Abstract
A fastidious, slowly growing, strictly aerobic, gram-negative bacterium was isolated from a culture of blood from a 25-year-old man with common variable immunodeficiency. The man had been admitted to hospital with febrile progressive cerebellar ataxia. The use of standard phenotypic schemes did not lead to identification, but sequence analysis demonstrated that the 16S rRNA gene of the isolate was most similar to those of the environmental bacteria Duganella zoogloeoides (formerly Zoogloea ramigera 115) and Telluria mixta. Further characterization of the bacterium by biochemical analysis, electron microscopy, G+C content estimation, and fatty acid analysis demonstrated significant differences between the bacterium and D. zoogloeoides and Telluria species; thus, we propose it as a new taxon with the name Massilia timonae gen. nov., sp. nov.
Children and young adults with severe antibody deficiencies such as X-linked gammaglobulinemia, common variable immunodeficiency, and X-linked hyper-immunoglobulin M syndrome are prone to recurrent infections which can affect different organ systems (17). Severe or recurrent pulmonary infections due to Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma spp., and Neisseria meningitidis are especially common, although gastrointestinal infections caused by Salmonella, Shigella, and Campylobacter spp.; joint infections caused by Mycoplasma and Ureaplasma spp.; and often fatal chronic meningitis caused by enterovirus are also encountered.
Here we report on the isolation of a novel gram-negative bacterium from the blood of an immunocompromised patient with meningoencephalitis. Polyphasic taxonomic analysis of this bacterium led to us to conclude that it warranted placement in a new genus, in line with contemporary taxonomic guidelines which reconcile the use of 16S rRNA gene sequence analysis with phenotypic and genotypic characterizations (13). The newly characterized organism has been named Massilia timonae gen. nov., sp. nov.
CASE REPORT
The patient from whom the strain was isolated was a 25-year-old man with common variable immunodeficiency (17). He was not human immunodeficiency virus positive but had been splenectomized when he was 10 years old and had subsequently been treated with gamma globulin infusions. He was admitted to hospital with progressive cerebellar ataxia and low-grade fever (temperature, 38.5°C). Brain imaging indicated a focal lesion in the right cerebellar white matter. The lesion was hypodense on a computed tomographic scan and hyperintense on a magnetic resonance imaging scan. The lesion had no obvious mass effect, and an intravenous injection of contrast material did not reveal any enhancement. Three sets of blood and cerebrospinal fluid (CSF) specimens for culture were collected prior to antibiotic therapy. CSF examination performed at admission yielded 4 cells/mm3 (mononuclear), a protein level of 0.46 g/liter, and a glucose level of 3.3 mmol/liter. Histologic examination of the CSF yielded no abnormal cells. Blood and CSF were negative for cryptococcal antigen. Direct examination of the CSF by Gram staining and Ziehl-Neelsen staining was negative. Cultures of CSF and blood were negative on standard media, mycobacterial media, mycoplasma media, and fungal media. Culture of the CSF for viruses was negative, as were PCR-based assays for the detection of enterovirus, herpes simplex virus, and JC virus. A few hours after admission, ceftriaxone and antimycobacterial chemotherapy were begun. Antimycobacterial chemotherapy was discontinued after 5 days, and he remained with ceftriaxone therapy for 3 weeks.
After 4 months, during which he remained afebrile and which saw a slow improvement of his ataxia and a regression of his cerebellar lesion, he suddenly developed a high-grade fever (temperature, 39.5°C) and renewed ataxia which led to his readmission to hospital 48 h later. A computed tomographic scan on admission revealed an enlargement of the cerebellar lesion. The bacterial and viral investigations carried out during his first admission were repeated with clinical specimens collected on the day of admission. In total, three separate collections of blood and CSF were done at approximately hourly intervals from the time of his admission. CSF examination yielded 2 cells/mm3 (mononuclear), a protein level of 0.57 g/liter, and a glucose level of 2.7 mmol/liter. Direct examination of the CSF by Gram staining and Ziehl-Neelsen staining was negative. The results of all bacterial and viral investigations remained negative with the exception of those for one blood culture, which led to the isolation of a gram-negative bacillus. Antibiotic therapy, initiated after collection of the third set of samples on the day of the patient’s admission, included imipenem and amikacin for 3 weeks. He became apyrexic within 3 days of the start of the treatment, and his ataxia continued to improve slowly. Two years later he remains well with only a cicatricial lesion in the cerebellar white matter.
