SUMMARY
mRNA in eukaryotic cells is packaged into highly compacted ribonucleoprotein particles (mRNPs) in the nucleus and exported to the cytoplasm for translation. mRNP packaging and export require the evolutionarily conserved transcription-export (TREX) complex. TREX facilitates loading of various RNA-binding proteins on mRNA through the action of its DDX39B subunit. SARNP (Tho1 [transcriptional defect of Hpr1 by overexpression 1] in yeast) is shown to interact with DDX39B and affect mRNA export. The molecular mechanism of how SARNP recognizes DDX39B and functions in mRNP assembly is unclear. Here, we determine the crystal structure of a Tho1/DDX39B/RNA complex, revealing a multivalent interaction mediated by tandem DDX39B interacting motifs in SARNP/Tho1. The high-order complex of SARNP and DDX39B is evolutionarily conserved, and human SARNP can engage with five DDX39B molecules. RNA sequencing (RNA-seq) from SARNP knockdown cells shows the most affected RNAs in export are GC rich. Our work suggests the role of the high-order SARNP/DDX39B/RNA complex in mRNP assembly and export.
In brief
Xie et al. describe a multivalent interaction between the mRNA export factors SARNP and DDX39B. They present the crystal structure of a ScTho1SARNP/DDX39B/RNA complex and show that SARNP features tandem DDX39B interacting motifs. This study highlights the role of DDX39B in regulating mRNP dynamics and assembly through high-order structures.
Graphical Abstract

INTRODUCTION
In eukaryotic cells, mRNA is transcribed in the nucleus. Newly synthesized mRNA is rapidly bound with protein factors to form a messenger ribonucleoprotein particle (mRNP) and is exported to the cytoplasm for translation.1–3 Although the principles of its assembly are not known, the mRNP is thought to exist in a highly compacted form. The exon junction complexes (EJCs) in humans multimerize with one another and with SR (serine- and arginine-rich) proteins to form high-order structures that are hypothesized to mediate mRNP compaction.1,2 In yeast, electron microscopy analysis of isolated mRNPs revealed 5- to 7-nm-wide structures, and the poly(A)-binding protein Nab2 may function in mRNP compaction.4 In general, mRNP maturation in the nucleus involves a large number of RNA-binding proteins.5 However, which protein factors are involved and how they mediate mRNP higher-order assembly remains elusive.
The transcription-export (TREX) complex serves as an mRNP assembly platform by recognizing various mRNA biogenesis factors such as the 5′ cap-binding complex and by facilitating loading of export factors including SR proteins and the mRNA export receptor NXF1/NXT1 onto mRNA.3,6,7 As the sole enzymatic component of the TREX complex, the evolutionarily conserved DEAD-box ATPase DDX39B (also called UAP56) is essential for mRNA export.8,9 In addition to DDX39B, TREX also contains a multisubunit transcriptional defect of Hpr1 by overexpression (THO) complex and an export adaptor protein ALYREF. In yeast, the DDX39B ortholog Sub2 is regulated by the THO complex.10–13 Recent structural and functional studies have elucidated the molecular mechanisms of how direct interactions with the Tho2 subunit of THO hold Sub2 in a half open conformation and prime it for RNA binding.12–15 Human THO regulates DDX39B activity with a similar mechanism.16
As a well-documented mRNP assembly factor, the EJC consists of the DEAD-box protein eIF4A3, as well as MAGOH, Y14, and MLN51. EJC forms a stable complex on RNA in the nucleus, and it accompanies mRNP to travel through the nuclear pore complex into the cytoplasm.17–20 Before translation, the ribosome-associated protein PYM disassembles the EJC from mRNP through direct interactions with MAGOH and Y14.21 Interestingly, interaction between eIF4A3 and the ALYREF subunit of TREX as well as the juxtaposition of EJC and ALYREF on mRNP suggest a possible coordination of the molecular events mediated by the two DEAD-box proteins eIF4A3 and DDX39B in nuclear mRNP assembly.22,23
The functions of DDX39B in mRNP assembly are facilitated by multiple mRNA export adaptor proteins, such as the ALYREF subunit of the TREX complex, UIF, CHTOP, and Luzp4. These adaptors all use a similar UAP56 binding motif (UBM) to recognize DDX39B.8,12,24–26 Our previous structural studies demonstrated that Yra1, the yeast ortholog of ALYREF, binds to the N-terminal RecA domain of Sub2 and that their interaction promotes Yra1 and Sub2 assembly into the mRNP.12
DDX39B also interacts with human SARNP (SAP domain-containing ribonucleoprotein), which is implicated in mRNA export.9 SARNP was initially identified from erythropoietin-stimulated UT7/Epo cells (thereby also known as cytokine-induced protein 29 kDa [CIP29]) and shows high expression levels in various cancer cells.27–30 SARNP does not have a UBM, and how SARNP recognizes DDX39B and facilitates mRNP assembly is not known. The role of SARNP in RNA metabolism is evolutionarily conserved. In Arabidopsis, mutations to the SARNP ortholog Mos11 show impaired mRNA export,31 and the function of Mos11 is also connected to the TREX complex.32 In yeast, the SARNP ortholog Tho1 is recruited to actively transcribed genes.33 Overexpression of Tho1 partially suppressed the mRNA export defect that results from a null mutation of the THO subunit Hpr1.34 Interestingly, Sub2 is also a multicopy suppressor for the hpr1 null mutant.33 We interpret these data to suggest functional links between Tho1/SARNP, Sub2/DDX39B, and the THO complex in mRNP assembly and export; however, the underlying mechanism is not clear.
