ABSTRACT
Mycobacterium abscessus biofilm aggregates have been shown in the lungs of cystic fibrosis patients and are often tolerant to drugs. Herein, we analyzed bi-dimensional images of either fluorescent or Congo red-stained M. abscessus colony-biofilms grown on a membrane to monitor growth and shape of M. abscessus smooth and rough variants. These colony-biofilms responded differently to rifabutin and bedaquiline, thus highlighting the importance of the morphotype to properly address antibiotic treatment in patients with biofilm-related infections.
KEYWORDS: Mycobacterium abscessus, biofilm, colony-biofilm, smooth, rough, morphotype, antibiotics
INTRODUCTION
Mycobacterium abscessus (Mabs) infections primarily affect vulnerable populations with underlying lung disorders such as cystic fibrosis (CF) and are particularly difficult to treat with very lengthy therapeutic options that are often associated with severe side effects (1, 2). The poor treatment outcomes are largely caused by the intrinsic resistance of Mabs to most antibacterials (3). Mabs presents smooth (S) and rough (R) morphotypes based on the presence (in S) or absence (in R) of glycopeptidolipids (GPL) (4), associated with different pathophysiological outcomes in macrophages (5) and patients (6). Mabs can also be associated with biomaterial-related infections (7), e.g., catheter-related or prosthetic joint infections, where the main pathological mechanism involves biofilm formation (8), a feature also encountered in CF patients (9). Biofilm development contributes to bacterial persistence in harsh environments (10), rendering eradication almost impossible. Several methodologies are available for studying and quantifying mycobacterial biofilms, including growth in 96-well plates and staining with crystal violet (11 - 14), tetrazolium salts (15, 16), and/or analysis by confocal laser microscopy (17 - 19). By requiring multiple washing/staining steps, these methods are difficult to apply to Mabs R biofilms due to the highly hydrophobic nature of this variant and the high propensity to aggregate, causing a loss of biofilm biomass recovery. Alternatively, the biofilm-colony model consists of inducing bacterial growth on a semipermeable membrane laying on an agar plate (20). Colony-biofilms can be fed with exogenous nutritional sources or antibiotics by simply relocating the biofilm-supporting membrane to a fresh agar plate, with the advantage of preventing washing steps (21). Herein, we developed a new technique for characterizing and comparing Mabs S and R colony-biofilms using two-dimensional (2D) images without the need for a complex equipment.
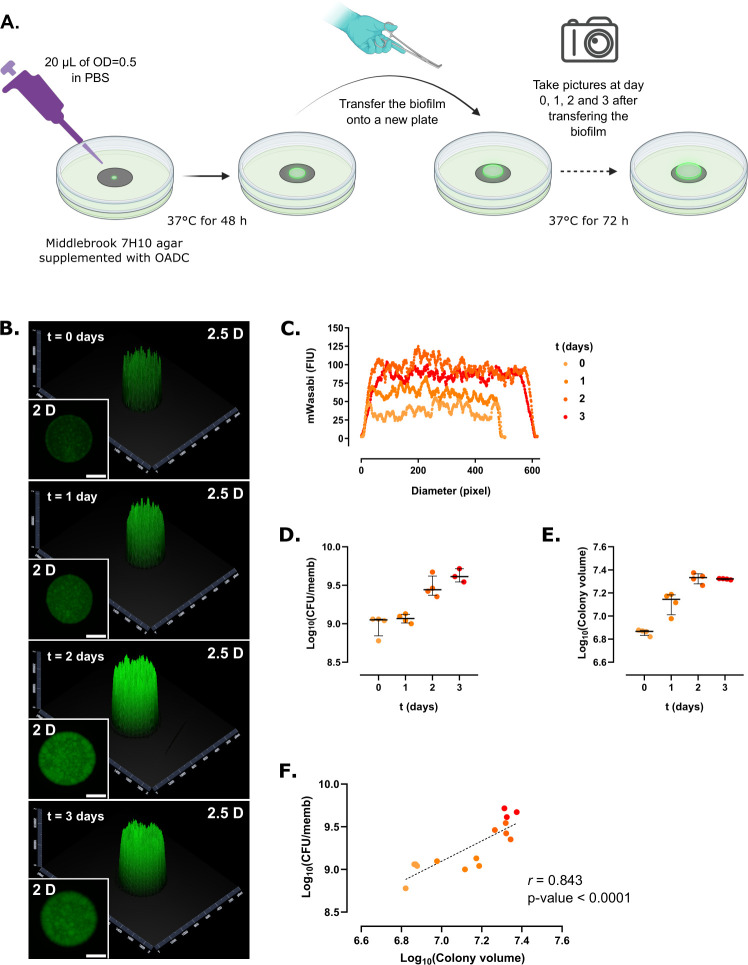
Mabs biofilms were grown according to the method previously described (22), with minor modifications. S and R cultures of Mabs CIP104536T expressing green fluorescent protein (mWasabi) were grown in Middlebrook 7H9 broth supplemented with oleic acid-albumin-dextrose-catalase (OADC) and tyloxapol (0.025%) at 37°C and 80 rpm for 72 h. Bacteria were collected by centrifugation at 3,500 rpm for 5 min, washed two times with sterile phosphate buffer saline (PBS) and then diluted to an optical density (OD600) of 0.5 [~1.5 × 108 colony-forming units (CFU)/mL]. Autoclaved black polycarbonate membranes (diameter, 25 mm; pore size, 0.2 µm, Whatman, Merck, Germany) were placed onto Middlebrook 7H10 plates supplemented with OADC and inoculated with 20 µL of the bacterial suspension (OD600 = 0.5). The membrane-supported biofilms were statically incubated for 48 h at 37°C, after which they were photographed every 24 h for 3 days (Fig. 1A). Pictures were taken using a binocular (ZEISS Axio Zoom.V16, Zeiss, Germany) equipped