Abstract
We evaluated the COBAS AMPLICOR PCR system (Roche Diagnostics) for the routine detection of Mycobacterium tuberculosis complex (MTBC) in clinical specimens. Diagnostic culture, considered as the reference method, was performed with BACTEC, Löwenstein-Jensen, Stonebrink, and Kirchner media. Occasionally MB-Redox, ESP, or MGIT medium was also used. A total of 643 respiratory and 506 nonrespiratory specimens collected from 807 patients were investigated. Of the 95 culture-positive specimens, 80 were COBAS AMPLICOR MTB positive, and of the 1,054 culture-negative specimens, 1,044 were COBAS AMPLICOR MTB negative. After resolving discrepancies by review of the medical history, the overall sensitivity, specificity, and positive and negative predictive values for the COBAS AMPLICOR MTB assay, respectively, were 83.5, 98.8, 86.7, and 98.6% compared to those of diagnostic culture. In smear-positive specimens, the sensitivity of the COBAS AMPLICOR MTB assay was 96%, versus 48% for smear-negative specimens. No significant differences in the test performance between respiratory and nonrespiratory specimens were observed. The overall inhibition rate was less than 2%, excluding stool specimens. The clear advantages of the COBAS AMPLICOR PCR system are standardized procedures and reagents for specimen processing as well as an internal control for reliable monitoring of PCR inhibitors. By simplifying the work flow through a completely automated amplification and amplicon detection procedure, the COBAS AMPLICOR PCR system proved itself as a very useful component for routine diagnostic procedures.
Clinical mycobacteriology laboratories play an important role in the control of the spread of tuberculosis (TB) through the timely detection, isolation, identification, and drug susceptibility testing of Mycobacterium tuberculosis. In the past, the mycobacteriology laboratory has had to rely on diagnostic methods that often do not rapidly give a definitive diagnosis of tuberculosis.
Conventional methods include the acid-fast stain, culture, and biochemical tests for detecting and identifying members of the M. tuberculosis complex (MTBC). Even with concentrated samples, the sensitivity of microscopy is not great, and M. tuberculosis cannot be reliably differentiated from other mycobacteria in acid-fast-stained smears. Although the use of fluorescent (auramine-rhodamine) stains increases the sensitivity and shortens the time required for reading of smears (6), microscopy, as a direct method, can provide at best only a preliminary diagnosis.
Cultural methods, properly applied, detect M. tuberculosis in clinical samples with reasonable sensitivity and provide accurate identification of the isolates. However, these methods are quite slow, requiring 3 to 8 weeks for completion, primarily because of the slow growth of the mycobacteria (13, 19). Once the presence of mycobacteria is indicated, additional testing is required to identify the species. There is thus an urgent need for a rapid, safe, and verifiable method to establish the diagnosis of TB.
Substantial changes in methodology have occurred with the widespread use of nonradioactive DNA probes for culture identification and, most recently, the application of highly sensitive in vitro nucleic acid amplification techniques for the direct detection and specific identification of mycobacteria in clinical specimens (3, 6, 25, 27). Although a number of amplification methods, such as ligase chain reaction, strand displacement amplification, and transcription-based amplification, have been developed in the emerging field of molecular diagnostics, the original and most widely applied amplification method is the PCR.
In the past few years, several research groups have described different PCR systems for the direct detection of MTBC or performed clinical studies based on PCR which had widely different results with respect to specificity and sensitivity (4, 7, 12, 15, 17, 21, 24, 27). Possible reasons for these differences may be the selection of the patient and specimen collection as well as methodological differences concerning sample preparation and DNA isolation. Moreover, different clinical parameters were used to evaluate PCR results in relation to the microbiological and clinical findings. This complicates the direct comparison of the results and an estimation of the true clinical value of PCR-based assay systems for the diagnosis of TB infections.
One aim of the present study was to evaluate the recently available COBAS AMPLICOR PCR system for the detection of MTBC in well-characterized clinical specimens and to compare the results with clinical classification and conventional culture and staining techniques. Other important goals were the assessment of the test performance for a broad collection of nonrespiratory specimens and the rating of the impact on laboratory management by integrating an automated test into the work flow of a routine diagnostic laboratory.
MATERIALS AND METHODS
Patients and clinical specimens.
In the scope of an open prospective study from February 1996 to March 1997, a total of 1,149 clinical specimens were selected from samples sent to our routine laboratory for MTBC testing. The specimens were collected from 807 patients with clinical signs or symptoms of pulmonary or extrapulmonary TB or in order to exclude the possibility of TB infection.
The specimens could be divided into two major groups: 643 respiratory specimens (sputa, bronchial and tracheal aspirates, bronchial secretions, bronchial washings, and bronchoalveolar lavages) and 506 specimens of nonrespiratory origin (ascitic fluids, biopsies, blood, bone marrow aspirates, cerebrospinal fluids, gastric secretions, aspirates, stool samples, urine samples, abscess fluids, and wound swabs).
Processing of specimens.
All specimens were decontaminated by the N-acetyl-l-cysteine (NALC)-NaOH method (11). Two volumes of NALC-NaOH solution (4% NaOH, 1.45% Na-citrate, 0.5% NALC) were mixed with the specimen on a test tube mixer for digestion, and the mixtures were allowed to stand for 15 min at room temperature. Ten volumes of 6.7 mM phosphate buffer (pH 7.4) were added for dilution, and the mixtures were centrifuged at 3,000 × g for 15 min. The sediment was resuspended in approximately 1.5 ml of the same phosphate buffer containing 0.5% Tween 80. Tween 80 was added to achieve better homogenization of the sediment and a more even distribution of the bacteria within the suspension. A 100-μl aliquot of the suspension was directly processed for PCR, and the remainder was inoculated onto culture media and used for acid-fast staining.