In this work, we carried out structural and functional studies of SARNP to provide mechanistic insights into its mode of action and into the interplay among SARNP, DDX39B, and TREX in mRNP assembly and export. We determined the crystal structure of a chimeric complex containing human DDX39B, yeast Tho1, and RNA. This structure reveals that SARNP facilitates the formation of DDX39B multimers on RNA, potentially covering longer stretches of RNA for more efficient mRNP packaging. The structure also indicates that the SARNP and THO binding sites on DDX39B partially overlap, suggesting they act at different steps during the functional cycle of the ATPase. We found that knockdown of SARNP led to poly(A) RNA accumulation in nuclear speckles. Furthermore, RNA sequencing (RNA-seq) analysis revealed that knockdown of SARNP affected mRNA export of a subset of genes, particularly GC-rich mRNAs, which is a feature also shared by mRNAs that are dependent on DDX39B and ALYREF for nuclear export.35 Together, our studies provide the structural basis for a multivalent interaction between SARNP and DDX39B on RNA, which we suggest facilitates high-order mRNP assembly that is important for mRNP export.
RESULTS
Evolutionarily conserved assembly of SARNP and DEAD-box protein DDX39B on RNA
DDX39B, like other DEAD-box proteins, is an RNA-dependent ATPase containing a functional core composed of two RecA-like domains, DDX39B-NTD (N-terminal domain) and DDX39B-CTD (C-terminal domain) (Figure 1A). Using an electrophoretic mobility shift assay (EMSA) with poly(U) RNA, we show that purified recombinant DDX39B forms a stable complex on RNA in the presence of the slowly hydrolysable ATP analog ATP-γ-S (Figures 1B and S1). We also found that SARNP has intrinsic RNA-binding ability (Figure 1B). When both DDX39B and SARNP were mixed with RNA, a slower migrating band appeared, suggesting the formation of a DDX39B/SARNP/RNA complex (Figure 1B). The yeast ortholog of SARNP, Tho1, also can bind to RNA (Figures 1C and S1). The major RNA-binding activity was shown to reside in the highly positively charged C-terminal region of Tho1.33 Like their human orthologs, yeast Tho1 and DEAD-box protein Sub2 also form a complex on RNA (Figure 1C). Of note, both SARNP and Tho1 facilitate RNA binding of the ATPase, as indicated by the lower amount of free RNA in the presence of SARNP and Tho1 (Figures 1B and 1C). Conceivably, the intrinsic RNA-binding abilities of SARNP and Tho1 contribute additional interactions to assemble the protein-RNA complex. Similar results were observed with Yra1 in our previous studies on the assembly of Yra1 and Sub2 on RNA.12 Simultaneous binding of RNA by DDX39B and SARNP could serve as a mechanism for concomitant loading of DDX39B and SARNP to remodel the mRNP. We also found that human DDX39B can cross species to form a chimeric complex with yeast Tho1 on RNA (Figure 1D). Therefore, the assembly of SARNP, DDX39B, and RNA is evolutionarily conserved.
Figure 1. SARNP and DEAD-box protein DDX39B assemble on RNA.
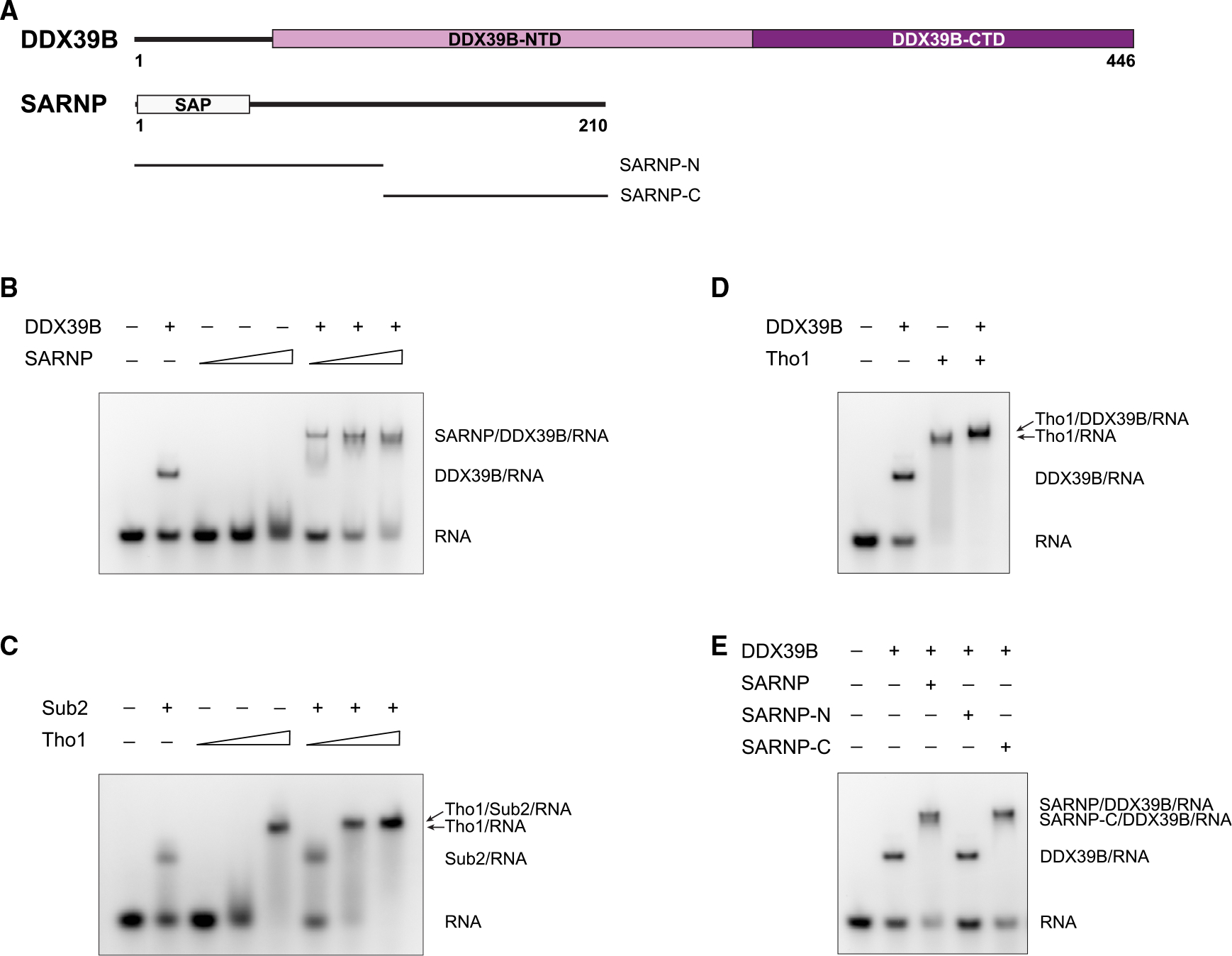
(A) Domain organization of human DDX39B and SARNP. DDX39B contains two RecA-like domains, DDX39B-NTD and DDX39B-CTD. SARNP contains a SAP domain.
