Y., Zhang C., Cao Y., Wu Y. Antibiotic resistance of Mycoplasma synoviae strains isolated in China from 2016 to 2019. BMC Vet. Res. 2022;18:1. doi: 10.1186/s12917-021-03104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.