Microscopy.
Fixed smears were stained with auramine-rhodamine fluorochrome as a screening method. Positive slides with acid-fast bacilli were confirmed to be positive by Ziehl-Neelsen staining (10).
Culture.
The major portion of the processed sediment (0.5 ml) was cultivated by the radiometric BACTEC technique with the BACTEC 460 instrument. Each vial containing BACTEC Middlebrook 7H12 medium (Becton Dickinson Diagnostic Instrument Systems, Sparks, Md.) was supplemented with 0.1 ml of PANTA (polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin). In addition, Kirchner medium was inoculated with 150 μl of the sediment, Löwenstein-Jensen medium was inoculated with 150 μl, and Stonebrink medium was inoculated with 100 μl. Occasionally, MB-Redox medium (Biotest AG, Dreieich, Germany), ESP MycoII medium (Difco GmbH, Augsburg, Germany), or MGIT medium (Becton Dickinson) was used instead of Kirchner medium. Slants and vials were incubated at 37°C for up to 8 weeks and inspected for growth twice a week for the first 3 weeks and weekly thereafter. The radiometric growth index of the 7H12 vials was recorded twice a week for 6 weeks. A growth index of >50 was considered positive. Isolates were identified by use of Accuprobe culture confirmation tests (Gen-Probe, Inc., San Diego, Calif.), 16S ribosomal DNA sequencing (22), or standard procedures (13).
Roche COBAS AMPLICOR PCR.
The Roche COBAS AMPLICOR PCR was done according to the manufacturer’s instructions. It consists of two steps: specimen preparation and combined amplification and detection. A detailed description of the technical procedure is given in reference 9. In brief, a 100-μl aliquot of the sediment sample was mixed with wash solution and centrifuged (14,000 × g) for 10 min. After centrifugation, the supernatant was removed and lysis reagent was added to the pellet. After being vortexed, the suspension was incubated at 60°C for 45 min to complete alkaline lysis of the mycobacteria. The lysed material was then neutralized by the addition of neutralizing reagent. For amplification, 50 μl of the neutralized specimen (200 μl) was added to 50 μl of the master mix reagent. The latter contains uracil N-glycosylase enzyme, which allows safe pre-PCR enzymatic decontamination of dU-containing PCR products (20), nucleotides, biotinylated primers, Taq DNA polymerase, and a synthetic internal control (IC). The IC nucleic acids (DNA plasmid) contain primer binding regions identical to those of the target sequence and a unique probe binding region that differentiates the IC from amplified mycobacterial target nucleic acid (18). In the COBAS AMPLICOR test, the IC is used at a concentration of 20 copies per test sample to indicate that amplification was sufficient to generate a positive signal from targets present at the limit of test sensitivity. It thereby increases sensitivity by enabling the user to identify and eventually retest samples inhibitory to PCR. During the course of the study, one M. tuberculosis-positive control and one M. tuberculosis-negative control were tested per 12-tube amplification ring. Following on-system amplification, the instrument added denaturation reagent to each PCR tube and a reagent containing an M. tuberculosis-specific oligonucleotide probe bound to paramagnetic microparticles to separate detection cups. After addition of denatured sample or control to the detection cups, reaction mixtures were washed four times and transferred to the incubator. Colorimetric detection of the amplicons was mediated by avidin-horseradish peroxidase. An absorbance reading greater than 0.350 optical density (OD) units was considered positive for the presence of MTBC DNA.
Analysis of discrepant results.
Diagnostic culture was considered as the “gold standard.” In those cases in which culture results were discrepant from the COBAS AMPLICOR MTB results, clinical data and other results obtained with additional specimens from the patient were considered. Assessment of each patient’s clinical picture included the patient’s history, symptoms, chest X ray, tuberculin skin test result, and history of drugs administered, whenever those data were available.
Statistical analysis.
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the COBAS AMPLICOR MTB assay were calculated by contrasting the PCR results with the culture results, which were considered as the reference. Statistical comparison was performed by using chi-square analysis.
RESULTS
The COBAS AMPLICOR MTB PCR system was evaluated for its ability to detect organisms of the MTBC in 643 respiratory and 506 nonrespiratory clinical specimens from 807 patients received in our routine laboratory for initial diagnosis or follow-up of mycobacterial infections. The distribution of the patients according to their clinical grouping and an initial comparison of PCR results versus clinical classification are depicted in Tables 1 and 2. Positive PCR results were obtained for only 14 of 705 patients who had no active TB disease (groups 1 and 2 in Table 2) but were obtained for 81 of 102 patients with a clinically active TB infection (groups 3 to 5 in Table 2).
TABLE 1.
Clinical classification of patients in this studya
| Group | Status | No. of patients |
|---|---|---|
| 1 | Exclusion of TB; negative tuberculin skin test; smear and culture negative; definitive other diagnosis obtained by bacteriological culture, histologically, or on the basis of clinical presentation | 521 |
| 2 | TB infection, smear and culture negative, not clinically active (positive tuberculin skin test and/or history of tuberculosis, definitive other diagnosis) | 184 |
| 3 | TB infection, smear and culture negative, clinically active (positive tuberculin skin test; history of TB; clinical, histological, or radiological signs of active disease; exclusion of other definitive diagnosis; improvement under treatment with antitubercular chemotherapy) | 15 |
| 4 | TB infection, smear negative, culture positive | 44 |
| 5 | TB infection, smear and culture positive | 43 |
The clinical classification of patients was carried out on the basis of information provided by the treating physicians, when available, according to the recommendations of the American Thoracic Society (presented in accordance with reference 2).