MATERIALS AND METHODS
Phenotypic study.
A sample of the patient’s blood was inoculated into a BACTEC aerobic bottle (NR 6-A*), and the bottle was incubated in a BACTEC NR-860 automated instrument (Becton Dickinson Diagnostic Instrument Systems, Sparks, Md.). Growth was detected 3 days later, blood culture broth was subcultured onto 5% sheep blood Columbia agar and chocolate agar plates, and these plates were incubated at 37°C. The morphological properties of the isolate were studied by Gram staining and Ziehl-Neelsen staining. For electron microscopy, bacteria were harvested from Trypticase soy medium (BioMerieux, Marcy l’Etoile, France), were pelleted by centrifugation, and were prefixed for 1 h at room temperature with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) containing 0.1 M sucrose. After washing overnight with the same buffer, the bacteria were fixed for 1 h at room temperature with 1% osmium tetroxide in 0.1 M cacodylate buffer, dehydrated through increasing concentrations (25 to 100%) of ethanol, and then embedded in Epon 812. Thin sections were cut out and poststained with a saturated solution of methanol-uranyl acetate and lead citrate in water before examination on a JEOL JEM 1200 EX electron microscope. The presence of flagella was assessed by depositing the bacteria on Formvar film and staining with a 0.33% solution of uranyl acetate.
Physiological characterization of the isolate, named the Timone isolate, was initially studied by plating onto Trypticase soy agar (BioMerieux), chocolate agar (BioMerieux), 5% sheep blood Columbia agar (BioMerieux), and MacConkey agar (BioMerieux). Growth was studied at 4 and 37°C. A motility test was performed, and respiratory type was assessed by incubation at 28°C in 0.2% agar Shaëdler broth (BioMerieux). Catalase activity was detected by emulsifying a colony in 30% hydrogen peroxide and checking for the presence of microscopic bubbles. Oxidase activity was detected with a dimethyl-para-phenylenediamine oxalate disk (Diagnostic Pasteur, Marnes, France). Further biochemical tests were performed by inoculation of API 20NE and API 20A systems (BioMerieux) according to the manufacturer’s instructions, but incubation was done under aerobic conditions at 28°C for 24 h. Carbon source utilization tests were performed by inoculation of the Biotype-99-carbon source strip (BioMerieux) with Biotype 2 assimilation medium (BioMerieux) and incubation at 28°C.
Cell wall fatty acid composition was analyzed by gas chromatography as described previously (12) with a 24-h-old culture of the Timone isolate grown on Trypticase soy agar (Becton Dickinson, Meylan, France).
Chromosomal DNA extraction, PCR, and sequencing of 16S rRNA gene.
DNA was extracted from a heavy suspension of the isolate with the QIAamp blood kit (Qiagen, Hilden, Germany) under the conditions stipulated by the manufacturer. The DNA extract was used as a template in a PCR incorporating the broad-spectrum eubacterial 16S rRNA gene primers fD1 and rP2 (purchased from Eurogentec, Seraing, Belgium) under previously described conditions (11, 21). The success of the amplification was determined by ethidium bromide staining, following the resolution of products by 1% agarose gel electrophoresis. Each experiment included sterile water (no DNA) as a negative control and Escherichia coli DNA as a positive control.
