Abstract
Introduction
Alpinetin is the bioactive component of a traditional Chinese medicine. This compound, one of the main constituents of the seeds of Alpinia katsumadai Hayata, is a member of the flavonoids, with anti-inflammatory, antibacterial, and other significant therapeutic activities of important potency and low systemic toxicity.
Material and methods
In our study, the inhibitory effect of isoliquiritigenin on HMG-CoA reductase showed a lower value of IC50 = 21.86 ±1.44 μg/ml. A molecular docking study was performed as a complementary study to provide additional data about the biological activities of alpinetin in the presence of urease. The docking calculations revealed that alpinetin with a docking score of –5.097 (kcal/mol) has an acceptable binding affinity to the enzyme, and because of various hydrophobic contacts and hydrogen bonds created by this chemical compound, alpinetin could be considered as an adequate inhibitor of urease.
Results
In the cellular and molecular part of the study, the cells treated with alpinetin were assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay for 48 h as regards the cytotoxicity and anti-human gastric carcinoma properties towards normal (human umbilical vein endothelial cells (HUVECs)) and gastric carcinoma cell lines, i.e. SNU-1, Hs 746T, and KATO III. The IC50 values of alpinetin were 426, 586, and 424 μg/ml against SNU-1, Hs 746T, and KATO III cell lines, respectively. The viability of the malignant gastric cell line decreased dose-dependently in the presence of alpinetin.
Conclusions
It seems that the anti-human gastric carcinoma effect of the investigated molecule is due to its antioxidant effects.
Keywords: alpinetin, anti-human gastric cancer, urease, molecular docking, enzyme inhibition
Introduction
Alternative drugs have become increasingly popular around the world. Containing a large family of polyphenolic secondary metabolites, flavonoids exhibit a wide variety of biological activities such as antioxidant, antiviral, antibacterial, antiprotozoal and antifungal effects. They also play an important role in plant growth and defense mechanisms against infection and injury. Alpinetin is a traditional Chinese medicine that is widely distributed in higher plants and prepared from its roots or seeds such as Alpinia katsumadai Hayata, Amomum, Alnus, Populus, Polygonum, and Scutellaria. It exhibits a wide range of biochemical activities and important therapeutic applications, such as significant anti-inflammatory, microbial resistance, and anti-hemostasis [1–3].
Urea is the most widely used fertilizer in agricultural applications. The use of urease inhibitors in agricultural applications has been studied for a long time as one of the strategies to ensure the food supply in sufficient quantities. This is because urea, one of the most widely used nitrogenous (N) fertilizers worldwide, is rapidly hydrolyzed by urease at the soil surface and is subject to hydrolysis with 70% N loss to the environment. Urease activity is an important viral marker in medicine as well as in the agricultural industry. Urease causes kidney stones and infections, as well as hepatic coma. In the treatment of such diseases, urease inhibitors that can inhibit the urease enzyme are being studied. Studies on urease inhibitors are very important in order to develop drugs to be used in the treatment of diseases caused by pathogens containing urease in humans and animals. In recent years, inhibitory effects of various plant extracts on urease enzyme have been investigated for this purpose [4–6].
Recently, theoretical investigation has become an integral part of the experimental evaluation of chemical compounds. Such investigation could provide more detailed insight into the biological activities and experimental outcomes [7]. Molecular docking studies have attracted considerable attention for biologists, since the results of docking calculation could give the researchers a comprehensive point of view for biological activities of compounds [8]. These data could help the researchers to identify the mechanisms in which the ligands and biological materials would interact with each other. There are a variety of parameters that will be obtained from docking calculations, such as binding affinity and the characteristics of interactions.
In the present study, the properties of alpinetin against common gastric carcinoma cell lines, i.e. SNU-1, Hs 746T, and KATO III, were evaluated, and also enzyme inhibition and molecular docking of it were investigated.