(B) DDX39B and SARNP assemble on RNA. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and SARNP at increasing concentrations (1, 3, and 10 μM) as indicated. Data are representative of three technical replicates.
(C) Evolutionarily conserved assembly as shown with yeast Sub2 and Tho1. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, Sub2 at 3 μM, and Tho1 at increasing concentrations (0.3, 1, and 3 μM) as indicated. Data are representative of three technical replicates.
(D) A chimeric assembly between human DDX39B and yeast Tho1 on RNA. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and Tho1 at 3 μM. Data are representative of three technical replicates.
(E) SARNP-C is sufficient to assemble with DDX39B on RNA. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and different SARNP proteins at 3 μM. Data are representative of three technical replicates.
SARNP contains an N-terminal SAP domain (a putative DNA-binding motif named after SAF-A/B, Acinus, and PIAS). The rest of the protein contains no known domains but is required in yeast Tho1 to suppress the defects of the hpr1 null mutant.33 To identify the DDX39B-binding region in SARNP, we dissected SARNP into a SAP domain containing SARNP-N and a C-terminal construct SARNP-C (Figure 1A). We found that SARNP-C mediates the interaction with DDX39B (Figure 1E).
Structural basis for the SARNP/DDX39B/RNA assembly
After extensive screening for DDX39B bound to different constructs from both human SARNP and yeast Tho1, we crystallized DDX39B (residues 44–428) in association with yeast Tho1 (residues 123–178) and poly(U) 15-mer RNA in the presence of the non-hydrolyzable ATP analog ADP-BeF3 and solved the structure at 2.5 Å resolution (Figures 2A and S2; Table S1). Intriguingly, the Tho1/DDX39B/RNA structure reveals a 1:2 ratio of Tho1 and DDX39B in the complex. The two DDX39B-Tho1 interfaces in the Tho1/DDX39B/RNA complex resemble each other. Tho1 contains two α-helices that are packed antiparallel. The C-terminal end of each α-helix recognizes the DDX39B-CTD of one DDX39B molecule (Figure 2B). We hereafter refer to this α-helical motif as DIM (DDX39B interacting motif); thus, Tho1 contains two DIMs, each bound to one DDX39B molecule.
Figure 2. Crystal structure of a Tho1/DDX39B/RNA complex at 2.5 Å resolution.
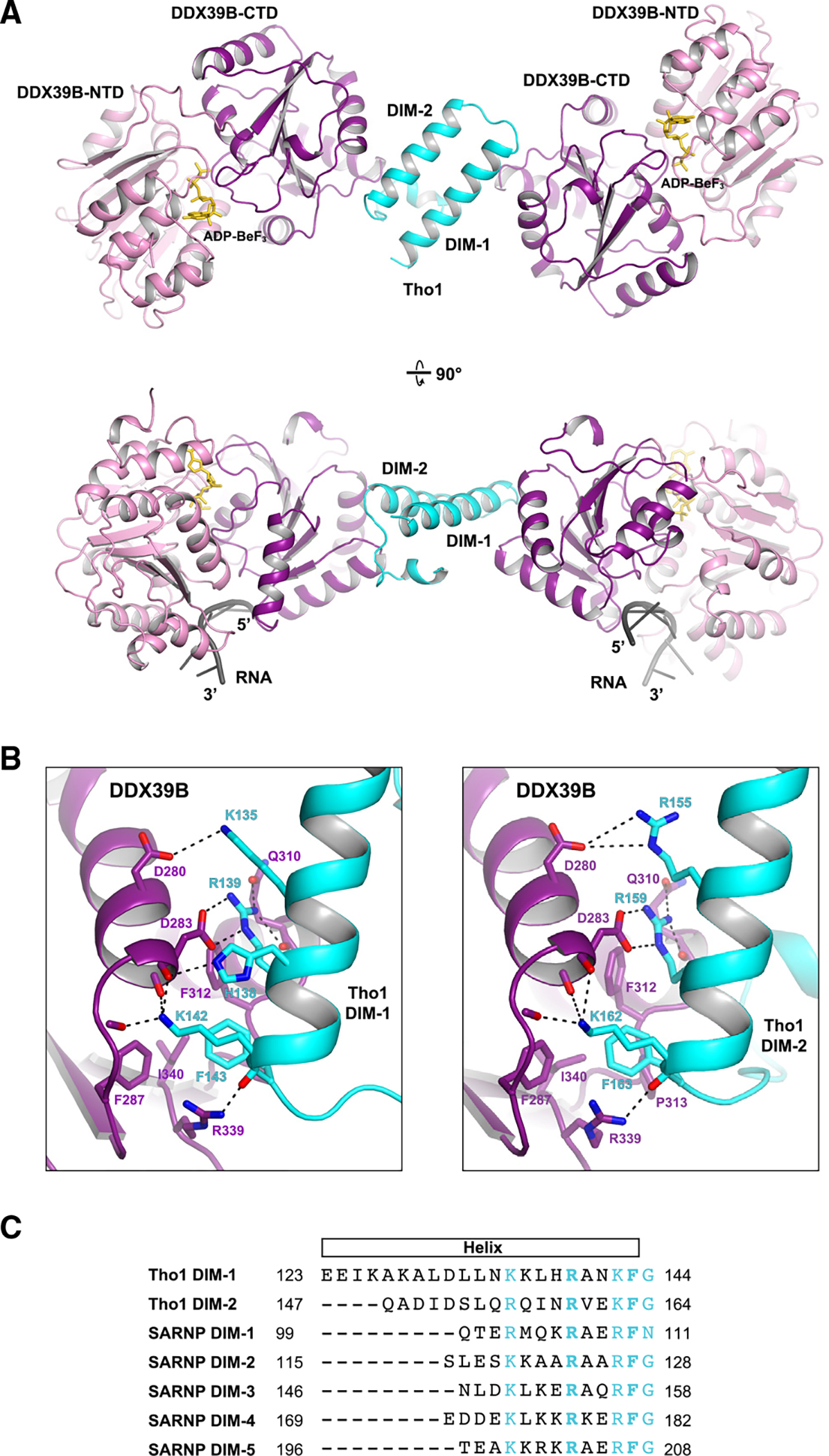
(A) DDX39B was crystallized with a C-terminal domain of yeast Tho1 in the presence of poly(U) 15-mer RNA and the non-hydrolyzable ATP analog ADP-BeF3. The model shown in two orientations features a 1:2 assembly of Tho1 and DDX39B. The structure reveals a DDX39B interacting motif (DIM) that recognizes the DDX39B-CTD domain.
(B) DDX39B interfaces with DIM-1 (left) and DIM-2 (right) of Tho1.
(C) Alignment of the two DIMs in yeast Tho1 and five DIMs in human SARNP. Conserved residues are highlighted in cyan. The two invariant residues (R5 and F9) are shown in bold.
