ABSTRACT
We describe the genome of a lytic phage EAb13 isolated from sewage, with broad activity against multidrug-resistant Acinetobacter baumannii. EAb13 is an unclassified siphovirus. Its genome consists of 82,411 bp, with 40.15% GC content, 126 protein-coding sequences, 1 tRNA, and 2,177 bp-long direct terminal repeats.
KEYWORDS: Acinetobacter baumannii, phage EAb13, broad host range, complete genome sequence, siphovirus, lytic phage, therapeutic candidate
ANNOUNCEMENT
Acinetobacter baumannii causes a growing number of severe nosocomial infections linked to extensively drug-resistant (XDR) and pandrug-resistant (PDR) strains (1). With the rise of XDR and PDR infections, phages have been successfully used as alternative therapeutics for the compassionate treatment of humans (2, 3). A challenge for phage therapy against A. baumannii infections is a typical narrow activity of phages (3, 4). Here, we describe the genome of phage EAb13 with an unusually broad host range that was able to lyse 91/105 (86.7%) of diverse drug-resistant A. baumannii isolates (5).
The EAb13 phage was isolated in 2020 from sewage collected in Washington, DC, using an XDR respiratory isolate of A. baumannii, MRSN 423159, for phage enrichment. Single plaque isolation was performed three times to ensure purity, the phage was propagated in broth, and its DNA was extracted with the QIAamp DNA Mini Kit (Qiagen, Germantown, MD), according to the manufacturer’s protocol. A library was constructed using the KAPA HyperPlus Kit (Roche Diagnostics, Indianapolis, IN), and sequencing was performed on an Illumina MiSeq (Illumina, Inc., San Diego, CA) with the MiSeq Reagent Kit v3 (600 cycles, 300 bp reads). The quality of 101,029 paired-end reads was evaluated with FastQC 0.11.9 (6), and the reads were trimmed with Trimmomatic (7), v0.39. The genome was assembled using Unicycler (8), its termini were identified with PhageTerm (9), and phage lifestyle was predicted with BACPHLIP (10). Protein coding sequences (CDSs) were annotated using Pharokka pipeline (11 - 21). Amino acid sequence similarity searches were performed using default parameters in DIAMOND (22, 23).
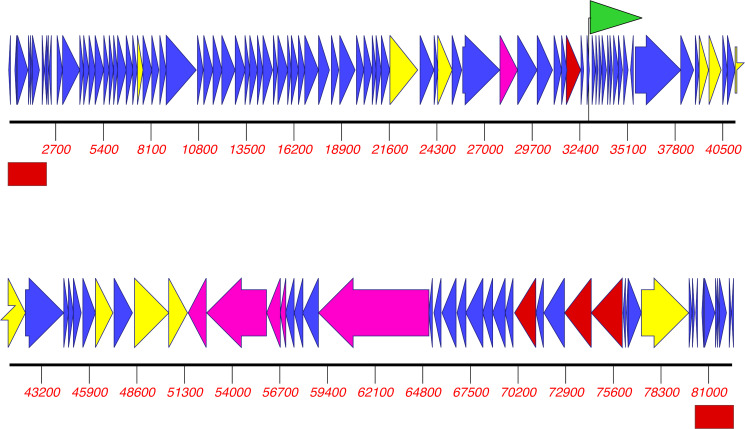
The average read coverage was 430×; EAb13 genome length was 82,411 bp, with a G + C content of 40.15%, 126 predicted CDSs, and direct terminal repeats of 2,177 bp (Fig. 1). MASH analysis (21) against the INPHARED database (20) identified one phage with significant DNA identity to EAb13 (88.0%) across the entire genome, TCUP2199 (GenBank Accession Number ON323491; 24). Their terminase large subunit sequences shared 95.4% amino acid and 88.02% nucleotide identity (22). Of the 91 proteins that matched sequences in the nr database, 89 had top hits within TCUP2199. TCUP2199 was similar to EAb13 in genome length and GC content, also showed a broad host range, and has a siphovirus morphology based on electron microscopy (EM) (24). EM found that EAb13 has a long, non-contractile tail (data not shown), suggesting that this is also an unclassified phage with a siphovirus morphology that belongs to the class Caudoviricetes.
Fig 1.
Genome organization of EAb13. Colored arrows denote the location of predicted coding sequences, transcriptional direction, and predicted function: unknown (blue), nucleotide metabolism (yellow), head and packing (red), and tail (pink). The green flag shows a tRNA sequence, and the red boxes mark terminal repeat regions.