TABLE 2.
Comparison of PCR results on initial testing and clinical classification
| Clinical groupa | No. (%) of patients with PCR resultb
|
|
|---|---|---|
| Positive | Negative | |
| 1 (no TB) | 10 (2) | 511 (98) |
| 2 (TB history) | 4 (2) | 180 (98) |
| 3 (TB positive) | 8 (50) | 8 (50) |
| 4 (TB, culture positive) | 31 (70) | 13 (30) |
| 5 (TB, culture and smear positive) | 43 (100) | |
The clinical classification is given in Table 1.
PCR was scored positive if the COBAS AMPLICOR reading was measured above the cutoff value.
Next to the classification obtained from the doctors in charge of the cases, the clinical performance of the COBAS AMPLICOR MTB assay was determined by comparison of the results of the assay with those of standard culture by using an expanded panel of culture media and acid-fast-staining methods.
Of the 1,149 specimens, 96 specimens were culture positive for M. tuberculosis. Of these, 69 specimens (71%) were smear positive, and 27 specimens (28%) were smear negative. Seventy-six of the 90 COBAS AMPLICOR MTB-positive specimens were culture positive for M. tuberculosis. Thirty-five specimens (18 smear-negative specimens) were culture positive for mycobacteria other than M. tuberculosis (MOTT). The species of MOTT identified from these specimens were M. avium-M. intracellulare complex (6 specimens), M. chelonae (4 specimens), M. haemophilum (3 specimens), M. interjectum (2 specimens), M. kansasii (3 specimens), and M. xenopi (17 specimens). A total of 1,054 specimens (12 smear positive) were culture negative.
The amplification results for all kinds of specimens are summarized in Table 3. Of the 1,149 specimens tested, 90 specimens (65 smear positive and 25 smear-negative) were COBAS AMPLICOR MTB assay positive, and 1,059 specimens (17 smear positive and 1,042 smear-negative) were COBAS AMPLICOR MTB assay negative, including 77 specimens showing an inhibition of the amplification reaction (5 specimens culture positive for M. tuberculosis). Of the 35 specimens with MOTT isolates, 33 were COBAS AMPLICOR MTB assay negative.
TABLE 3.
Distribution of species and comparison of the initial COBAS AMPLICOR MTB results with culture results
| Type of specimen | No. of specimens | No. of specimens with result
|
||||
|---|---|---|---|---|---|---|
| PCR Positive | PCR Negative | Inhibitiona | False positiveb | False negativec | ||
| Bronchoalveolar lavage | 383 | 17 | 366 | 3 | 3 | 2 |
| Bronchial secretion | 32 | 3 | 29 | 1 | 1 | |
| Bronchial washing | 8 | 0 | 8 | 0 | ||
| Tracheal aspirate | 15 | 2 | 13 | 0 | 1 | |
| Sputum | 205 | 31 | 174 | 4 | 7 | |
| Abscess fluid | 4 | 1 | 3 | 1 | ||
| Ascitic fluid | 25 | 1 | 24 | 1 | 1 | 1 |
| Aspirate | 19 | 4 | 15 | 0 | 1 | 1 |
| Biopsy | 35 | 3 | 32 | 2 | ||
| Blood | 71 | 5 | 66 | 4 | 3 | 1 |
| Bone marrow aspirate | 6 | 1 | 5 | 0 | ||
| Cerebrospinal fluid | 77 | 3 | 74 | 0 | 1 | 1 |
| Gastric secretion | 55 | 6 | 49 | 4 | 1 | |
| Pleural exudate | 69 | 3 | 66 | 1 | 1 | 1 |
| Stool | 69 | 0 | 69 | 54 | ||
| Urine | 57 | 6 | 51 | 1 | 1 | |
| Wound swab | 19 | 4 | 15 | 1 | 1 | |
| Total | 1,149 | 90 | 1,059 | 77 | 14 | 15 |
Reading of the built-in PCR inhibition control was negative.
PCR-positive, culture-negative specimens were initially classified as false positives.
PCR-negative, culture-positive specimens were initially classified as false negatives.
As depicted in Table 3, 29 discrepant results were observed. Fourteen M. tuberculosis culture-negative specimens (1 smear positive and 13 smear negative) were COBAS AMPLICOR MTB assay positive and thus were initially considered as false positives. A detailed analysis of the patients clinical history revealed that nine of these specimens (smear negative) came from tumor patients who underwent X-ray therapy or chemotherapy. Two specimens (one smear positive and one smear negative) with a weak positive COBAS reading had a positive culture for M. xenopi. Of the remaining three specimens (smear negative), two specimens were obtained from patients who had subsequently positive cultures for M. tuberculosis and had received antibiotic treatment for 6 months or had been previously treated 1 year ago, respectively. Based on these findings, the latter were reclassified as true-positive PCR results (Table 4).
TABLE 4.