Alignment of the DIM-1 and DIM-2 sequences (Figure 2C) reveals the conserved residues in this motif, all of which situate on one side of the α-helix interfacing with DDX39B (Figure 2B). Both DIM-1 and DIM-2 contain arginine residues (R139 and R159) that form two salt bridges with DDX39B D283 and two hydrogen bonds with the main-chain and side-chain carbonyl groups of DDX39B Q310. Both DIMs also contain phenylalanine residues (F143 and F163) that are buried in a hydrophobic pocket formed by F287, F312, P313, and I340 of DDX39B. In addition, lysine residues (K142 in DIM-1 and K162 in DIM-2) form hydrogen bonds with three main-chain carbonyl groups in DDX39B. There are modest differences between DIM-1 and DIM-2 binding to DDX39B. For example, DIM-1 features an additional interaction mediated by H138 and the main-chain carbonyl group of DDX39B D283.
Within the Tho1/DDX39B/RNA complex, each DDX39B molecule binds to the phosphate-ribose backbone of an RNA molecule. Like other DEAD-box proteins, RNA binding of DDX39B involves the conserved motifs from both the NTD and the CTD (Figure S3).36 The two RNA molecules in this complex both face the same side of the complex. The footprint of DDX39B is 6 nucleotides, and the distance between the 3′ end of one RNA molecule to the 5′ end of the other RNA molecule is 7 nm. Although the U15 RNA used in crystallization cannot bridge two DDX39B molecules, the structure suggests that Tho1/DDX39B could assemble on a single RNA molecule with sufficient length.
Evolutionarily conserved multivalent assembly of SARNP and DDX39B
Intriguingly, analysis of the human SARNP protein sequence reveals the presence of five DIMs (Figure 2C). This observation prompted us to analyze the orthologous SARNP sequences from a variety of eukaryotes from yeast to humans. We found that SARNP orthologs contain various numbers of DIMs, ranging from two as in S. cerevisiae Tho1 to as many as six as in C. elegans SARNP (Figure 3A). In humans, SARNP contains five DIMs; thus, one human SARNP potentially can engage with five DDX39B molecules. SARNP and DDX39B together can bind to a long stretch of RNA, forming a high-order mRNP assembly. Analysis of all 37 DIMs in representative species shown in Figure 3A reveals the consensus sequence of DIM (K1×2×3×4R5×6×7R/K8F9G10) (Figure 3B). The most conserved residues are residues 5 (arginine) and 9 (phenylalanine), which are key residues mediating DDX39 binding (Figure 2B) and are invariable in all 37 DIMs. Residue 8 is also highly conserved, featuring either arginine or lysine residues. In addition, residues 1 and 10 have preferences for lysine/arginine and glycine, respectively.
Figure 3. Evolutionarily conserved multivalent assembly between SARNP and DDX39B.
