Abstract
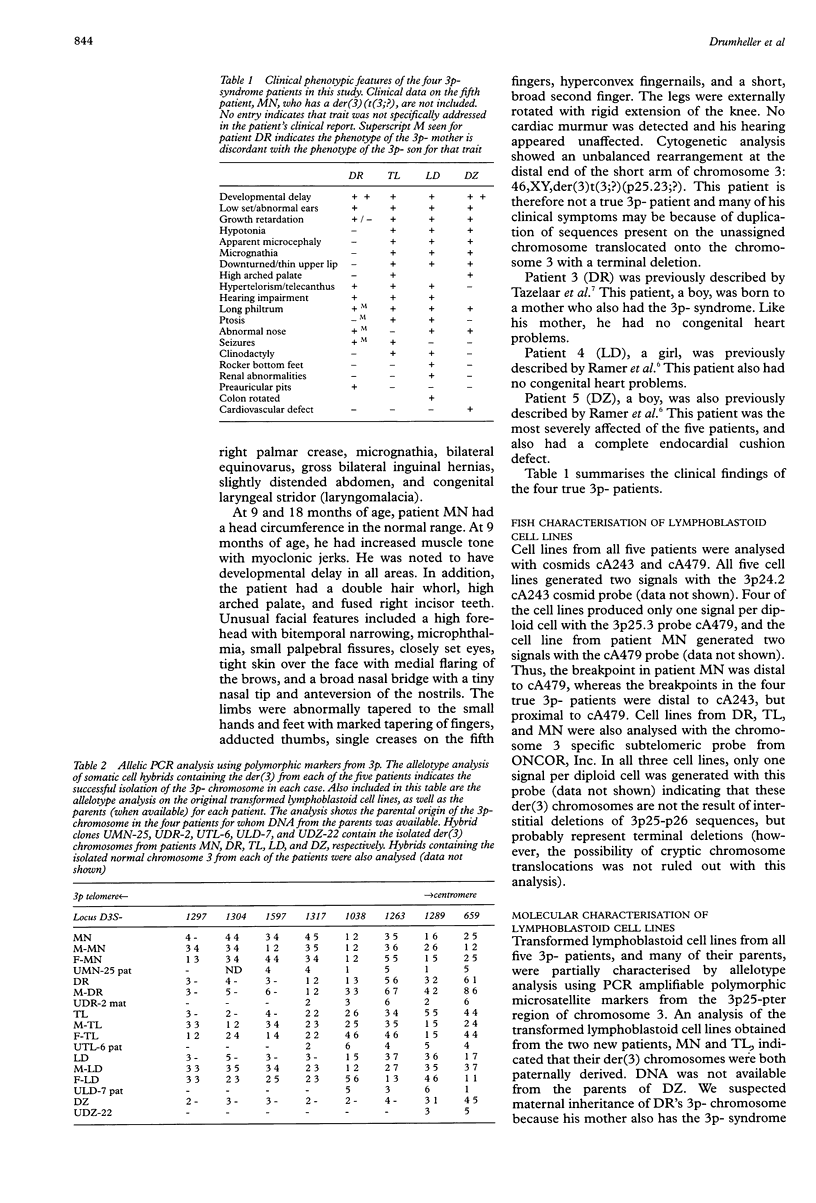
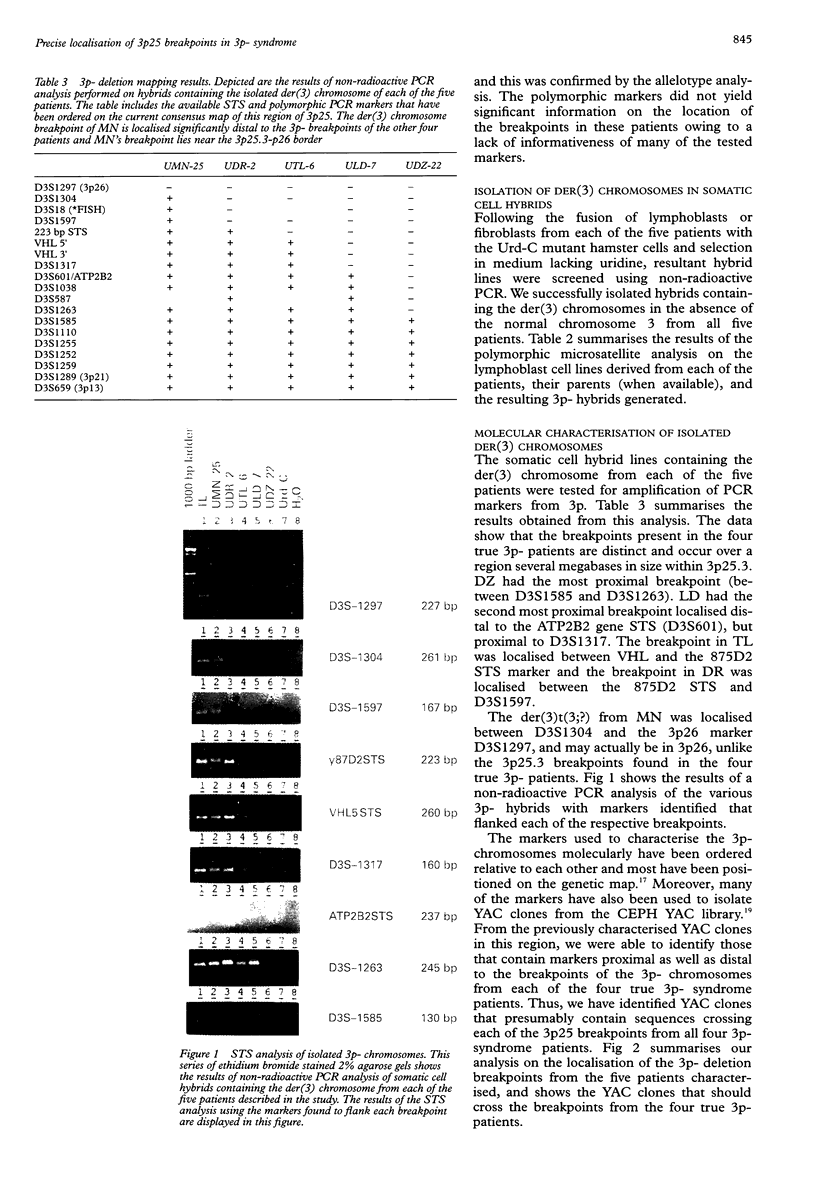
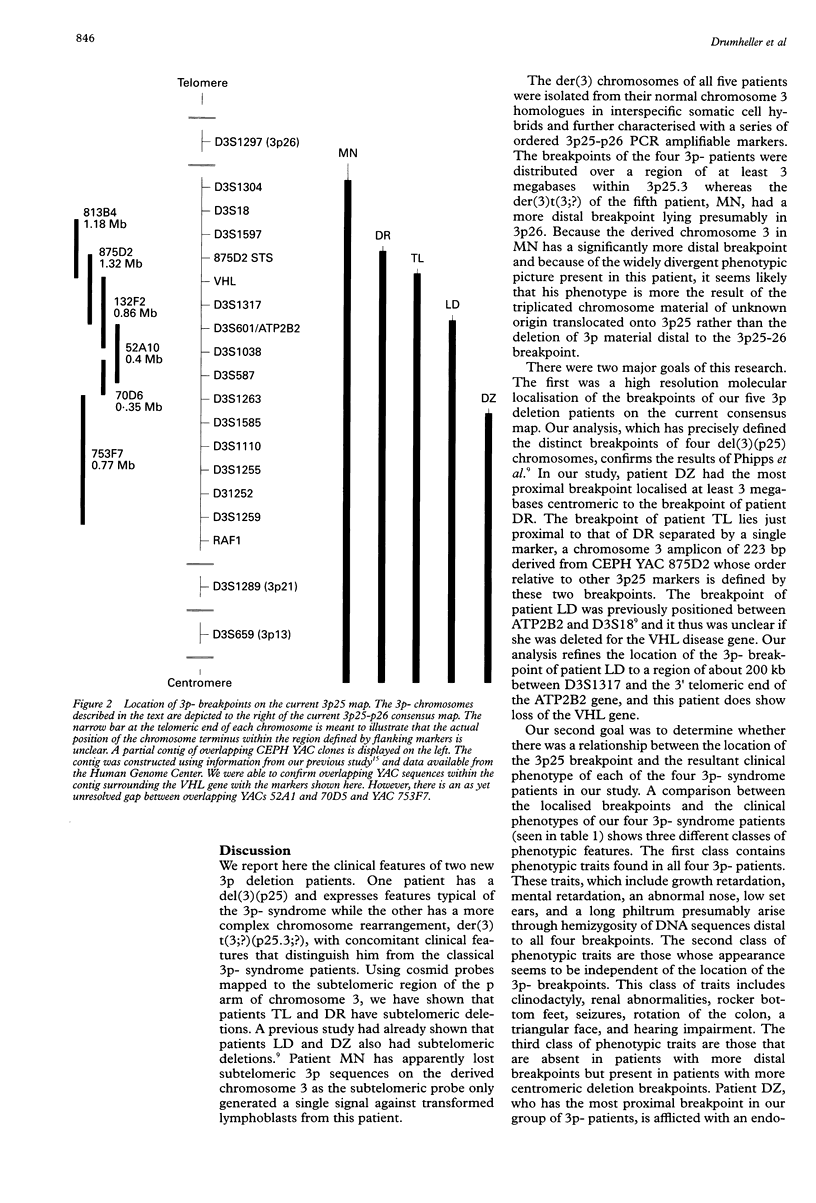
In patients with the 3p-syndrome, hemizygous deletion of 3p25-pter is associated with profound growth failure, characteristic facial features, and mental retardation. We performed a molecular genetic analysis of 3p25 breakpoints in four patients with the 3p- syndrome, and a fifth patient with a more complex abnormality, 46,XY,der(3)t(3;?)(p25.3;?). EBV transformed lymphoblasts from each of the patients were initially characterised using fluorescent in situ hybridisation (FISH) and polymorphic microsatellite analyses. The 3p-chromosome from each patient was isolated from the normal chromosome 3 in somatic cell hybrid lines and subsequently analysed with polymorphic and monomorphic PCR amplifiable markers from 3p25. The analysis clearly shows that all five breakpoints are distinct. Furthermore, we have identified yeast artificial chromosomes that cross the 3p25 breakpoints of all four 3p-patients. Two of the patients were deleted for the von Hippel-Lindau (VHL) tumour suppressor gene, although neither has yet developed evidence of VHL disease. The patient with the most centromeric breakpoint, between D3S1585 and D3S1263, had the most severe clinical phenotype including an endocardial cushion defect that was not observed in any of the four patients who had more telomeric breakpoints. This study should provide useful insights into critical regions within 3p25 that are involved in normal human growth and development.