Material and methods
Anti-human gastric carcinoma properties of alpinetin
SNU-1, Hs 746T, and KATO III cells were used to evaluate the anticancer effect of alpinetin on cell culture. For this purpose, each cell line was placed separately in T25 flasks with a complete culture medium (including DMEM (Dulbecco’s Modified Eagle Medium), 10% complementary bovine fetal serum, and 1% penicillin-streptomycin solution) and at 37°C in the incubator, the cell culture was incubated with 5% CO2. After obtaining 80% cell density, the sample was exposed to 1% trypsin-EDTA solution and after 3 min of incubation at 37°C in a cell culture incubator with 5% CO2 and observation of cells removed from the bottom of the plate, the sample was centrifuged at 5000 rpm for 5 min and then the cell precipitate was decrypted digested by adding trypsin culture medium. Then, the cell suspensions after adding trypan blue dye were counted by a neobar slide and a cytotoxicity test was performed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) method. For this purpose, in each well of a 98-cell culture plate, 10000 SNU-1, Hs 746T, and KATO III cells were introduced with 200 μl from the complete cell culture medium and to achieve the cell monolayer density, the plate was re-exposed to 5% CO2 at 37°C. After reaching 80% cell growth, the culture medium was removed and the cell surface was first washed with PBS buffer; again, in all wells, a complete two-concentration culture medium of 100 μl was introduced and 100 μl of a solution of alpinetin dissolved in PBS (mg/ml) was introduced into well No. 1. After mixing the molecule in the culture medium, 100 μl of it was removed and added to the second well. In the next step, 100 μl of the second well was removed after stirring the medium and added to well 3. This operation was performed up to well 11 and thus the amount of molecule in each well was halved. Well No. 12 contained only one cell and complete culture medium of one concentration and was used as a control. The plate was again exposed to 5% CO2 at 37°C for 24 h and after 24 h the cytotoxicity was determined using tetrazolium dye. 10 μl of tetrazolium dye (5 mg/ml) was added to all wells, including the control, and the plate was exposed to 5% CO2 at 37°C for 2 h. The dye was then removed from the wells and 100 μl of DMSO (dimethyl sulfoxide) was added to the wells. The plate was wrapped in aluminum foil and shaken thoroughly in a shaker for 20 min. Finally, cell survival was recorded in an ELISA reader at 540 nm [9]: Cell viability (%) = (Sample A/Control A) × 100.
Enzyme study
Mobley’s method [10] was used to determine the urease inhibition activity. For the urease enzyme inhibition activity, urea was used as a substrate after the extracts were interacted with urease enzyme and the ammonia formed as a result of the reaction was measured spectroscopically [11]. Thiourea was used as a positive control. The solutions required for analysis were prepared as follows: 1) 0.01 M phosphate buffer (pH = 8.2): the pH was adjusted to 8.2 by mixing 0.01 M NaH2PO4 and 0.01 M Na2HPO4 buffer solutions in appropriate proportions. 2) Urease enzyme: 1 mg urease enzyme was dissolved in 1 ml of buffer solution. When used by dividing into 50 μl portions, appropriate dilution was made in the range of 2000–3000 μl. Substrate: A 0.1 M urea solution (in pH = 8.2 phosphate buffer) was used as the substrate [12]. Phenol reagent: For 1% phenol reagent, 200 mg of phenol was weighed and dissolved in 10 ml of distilled water and 2% phenol reagent was obtained. With subsequent dilutions, the final concentration was 1%. For 0.005% sodium nitroprusside, 1 mg of sodium nitroprusside was weighed and dissolved in 10 ml of distilled water and a solution of 0.01% was obtained. With subsequent dilutions, the final concentration was 1%. Two prepared solutions were mixed in a one-to-one (1 : 1) ratio [13]. Alkaline reagent: for 0.5% NaOH; 100 mg of NaOH was weighed and dissolved in 10 ml of water. For 0.1% NaOCl, 0.1105 ml of 10% NaOCl was taken and added to 10 ml of distilled water. Two prepared solutions were mixed in a one-to-one (1 : 1) ratio. We pipetted all of them into cuvettes sequentially with different pipettes and measured the activities at 630 nm wavelength. After finding a control value, we started the IC50 study. We found that their activities were decreased by using different inhibitor concentrations. We determined that the activities decreased as the amount of inhibitor increased [14].