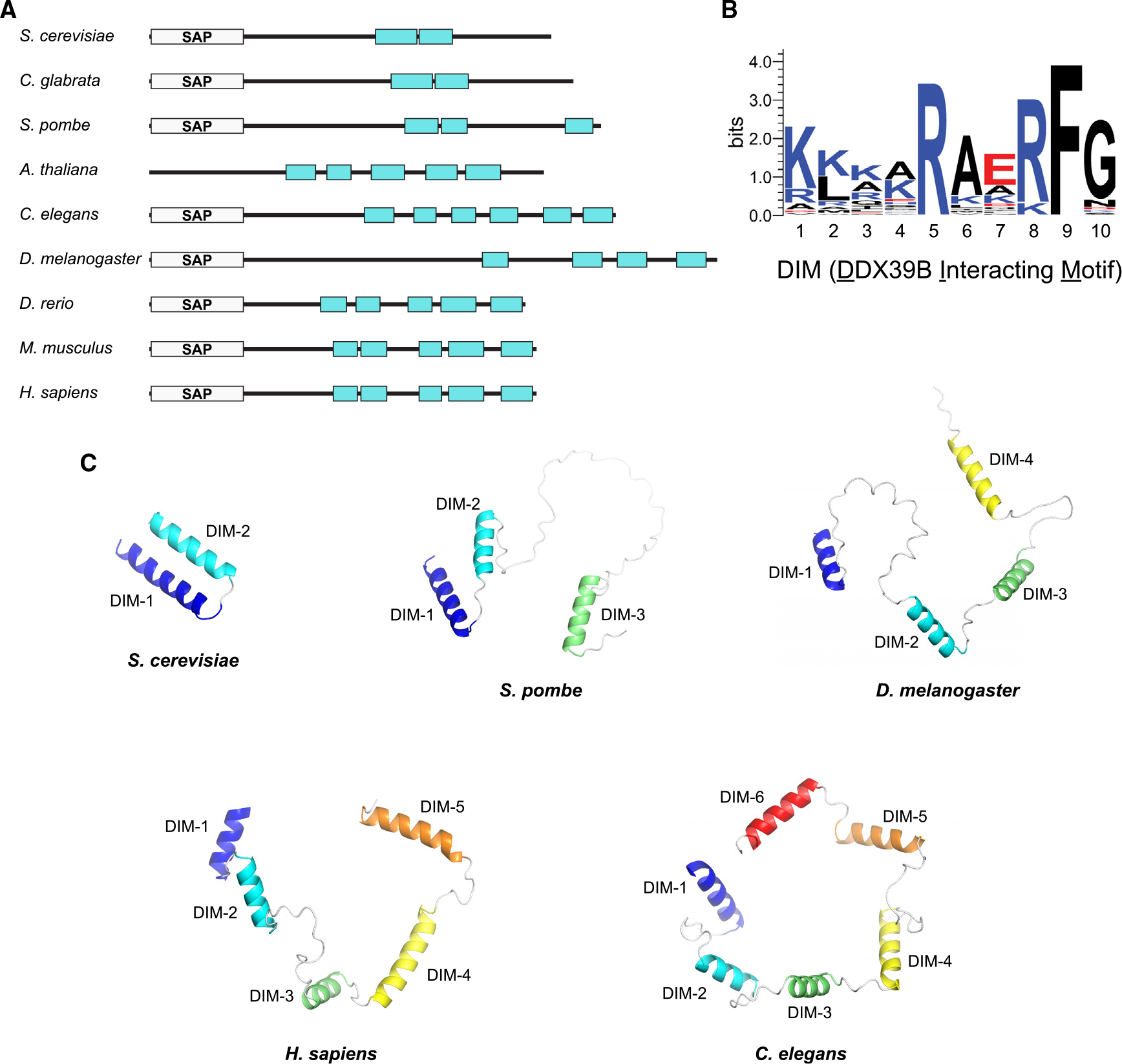
(A) Domain organization of SARNP in different organisms. DIM is represented as cyan rectangles.
(B) Sequence logo of the DIM motif generated with WebLogo.
(C) Comparison of SARNP structures in different organisms featuring 2–6 DIMs. For simplicity, only the DIM-containing C-terminal region is shown. Except for S. cerevisiae protein, which uses the structure reported here, all others use AlphaFold-predicted structures (AF-O74871-F1 for S. pombe, AF-Q9VHC8-F1 for D. melanogaster, AF-P82979-F1 for H. sapiens, and AF-Q9N3G0-F1 for C. elegans).
We compared the predicted structures of SARNP in various species by AlphaFold (Figure 3C). The ortholog in C. glabrata, like S. cerevisiae Tho1, contains two DIMs that interact with each other. Interestingly, in higher organisms, the multiple DIMs in SARNP do not interact with each other and are spaced out. These DIMs are connected by linkers with various lengths. Such versatile arrangement conceivably can accommodate a more flexible configuration of SARNP/DDX39B on RNA to facilitate high-order mRNP assembly.
To validate the DDX39-DIM interaction observed in the crystal structure, we generated a Tho1 mutant (Tho1-mut) in which the invariant residues R5 and F9 are mutated to alanine in both DIM-1 and DIM-2. We performed EMSAs using poly(U) RNA with DDX39B and Tho1 (Figures 4A and S1). In contrast to wild-type Tho1, which showed a slower migrating band indicating the Tho1/DDX39B/RNA complex, Tho1-mut with both crucial DIM residues altered did not produce a new band. Of note, Tho1-mut alone showed reduced RNA interaction compared with wild-type Tho1, suggesting that the positively charged residues of DIMs contribute to the intrinsic RNA-binding ability of Tho1. We also mutated residues R5 and F9 in all five DIMs in human SARNP. The corresponding SARNP-mut1 protein did not form a complex with DDX39B on RNA (Figures 4B and S1).
Figure 4. Characterization of DIM interaction with Sub2/DDX39B.