with a lighting device (Zeiss HXP 200C) at 7× zoom for detecting the mWasabi fluorescence. The spatial fluorescence over time was measured from the pictures and processed using ZEN 2 (Blue Edition) and covering only one whole diameter per colony. The colony volume was calculated by using the half-volume of an ellipsoid: 2/3 × π × FIM × (CD/2)2, where FIM is the fluorescence intensity maximum in fluorescence intensity units (FIU) and CD is the colony diameter in pixels. After taking pictures, each membrane-supported biofilm was processed for quantifying the number of CFU per membrane. For that, each biofilm was removed from the plates with sterile clamps, deposited in a 50-mL tube containing 10 mL of PBS supplemented with 0.025% tyloxapol and a mixture of 4 mm (five to six beads) and 1 mm diameter (~1 mL) glass beads, vortexed for 15 s, and sonicated in an ultrasonic bath (Bandelin Sonorex Super RK 255 H, Sigma Aldrich) for 5 min once for Mabs S and twice for Mabs R. The resulting homogenous bacterial suspensions were diluted in a 10-fold bank dilution, plated on Luria-Bertoni (LB) agar, and incubated at 37°C for 4 days prior to CFU counting.
Fig 1.
Colony-biofilm growth of M. abscessus S expressing mWasabi. (A) Schematic representation of the experimental protocol used to monitor Mabs biofilm growth. (B) Representative 2- and 2.5-dimensional pictures of a colony-biofilm taken at 0, 1, 2, and 3 days after transferring the biofilm-supporting membrane onto a fresh plate. White bar represents 2 mm. (C) 2D profile of a colony-biofilm estimated from the fluorescence intensity profile of the colony over time. (D) Colony-forming units (CFU) per membrane (CFU/memb) over time. (E) Colony volume over time. (F) Correlation between CFU/memb and colony volume. The discontinuous line represents the linear tendency correlation. Experiments were done using four biological replicates per strain and per time point.
Although the colony-biofilm model was to study the effects of antibiotic treatment (21), to evaluate the biomechanical characteristics of Mabs biofilms (22) or growth in the presence of Pseudomonas aeruginosa in dual biofilms (23, 24), we have implemented a new set-up to monitor and compare the growth of fluorescent Mabs CIP104536T S and R variants over time. Strains were first transformed with pMV306::Pleft*mWasabi, generated by cloning the Pleft*mWasabi insert from plasmid L5 attB::Pleft*mWasabi (AddGene number #169409) (25) into the EcoRV-restricted integrative pMV306 (26). Fluorescent strains were selected Middlebrook 7H10 supplemented with 10% OADC enrichment with 0.2% glycerol and 250 µg/mL kanamycin. Representative 2- and 2.5-dimensional pictures of a colony-biofilm of Mabs S overexpressing mWasabi were imaged after 1, 2, or 3 days after transferring it to a new agar plate (Fig. 1B). The 2.5-dimensional pictures represent intensity values in a two-dimensional image as a height map. This strain tended to grow mainly vertically as evidenced by measuring the fluorescence intensity of the biofilms over time, although the optimal size was reached on day 2 (Fig. 1C). To address whether the colony volume estimated from fluorescence intensity increase correlated with the number of bacilli in the growing colony-biofilm, biofilms were detached from the membranes and subjected to physical dissociation prior to CFU determination. Both the CFU per membrane (Fig. 1D) and colony volume (Fig. 1E) increased over time. Plotting the number of CFU in the function of the colony volume showed a very high positive correlation (r = 0.843, P-value <0.0001) (Fig. 1F). Our results suggest that the Mabs S forms a biofilm-colony which hardly grows beyond day 2.