Molecular docking study
The biological activities of alpinetin as an inhibitor for urease were investigated using molecular docking calculations. For this purpose, the crystal structure of urease from Jack bean (Canavalia ensiformis) at 1.49 Å resolution with the X-ray diffraction method was downloaded from the protein data bank (http://www.rcsb.org/pdb). The enzyme structure was prepared with the protein preparation module of the Schrödinger Suite [15] before using for docking study. Some of the major jobs in this step are the addition of hydrogen bonds, removing water molecules beyond 5 Å, and creating an H-bond network with the optimization module. Finally, the structure was subjected to minimization using the OPLS3e force field, and the prepared enzyme was used for the docking study. The binding site prediction was performed utilizing SiteMap of Schrödinger [16] to predict the active sites of the enzyme. After that, a grid box (20 × 20 × 20 Å3) was generated at the centroid of the predicted active site. The SDF format of alpinetin was obtained from the PubChem database and prepared with the LigPrep module of Schrödinger [17] to produce correct molecular geometric and protonation states. Eventually, the calculations of molecular docking were conducted using Glide of the Schrödinger suite.
Results and discussion
Enzyme results
In our study, the inhibitory effect of isoliquiritigenin on HMG-CoA reductase showed a lower value, IC50 = 21.86 ±1.44 μg/ml. Urease as a key enzyme plays a key role in peptic ulcers and pathogenesis of gastritis. Indeed, its inhibition protects our bodies from many disturbances including creation of urinary calculi. In agricultural studies, high urease enzyme content causes intense environmental and hence economic issues. Due to the lack of effective and safe drugs to tackle the aforementioned disturbances, the quest for new scaffolds is becoming mandatory in the field of medicinal chemistry [18]. The diverse and important roles of urease support this enzyme to be the focus of researchers around the world in the fields of biochemistry, genetics, and physiology. Strategies based on urease inhibition are considered promising tools for treating diseases caused by urease-synthesizing bacteria and reducing the loss of nitrogen from urea used as a fertilizer [19]. Therefore, it is not surprising that research on urease inhibitors has increased recently. Studies on urease inhibitors are very important in order to develop drugs to be used in the treatment of diseases caused by pathogens containing urease in humans and animals and to repair these negative effects on the environment. Until studies on urease inhibition guide the use of drugs used according to the variety of physiological conditions, their importance in medical research will continue to be ignored [20].
Molecular docking results
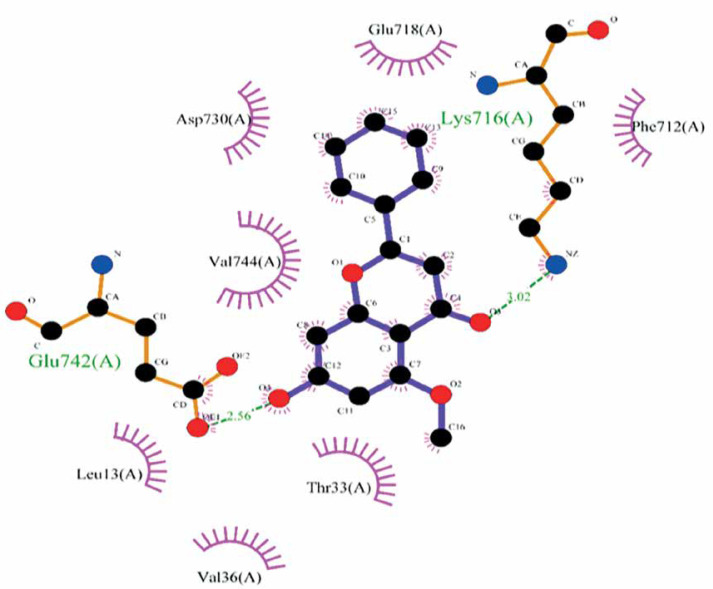
The molecular docking calculation as a versatile theoretical study was used for the evaluation of alpinetin biological activities. The predicted docking pose of alpinetin among the residues of urease is presented in Figure 1, and the interactions between the ligand and enzyme are presented in Figure 2. As can be seen, Lys716 and Glu742 have created two hydrogen bonds with alpinetin. NH of Lys716 and oxygen of Glu742 from the peptide backbone have created these H-bonds with two oxygens of the ligand. There are also seven hydrophobic contacts between alpinetin and residues of urease. These amino acids are Leu13, Thr33, Val36, Phe712, Glu718, Asp730, and Val744. The oxygen atom of alpinetin, which has created a hydrogen bond with Lys716, has emerged as a hydrogen bond acceptor, and the oxygen that has created a hydrogen bond with Glu742 is an H-bond donor. Therefore, it is vividly clarified that alpinetin could create hydrogen bonds in both the hydrogen bond acceptor and hydrogen bond donor area of the enzyme. Some of the calculated parameters from molecular docking are presented in Table I. The docking score, which is the most important parameter [21], has a value of –5.097 kcal/mol. This parameter shows the binding affinity of the ligand to the enzyme. The Glide ligand efficiency is a parameter to indicate the binding energy between the atoms of the molecule and their binding