Analysis of 14 specimens from 13 patients culture negative for M. tuberculosis and positive by COBAS AMPLICOR MTBc
| Type of specimen | Clinical groupa | Smear result | Comments | Final interpretation of PCR result |
|---|---|---|---|---|
| Bronchoalveolar lavage | 1 | Positive | No evidence of TB, grew M. xenopi, OD of 0.5b | False positive |
| 1 | Negative | No evidence of TB, nasopharyngeal lymphoma | False positive | |
| 1 | Negative | No evidence of TB, positive tuberculin skin test, bronchogenic carcinoma | False positive | |
| Bronchial secretion | 1 | Negative | No evidence of TB, grew M. xenopi, OD of 0.6b | False positive |
| Tracheal aspirate | 1 | Negative | No evidence of TB, positive tuberculin skin test, bronchogenic carcinoma, OD of 4.0b | False positive |
| Ascitic fluid | 3 | Negative | On treatment for 26 wk, severe lung disease, subsequent specimen culture positive, OD of 1.3b | True positive |
| Aspirate | 1 | Negative | No evidence of TB, bronchogenic carcinoma | False positive |
| Blood | 1 | Negative | No evidence of TB, liver carcinoma, also urine specimen false positive (see below) | False positive |
| 1 | Negative | No evidence of TB, bronchogenic carcinoma | False positive | |
| 1 | Negative | No evidence of TB, bronchogenic carcinoma | False positive | |
| Cerebrospinal fluid | 1 | Negative | No evidence of TB, clinical symptoms of meningitis | False positive |
| Gastric secretion | 3 | Negative | Previously treated for TB 1 yr ago, persistent cavity, subsequent culture positive | True positive |
| Pleural exudate | 1 | Negative | No evidence of TB, bronchogenic carcinoma | False positive |
| Urine | 1 | Negative | No evidence of TB, liver carcinoma, also blood specimen false positive (see above) | False positive |
The clinical classification is given in Table 1.
COBAS AMPLICOR reading.
The interpretive criteria for the COBAS AMPLICOR MTB assay reading were as follows: OD of ≥0.35, positive; OD of <0.35, negative.
Twenty M. tuberculosis culture-positive specimens (5 smear positive and 15 smear negative) were COBAS AMPLICOR MTB assay negative after repeated testing, and thus they were initially considered as false negatives. Of these, the DNA preparation of five specimens (three smear positive and two smear negative) showed an inhibition of the amplification reaction after repeated testing, and hence these results were not considered in the statistical analysis. Of the remaining 15 specimens (13 smear negative and 2 smear positive), 12 specimens exhibited only positive culture results after a period of ≥20 days of incubation, which indicates the presence of a small number of mycobacteria in the clinical specimen examined (Table 5). This is in agreement with the patients’ clinical history, indicating that 9 of the 13 patients are currently receiving or have previously received antibiotic treatment. The results for two specimens originating from patients without any clinical signs of TB infection (clinical group 1 in Table 5) were reclassified as true negatives. In one case, the AccuProbe M. tuberculosis test (Gen-Probe, Inc.) for culture confirmation was negative, and since subsequent specimens received from both patients remained culture negative, culture contaminations could be suspected.
TABLE 5.
Analysis of 15 specimens from 13 patients culture positive for M. tuberculosis and negative for COBAS AMPLICOR MTB
| Type of specimen | Clinical groupa | Smear result | No. of days to positive cultureb | Comments | Final interpretation of PCR result |
|---|---|---|---|---|---|
| Bronchoalveolar lavage | 4 | Negative | 22 (B) | On treatment for 14 wk, 2 of 3 specimens PCR positive | False negative |
| 4 | Negative | 22 (B) | Previously treated for TB 2 yr ago, clinical signs of reactivated disease, subsequent specimens PCR positive | False negative | |
| Sputum | 4 | Positive | 35 (B) | On treatment for 4 wk, active TB | False negative |
| 4 | Negative | 20 (B) | On treatment for 12 wk, active TB | False negative | |
| 1 | Negative | 32 (B) | No evidence of TB, presence of MOTT excluded, possible culture contamination, subsequent specimens culture negative | True negative | |
| 4 | Negative | 14 (B) | On treatment for 12 wk, clinical data unavailable | False negative | |
| 4 | Negative | 18 (B) | On treatment for 3 wk, radiological and clinical signs of active TB | False negative | |
| 4 | Negative | 30 (ESP) | Previously treated for TB 3 yr ago, human immunodeficiency virus positive, clinical signs of active TB | False negative | |
| 4 | Positive | 45 (B) | On treatment for 8 wk, active TB | False negative | |
| Ascitic fluid | 4 | Negative | 24 (B) | On treatment for 16 wk, severe lung disease | False negative |
| Aspirate | 4 | Negative | 21 (B) | On treatment for 9 wk, clinical signs of active TB | False negative |
| Blood | 4 | Negative | 35 (B) | On treatment for 14 wk, subsequent specimens PCR positive | False negative |
| Cerebrospinal fluid | 4 | Negative | 7 (Ki) | Clinical data unavailable | False negative |
| Pleural exudate | 1 | Negative | 33 (B) | No evidence of TB, APr negativec, presence of MOTT suspected, subsequent cultures negative | True negative |
| Wound swab | 4 | Negative | 26 (B) | Skin TB, two consecutive specimens culture positive | False negative |
The clinical classification is given in Table 1. Four culture-positive stool specimens and one pleural exudate which showed an inhibition in the PCR assay are not listed.
B, BACTEC; Ki, Kirchner medium; ESP, ESP Myco II medium.
APr, Accuprobe M. tuberculosis test (Gen-Probe, Inc., San Diego, Calif.).