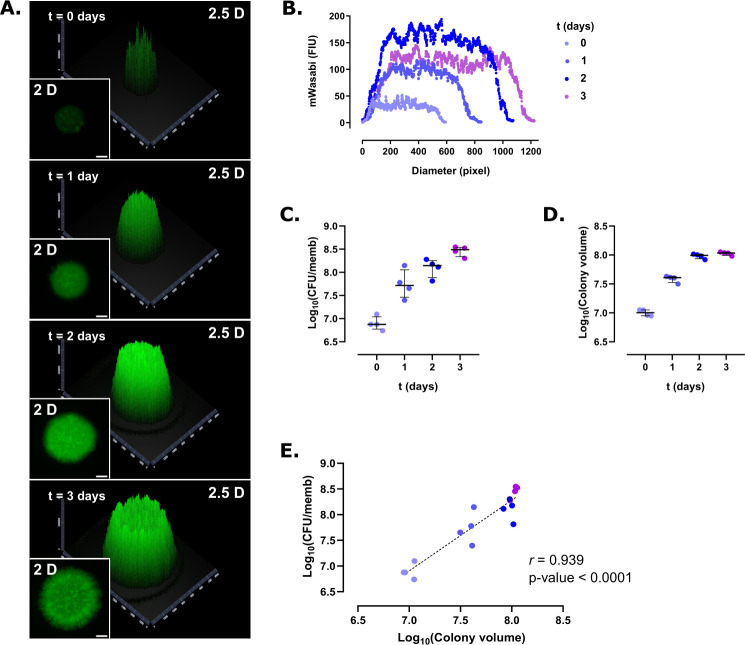
Whether Mabs R forms colony-biofilms was next investigated using the same set-up. Two-dimensional and 2.5D images showed that the spatial shape and growth of R biofilms differ from those of S biofilms and tend to grow more horizontally (Fig. 2A), which was confirmed by measuring the diameter of the colony-biofilms over time (Fig. 2B). While the diameter of the S variant biofilm reached 600 pixels in 3 days, this value increased up to 1,200 pixels for the R biofilm. Noteworthy, R biofilms continue to grow beyond day 2. Both the CFU per membrane (Fig. 2C) and colony volume (Fig. 2D) increased over time and a high correlation was found between the CFU number in the function of the colony volume (r = 0.939, P-value <0.0001) (Fig. 2E). Overall, this indicates that the colony volume mirrors the CFU in S and R colony-biofilm and that determining the colony volume by measuring the intensity of fluorescence allows to rapidly estimate number of bacilli, a procedure particularly suited for the screening of multiple growth conditions in the same study.
Fig 2.
Colony-biofilm growth of M. abscessus R expressing mWasabi. (A) Representative 2- and 2.5-dimensional pictures of a colony-biofilm taken at 0, 1, 2, and 3 days after transferring the biofilm-supporting membrane onto a fresh plate. White bar represents 2 mm. (B) 2D profile of a colony-biofilm estimated from the fluorescence intensity profile of the colony over time. (C) Colony-forming units per membrane (CFU/memb) over time. (D) Colony volume over time. (E) Correlation between CFU/memb and colony volume. The discontinuous line represents the linear tendency correlation. Experiments were done using four biological replicates per strain and per time point.
We next evaluated the influence of antibiotics in colony-biofilm development by determining the activity of rifabutin, known to inhibit Mabs growth in vitro, in macrophages and animal models (27 - 29). The experimental design was similar to the one described above except that, after a 48-h growth, the biofilm-supporting membranes were transferred to 7H10 agar supplemented with 10% OADC and containing 75 µg/mL rifabutin, a concentration based on previous MIC determinations (12.5 µg/mL and 50 µg/mL against R and S strains, respectively) (29) (Fig. 3A). Rifabutin treatment was associated with a clear drop in fluorescence intensity at day 3 (Fig. 3B), suggesting that the antibiotic stops the colony-biofilm growth, translating into a 68% decrease in the CFU per membrane (Fig. 3C and J) and a marked reduction (73%) in the colony volume (Fig. 3D). Considering the starting number of CFU per membrane, one can propose that the rifabutin effect on both M. abscessus S and R biofilms is inhibitory (Fig. S1). A high positive correlation was observed between the CFU per membrane and the colony volume in the rifabutin-treated versus untreated biofilms (r = 0.899, P-value <0.001) (Fig. 3E). Similarly, the 72-h rifabutin treatment severely reduced the fluorescence signal in R biofilms (Fig. 3F), accompanied by an 89% drop in the CFU numbers (Fig. 3G and J) and an 81% reduction in the colony volume (Fig. 3H). A high positive correlation was found between the number of viable bacilli and the colony volume in the rifabutin-treated versus untreated biofilms (r = 0.907, P-value <0.001) (Fig. 3I). This indicates that rifabutin is more active in R biofilms than in S biofilms, in line with the higher susceptibility of R than S bacilli in planktonic state (29). This differential susceptibility of Mabs S and R to rifabutin (Fig. 3J) might be explained by the presence or absence of the GPL layer, which modifies the mycobacterial surface properties. Whereas the S variant displays nanoscopic surface areas with varying degrees of hydrophobicity, the R variant is homogenously hydrophobic (30), possibly favoring direct hydrophobic interaction between rifabutin and the Mabs R surface (31). While these results imply that the determination of the colony volume estimated from fluorescence intensity measurement allows monitoring the evolution of biofilm growth in the presence of an antibiotic, this method, however, pre-requires to generate a fluorescent strain. Although easily achievable in type strains, this may not be the case in clinical isolates, often refractory to DNA transformation, thus adding an additional delay when one wants to rapidly compare biofilm growth of different strains. Therefore, we thought of replacing the fluorescence marker in the bacterial strain with a non-fluorescent dye directly added into the agar and able to diffuse homogenously inside the biofilm.