In preparation for base sequence determination, the amplification product was purified with Microspin S-400 HR columns (Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer’s instructions. Cycle sequencing reactions were prepared with the Amplicycle sequencing kit (Perkin-Elmer), according to the manufacturer’s instructions, and incorporated 1 of the 10 primers listed in Table 1. If the estimated annealing temperature of the primer was ≥50°C, the thermal cycle used consisted of an initial denaturation step at 95°C for 1 min, which was followed by 25 cycles of denaturation at 95°C for 30 s, annealing for 30 s, and extension at 72°C for 1 min. The amplification was completed by extension for 5 min at 72°C to allow full extension of the amplified products. If the estimated annealing temperature of the primer was <50°C, an initial denaturation step at 95°C for 1 min was followed by 30 cycles of denaturation at 95°C for 30 s, annealing for 30 s, and extension at 60°C for 2 min and then 10 supplementary cycles of denaturation at 95°C for 10 s and extension at 60°C for 90 s. A Perkin-Elmer 9600 thermal cycler was used for the sequencing reactions. Reaction products were resolved by electrophoresis on 6% READIMIX polyacrylamide gels (Pharmacia Biotech), and sequence data were determined with an ALF Automated Sequencer and related software (Pharmacia Biotech).
TABLE 1.
Summary of primers used for sequencing in this study
| Sequencing primers | Sequences (5′ to 3′) | Hybridization temp (°C) |
|---|---|---|
| fD1 | -AGAGTTTGATCCTGGCTGAG- | 55 |
| rP2 | -ACGGCTACCTTGTTACGACTT- | 55 |
| 200fti | -GCTAGTTGGTGAGGTAAAGC- | 55 |
| 200rti | -CCTTTACCTCACCAACTAGC- | 55 |
| 500rti | -CTTTACGCCCAGTAATTCCG- | 55 |
| 800f | -ATTAGATACCCTGGTAG- | 50 |
| 800r | -CTACCAGGGTATCTAAT- | 50 |
| 1050f | -TGTCGTCAGCTCGTG- | 43 |
| 1050r | -CACGAGCTGACGACA- | 43 |
| 1200f | -TATGTCCTGGGCTACACACG- | 57 |
Sequence analysis.
Sequence data derived from each primer were compared, aligned, and combined into a single, almost complete 16S rRNA gene sequence by using the program PC gene (Intelligenetics, Geneva, Switzerland). The validity of the primary sequence obtained was assessed by reference to theoretical base pairing in the stems of a standard eubacterial secondary structure model. The sequence was compared with all eubacterial 16S rRNA sequences available in the GenBank database by using the multisequence comparison program FASTA, which is part of the BISANCE software package (6). The 16S rRNA gene sequence of the Timone isolate, together with those found to be most similar in the FASTA analysis, and sequences of other members of the class Proteobacteria were then aligned by using CLUSTAL W (9) supported in BISANCE. The resulting alignment was first edited by removal of sequences at the 5′ and 3′ ends of longer sequences so that their lengths matched that of the shortest sequence. Next, ambiguous base positions were taken out, and finally, any gaps in the alignment, which corresponded to recognized highly variable loop regions in the 16S rRNA secondary structure model (data not shown), were removed to yield a final alignment suitable for use in phylogenetic reconstructions. The data were analyzed by using the distance matrix, parsimony, and maximum likelihood programs (DNADIST/NEIGHBOUR JOINING, DNAPARS, and DNAMLK, respectively) of the PHYLIP (8) package, again supported in BISANCE. The stability of reconstructions inferred by each method were assessed by generation of 100 bootstrap samples (DNABOOT) and construction of strict majority rule consensus trees (CONSENSE).
G+C content analysis.
Bacterial colonies grown on Trypticase soy agar were gently scraped from the plate, and DNA was extracted as described previously (2). Estimations were performed by high-pressure liquid chromatography with a model 46200A system pump (Merck Clevenot, Nogent sur Marne, France). An aliquot of 5 μl of the hydrolysate was applied onto the Nucleosil 5C18 Lichrocart column (4 by 250 mm; Merck). Elution was carried out at room temperature with a mixture of 0.2 M NH4H2PO4 (pH 4.5) and acetonitrile (96:4 [vol/vol]). A flow rate of 1 ml/min was used, and the absorbance was monitored at 270 nm. The calibration curve was obtained from a mixture of four standard nucleotides (5 nmol/ml in distilled water; Sigma Chemical Co., St. Louis, Mo.). After chromatography, the relative concentration of each nucleotide was calculated on the basis of the peak area in the high-pressure liquid chromatography elution profile and was corrected as described previously (18). Determinations of G+C contents were repeated five times.
Antibiotic susceptibility tests.