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Higginbottom M. C., Mascarello J. T., Hassin H., McCord W. K. A second patient with partial deletion of the short arm of chromosome 3: karyotype 46,XY,del(3)(p25). J Med Genet. 1982 Feb;19(1):71–73. doi: 10.1136/jmg.19.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif F., Tory K., Gnarra J., Yao M., Duh F. M., Orcutt M. L., Stackhouse T., Kuzmin I., Modi W., Geil L. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993 May 28;260(5112):1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- Liu W., Vance J., Smith D. I. cA479 (D3S719): a cosmid mapped telomeric of the Von Hippel Lindau disease gene contains the D3S18 locus. Hum Mol Genet. 1992 Jun;1(3):201–201. doi: 10.1093/hmg/1.3.201. [DOI] [PubMed] [Google Scholar]
- Mowrey P. N., Chorney M. J., Venditti C. P., Latif F., Modi W. S., Lerman M. I., Zbar B., Robins D. B., Rogan P. K., Ladda R. L. Clinical and molecular analyses of deletion 3p25-pter syndrome. Am J Med Genet. 1993 Jul 1;46(6):623–629. doi: 10.1002/ajmg.1320460604. [DOI] [PubMed] [Google Scholar]
- Naylor S. L., Buys C. H., Carritt B. Report and abstracts of the Fourth International Workshop on Human Chromosome 3 Mapping. Cytogenet Cell Genet. 1994;65(1-2):2–50. [PubMed] [Google Scholar]
- Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986 Aug;73(4):320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- Patterson D., Jones C., Morse H., Rumsby P., Miller Y., Davis R. Structural gene coding for multifunctional protein carrying orotate phosphoribosyltransferase and OMP decarboxylase activity is located on long arm of human chromosome 3. Somatic Cell Genet. 1983 May;9(3):359–374. doi: 10.1007/BF01539144. [DOI] [PubMed] [Google Scholar]
- Ramer J. C., Ladda R. L., Frankel C. Two infants with del(3)(p25pter) and a review of previously reported cases. Am J Med Genet. 1989 May;33(1):108–112. doi: 10.1002/ajmg.1320330115. [DOI] [PubMed] [Google Scholar]
- Reifen R. M., Gale R., Kerem E., Armon Y., Brand A., Dagan J., Kohn G. Partial deletion of the short arm of chromosome 3: further delineation of the 3p25-3pter syndrome. Clin Genet. 1986 Aug;30(2):127–130. doi: 10.1111/j.1399-0004.1986.tb00581.x. [DOI] [PubMed] [Google Scholar]
- Smith D. I., Glover T. W., Gemmill R., Drabkin H., O'Connell P., Naylor S. L. Report and abstracts of the fifth international workshop on human chromosome 3 mapping 1994. Ann Arbor, Michigan, May 8-9, 1994. Cytogenet Cell Genet. 1995;68(3-4):125–146. doi: 10.1159/000133907. [DOI] [PubMed] [Google Scholar]