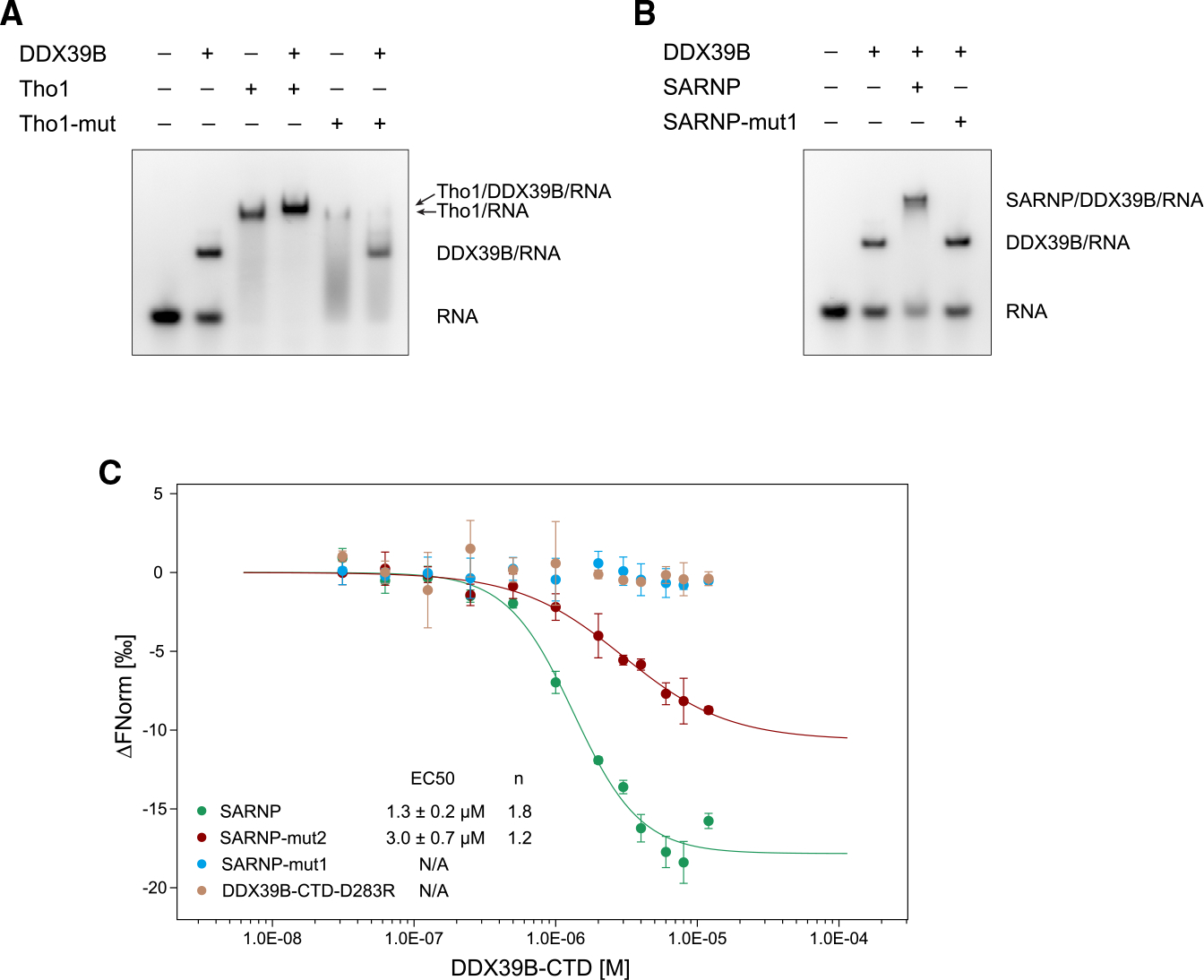
(A) Mutation of the R5 and F9 residues in yeast Tho1 DIMs (Tho1-mut) disrupted binding of DDX39B. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and Tho1 or Tho1-mut (R139A/F143A/R159A/F163A) at 3 μM. Data are representative of three technical replicates.
(B) Mutation of the R5 and F9 residues in all SARNP DIMs (SARNP-mut1) disrupted binding of DDX39B. EMSA was carried out with poly(U) 15-mer RNA at 100 nM, DDX39B at 0.3 μM, and SARNP or SARNP-mut1 (R106A/F110A/R123A/F127A/R153A/F157A/R177A/F181A/R203A/F207A) at 3 μM. Data are representative of three technical replicates.
(C) Microscale thermophoresis (MST) analysis of the SARNP-DDX39B interaction. Measurements of SARNP, SARNP-mut2 (R153A/F157A/R177A/F181A/R203A/F207A, DIM3–5 mutated), or SARNP-mut1 binding to DDX39B-CTD are colored in green, red, and blue, respectively. Measurements of SARNP binding to DDX39B-CTD-D283R are colored in brown. Data were fitted using the Hill equation. EC50 and Hill coefficient (n) are shown. Error bars represent SD from three technical replicates.
We next sought to examine the role of the multiple DIMs of SARNP in DDX39B interaction. We generated a SARNP mutant (SARNP-mut2, R153A/F157A/R177A/F181A/R203A/F207A) that contains only two functional DIMs (DIM-1 and DIM-2). Microscale thermophoresis (MST) was used to compare the binding of wild-type SARNP, SARNP-mut1, and SARNP-mut2 to the DDX39B-CTD (Figures 4C and S1). Wild-type SARNP showed an n and an EC50 of 1.8 and 1.3 μM, respectively, with a Hill model fitting, suggesting that multiple DDX39B-CTD molecules bind to SARNP. SARNP-mut2 showed an n and an EC50 of 1.2 and 3 μM, respectively, suggesting that the remaining two DIMs in SARNP-mut2 are capable of binding DDX39B and that the lack of functional DIM-3 to DIM-5 reduced DDX39B interaction. In line with the EMSA experiments, mutations of all DIMs abolished DDX39B interaction (Figure 4B). In addition, mutation of the D283 residue on DDX39B, a key residue at the DDX39B-DIM interface (Figure 2B), abolished SARNP binding. Together, our biochemical and structural studies reveal a multivalent interaction between SARNP and DDX39B.
Comparison of SARNP, ALYREF, and THO complex binding to DDX39B
We previously determined a crystal structure of the yeast Sub2-Yra1-RNA complex.12 Yra1 recognizes the NTD of the ATPase as opposed to SARNP, which binds to the CTD (Figures 5A and S3). Therefore, ALYREF and SARNP can function together to assemble with DDX39B on RNA.