partners. There are some parameters, such as Glide Evdw and Glide Ecoul, that are interaction-related parameters. Glide Evdw shows the Van der Waals energy, and Glide Ecoul is the value of Coulomb energy. The Glide energy is the modification of Coulomb-van der Waals interaction energy. The amount of interaction pose is determined and performed with Glide Emodel [22]. The position of the first predicted active site is presented in Figure 3. This site has a site score of 1.02 and a Dscore of 1.05. Both of these values are greater than one, which indicates the potential druggability of the active site. This druggability shows the potential of this site to construct various contacts with inhibitor compounds. The areas with red color are hydrogen acceptor areas, and the blue areas represent the hydrogen bond donor spaces. Those areas presented with yellow color are hydrophobic. The residues that are engaged in the first active site are shown in Table II. Based on the molecular docking calculation results, alpinetin has the potential to be considered as an inhibitor of urease. In general, alpinetin is able to create various hydrophobic contacts and hydrogen bonds, both in the H-bond acceptor domain and the H-bond donor area of the biological materials. These characteristics make this compound a compatible inhibitor of urease.
Figure 1.
Docking pose of alpinetin among the urease residues
Figure 2.
Interactions of alpinetin and urease. Green dashed lines indicate the hydrogen bonds, and semicircles show the hydrophobic contacts
Table I.
Parameters obtained from the molecular docking calculations
| Variable | Alpinetin |
|---|---|
| IC50 [mM] | 21.86 |
| Docking score [kcal/mol] | –5.097 |
| Glide ligand efficiency [kcal/mol] | –0.255 |
| Glide Ecoul [kcal/mol] | –5.014 |
| Glide Evdw [kcal/mol] | –24.160 |
| Glide Emodel [kcal/mol] | –36.006 |
| Glide energy [kcal/mol] | –29.174 |
Figure 3.
First predicted active site of urease. Red areas are hydrogen acceptors, blue areas are hydrogen donors, and yellow areas are hydrophobic
Table II.
Residues of the first active site with Dscore of 1.05 and site score of 1.02
| Predicted residues of the first active site | |
|---|---|
| Chain A | 13,16,23,26,28,29,31,32,33,34,36,37,71,72,73,76,77,78,79,80,81,82,95,131,132,133,134,135,136,137, 277,280,281,285,291,295,296,297,345,392,396,397,414,415,418, 421,422,573,576,578,579,580,581,584,634,635,639,640,641,642,643,644,645,646, 649,652,653,656,707,709,710,711,712,714,715,716,717,718,719,728,729,730,740, 741,742,743,744,745,746,828,830,831,832,833,834,835,836,838,839,840,905,1122, 1552 |
Cancer results
Cancer is now one of the leading causes of death worldwide. Existing treatments have not been able to meet the treatment needs for various types of cancer. Therefore, the use of new technologies in the prevention and treatment of cancer can be helpful. Extensive research on molecules has been conducted in recent years [23, 24]. The advent of biotechnology has had a profound effect on many areas of healthcare and scientific research. Common cancer treatments, including chemotherapy, radiation and surgery, may reduce the size of the tumor, but the effect of these methods is transient and has no positive effect on patient survival. Therefore, replacing more effective, more specific therapies with fewer side effects with higher anti-cancer activity is a dominant issue in clinical oncology [25–27]. The gradual maturation of biotechnology has been considered not only for treating cancer but also for a wide variety of applications, especially for drug delivery and diagnostic and imaging cases. There are many types of molecules available, and choosing the right carriers according to demand is a key issue [28–30]. Molecules are very close in size to biological molecules in terms of size and can easily penetrate the cell, for this reason, one of the goals of biotechnology is to mount molecules and drugs on molecules and transfer them to the target cell [31, 32]. It is also possible to create different surface properties for molecules by attaching protective ligands to increase the molecules’ resistance to the immune system and increase their presence in the bloodstream, and even binding ligands to specifically bind the molecules to the target tissue [33–36].