Fig 3.
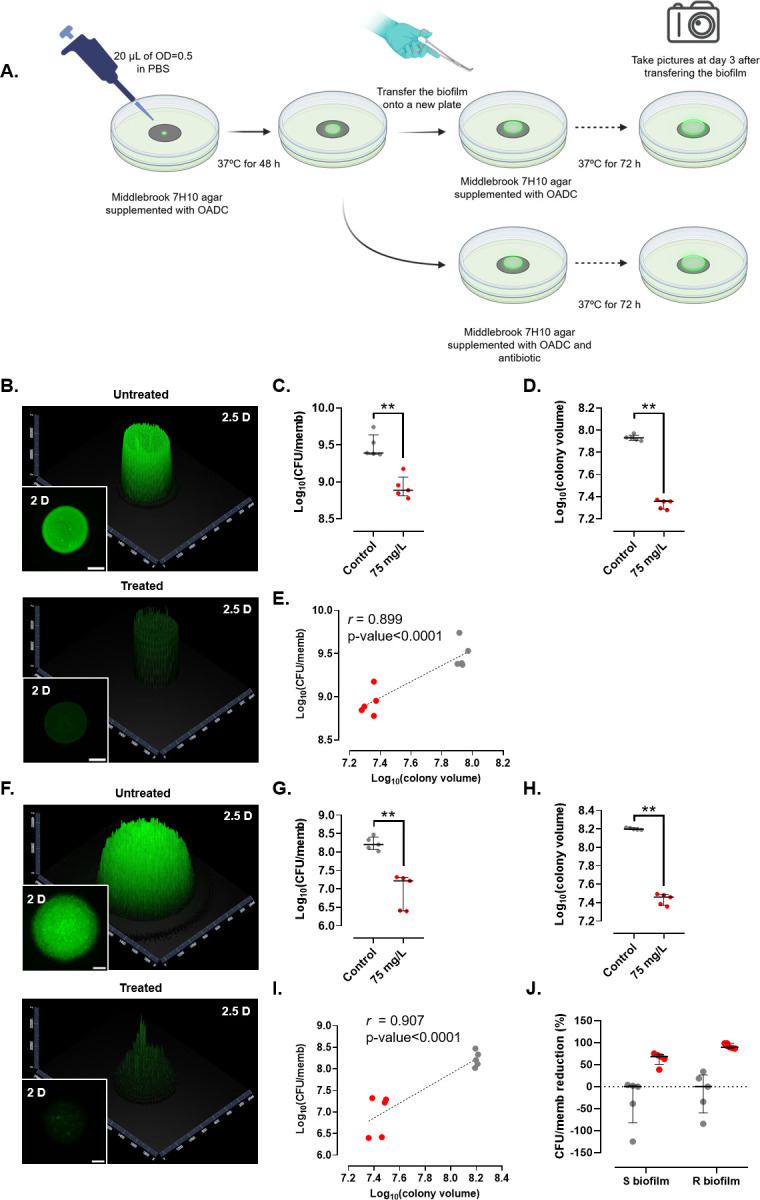
Effect of rifabutin on fluorescent M. abscessus S (B–E) and R (F–I) biofilms. (A) Schematic representation of the experimental protocol used to monitor the effect of drugs on biofilm growth. (B, F) Representative 2- and 2.5-dimensional pictures of a colony-biofilm in the absence (upper panels) or presence (lower panels) of rifabutin, 3 days after transferring the biofilm-supporting membrane onto a fresh plate. White bar represents 2 mm. (C, G) Colony-forming units per membrane (CFU/memb) with or without treatment. (D, H) Colony volume with or without treatment. (E, I) Correlation between CFU/memb and colony volume with or without treatment. The discontinuous line represents the linear tendency correlation. (J) CFU/memb reduction in S and R biofilms. Untreated biofilms are in gray and rifabutin-treated biofilms are in red. **, P-value <0.01 for Wilcoxon test. Experiments were done using five biological replicates per strain and condition.
