ABSTRACT
We report the draft genome sequences of Pseudomonas extremaustralis NQ5, Arthrobacter strain NQ4, and Arthrobacter strain NQ7 isolated from a laboratory-scale membrane bioreactor, soils from San Antonio, TX, USA and sediments from Galveston Bay, TX, USA, respectively. These bacteria degrade the explosive compound nitroguanidine, which is present in some insensitive munitions.
KEYWORDS: biodegradation, insensitive munitions, nitroguanidine
ANNOUNCEMENT
Nitroguanidine (NQ) is an explosive compound used in a variety of insensitive munitions developed to resist impact, friction, and heat (1). NQ is also used in the synthesis of some organic compounds, such as herbicides, and documented to cause acute and/or chronic toxicity in mice, aquatic organisms, and plants (2 - 4). Pseudomonas extremaustralis NQ5 was isolated from a laboratory-scale membrane bioreactor treating various explosive compounds, including NQ. Arthrobacter strain NQ4 was isolated from soil collected from a per- and polyfluoroalkyl substances-contaminated site in San Antonio, TX, USA. Arthrobacter strain NQ7 was isolated from sediments collected from Galveston Bay, TX, USA. Sample matrices were suspended and enriched in nitrogen-free mineral salts medium (N-free MSM) containing 2 mM NQ as sole nitrogen source and 11 mM glucose as sole carbon source for 3 weeks. The strains were isolated from N-free MSM agar plates with the same amendments by picking individual colonies. The colonies grew within a day (NQ5) or more than a week (NQ4 and NQ7) in N-free MSM with NQ and glucose as sole nitrogen and carbon sources, respectively (Fig. 1). NQ concentrations were analyzed by high-performance liquid chromatography using the method in Fuller et al. (5). Genomic DNAs (gDNA) were extracted using the FastDNA SPIN kit for soil (MP biomedical, Irvine, CA, USA) using the provided method. 16S rRNA genes were amplified using the 24F/1532R primer set. Sanger sequencing was conducted on both ends of the amplicons by Eton Biosciences (San Diego, CA, USA), and the trimmed paired-end reads were merged through VSearch (5). Through BLAST, NQ5, NQ4, and NQ7 were identified as Pseudomonas extremaustralis strain BF11 (MT441542.1), Arthrobacter strain AFS039412 (OP986498.1), and Arthrobacter strain SE3A52 (OQ121141.1), respectively, with 100% identity.
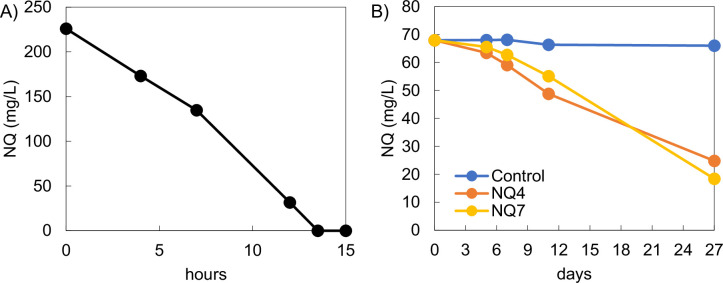
Fig 1.
Time course of NQ degradation by (A) strain NQ5 and (B) strains NQ4 and NQ7, when NQ was supplied as sole nitrogen source and glucose was supplied as sole carbon source.
The same gDNAs used for 16S rRNA gene sequencing were quantified using a Qubit High Sensitivity (HS) Assay Kit on the Qubit Fluorometer 4.0 (Invitrogen, Waltham, MA, USA). The gDNA integrities were checked with the Genomic DNA ScreenTape Assay Kit (Agilent, Santa Clara, CA, USA) on a 4200 TapeStation (Agilent). Libraries were prepared with the Illumina DNA Prep Kit and IDT for Illumina UD indexes (Plate A) (Illumina, San Diego, CA, USA). Post-library quantification was conducted with the HS Assay Kit. Quality was checked with the D1000 ScreenTape Assay Kit (Agilent) on the 4200 TapeStation. Sequencing was performed through the MiSeq platform (Illumina) with the paired-end 2 × 250 bp strategy. Reads were trimmed, adapter removed, and quality controlled by FastQC within Trim Galore version 0.6.7 (6). The genomes were de novo assembled using SPAdes version 3.15.3 (7) on the Grace computing cluster at Texas A&M University. Annotated genomes of Pseudomonas sp. 02C 26 (CP025262.1), Arthrobacter crystallopoietes strain DSM 20117 (CP018863.1), and Arthrobacter phenanthrenivorans strain SWC37 (JWTB00000000) were used as references to identify contaminants within the NQ5, NQ4, and NQ7 assemblies that ran through the RASTtk-based custom annotation pipeline at BV-BRC (8, 9) (https://www.bv-brc.org/app/Annotation). Contaminants were removed using the BV-BRC Metagenomic Binning Tool (https://www.bv-brc.org/app/MetagenomicBinning). The filtered contigs were submitted and annotated through NCBI PGAP version 4.6 (10). Genome coverages were calculated with samtools version 1.16.1 (11) after mapping paired-end reads to the assemblies using BWA-MEM2 version 2.2.1 (12). Genomic features are provided in Table 1.
TABLE 1.