Antibiotic susceptibility tests were performed by the disk diffusion method on Mueller-Hinton II agar (BioMerieux) after a 24-h incubation at 37°C under aerobic conditions (1). The MICs of the antibiotics listed in Table 2 were determined.
TABLE 2.
MICs of selected antibiotics for the Timone isolate as determined by the disk diffusion method
| Antibiotic | MIC (μg/ml) |
|---|---|
| Penicillin G | 8 |
| Amoxicillin | 4 |
| Ticarcillin | 2 |
| Piperacillin | 4 |
| Amoxicillin-clavulanate | 4 |
| Ticarcillin-clavulanate | 2 |
| Imipenem | 0.5 |
| Cephalothin | 16 |
| Cefotaxime | 0.125 |
| Ceftazidime | 1 |
| Gentamicin | 0.5 |
| Tobramycin | 2 |
| Netilmicin | 0.25 |
| Amikacin | 1 |
| Trimethoprim-sulfamethoxazole | <0.03 |
| Pefloxacin | <0.125 |
| Ofloxacin | <0.125 |
| Ciprofloxacin | <0.125 |
| Rifampin | 0.25 |
| Fosfomycin | <32 |
| Colistin | <2 |
Nucleotide sequence accession number.
The 16S rRNA gene sequence of Massilia timonae has been deposited in GenBank under accession no. U54470.
RESULTS
Phenotypic study.
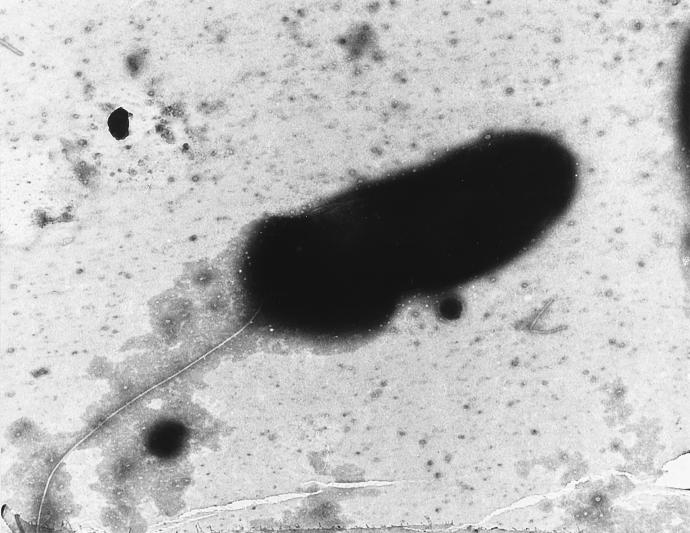
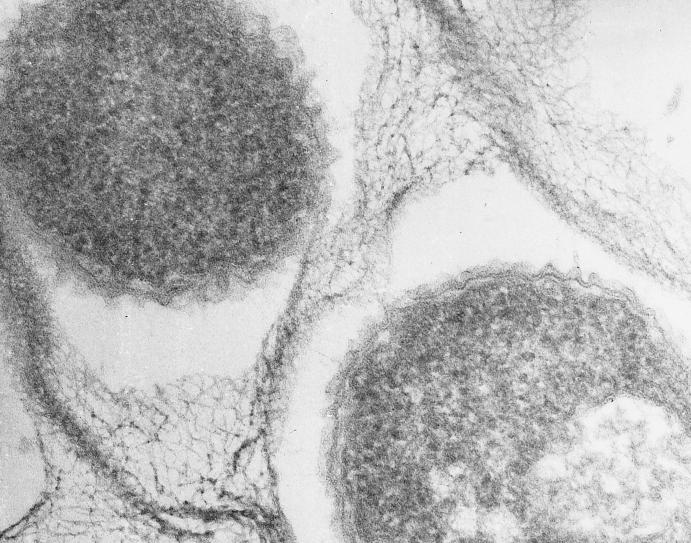
The Timone isolate was found to be a gram-negative, rod-shaped organism with a single polar flagellum (Fig. 1). The tendency of this bacterium to aggregate, its morphology, and the structure of its cell wall, which was typical of that of gram-negative bacteria, were confirmed by electron microscopy (Fig. 2). Growth was first detected in an automated blood culture system after 3 days. Colonies were detected on subculture after 48 h. On subsequent passage, the colonies reached 1 mm in diameter after 24 h of incubation and appeared circular, completely opaque, and pale yellow. Straw-colored, mature colonies are distinctively tenacious and could be lifted intact from the agar surface with a needle. Colonies of the isolate were apparent after 24 h at 28 and 37°C on Trypticase soy agar, 5% sheep blood Columbia agar (best growth), and MacConkey agar, incubated with or without 5% CO2. No growth was obtained at 4°C. Bacterial growth had a tendency to form pellicles at the surface of liquid medium, with the bacteria embedded in a gelatinous matrix (Fig. 2), but never strictly finger-like formations. The Timone isolate was only actively motile in young cultures cultivated in liquid media. Growth only occurred aerobically. The results of biochemical and physiological assessments are presented in Table 3. Because phylogenetic analysis of the Timone isolate demonstrated that it shares the most evolutionary relatedness with environmental organisms which grow optimally at 28°C, all phenotypic tests of the Timone isolate were carried out with strains grown at this temperature to ensure that the observed differences were not the result of physiological variation induced by different growth temperatures.