Congo red (CR) is a hydrophobic diazo dye that binds to lipids and lipoproteins (32), widely used to evaluate mycobacterial surface hydrophobicity (32 - 35) and in biofilm-forming bacteria such as staphylococci (36, 37). We monitored the efficacy of bedaquiline using non-fluorescent S and R biofilms. White polycarbonate membranes (diameter, 13 mm; pore size, 0.2 µm, Whatman, Merck, Germany) supporting 48-h biofilms of non-fluorescent S and R variants were transferred onto 7H10 plates supplemented with 10% OADC, containing 100 µg/mL CR (Sigma-Aldrich, Merck, United States) and supplemented with or without 1 µg/mL bedaquiline and incubated at 37°C for 72 h. After incubation, each biofilm was photographed using the light shutter completely open with an exposure time of 1 ms. Following image processing, the colony volume was estimated as described above, except that the FIM was replaced by the difference between the maximum and minimum intensity of the gray level intensity (GLI, in GLI units or GLIU). Each membrane-supported biofilm was then processed for quantifying the number of CFU per membrane. Experiments were performed five times per strain and per condition using five biological replicates per strain and per time point.
CR diffuses through the membrane and is incorporated inside the growing biofilms while providing the biofilm a reddish color that allows the gray level in black-and-white photography to be assessed. Bedaquiline inhibits the ATP synthase (38) and is bacteriostatic against Mabs in vitro (MIC of 0.12 µg/mL) (39) but, to our knowledge, has not been evaluated in Mabs biofilms. Bedaquiline reduced growth of S biofilms (Fig. 4A through D and I) but not of R biofilms (Fig. 4 E through I), causing a 29% decrease in the CFU per membrane (P-value = 0.0317) (Fig. 4B) and a 27.5% decrease in the colony volume (P-value = 0.0317) (Fig. 4C) for the S strain. The R biofilm appeared more tolerant to bedaquiline as judged by the CFU counts (Fig. 4F), although a significant 28% decrease in the colony volume was noticed (P-value = 0.0317) (Fig. 4G). Only S biofilms showed a high correlation between the CFU per membrane and colony volume (r = 0.639, P-value = 0.0467) (Fig. 4D and H). The anti-biofilm effect of bedaquiline observed on S biofilms supports an inhibitory effect on Mabs S proliferation, in line with the observation showing that bedaquiline inhibits the growth and biofilm formation of oral streptococci (40). Overall, it can be inferred that the CR-derived method can be applied to non-fluorescent mycobacterial strains, e.g., clinical strains, to rapidly determine the effect of various antibiotics on biofilms. Importantly, to demonstrate that the same results generated using the CR-based methodology are comparable to those using the fluorescence assay, we repeated the experiment demonstrating the efficacy of rifabutin against the S and R biofilms with CR. The results obtained are essentially similar to those obtained by monitoring fluorescence (Fig. S2), thus validating the usefulness of the CR-based approach.
Fig 4.
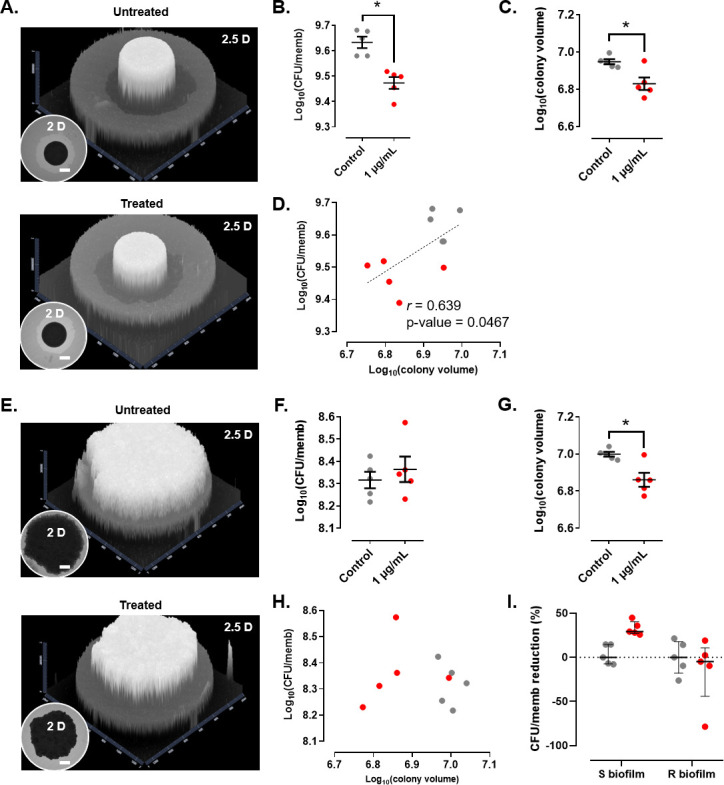
Effect of bedaquiline on non-fluorescent M. abscessus S (A–D) and R (E–H) biofilms. (A, E) Representative 2- and 2.5-dimensional pictures of a colony-biofilm in the absence (upper panels) or presence (lower panels) of bedaquiline, 3 days after transferring the biofilm-supporting membrane onto a fresh plate. White bar represents 2 mm. Black and white must be inverted to represent the 2.5D figures. (B, F) CFU/memb with or without treatment. (C, G) Colony volume with or without treatment. (D, H) Correlation between CFU/memb and colony volume with or without treatment. The discontinuous line represents the linear tendency correlation. (I) CFU/memb reduction in S and R biofilms. Untreated biofilms are in gray and bedaquiline-treated biofilms are in red. *, P-value <0.05 for Wilcoxon test. Experiments were done using five biological replicates per strain and condition.