In summary, given that the overall positivity rate of the study collection was 11.9% (96 of 1,149 specimens), and omitting the 5 culture-positive specimens showing an inhibition in the PCR assay, the sensitivity, specificity, PPV, and NPV were 83.5, 98.8, 86.7, and 98.6%, respectively, for the COBAS AMPLICOR MTB assay. Divided up by respiratory, nonrespiratory, smear-positive, and smear-negative specimens, the values were calculated as 95.4, 90.9, 97.7, and 83.3%, respectively, for smear-positive respiratory specimens; 50.0, 99.3, 60.0, and 98.9%, respectively, for smear-negative respiratory specimens; 100, 100, 100, and 100%, respectively, for smear-positive nonrespiratory specimens; and 61.5, 98.4, 53.3, and 98.4%, respectively, for smear-negative nonrespiratory specimens (Table 6).
TABLE 6.
Comparison of confirmed results by the COBAS AMPLICOR MTB assay and culture in 643 respiratory and 506 nonrespiratory specimens
| COBAS AMPLICOR MTB specimen group | No. of specimens with culture result
|
% Sensitivitya | % Specificitya | % PPVa | % NPVa | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | |||||
| All | |||||||
| Positive | 76 | 14 | 90 | ||||
| Negative | 20b | 1,039 | 1,059 | 83.5 | 98.8 | 86.7/96.7c | 98.6 |
| Total | 96b | 1,053 | 1,149 | ||||
| Respiratory | |||||||
| Smear positive | |||||||
| Positive | 42 | 1 | 43 | ||||
| Negative | 2 | 10 | 12 | 95.4 | 90.9 | 97.7 | 83.3 |
| Total | 44 | 11 | 55 | ||||
| Smear negative | |||||||
| Positive | 6 | 4 | 10 | ||||
| Negative | 7 | 571 | 578 | 50.0 | 99.3 | 60.0/85.7c | 98.9 |
| Total | 13 | 575 | 588 | ||||
| Nonrespiratory | |||||||
| Smear positive | |||||||
| Positive | 22 | 0 | 22 | ||||
| Negative | 3b | 2 | 5 | 100 | 100 | 100 | 100 |
| Total | 25b | 2 | 27 | ||||
| Smear negative | |||||||
| Positive | 6 | 9 | 15 | ||||
| Negative | 8b | 456 | 464 | 61.5 | 98.4 | 53.3/88.8c | 98.4 |
| Total | 14b | 465 | 479 | ||||
Considering the final interpretation of PCR results (Tables 4 and 5) and excluding five culture-positive specimens which showed an inhibition in the PCR assay.
Including three smear-positive and two smear-negative, culture-positive specimens which showed an inhibition in the PCR assay.
Excluding culture-negative specimens originating from tumor patients.
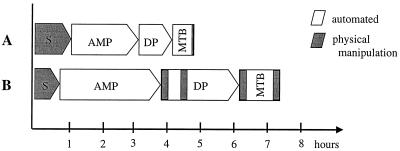
A workload assessment clearly demonstrates the savings in both hands-on time and total assay time by using the automated COBAS AMPLICOR PCR system instead of manual or semiautomated assay formats. A comparison of a typical in-house PCR assay with an enzyme-linked oligosorbent assay-based method of detection of specific amplification products (16) is depicted in Fig. 1.
FIG. 1.
Comparison of the workload organization of the COBAS AMPLICOR MTB assay (A) and a conventional PCR assay with that of an enzyme-linked oligosorbent assay-based amplicon detection (B). S, sample preparation; AMP, amplification; DP, detection process; MTB, analytic run.
DISCUSSION
The laboratory diagnosis of mycobacterial infections by culture of the organisms usually requires 1 to 8 weeks. The increased incidence of tuberculosis has stimulated the development of rapid assays to detect M. tuberculosis directly in clinical samples. Since M. tuberculosis is usually not found in clinical samples in the absence of infection, the detection of M. tuberculosis nucleic acids should be diagnostic as well. In recent years, various in vitro nucleic acid amplification systems for the highly sensitive detection and specific identification of mycobacterial species have been reported. Clinical evaluations of the Roche AMPLICOR MTB PCR assay have shown high specificity for direct detection of MTBC in respiratory specimens (4, 5, 9, 15, 21, 24, 27), smear-positive nonrespiratory specimens (5, 15, 24), and BACTEC 12B cultures (23, 27). In addition, the successful application of several other standardized commercial detection assays for the detection of MTBC has been reported (1, 2, 8, 14, 25, 28). Following resolution of discrepant results, the average sensitivity and specificity of these assays were about 82 and 99%, respectively.
Most of these assays were shown to be rapid and specific, but their methodologies are not well suited for use in routine clinical laboratories. In principle, every in vitro nucleic acid amplification test format consists of three typical steps: (i) nucleic acid preparation from the clinical specimen, (ii) amplification of the target sequence, and (iii) the specific detection of the amplicons. Although most of the currently available commercial assays rely on special devices and contain prefabricated buffer and reagent solutions, most of the manual work has to be performed stepwise, and the work flow is repeatedly interrupted by a number of necessary incubation and washing steps. With respect to the hands-on time and the laboratory’s work flow, an automated device would be highly desirable. The analytical concept behind the COBAS AMPLICOR PCR system is based on the manual AMPLICOR PCR system, but both amplification and detection were fully automated (9). The COBAS AMPLICOR PCR system is the first automated nucleic acid amplification test in which the laboratory technician was able to walk away after loading the detection reagents and placing the specimens in the thermal cycler module.