- Swaroop A., Yang-Feng T. L., Liu W., Gieser L., Barrow L. L., Chen K. C., Agarwal N., Meisler M. H., Smith D. I. Molecular characterization of a novel human gene, SEC13R, related to the yeast secretory pathway gene SEC13, and mapping to a conserved linkage group on human chromosome 3p24-p25 and mouse chromosome 6. Hum Mol Genet. 1994 Aug;3(8):1281–1286. doi: 10.1093/hmg/3.8.1281. [DOI] [PubMed] [Google Scholar]
- Tazelaar J., Roberson J., Van Dyke D. L., Babu V. R., Weiss L. Mother and son with deletion of 3p25-pter. Am J Med Genet. 1991 May 1;39(2):130–132. doi: 10.1002/ajmg.1320390203. [DOI] [PubMed] [Google Scholar]
- Verjaal M., De Nef M. B. A patient with a partial deletion of the short arm of chromosome 3. Am J Dis Child. 1978 Jan;132(1):43–45. doi: 10.1001/archpedi.1978.02120260045012. [DOI] [PubMed] [Google Scholar]
- Warburg M., Bugge M., Brøndum-Nielsen K. Cytogenetic findings indicate heterogeneity in patients with blepharophimosis, epicanthus inversus, and developmental delay. J Med Genet. 1995 Jan;32(1):19–24. doi: 10.1136/jmg.32.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt D. R., Biedermann B., Hall J. G. Partial deletion of the short arm of chromosome 3 (3p25----3pter). Further delineation of the clinical phenotype. Clin Genet. 1985 Apr;27(4):402–407. doi: 10.1111/j.1399-0004.1985.tb02283.x. [DOI] [PubMed] [Google Scholar]