Genomic features of strains NQ5, NQ4, and NQ7
| Features | NQ5 | NQ4 | NQ7 | |||
|---|---|---|---|---|---|---|
| GenBank accession no. | JARBJR010000000 | JARETC010000000 | JARFXU020000000 | |||
| Assembly accession no. | GCA_029581615.1 | GCA_029077345.1 | GCA_029581595.2 | |||
| SRA a accession no. | SRR23696550 | SRR23696941 | SRR23692575 | |||
| No. of paired-end reads | 489,473 | 511,004 | 467,527 | |||
| No. of contigs | 95 | 37 | 78 | |||
| Assembly length (bp) | 6,533,326 | 4,483,921 | 4,874,196 | |||
| Genome coverage (×) | 33 | 50 | 40 | |||
| Contig N50 (bp) | 205,006 | 298,154 | 145,272 | |||
| Contig L50 (bp) | 13 | 5 | 9 | |||
| GC content (%) | 60.5 | 66.5 | 66.5 | |||
| No. of CDS b | 6,038 | 4,074 | 4,560 | |||
| No. of CDS with proteins | 5,908 | 4,052 | 4,522 | |||
| No. of complete rRNAs (5S, 16S, 23S) | 2, 1, 1 | 2, 1, 1 | 3, 1, 0 | |||
| No. of predicted tRNAs | 59 | 51 | 53 | |||
Sequence Read Archive (SRA).
Coding sequences (CDS).
ACKNOWLEDGMENTS
This work was supported by the Strategic Environmental Research and Development Program (SERDP; Project ER19-1198), under contract number W912HQ-19-C-0016.
We acknowledge Dr. John C. Blazier and staff members at the Institute for Genome Sciences and Society at Texas A&M University for their assistance in sequencing and sequence data processing and Paul Hedman at Aptim Federal Services for analytical support.
Contributor Information
Kung-Hui Chu, Email: kchu@civil.tamu.edu.
Irene L. G. Newton, Indiana University, Bloomington, Bloomington, Indiana, USA
DATA AVAILABILITY
Whole Genome Shotgun projects have been deposited in DDBJ/ENA/GenBank under accession numbers JARBJR000000000 (NQ5), JARETC000000000 (NQ4), and JARFXU000000000 (NQ7). The versions described in this paper are versions JARBJR010000000 (NQ5), JARETC010000000 (NQ4), and JARFXU020000000 (NQ7). BioProject accession numbers are PRJNA934168 (NQ5), PRJNA934170 (NQ4), and PRJNA934176 (NQ7). BioSample accession numbers are SAMN33268489 (NQ5), SAMN33268523 (NQ4), and SAMN33268549 (NQ7). The Sequence Read Archive (SRA) accession numbers are SRR23696550 (NQ5), SRR23696941 (NQ4), and SRR23692575 (NQ7).
REFERENCES
- 1. Kaplan DL, Cornell JH, Kaplan AM. 1982. Decomposition of nitroguanidine. Environ Sci Technol 16:488–492. doi: 10.1021/es00102a012 [DOI] [Google Scholar]
- 2. Schalie WH. 1985. The toxicity of nitroguanidine and photolyzed nitroguandine to freshwater aquatic organisms. Army Medical Bioengineering Research and Development Lab, Fort Detrick, MD. [Google Scholar]
- 3. Heitholt JJ, Hodgson RH, Tworkoski TJ. 1990. Toxicity and uptake of nitroguanidine in plants. Bull Environ Contam Toxicol 44:751–758. doi: 10.1007/BF01701798 [DOI] [PubMed] [Google Scholar]
- 4. Kanne DB, Dick RA, Tomizawa M, Casida JE. 2005. Neonicotinoid nitroguanidine insecticide metabolites: synthesis and nicotinic receptor potency of guanidines, aminoguanidines, and their derivatives. Chem Res Toxicol 18:1479–1484. doi: 10.1021/tx050160u [DOI] [PubMed] [Google Scholar]
- 5. Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krueger F 2015. Trim Galore! a wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data.
- 7. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olson RD, Assaf R, Brettin T, Conrad N, Cucinell C, Davis JJ, Dempsey DM, Dickerman A, Dietrich EM, Kenyon RW, Kuscuoglu M, Lefkowitz EJ, Lu J, Machi D, Macken C, Mao C, Niewiadomska A, Nguyen M, Olsen GJ, Overbeek JC, Parrello B, Parrello V, Porter JS, Pusch GD, Shukla M, Singh I, Stewart L, Tan G, Thomas C, VanOeffelen M, Vonstein V, Wallace ZS, Warren AS, Wattam AR, Xia F, Yoo H, Zhang Y, Zmasek CM, Scheuermann RH, Stevens RL. 2023. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 51:D678–D689. doi: 10.1093/nar/gkac1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vasimuddin M, Misra S, Li H, Aluru S. 2019. Efficient architecture-aware acceleration of BWA-MEM for multicore systems 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS); Rio de Janeiro, Brazil. doi: 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole Genome Shotgun projects have been deposited in DDBJ/ENA/GenBank under accession numbers JARBJR000000000 (NQ5), JARETC000000000 (NQ4), and JARFXU000000000 (NQ7). The versions described in this paper are versions JARBJR010000000 (NQ5), JARETC010000000 (NQ4), and JARFXU020000000 (NQ7). BioProject accession numbers are PRJNA934168 (NQ5), PRJNA934170 (NQ4), and PRJNA934176 (NQ7). BioSample accession numbers are SAMN33268489 (NQ5), SAMN33268523 (NQ4), and SAMN33268549 (NQ7). The Sequence Read Archive (SRA) accession numbers are SRR23696550 (NQ5), SRR23696941 (NQ4), and SRR23692575 (NQ7).