In our study, we evaluated the performance of the COBAS AMPLICOR PCR system over a 1-year period for the detection of MTBC in respiratory and nonrespiratory clinical specimens sent to our laboratory for routine MTBC testing. In contrast to hospitals specializing in pulmonary disorders, a considerable portion of our specimens are of nonrespiratory origin. Therefore, we decided to include every kind of specimen in the study. A total of 643 respiratory and 506 nonrespiratory specimens collected from 807 patients were investigated. Every sample was subjected to standard NACL decontamination, and a 100-μl aliquot of the 1.5-ml sediment suspension was transferred to PCR. Some studies with clinical specimens suffer from sampling problems commonly encountered with small loads of mycobacteria as a result of their tendency to clump together. We found that addition of 0.5% Tween 80 to the phosphate buffer was advantageous in achieving much better homogenization of the sediment and a more even distribution of the bacteria within the suspension.
Diagnostic culture, considered as the reference method, was performed with well-established BACTEC, Löwenstein-Jensen, Stonebrink, and Kirchner media. In the course of a clinical evaluation, the recently developed MB-Redox, ESP, and MGIT media were occasionally used as well. Although we routinely used two liquid and two solid media for diagnostic culture (the use of one liquid medium and two solid media is recommended in Germany), a good overall correlation of PCR results with culture results was observed. Based on the culture results, and excluding five culture-positive specimens which showed an inhibition of the PCR assay, the diagnostic sensitivity and specificity of the COBAS AMPLICOR MTB assay were determined to be 84.2 and 99.1%, respectively, for respiratory specimens and 82.3 and 98.0%, respectively, for nonrespiratory specimens. Previously published data on respiratory specimens are in agreement with our findings: the sensitivities and specificities of the manual AMPLICOR MTB assay ranged from 66.7 to 86% and from 97 to 99.6%, respectively (5, 7, 12, 21). Conclusively, the results obtained by the COBAS AMPLICOR MTB assay with nonrespiratory specimens are in good accordance with those obtained with respiratory specimens.
As with each of the commercially available amplification tests for direct detection of MTBC from clinical specimens, the COBAS AMPLICOR MTB assay showed reduced performance with smear-negative specimens.
In comparison with diagnostic culture, 14 false-positive results and 15 false-negative results were observed with the COBAS AMPLICOR MTB assay. Of the false positives, a strikingly high number came from tumor patients who underwent X-ray therapy or chemotherapy (9 of 14 specimens). Here it may be speculated that the radical treatment partially destroys the lung tissue, and as a consequence, mycobacteria (or mycobacterial nucleic acids) originating from primary complexes caused by an early MTBC infection of the patients are dislocated throughout the organism and are also circulating in the blood (three PCR-positive blood specimens and one urine specimen). Unfortunately there were no previous clinical data available to support an MTBC history for these patients. Because of the exceptionally high sensitivity of the PCR, mycobacterial DNA could be detected and may give rise to positive PCR results while culture fails. Automated DNA sequencing of the amplification products confirmed the presence of MTBC DNA in these cases. An alternative interpretation would be a therapy-induced damage of the proliferation activity of the mycobacteria (like in the case of antibiotic treatment), leading to negative culture results in these cases. Positive results obtained by MTB PCR for culture-negative blood samples have been reported previously (17). Since all of these specimens were smear negative and the patients have not developed clinical signs of MTBC infection, we considered them true false-positive PCR results. They could, however, represent false-negative culture results. This observation should be considered for the clinical interpretation of positive PCR results obtained for specimens originating from tumor patients. They may be due to nonviable mycobacteria or the presence of mycobacterial DNA in specimens from patients without clinical symptoms of an MTBC infection.
Two PCR-positive specimens had a positive culture only for M. xenopi. The culture result was confirmed by 16S ribosomal DNA sequencing, and the absence of M. tuberculosis DNA in these specimens was confirmed by an in-house PCR based on the IS6110 target sequence. Thus it is unlikely that these patients were dually infected with M. xenopi and M. tuberculosis. False-positive results with the Amplicor MTB assay on specimens culture positive for M. xenopi have also been reported by Wobeser et al. (27). The ODs of these specimens tended to be lower than those of specimens containing M. tuberculosis but were measured above the assay cutoff value. Since our M. xenopi-positive specimens had COBAS AMPLICOR MTB readings of 0.5 and 0.6, respectively, this lack of specificity could initially be overcome by raising the assay cutoff value before the biological basis for this potential cross-reactivity is investigated in more detail. Considering that 15 M. xenopi-positive specimens gave negative PCR results, a general cross-reactivity of the M. tuberculosis-specific assay with M. xenopi is thus unlikely.
Of the remaining three false-positive specimens, two specimens originated from patients with a history of anti-TB therapy for periods of 4 and 6 months, respectively. It is well-known that antibiotic treatment interferes with culture but not with nucleic acid analysis, and these specimens cannot be considered as true false positives. Taking these clinical facts into account, there are three specimens remaining which can be considered as true PCR false positives.
We obtained false-negative PCR results for 20 specimens, which remained PCR negative even after repeated testing. Of these, the IC of the COBAS AMPLICOR MTB assay had a negative reading with the DNA preparations of five specimens (four stool specimens and one pleural exudate), indicating the presence of inhibitory substances for the amplification process. Of the remaining 15 specimens (13 smear negative and 2 smear positive), 13 specimens exhibited only positive culture results after a period of more than 20 days of incubation, indicating a small number of mycobacteria in the clinical specimen examined. Negative results obtained by amplification assays for culture-positive samples are usually explained by the presence of inhibitors of enzymatic amplification, a low number of mycobacteria, and/or an unequal distribution in the test suspension. Because we are routinely adding Tween 80 to the suspension buffer to obtain an as even as possible distribution of the bacteria within the suspension and because the COBAS AMPLICOR MTB assay has an internal control for monitoring the presence of PCR inhibitors, the failure of PCR to detect the mycobacteria in the specimens mentioned above is probably due to their small number in the corresponding specimens. This supposition is supported by the facts that (i) 9 of the 13 patients are currently under treatment or have received antitubercular chemotherapy and (ii) 13 of the 15 specimens were smear negative.