This study presents, however, a few limitations: (i) the starting time point to monitor the biofilm growth corresponds to 48 h after inoculation of the bacterial suspension onto the membrane, implying that potential differences in growth/drug activity may be overlooked during the first 48 h; (ii) the OD/CFU ratio between the S and R morphotypes in the initial inoculum onto the membranes varies, implying that the CFU per membrane of each morphotype should be compared; (iii) units (FIU × pixel2 or GLIU × pixel2) may not be comparable between studies since both factors are depending on the fluorescent reporter used or on the software from which images are acquired and processed; (iv) the use of the FIM to calculate the volume of a colony may oversimplify the differences in the colony contours between both Mabs variants since, although the S morphotype shows a more uniform dome-shaped morphology, the R morphotype has an irregular shape with ridges; (v) although the fluorescent and non-fluorescent Mabs strains used in this study possess the same genetic background, high levels of expression of the fluorescent protein may affect the growth rates and the biomass yield (41, 42), thus influencing biofilm development; (vi) the percentage of agarose in the medium onto which the colony-biofilm is growing modifies the biofilm characteristics in both variants (Fig. S3). While Mabs S showed no changes related to CFU per membrane, the colony volume dropped significantly with a high agarose concentration (1.5%). In contrast, for Mabs R, this high agarose concentration showed an increase in the CFU per membrane and a decrease in colony volume.
In summary, the methods described here allow to rapidly estimate the volume associated with each biofilm, which may be very helpful to characterize growth evolution and/or the potential impact of drugs on both fluorescent and non-fluorescent Mabs colony-biofilms. This study also reveals that S and R biofilms show distinct tolerance to bedaquiline, thus underscoring the importance of the morphotype to adequately address antibiotic treatment in patients with biofilm-related infections.
ACKNOWLEDGMENTS
This project has been funded by the French National Research Agency and is supported by the ANR grants ILIome-20-CE44-0019 and SUNLIVE-19-CE15-0012-01 (SUNLIVE). Figure 1A and 3A have been created with BioRender.com.
The authors report there are no competing interests to declare.
Contributor Information
Laurent Kremer, Email: laurent.kremer@irim.cnrs.fr.
Kelly E. Dooley, Vanderbilt University Medical Center, Nashville, Tennessee, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00402-23.
Additional experimental details, additional results.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Martiniano SL, Nick JA, Daley CL. 2016. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med 37:83–96. doi: 10.1016/j.ccm.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 2. Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ Jr, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. 2020. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J 56:2000535. doi: 10.1183/13993003.00535-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johansen MD, Herrmann J-L, Kremer L. 2020. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol 18:392–407. doi: 10.1038/s41579-020-0331-1 [DOI] [PubMed] [Google Scholar]
- 4. Gutiérrez AV, Viljoen A, Ghigo E, Herrmann J-L, Kremer L. 2018. Glycopeptidolipids, a double-edged sword of the Mycobacterium abscessus complex. Front Microbiol 9:1145. doi: 10.3389/fmicb.2018.01145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roux A-L, Viljoen A, Bah A, Simeone R, Bernut A, Laencina L, Deramaudt T, Rottman M, Gaillard J-L, Majlessi L, Brosch R, Girard-Misguich F, Vergne I, de Chastellier C, Kremer L, Herrmann J-L. 2016. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol 6:160185. doi: 10.1098/rsob.160185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Catherinot E, Roux A-L, Macheras E, Hubert D, Matmar M, Dannhoffer L, Chinet T, Morand P, Poyart C, Heym B, Rottman M, Gaillard J-L, Herrmann J-L. 2009. Acute respiratory failure involving an R variant of Mycobacterium abscessus. J Clin Microbiol 47:271–274. doi: 10.1128/JCM.01478-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esteban J, García-Coca M. 2018. Mycobacterium biofilms. Front. Microbiol 8:2651. doi: 10.3389/fmicb.2017.02651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martins N, Rodrigues CF. 2020. Biomaterial-related infections. J Clin Med 9:722. doi: 10.3390/jcm9030722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hunt-Serracin AC, Parks BJ, Boll J, Boutte CC. 2019. Mycobacterium abscessus cells have altered antibiotic tolerance and surface glycolipids in artificial cystic fibrosis sputum medium. Antimicrob Agents Chemother 63:e02488-18. doi: 10.1128/AAC.02488-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson LR. 2008. Microcolony and biofilm formation as a survival strategy for bacteria. J Theor Biol 251:24–34. doi: 10.1016/j.jtbi.