FIG. 1.
Electron micrograph of M. timonae showing the polar flagellum. Magnification, ×12,000.
FIG. 2.
Electron micrograph of M. timonae embedded in an exopolymer matrix (arrows) showing the cell wall of a gram-negative-type organism. Magnification, ×60,000.
TABLE 3.
Characteristics of M. timonae, T. mixta, and D. zoogloeoides
| Characteristic | M. timonae | T. mixta | D. zoogloeoides |
|---|---|---|---|
| Flagellar arrangement | |||
| Monotrichous only | + | + | + |
| Monotrichous and polytrichous | |||
| Zoogloeae are produced | − | − | + |
| Straw-colored colonies | + | − | + |
| Oxidase production | − | + (weak) | + |
| Catalase production | + | + | + |
| Hydrolysis of gelatin | + | + | + |
| Hydrolysis of esculin | + | + | + |
| Arginine dihydrolase activity | + | − | − |
| Growth in 3% NaCl | − | − | − |
| Growth on MacConkey agar | + | − | + (weak) |
| Denitrification | − | − | − |
| Urease activity | − | + | + |
| Hydrolysis of starch | + | + | + |
| Acid formed from rhamnose and mannitol | − | − | − |
| Acid formed from glucose, xylose, arabinose, sucrose, maltose, cellobiose, glycogen, lactose, and mannose | − | − | + |
| Utilized as sole carbon sources: | |||
| α-Lactose, d-galacturonate, protocatechuate, gentisate, α-ketoglutarate d-mannose, maltose, d-cellobiose, l-arabinose, d-xylose, l-malate, fumarate, malonate | + | + | + |
| α-l-Rhamnose, d-tagatose, d-malate, d-glucuronate, succinate, l-alanine | + | + | + |
| α-Hydroxybutyrate, citrate, l-aspartate, d-glucose, d-fructose, d-galactose, sucrose, l-tartrate, l-aspartate, l-glutamate, d-alanine | − | + | + |
| d-Trehalose, d-melezitose, d-saccharate, mucate, dl-lactate, dl-glycerate, l-serine | − | + | − |
| G+C DNA content (mol%) | 64.61 ± 1.78 | 64–70 | 63–64 |
The cell wall fatty acid pattern was C10:0 (0.68%), C11:0 (0.28%), C10:0 3OH (5.66%), C12:0 (6.49%), C13:0 (0.27%), C12:0 2OH (2.77%), C14:0 (3.3%), C15:0 (0.96%), C16:1 ω7c (48.28%), C16:0 (21.27%), C17:1 ω8c (0.48%), C17:1 ω6c (0.52%), C17:0 (0.39%), C18:1 (8.14%).
PCR amplification and sequence analysis of a 16S rRNA gene of the Timone isolate.