Taking into account that the portion (0.5 ml) of the processed sediment transferred to the BACTEC Middlebrook 7H12 medium was five times higher than the aliquot processed for PCR, “aliquotation effects” may give rise to false-negative PCR results. Here the number of false negatives could be further reduced by raising the portion of the original sediment introduced to PCR testing.
Although we used a variety of clinical specimens as starting material, the portion of inhibited amplification reactions was low. Except for stool specimens, which showed an inhibition rate of 78%, an overall inhibition rate of less than 2% was observed. There was no significant difference between the inhibition rates of respiratory specimens (1%) and those of nonrespiratory specimens (2.9%). With respect to several in-house PCR assays, it is important to mention that the sensitivity of the assay would have been considerably lower without having a built-in inhibition control available. Use of the internal control increases sensitivity, because repeatedly inhibitory specimens are uninterpretable and were therefore excluded from the sensitivity calculations. When the internal control results were ignored, these specimens were classified as false negatives. Furthermore, a positive IC result indicates that amplification has occurred and thus provides assurance that negative test results are truly negative. This is also an important feature when testing nonvalidated types of specimens. Because the COBAS AMPLICOR MTB assay exhibited low rates of inhibition (with the exception of stool samples), consideration of the internal control resulted, at least in our study, in very moderate improvements in test sensitivity and positive predictive value. The excellent performance with respect to inhibition may be partly due to an especially careful execution of the decontamination procedure in our routine laboratory practice. However, removal of inhibitory substances should be subject to further improvement of the assay procedure before the specimen range of the test kit can be extended to stool samples. Various inhibitors in fecal specimens have been suspected to produce false-negative results. Such inhibitors include heme and its metabolic products, bile salts, bilirubin, and polysaccharides (26). There are several different strategies that claim to be able to remove these inhibitors from DNA preparations, including solvent extractions, protein salting-out, and solid-phase DNA binding. Which one of these strategies will turn out to be efficient and can be optionally introduced into the protocol of the test kit has to be determined.
Since the appearance of nucleic acid-based test systems, the primary question has been the sensitivity of these assays, which with smear-negative samples has been less than that of culture. Because of the necessity for susceptibility testing and epidemiological characterization of isolates, it is beyond any doubt that the current amplification assays cannot substitute for culture. However, the ability to detect MTBC in most samples within a few hours rather than in 2 to 6 weeks seems to be a considerable advance from the practical point of view. Samples that are negative by the direct test may still prove positive by culture, and a negative test result should not be interpreted as ruling out TB. Although increased sensitivity would be desirable, the clinical studies demonstrate that some of the currently available amplification assays are rapid and specific enough to reliably detect those patients who are most likely to be infectious.
In summary, although the exact role of amplification assays for diagnosis of MTBC infections has yet to be determined, the COBAS AMPLICOR MTB assay was shown to be rapid, sensitive, and specific for the detection of MTBC from smear-positive specimens originating from a variety of respiratory as well as nonrespiratory sources. Furthermore, our results suggest a potential role for the COBAS AMPLICOR MTB assay in evaluating smear-negative specimens, since PPVs of above 85% could be observed with smear-negative specimens if tumor patient specimens were excluded. Clear advantages of the prefabricated test kit are the standardized procedures and reagents for specimen processing and amplification (reproducibility) as well as an internal control for monitoring the presence of PCR inhibitors (reliability). From the point of view of a technician, a significant reduction in hands-on time can be experienced in addition to the reduction in total assay time by the truly automated amplification and detection procedure. In cases in which emergency testing is requested, reaction mixtures can be completed in the late afternoon and results are available the next morning. Because of its completely automated amplification and detection procedure, the COBAS AMPLICOR MTB assay proved to be well suited for a clinical microbiology laboratory’s work flow.
ACKNOWLEDGMENTS
We thank Stefan Emler for active support and Sabine Philippi-Schulz for critically reading the manuscript, and we gratefully acknowledge the excellent technical assistance of Birgit Haber, Eveline Lang, and Michaela Pöhlmann during the study.