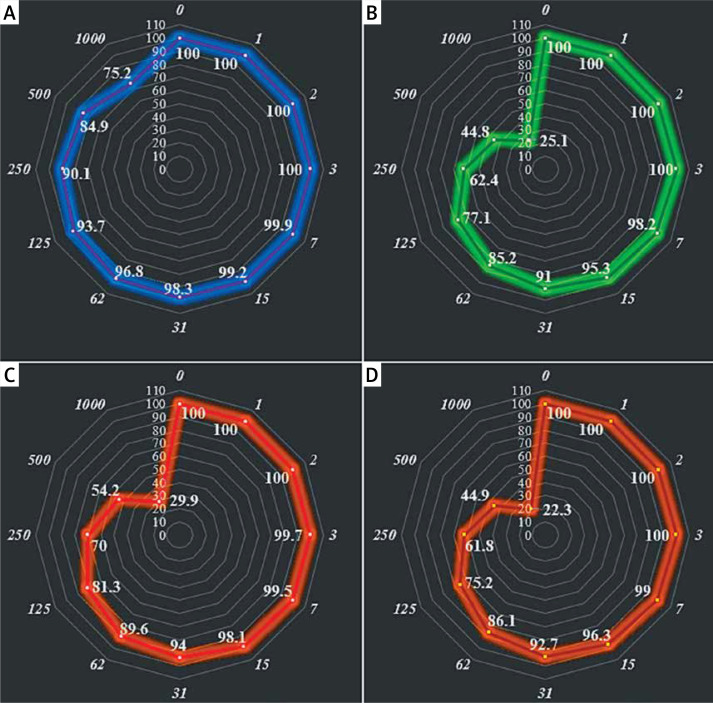
In this investigation, the cells treated with different concentrations of alpinetin were assessed by MTT assay for 48 h as regards the cytotoxic properties towards normal (HUVEC) and gastric malignancy cell lines, i.e. SNU-1, Hs 746T, and KATO III (Figure 4).
Figure 4.
Anti-human gastric carcinoma properties (cell viability (%)) of alpinetin (concentrations of 0–1000 μg/ml) against normal (HUVEC: I) and human gastric carcinoma (SNU-1 (A), Hs 746T (B), and KATO III (C, D)) cell lines. The numbers indicate the percent of cell viability at the concentrations of 0–1000 μg/ml of alpinetin against several human gastric carcinoma cell lines
The viability of the malignant gastric cell line decreased dose-dependently in the presence of alpinetin. The IC50 values of alpinetin were 426, 586, and 424 μg/ml against SNU-1, Hs 746T, and KATO III cell lines, respectively (Table I).
The absorbance rate was evaluated at 570 nm, which represented viability on the normal cell line (HUVEC) even up to 1000 μg/ml for alpinetin (Table III, Figure 4). onclusion, the theoretical studies are attractive approaches that can be conducted as fascinating investigations of experimental studies. The molecular docking study is one of the most interesting methods in this area and has attracted a lot of attention in recent years. In this study, the biological activities of alpinetin were assessed against urease using the molecular docking method. The results of this study revealed that this chemical compound has acceptable inhibitor activity against urease. This inhibitory activity could be related to the various hydrophobic contacts and H-bonds created by alpinetin. These chemical structures have attracted the attention of many researchers because of the advantages of the molecules such as controlling drug release, carrying drugs, reducing toxicity and delivering specific drugs to the target tissue. As a result of these properties, biochemistry and biotechnology have great potential for cancer treatment that can pass from a laboratory study to the patient’s bed. A possible concern limiting the application of some compounds in cancer therapy is their toxicity, which should be investigated further. However, molecule-based cancer treatments will continue to be developed to improve treatment outcomes. The viability of the malignant gastric cell line decreased dose-dependently in the presence of alpinetin. The IC50 values of alpinetin were 426, 586, and 424 μg/ml against SNU-1, Hs 746T, and KATO III cell lines, respectively. After clinical study, alpinetin can be utilized as an efficient drug in the treatment of gastric carcinoma in humans.