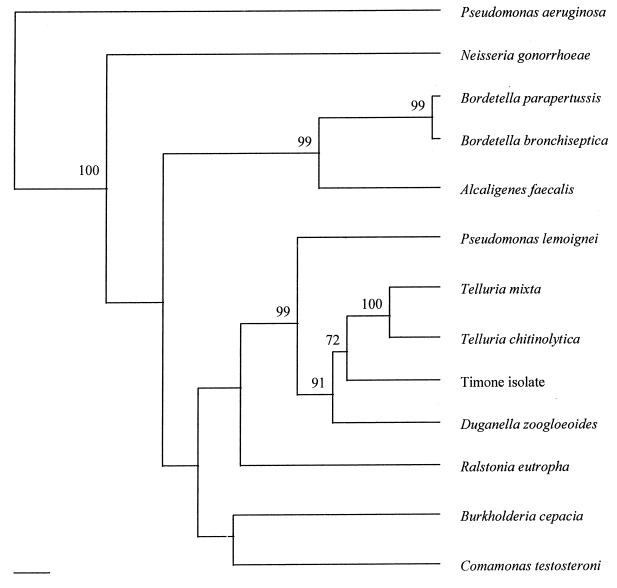
Amplification with the fD1 and rP2 primer set yielded a product of approximately 1,500 bp. The sequencing primers used in this study allowed determination of 1,494 bp of the sequence. When aligned and compared with 16S rRNA gene sequences available in the GenBank database, the organisms with the most closely related 16S rRNA sequences were Duganella zoogloeoides (94.6%), Telluria mixta (94.4%), and Telluria chitinolytica (92.9%). Following construction and editing of the 16S rRNA sequence alignment, 1,382 bp remained for phylogenetic analysis. Parsimony and maximum likelihood inferral methods yielded trees on which the proposed branching orders of the β subdivision of the class Proteobacteria around the Timone isolate were indistinguishable (Fig. 3). Neighbor joining analysis varied slightly, clustering the Timone isolate with D. zoogloeoides. However, whereas the branching orders proposed by the former two analyses had strong support, as demonstrated by high bootstrap values at the relevant nodes (Fig. 3), that proposed by neighbor joining was only weakly supported. As a result of these observations, we considered the architecture of the parsimony- and maximum likelihood-derived trees to be more reliable than that of the distance matrix tree, and our conclusions were based on that consideration.
FIG. 3.
Unrooted phylogenetic tree based on comparison of 16S rRNA gene sequences demonstrating the evolutionary relationships between M. timonae and closely related members of the β subdivision of the class Proteobacteria. The tree was inferred by a maximum likelihood method with molecular clock (DNAMLK). Numbers are percent probabilities obtained with 100 bootstrapped runs for individual nodes. The scale bar represents 1% divergence.
G+C content analysis.
The mean ± standard deviation moles percent G+C content of the DNA of the Timone isolate was 64.61 ± 1.78.
Antibiotic susceptibility tests.
The results of antibiotic susceptibility tests are presented in Table 2. The MICs of cefotaxime, the fluoroquinolones, and trimethoprim-sulfamethoxazole were especially low.
DISCUSSION
We isolated a previously undescribed member of the β subdivision of the class Proteobacteria from the blood of a French man with a common variable immunodeficiency who had been admitted to hospital with acute-onset fever and progressive cerebellar ataxia associated with cerebellar lesions. The clinical presentation of the syndrome suggested the involvement of an infectious agent, and there was strong evidence that the agent involved was the Timone isolate; (i) recovery of the Timone isolate from the patient’s blood coincided with an acute, high fever, (ii) despite thorough efforts, no other organism could be recovered or detected in appropriately timed blood and CSF specimens, and (iii) pre- and postrelapse antibiotic prescriptions were effective in treating the syndrome. Unfortunately, due to his agammaglobulinemic state, the patient was unable to mount an immune response; thus, we were unable to verify the involvement of the Timone isolate by serological methods.