REFERENCES
- 1.Ausina V, Gamboa F, Gazapo E, Manterola J M, Lonca J, Matas L, Manzano J R, Rodrigo C, Cardona P J, Padilla E. Evaluation of the semiautomated Abbott LCx Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis in respiratory specimens. J Clin Microbiol. 1997;35:1996–2002. doi: 10.1128/jcm.35.8.1996-2002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beige J, Lokies J, Schaberg T, Finckh U, Fischer M, Mauch H, Lode H, Köhler B, Rolfs A. Clinical evaluation of a Mycobacterium tuberculosis PCR assay. J Clin Microbiol. 1995;33:90–95. doi: 10.1128/jcm.33.1.90-95.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodmer T, Gurtner A, Scholkmann M, Matter L. Evaluation of the COBAS AMPLICOR MTB system. J Clin Microbiol. 1997;35:1604–1605. doi: 10.1128/jcm.35.6.1604-1605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpentier E, Drouillard B, Dailloux M, Moinard D, Vallee E, Dutilh B, Maugein J, Bergogne-Berezin E, Carbonnelle B. Diagnosis of tuberculosis by Amplicor Mycobacterium tuberculosis test: a multicenter study. J Clin Microbiol. 1995;33:3106–3110. doi: 10.1128/jcm.33.12.3106-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford J T. New technologies in the diagnosis of tuberculosis. Semin Respir Infect. 1994;9:62–70. [PubMed] [Google Scholar]
- 7.D’Amato R F, Wallmann A A, Hochstein L H, Colaninno P M, Scardamaglia M, Ardila E, Ghouri M, Kim K, Patel R C, Miller A. Rapid diagnosis of pulmonary tuberculosis by using Roche AMPLICOR Mycobacterium tuberculosis PCR test. J Clin Microbiol. 1995;31:1832–1834. doi: 10.1128/jcm.33.7.1832-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamboa F, Manterola J M, Viñado B, Matas L, Giménez M, Lonca J, Manzano J R, Rodrigo C, Cardona P J, Padilla E, Dominguez J, Ausina V. Direct detection of Mycobacterium tuberculosis complex in nonrespiratory specimens by Gen-Probe Amplified Mycobacterium tuberculosis Direct Test. J Clin Microbiol. 1997;35:307–310. doi: 10.1128/jcm.35.1.307-310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jungkind D, DiRenzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent P T, Kubica G. Public health mycobacteriology. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 11.Kubica G P W, Dye E, Cohn M L, Middlebrook G. Sputum digestion and decontamination with N-acetyl-L-cysteine-sodiumhydroxide for culture of mycobacteria. Am Rev Respir Dis. 1993;87:775–779. doi: 10.1164/arrd.1963.87.5.775. [DOI] [PubMed] [Google Scholar]
- 12.Moore D F, Curry J I. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by Amplicor PCR. J Clin Microbiol. 1995;33:2686–2691. doi: 10.1128/jcm.33.10.2686-2691.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolte F S, Metchock B. Mycobacterium. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 400–437. [Google Scholar]
- 14.Pfyffer G E, Kissling P, Jahn E M I, Welscher H-M, Salfinger M, Weber R. Diagnostic performance of Amplified Mycobacterium tuberculosis Direct Test with cerebrospinal fluid, other nonrespiratory, and respiratory specimens. J Clin Microbiol. 1996;34:834–841. doi: 10.1128/jcm.34.4.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piersimoni C, Callegaro A, Nista D, Bornigia S, De Conti F, Santini G, De Sio G. Comparative evaluation of two commercial amplification assays for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 1997;35:193–196. doi: 10.1128/jcm.35.1.193-196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reischl U, Kochanowski B. Quantitative PCR—a survey of the present technology. Mol Biotechnol. 1995;3:55–71. doi: 10.1007/BF02821335. [DOI] [PubMed] [Google Scholar]
- 17.Rolfs A, Beige J, Finckh U, Köhler B, Schaberg T, Lokies J, Lode H. Amplification of Mycobacterium tuberculosis from peripheral blood. J Clin Microbiol. 1995;33:3312–3314. doi: 10.1128/jcm.33.12.3312-3314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenstraus M, Wang Z, Chang S-Y, DeBonville D, Spadoro J P. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J Clin Microbiol. 1998;36:191–197. doi: 10.1128/jcm.36.1.191-197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Runyon E H, Karlson A G, Kubica G P, Wayne L G. Mycobacterium. In: Lennette E H, Balows A, Hausler W J Jr, Truant J P, editors. Manual of clinical microbiology. 3rd ed. Washington, D.C: American Society for Microbiology; 1980. pp. 150–179. [Google Scholar]
- 20.Rys P N, Persing D H. Preventing false positives: quantitative evaluation of three protocols for inactivation of polymerase chain reaction amplification products. J Clin Microbiol. 1993;31:2356–2360. doi: 10.1128/jcm.31.9.2356-2360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirm J, Oostendorp L A B, Mulder J G. Comparison of AMPLICOR, in-house PCR, and conventional culture for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1995;33:3221–3224. doi: 10.1128/jcm.33.12.3221-3224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schröder K-H, Naumann L, Kroppenstedt R M, Reischl U. Mycobacterium hassiacum sp. nov., a new rapidly growing thermophilic mycobacterium. Int J Syst Bacteriol. 1997;47:86–91. doi: 10.1099/00207713-47-1-86. [DOI] [PubMed] [Google Scholar]
- 23.Smith M B, Bergmann J S, Woods G L. Detection of Mycobacterium tuberculosis in BACTEC 12B broth cultures by the Roche Amplicor PCR assay. J Clin Microbiol. 1997;35:900–902. doi: 10.1128/jcm.35.4.900-902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stauffer F, Mutschlechner R, Hasenberger P, Stadlbauer S, Schinko H. Detection of Mycobacterium tuberculosis complex in clinical specimens by a commercial polymerase chain reaction kit. Eur J Clin Microbiol Infect Dis. 1995;14:1046–1051. doi: 10.1007/BF01590937. [DOI] [PubMed] [Google Scholar]
- 25.Vlaspolder F, Singer P, Roggeveen C. Diagnostic value of an amplification method (Gen-Probe) compared with that of culture for diagnosis of tuberculosis. J Clin Microbiol. 1995;33:2699–2703. doi: 10.1128/jcm.33.10.2699-2703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wobeser W L, Krajden M, Conly J, Simpson H, Yim B, D’Costa M, Fuksa M, Hian-Cheong C, Patterson M, Phillips A, Bannatyne R, Haddad A, Brunton J L, Krajden S. Evaluation of Roche Amplicor PCR assay for Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:134–139. doi: 10.1128/jcm.34.1.134-139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuen K-Y, Yam W-C, Wong L-P, Seto W-H. Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J Clin Microbiol. 1997;35:1385–1389. doi: 10.1128/jcm.35.6.1385-1389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]