Figure 5. Comparison of SARNP, Yra1/ALYREF, and THO complex binding to DDX39B.
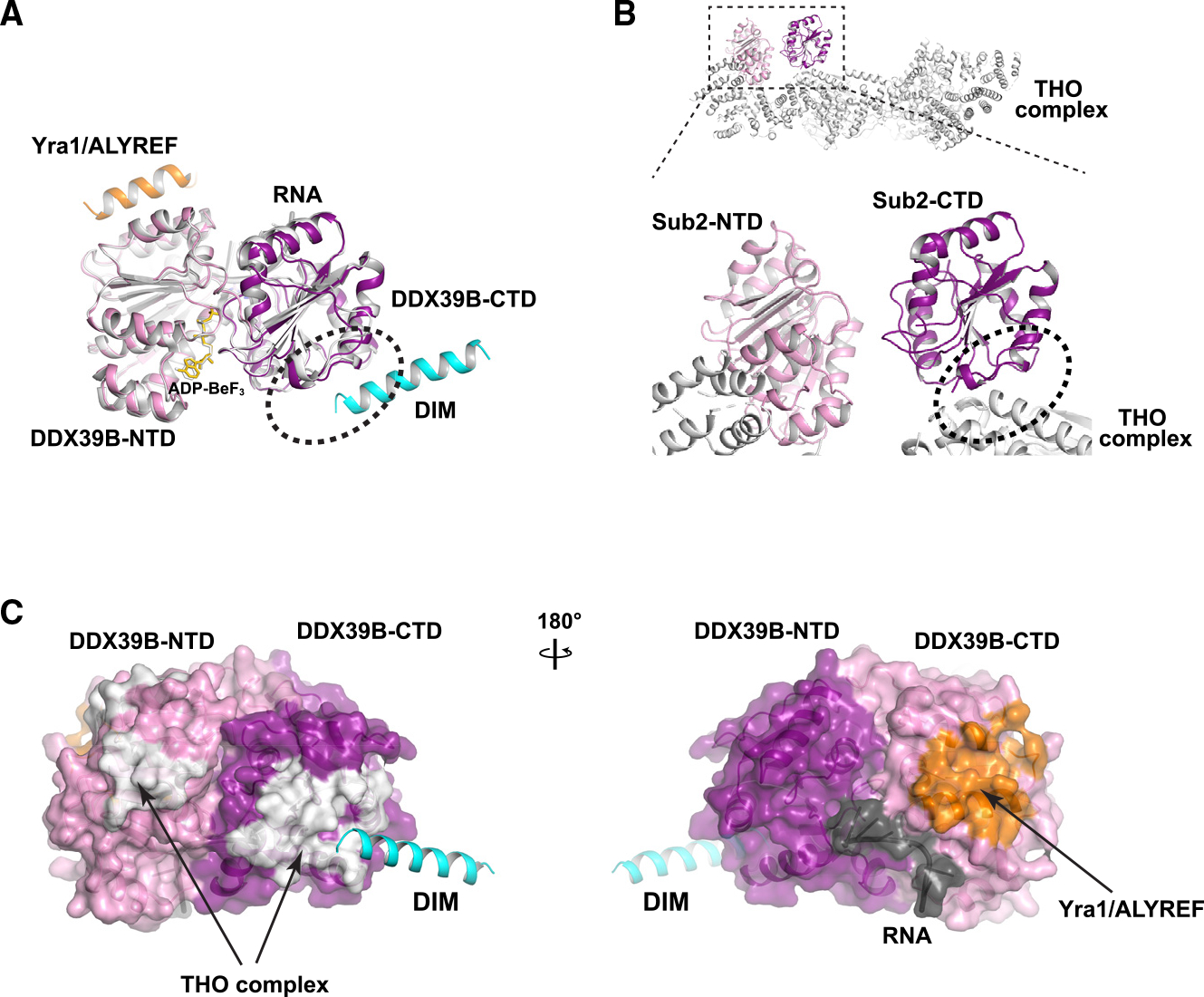
(A) Yra1/ALYREF and DIM bind to different domains of DDX39B. The DDX39B/Tho1/RNA structure, colored as in Figure 2A, is overlayed on the Sub2/Yra1/RNA structure (PDB: 5SUP), colored in gray except for Yra1 (orange).
(B) THO complex (PDB: 7LUV) and DIM bind to overlapping regions (indicated by an oval) on DDX39B-CTD.
(C) Comparison of Tho1/SARNP, Yra1/ALYREF, and THO binding interfaces on DDX39B. DDX39B is shown in surface model. Yra1/ALYREF and THO binding interfaces are analyzed based on their structures in complex with Sub2 shown in (A) and (B); corresponding residues in DDX39B are colored in white and orange, respectively.
Sub2/DDX39B is recruited to the transcription machinery via the THO complex. Cryoelectron microscopy (cryo-EM) structures of Sub2/DDX39B in complex with THO determined by us and others reveal that THO recognizes both the NTD and the CTD of the ATPase.12–16 Intriguingly, DIMs and THO bind to overlapping regions on the CTD of the ATPase (Figures 5B and 5C). This observation suggests that SARNP and THO act at different steps during the ATPase reaction cycle. The THO-Sub2 complex structure captures a snapshot in which the ATPase is “primed” for later engagement with RNA. The crystal structures of Tho1/SARNP and Yra1 in association with the ATPase represent the “engaged” state in which the ATPase binds to RNA and concomitantly loads Tho1/SARNP and Yra1 onto RNA. The overlapping binding of THO and SARNP on DDX39B suggests that some conformational changes have to occur for the THO-Sub2/DDX39B complex as the ATPase transitions from the “primed” state to the “engaged” state.