Table III.
IC50 of alpinetin in the anti-human gastric carcinoma test
| HUVEC | SNU-1 | Hs 746T | KATO III | |
|---|---|---|---|---|
| IC50 [μg/ml] | – | 426 ±0 | 586 ±0 | 424 ±0 |
Acknowledgments
Hui Zhang and Qian Jiang are co-first authors, they contributed equally to this work.
We acknowledge Taif University for Researchers Supporting Project (TURSP – number 2020/83), Taif University, Taif, Saudi Arabia.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Cody V, Middleton E, Harborne JB. Plant Flavonoids in Biology and Medicine. Alan R. Liss, New York: (1986). [Google Scholar]
- 2.Havsteen B. Flavonoids, a class of natural products of high pharmacological potency. Biochem Pharmacol 1983; 32: 1141-8. [DOI] [PubMed] [Google Scholar]
- 3.Molecular Luminescence Spectroscopy: Methods and Applications. Part I. Wolfbeis OS. Schulman SG (eds.). Wiley, New York; 1985; 167. [Google Scholar]
- 4.Mira AB, Cantarella H, Souza-Netto GJM, Moreira LA, Kamogawa MY, Otto R. Optimizing urease inhibitor usage to reduce ammonia emission following urea application over crop residues. Agriculture Ecosystems Environment 2017; 248: 105-12. [Google Scholar]
- 5.Drury CF, Yang X, Dan Reynolds W, Calder W, Oloya TO, Woodley AL. Combining urease and nitrification inhibitors with incorporation reduces ammonia and nitrous oxide emissions and increases corn yields. J Environm Qual 2017; 46: 939-49. [DOI] [PubMed] [Google Scholar]
- 6.Lam SK, Suter H, Bai M, et al. Using urease and nitrification inhibitors to decrease ammonia and nitrous oxide emissions and improve productivity in a subtropical pasture. Sci Total Environ 2018; 644: 1531-5. [DOI] [PubMed] [Google Scholar]
- 7.Kaczmarczyk-Sedlak I, Folwarczna J, Sedlak L, et al. Effect of caffeine on biomarkers of oxidative stress in lenses of rats with streptozotocin-induced diabetes. Arch Med Sci 2019; 15: 1073-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jhong CH, Riyaphan J, Lin SH, Chia YC, Weng CF. Screening alpha-glucosidase and alpha-amylase inhibitors from natural compounds by molecular docking in silico. BioFactors 2015; 41: 242-51. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y, Wan X, Li L, Sun P, Liu G. Synthesis of a reusable composite of graphene and silver nanoparticles for catalytic reduction of 4-nitrophenol and performance as anti-colorectal carcinoma. J Materials Res Technol 2021; 12: 1832-43. [Google Scholar]
- 10.Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev 1995; 59: 451-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modolo LV, da-Silva CJ, Brandão DS, Chaves IS. A minireview on what we have learned about urease inhibitors of agricultural interest since mid-2000s. J Adv Res 2018; 13: 29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sha Z, Lv T, Staal M, et al. Effect of combining urea fertilizer with P and K fertilizers on the efficacy of urease inhibitors under different storage conditions. J Soils Sediments 2020: 20: DOI: 10.1007/s11368-019-02534-w. [DOI] [Google Scholar]
- 13.Fu Q, Abadie M, Blaud A, et al. Effects of urease and nitrification inhibitors on soil N, nitrifier abundance and activity in a sandy loam soil. Biol Fertil Soils 2020: 56: 185-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Zhang M, Li Z, et al. Inhibition of urease activity by humic acid extracted from sludge fermentation liquid. Bioresource Technol 2019; 290: 121767. [DOI] [PubMed] [Google Scholar]
- 15.“Schrödinger Release 2020-4: Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY: 2016; Impact, Schrödinger, LLC, New York, NY 2016; Prime, Schrödinger, LLC, New York, NY 2020. [Google Scholar]