Analysis of the isolate based on comparison of 16S rRNA gene sequences demonstrated that it shares the highest degree of similarity with members of the genus Telluria and D. zoogloeoides (formerly Zooglea ramigera 115). In order to assess the taxonomic relationship between the Timone isolate and these species, we subjected it to polyphasic characterization on the basis of phylogenetic, phenotypic, and genotypic assessments. Our phylogenetic reconstructions demonstrate that the Timone isolate is a previously unrecognized organism sharing a line of common evolutionary descent with members of the genera Telluria and Duganella. The evolutionary homology of these organisms is reflected in genotypic and phenotypic similarities. All have high moles percent G+C contents (63 to 70) and possess similar physiologies, as demonstrated by their performances in biochemical tests, their growth requirements, and their microscopic appearances. However, although closely related, Telluria spp. and D. zoogloeoides and the Timone isolate have diverged sufficiently to exhibit phenotypic differences which warrant their allocation to distinct taxa (4, 5, 10). The Timone isolate is therefore best accommodated as a new species within a new genus, and we proposed it as the taxon Massilia timonae.
Telluria and Duganella species have never been implicated as causes of human infections. Both genera have been encountered only environmentally, being isolated from soil. The phylogenetic position of the Timone isolate within a cluster of soil-living bacteria suggests that it, too, may naturally reside in such an environment; however, the fact that the Timone isolate is within this cluster is not sufficient to deduce with any certainty that it really does inhabit such a habitat. As elsewhere among members of the class Proteobacteria, organisms adapted to markedly different environments can lie in very close evolutionary proximity. Bartonella and Brucella species, for example, lie among a large cluster of plant-associated bacteria within the α2 subdivision of the class Proteobacteria (3). We therefore know nothing about the source of this infection or the basis of its pathogenicity.
During the last two decades, the increasing number of immunocompromised patients who, following improved methods of treatment and prophylaxis, now survive longer has created a subpopulation which is particularly susceptible to numerous opportunistic pathogens. Improvements in the techniques for bacterial isolation and identification have also led to the description of new pathogens that previously have been uncultivable or unidentifiable by standard microbiological procedures (14, 15). M. timonae is one such organism, and it is likely that many more will be identified over the next few years.
Although patients with severe common variable immunodeficiency have been successfully treated with intramuscular immunoglobulin for many years, many still suffer opportunistic infections, especially when they have been splenectomized (as was the case for our patient). We can hypothesize that the common preparation of immunoglobulin used for substitutive therapy is probably devoid of antibodies against rarely opportunistic pathogens, thus leaving the recipient susceptible to organisms which are only rarely encountered elsewhere in medical microbiology (17).
The improved quality of the PCR reagents and equipment and the automation of DNA sequencing methodologies have greatly enhanced the user friendliness of these techniques and thus their suitability for more routine applications. These methods, together with the availability of large, public-domain molecular sequence databases, have permitted the development of a new approach to the identification of disease-associated microorganisms, based on analysis of amplified 16S rRNA gene sequences. Such an approach has already been used in the identification of both uncultured pathogens and less fastidious but unexpected organisms which fail to be identified or detected by commonly used phenotypic schemes (11, 14, 15, 19). By molecular methods, a taxonomically accurate identity for this isolate was quickly obtained. It is unlikely that such an achievement would have been straightforward and quick by traditional phenotypic methods. A more widespread introduction of these new methods to complement existing schemes is likely to help in the identification of numerous other unexpectedly pathogenic organisms.
Description of Massilia timonae gen. nov., sp. nov.
Massilia is taken from the old Greek and Roman name for Marseille, Massilia, where our hospital is situated; timonae is taken from the name Hôpital de la Timone. Gram-negative cells are straight rods that are 1.0 μm wide and 3.0 μm long. Occasionally, filamentous cells are formed in old cultures and are especially formed at 37°C. Growth occurs on MacConkey agar. The organism does not form spores. A single polar flagellum is formed on each cell. The organism is actively motile, especially in young cultures. It is strictly aerobic and forms straw-colored colonies. It has a tendency to form flocs and films in liquid medium without finger-like projections. It is oxidase negative and catalase positive. Acid is not formed from carbohydrates. It is arginine dihydrolase positive, urease negative, esculin positive, and proteolytic on gelatin. Denitrification does not occur. Additional characteristics are listed in Table 2. The moles percent G+C content of the DNA of the strain is 64.61 ± 1.78. The organism was isolated from a culture of blood from a febrile patient with common variable immunodeficiency. The type strain of M. timonae is strain UR/MT95, which has been deposited in the Collection de l’Institut Pasteur, Paris, France, as strain CIP 105350. Its 16S rRNA gene sequence has been lodged within the GenBank sequence database under accession no. U54470.