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 11. Belardinelli JM, Li W, Avanzi C, Angala SK, Lian E, Wiersma CJ, Palčeková Z, Martin KH, Angala B, de Moura VCN, Kerns C, Jones V, Gonzalez-Juarrero M, Davidson RM, Nick JA, Borlee BR, Jackson M. 2021. Unique features of Mycobacterium abscessus biofilms formed in synthetic cystic fibrosis medium. Front Microbiol 12:743126. doi: 10.3389/fmicb.2021.743126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belardinelli JM, Li W, Martin KH, Zeiler MJ, Lian E, Avanzi C, Wiersma CJ, Nguyen TV, Angala B, de Moura VCN, Jones V, Borlee BR, Melander C, Jackson M. 2022. 2-Aminoimidazoles inhibit Mycobacterium abscessus biofilms in a zinc-dependent manner. Int J Mol Sci 23:2950. doi: 10.3390/ijms23062950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clary G, Sasindran SJ, Nesbitt N, Mason L, Cole S, Azad A, McCoy K, Schlesinger LS, Hall-Stoodley L. 2018. Mycobacterium abscessus smooth and rough morphotypes form antimicrobial-tolerant biofilm phenotypes but are killed by acetic acid. Antimicrob Agents Chemother 62:e01782-17. doi: 10.1128/AAC.01782-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malcolm KC, Nichols EM, Caceres SM, Kret JE, Martiniano SL, Sagel SD, Chan ED, Caverly L, Solomon GM, Reynolds P, Bratton DL, Taylor-Cousar JL, Nichols DP, Saavedra MT, Nick JA. 2013. Mycobacterium abscessus induces a limited pattern of neutrophil activation that promotes pathogen survival. PLoS One 8:e57402. doi: 10.1371/journal.pone.0057402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richards JP, Cai W, Zill NA, Zhang W, Ojha AK. 2019. Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrob Agents Chemother 63:e01213-19. doi: 10.1128/AAC.01213-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marini E, Di Giulio M, Ginestra G, Magi G, Di Lodovico S, Marino A, Facinelli B, Cellini L, Nostro A. 2019. Efficacy of carvacrol against resistant rapidly growing mycobacteria in the planktonic and biofilm growth mode. PLoS One 14:e0219038. doi: 10.1371/journal.pone.0219038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pradal I, Esteban J, Mediero A, García-Coca M, Aguilera-Correa JJ. 2020. Contact effect of a Methylobacterium sp. extract on biofilm of a Mycobacterium chimaera strain isolated from a 3t heater-cooler system. Antibiotics 9:474. doi: 10.3390/antibiotics9080474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muñoz-Egea M-C, García-Pedrazuela M, Mahillo-Fernandez I, Esteban J. 2016. Effect of antibiotics and antibiofilm agents in the ultrastructure and development of biofilms developed by nonpigmented rapidly growing mycobacteria. Microb Drug Resist 22:1–6. doi: 10.1089/mdr.2015.0124 [DOI] [PubMed] [Google Scholar]
- 19. García-Coca M, Rodríguez-Sevilla G, Pérez-Domingo A, Aguilera-Correa J-J, Esteban J, Muñoz-Egea M-C. 2020. Inhibition of Mycobacterium abscessus, M. chelonae, and M. fortuitum biofilms by Methylobacterium sp. J Antibiot (Tokyo) 73:40–47. doi: 10.1038/s41429-019-0232-6 [DOI] [PubMed] [Google Scholar]
- 20. Anderl JN, Franklin MJ, Stewart PS. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 44:1818–1824. doi: 10.1128/AAC.44.7.1818-1824.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merritt JH, Kadouri DE, O’Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gloag ES, Wozniak DJ, Stoodley P, Hall-Stoodley L. 2021. Mycobacterium abscessus biofilms have viscoelastic properties which may contribute to their recalcitrance in chronic pulmonary infections. Sci Rep 11:5020. doi: 10.1038/s41598-021-84525-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodríguez-Sevilla G, García-Coca M, Romera-García D, Aguilera-Correa JJ, Mahíllo-Fernández I, Esteban J, Pérez-Jorge C. 2018. Non-tuberculous mycobacteria multispecies biofilms in cystic fibrosis: development of an in vitro Mycobacterium abscessus and Pseudomonas aeruginosa dual species biofilm model. Int J Med Microbiol 308:413–423. doi: 10.1016/j.ijmm.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 24. Rodríguez-Sevilla G, Crabbé A, García-Coca M, Aguilera-Correa JJ, Esteban J, Pérez-Jorge C. 2019. Antimicrobial treatment provides a competitive advantage to Mycobacterium abscessus in a dual-species Biofilm with Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e01547-19. doi: 10.1128/AAC.01547-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolbe K, Bell AC, Prosser GA, Assmann M, Yang H-J, Forbes HE, Gallucci S, Mayer-Barber KD, Boshoff HI, Barry III CE. 2020. Development and optimization of chromosomally-integrated fluorescent Mycobacterium tuberculosis reporter constructs. Front. Microbiol 11:591866. doi: 10.3389/fmicb.2020.591866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0 [DOI] [PubMed] [Google Scholar]