- 16.Poustforoosh A, Hashemipour H, Tüzün B, Pardakhty A, Mehrabani M, Nematollahi MH. Evaluation of potential anti-RNA-dependent RNA polymerase (RdRP) drugs against the newly emerged model of COVID-19 RdRP using computational methods. Biophys Chem 2021; 272: 106564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokalı FS, Taslimi P, Usanmaz H, Karaman M, Şendil K. Synthesis, characterization, biological activity and molecular docking studies of novel schiff bases derived from thiosemicarbazide: biochemical and computational approach. J Mol Structure 2021; 1231: 129666. [Google Scholar]
- 18.Galindo FS, Carvalho Minhoto Teixeira Filho M, Buzetti S, Pagliari PH, Kondo Santini JM. Can NBPT urease inhibitor in combination with Azospirillum brasilense inoculation improve wheat development? Nutrient Cycling in Agroecosystems 2020; 117: 131-43. [Google Scholar]
- 19.Hartmann TE, Guzman-Bustamante I, Ruser R, Müller T. Turnover of urea in a soil from the North China plain as affected by the urease inhibitor NBPT and wheat straw. Agronomy 2020; 10: 857. [Google Scholar]
- 20.Yang G, Ji H, Sheng J, et al. Combining Azolla and urease inhibitor to reduce ammonia volatilization and increase nitrogen use efficiency and grain yield of rice. Sci Total Environ 2020; 743: 140799. [DOI] [PubMed] [Google Scholar]
- 21.Schrödinger Release 2020-4: LigPrep, Schrödinger, LLC, New York, NY: 2020. [Google Scholar]
- 22.Eckhard U, Schönauer E, Brandstetter H. Structural basis for activity regulation and substrate preference of clostridial collagenases G, H, and T. J Biol Chem 2013; 288: 20184-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Wang Z, Liu H, Wang L, Huang G. Liposome encapsulated of temozolomide for the treatment of glioma tumor: preparation, characterization and evaluation. Drug Discov Ther 2015; 9: 205-12. [DOI] [PubMed] [Google Scholar]
- 24.Mohammed MI, Makky AM, Teaima MH, Abdellatif MM, Hamzawy MA, Khalil MA. Transdermal delivery of van-comycin hydrochloride using combination of nanoethosomes and iontophoresis: in vitro and in vivo study. Drug Deliv 2016; 23: 1558-64. [DOI] [PubMed] [Google Scholar]
- 25.Zhou YX, Quan GL, Wu QL, Zhang XX, Niu BY, Wu BY. Mesoporous silica nanoparticles for drug and gene delivery. Acta Pharm Sin B 2018; 8: 165-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F, Jin C, Jiang Y, et al. Liposome based delivery systems in pancreatic cancer treatment: from bench to bedside. Cancer Treat Rev 2011; 37: 633-42. [DOI] [PubMed] [Google Scholar]
- 27.Xinli DHZS. Applications of nanocarriers with tumor molecular targeted in chemotherapy. Chemistry 2012; 75: 621-7. [Google Scholar]
- 28.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer 2002; 2: 750-63. [DOI] [PubMed] [Google Scholar]
- 29.Byrne JD, Betancourt T, Brannon-Peppas L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv Drug Del Rev 2008; 60: 1615-26. [DOI] [PubMed] [Google Scholar]
- 30.Torchilin VP. Targeted pharmaceutical nanocarriers for cancer therapy and imaging. AAPS J 2007; 9: E128-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshpande PP, Biswas S, Torchilin VP. Current trends in the use of liposomes for tumor targeting. Nanomedicine 2013; 8: 1509-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Huang Y, Li S. Polymeric micelles: nanocarriers for cancer-targeted drug delivery. AAPS Pharm Sci Tech 2014; 15: 862-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumura Y, Hamaguchi T, Ura T, et al. Phase I clinical trial and pharmacokinetic evaluation of NK911, a micelleencapsulated doxorubicin. Br J Cancer 2004; 91: 1775-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie S, Xing Y, Kim GJ, Simons JW. Nanotechnology applications in cancer. Annu Rev Biomed Eng 2007; 9: 257-88. [DOI] [PubMed] [Google Scholar]
- 35.Gao Z, Lukyanov AN, Singhal A, Torchilin VP. Diacyllipid-polymer micelles as nanocarriers for poorly soluble anticancer drugs. Nano Lett 2002; 2: 979-82. [Google Scholar]
- 36.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov 2008; 7: 771-82. [DOI] [PubMed] [Google Scholar]