REFERENCES
- 1.Acar J F, Goldstein F W. Disk susceptibility test. In: Lorian V, editor. Antibiotics in laboratory medecine. 4th ed. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 1–51. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 2.4.1–2.4.5. [Google Scholar]
- 3.Birtles R J, Harrison T G, Saunders N A, Molyneux D H. Proposals to unify the genera Grahamella and Bartonella, with description of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Bowman J P, Hayward A C, Sly L I. Pseudomonas mixta sp. nov., a bacterium from soil with degradative activity on a variety of complex polysaccharides. Syst Appl Microbiol. 1988;11:53–59. [Google Scholar]
- 5.Bowman J P, Sly L I, Hayward A C, Spiegel Y, Stackebrandt E. Telluria mixta (Pseudomonas mixta Bowman, Sly, and Hayward 1988) gen. nov., comb. nov., and Telluria chitinolytica sp. nov., soil-dwelling organisms which actively degrade polysaccharides. Int J Syst Bacteriol. 1993;43:120–124. doi: 10.1099/00207713-43-1-120. [DOI] [PubMed] [Google Scholar]
- 6.Dessen P, Frondat C, Valencien C, Munier G. BISANCE: a French service for access to biomolecular sequences databases. Comput Appl Biosci. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- 7.Dugan P R, Stoner D L, Pickrum H M. The genus Zoogloea. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 3952–3964. [Google Scholar]
- 8.Felsenstein J. PHYLIP: phylogeny inference package, version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 9.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 10.Hiraishi A, Shin Y K, Sugiyama J. Proposal to classify Zoogloea ramigera IAM 12670 (P. R. Dugan 115) as Duganella zoogloeoides gen. nov., sp. nov. Int J Syst Bacteriol. 1997;47:1246–1248. doi: 10.1099/00207713-47-4-1249. [DOI] [PubMed] [Google Scholar]
- 11.La Scola B, Michel G, Raoult D. Use of amplification and sequencing of the 16S rRNA gene to diagnose Mycoplasma pneumoniae osteomyelitis in a patient with hypogammaglobulinemia. Clin Infect Dis. 1997;24:1161–1163. doi: 10.1086/513631. [DOI] [PubMed] [Google Scholar]
- 12.Miller L, Berger T. Bacterial identification by gas chromatography of whole cell fatty acids. Hewlett-Packard application note 228-241. Avondale, Pa: Hewlett-Packard; 1985. [Google Scholar]
- 13.Murray R G E, Brenner D J, Colwell R R, De Vos P, Goodfellow M, Grimont P A D, Pfennig N, Stackebrandt E, Zavarzin G A. Report of the Ad Hoc Committee on Approaches to Taxonomy within the Proteobacteria. Int J Syst Bacteriol. 1990;40:213–215. [Google Scholar]
- 14.Relman D A, Loutit J S, Schmidt T M, Falkow S, Thompkins L S. The agent of bacillary angiomatosis: an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 15.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 16.Shin Y K, Hiraishi A, Sugiyama J. Molecular systematics of the genus Zoogloea and emendation of the genus. Int J Syst Bacteriol. 1993;43:826–831. doi: 10.1099/00207713-43-4-826. [DOI] [PubMed] [Google Scholar]
- 17.Spickett G P, Misbah S A, Chapel H M. Primary antibody deficiency in adults. Lancet. 1991;337:281–284. doi: 10.1016/0140-6736(91)90882-p. [DOI] [PubMed] [Google Scholar]
- 18.Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–128. [Google Scholar]
- 19.Tee W, Dyall-Smith M, Woods W, Eisen D. Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J Clin Microbiol. 1996;34:1760–1764. doi: 10.1128/jcm.34.7.1760-1764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unz R F. Genus IV. Zoogloea Itzigsohn 1868, 30AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 214–219. [Google Scholar]
- 21.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]