- 27. Aziz DB, Low JL, Wu M-L, Gengenbacher M, Teo JWP, Dartois V, Dick T. 2017. Rifabutin is active against Mycobacterium abscessus complex. Antimicrob Agents Chemother 61:e00155-17. doi: 10.1128/AAC.00155-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dick T, Shin SJ, Koh W-J, Dartois V, Gengenbacher M. 2020. Rifabutin is active against Mycobacterium abscessus in mice. Antimicrob Agents Chemother 64:e01943–19. doi: 10.1128/AAC.01943-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johansen MD, Daher W, Roquet-Banères F, Raynaud C, Alcaraz M, Maurer FP, Kremer L. 2020. Rifabutin is bactericidal against intracellular and extracellular forms of Mycobacterium abscessus. Antimicrob Agents Chemother 64:e00363-20. doi: 10.1128/AAC.00363-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Viljoen A, Viela F, Kremer L, Dufrêne YF. 2020. Fast chemical force microscopy demonstrates that glycopeptidolipids define nanodomains of varying hydrophobicity on mycobacteria. Nanoscale Horiz 5:944–953. doi: 10.1039/c9nh00736a [DOI] [PubMed] [Google Scholar]
- 31. Heipieper HJ, Cornelissen S, Pepi M. 2010. Surface properties and cellular energetics of bacteria in response to the presence of hydrocarbons, p 1615–1624. In Timmis KN (ed), Handbook of hydrocarbon and lipid Microbiology. Springer, Berlin Heidelberg, Berlin, Heidelberg. doi: 10.1007/978-3-540-77587-4 [DOI] [Google Scholar]
- 32. Cangelosi GA, Palermo CO, Laurent J-P, Hamlin AM, Brabant WH. 1999. Colony morphotypes on Congo red agar segregate along species and drug susceptibility lines in the Mycobacterium avium-intracellulare complex. Microbiology (Reading) 145 (Pt 6):1317–1324. doi: 10.1099/13500872-145-6-1317 [DOI] [PubMed] [Google Scholar]
- 33. Miyamoto Y, Mukai T, Takeshita F, Nakata N, Maeda Y, Kai M, Makino M. 2004. Aggregation of mycobacteria caused by disruption of fibronectin-attachment protein-encoding gene. FEMS Microbiol Lett 236:227–234. doi: 10.1016/j.femsle.2004.05.047 [DOI] [PubMed] [Google Scholar]
- 34. Singh P, Rao RN, Reddy JRC, Prasad RBN, Kotturu SK, Ghosh S, Mukhopadhyay S. 2016. PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci Rep 6:21624. doi: 10.1038/srep21624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Etienne G, Villeneuve C, Billman-Jacobe H, Astarie-Dequeker C, Dupont M-A, Daffé M. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis . Microbiology (Reading) 148:3089–3100. doi: 10.1099/00221287-148-10-3089 [DOI] [PubMed] [Google Scholar]
- 36. de Castro Melo P, Ferreira LM, Filho AN, Zafalon LF, Vicente HIG, de Souza V. 2013. Comparison of methods for the detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Braz J Microbiol 44:119–124. doi: 10.1590/S1517-83822013005000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. 2006. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol 24:25–29. doi: 10.4103/0255-0857.19890 [DOI] [PubMed] [Google Scholar]
- 38. Deshkar AT, Shirure PA. 2022. Bedaquiline: a novel diarylquinoline for multidrug-resistant pulmonary tuberculosis. Cureus 14:e28519. doi: 10.7759/cureus.28519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dupont C, Viljoen A, Thomas S, Roquet-Banères F, Herrmann J-L, Pethe K, Kremer L. 2017. Bedaquiline inhibits the ATP synthase in Mycobacterium abscessus and is effective in infected zebrafish. Antimicrob Agents Chemother 61:e01225-17. doi: 10.1128/AAC.01225-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang M, Yu W, Zhou S, Zhang B, Lo ECM, Xu X, Zhang D. 2021. In vitro antibacterial activity of an FDA-approved H+-ATPAse inhibitor, bedaquiline, against Streptococcus mutans in acidic milieus. Front. Microbiol 12:647611. doi: 10.3389/fmicb.2021.647611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoffmann F, Rinas U. 2004. Stress induced by recombinant protein production in Escherichia coli. Adv Biochem Eng Biotechnol 89:73–92. doi: 10.1007/b93994 [DOI] [PubMed] [Google Scholar]
- 42. Soares A, Azevedo A, Gomes LC, Mergulhão FJ. 2019. Recombinant protein expression in biofilms. AIMS Microbiol 5:232–250. doi: 10.3934/microbiol.2019.3.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional experimental details, additional results.