In contrast to TCUP2199, EAb13 encodes one tRNA, tRNA-Asn. Both phages encode several genes related to DNA synthesis (Fig. 1). The high (95.2%) BACPHLIP score suggested a virulent lifestyle of EAb13 (10). Its putative proteins did not show significant similarities to drug resistance or virulence determinants in the databases CARD (16) and VFDB (17), any other bacterial proteins, or phage products responsible for lysogenic lifestyle or gene transfer. Therefore, EAb13 appears to be a lytic phage and a promising candidate for therapeutic applications.
ACKNOWLEDGMENTS
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense.
This study was supported by the Military Infectious Diseases Research Program, grant W0330_19_WR, and Peer Reviewed Medical Research Program, Focused Program Award PR182667. The Multidrug Resistant Organism Repository and Surveillance Network (MRSN) at the Walter Reed Army Institute of Research provided the strain MRSN 423159 used for phage isolation and diverse clinical isolates of A. baumannii for phage host range testing.
We thank Yunxiu He for her technical assistance.
Contributor Information
Andrey A. Filippov, Email: andrey.a.filippov.ctr@health.mil.
Jelle Matthijnssens, Katholieke Universiteit Leuven, Leuven, Belgium .
DATA AVAILABILITY
The EAb13 genome BioProject, BioSample, GenBank, and NCBI Sequence Read Archive accession numbers are PRJNA948145, SAMN33875313, OQ717042, and SRX19782736, respectively.
REFERENCES
- 1. Ayoub Moubareck C, Hammoudi Halat D. 2020. Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 9:119. doi: 10.3390/antibiotics9030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dehbanipour R, Ghalavand Z. 2022. Acinetobacter baumannii: pathogenesis, virulence factors, novel therapeutic options and mechanisms of resistance to antimicrobial agents with emphasis on tigecycline. J Clin Pharm Ther 47:1875–1884. doi: 10.1111/jcpt.13787 [DOI] [PubMed] [Google Scholar]
- 3. Strathdee SA, Hatfull GF, Mutalik VK, Schooley RT. 2023. Phage therapy: from biological mechanisms to future directions. Cell 186:17–31. doi: 10.1016/j.cell.2022.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lai M-J, Chang K-C, Huang S-W, Luo C-H, Chiou P-Y, Wu C-C, Lin N-T. 2016. The tail associated protein of Acinetobacter baumannii phage ΦAB6 Is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS One 11:e0153361. doi: 10.1371/journal.pone.0153361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Supplementary table S1 . n.d. Comparative activity of Acinetobacter baumannii phages EAb13 and EAb1. Available from: 10.6084/m9.figshare. https://doi.org/10.6084/m9.figshare [DOI]
- 6. Andrews S. 2010. Fastqc: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 7. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garneau JR, Depardieu F, Fortier L-C, Bikard D, Monot M. 2017. Phageterm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292. doi: 10.1038/s41598-017-07910-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hockenberry AJ, Wilke CO. 2021. BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ 9:e11396. doi: 10.7717/peerj.11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald P-J, Vreugde S. 2023. Pharokka: a fast Scalable bacteriophage annotation tool. Bioinformatics 39:btac776. doi: 10.1093/bioinformatics/btac776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly Interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steinegger M, Söding J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35:1026–1028. doi: 10.1038/nbt.3988 [DOI] [PubMed] [Google Scholar]
- 15. McNair K, Zhou C, Dinsdale EA, Souza B, Edwards RA. 2019. PHANOTATE: a novel approach to gene identification in phage genomes. Bioinformatics 35:4537–4542. doi: 10.1093/bioinformatics/btz265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–8. doi: 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49:9077–9096. doi: 10.1093/nar/gkab688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Terzian P, Olo Ndela E, Galiez C, Lossouarn J, Pérez Bucio RE, Mom R, Toussaint A, Petit M-A, Enault F. 2021. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genom Bioinform 3:lqab067. doi: 10.1093/nargab/lqab067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cook R, Brown N, Redgwell T, Rihtman B, Barnes M, Clokie M, Stekel DJ, Hobman J, Jones MA, Millard A. 2021. INfrastructure for a PHAge REference database: identification of large-scale biases in the current collection of cultured phage genomes. Phage 2:214–223. doi: 10.1089/phage.2021.0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using minhash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 23. Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. doi: 10.1038/s41592-021-01101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mardiana M, Teh S-H, Lin L-C, Lin N-T. 2022. Isolation and characterization of a novel Siphoviridae phage, vB_AbaS_TCUP2199, infecting multidrug-resistant Acinetobacter baumannii. Viruses 14:1240. doi: 10.3390/v14061240 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The EAb13 genome BioProject, BioSample, GenBank, and NCBI Sequence Read Archive accession numbers are PRJNA948145, SAMN33875313, OQ717042, and SRX19782736, respectively.