Function of SARNP in mRNA nuclear export
To determine the effect of SARNP on mRNA export in situ, we knocked down SARNP levels using small interfering RNAs (siRNAs) and assessed its protein levels, localization, and impact on the intracellular distribution of mRNAs. SARNP levels were efficiently knocked down, as observed by the robust decrease in its intranuclear pool colocalized with the nuclear speckle marker SC35 (Figure 6A) and its low levels detected by western blot (Figure 6B). For clarification, the SC35 antibody used in this study recognizes the SC35 protein and not the SRRM2 protein, as the latter is known to be recognized by another antibody previously characterized as an SC35 antibody. The low levels of SARNP resulted in inhibition of nuclear export of a subset of poly(A) RNA at nuclear speckles, which is shown by RNA fluorescence in situ hybridization (RNA-FISH) combined with immunofluorescence to label the nuclear speckle assembly factor SON (Figures 6C and 6D).
Figure 6. SARNP knockdown inhibits nuclear export of a subset of mRNAs with high GC content.
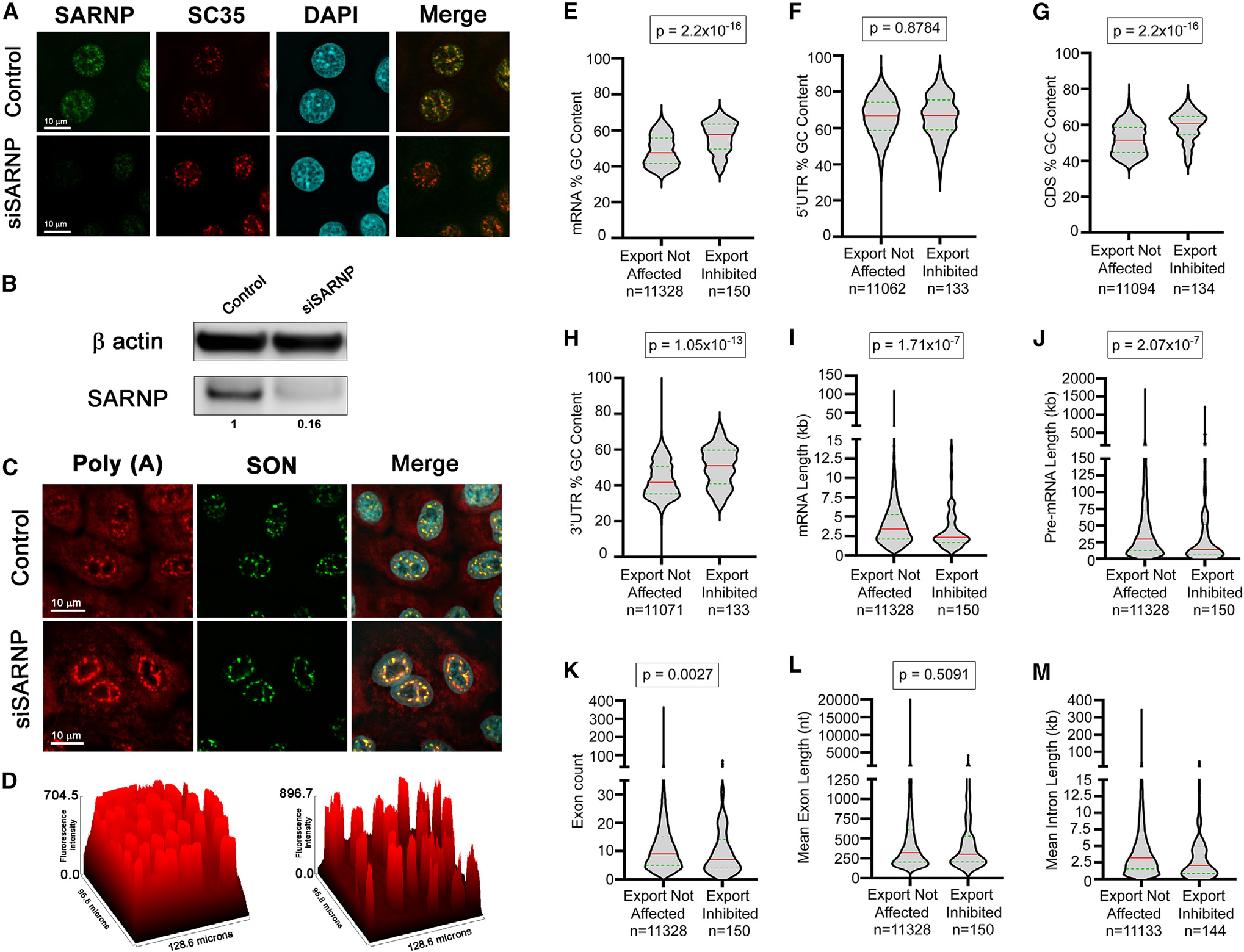
(A–D) A549 cells were transfected with nontargeting siRNAs or with siRNAs that target SARNP. After 48 h, cells were subjected to immunofluorescence microscopy (A) or western blot analysis (B) with the depicted antibodies or were subjected to RNA-FISH to detect the intracellular distribution of poly(A) RNA in control cells and in cells depleted of SARNP (C). The nuclear speckle assembly and splicing factor SON is used as a nuclear speckle marker. The graph in (D) depicts accumulation of poly(A) RNA at nuclear speckles upon SARNP depletion (right) compared to control cells (left). The surface plot tool of Fiji (ImageJ) was used to calculate the fluorescence intensity of poly(A) RNA.
(E–M) A549 cells were transfected with non-targeting siRNAs or with siRNAs that target SARNP. After 48 h, RNA was isolated from whole-cell, cytoplasmic, and nuclear fractions. RNA-seq data obtained from these samples in two biological replicates were analyzed to identify RNA features associated with cellular mRNAs that are dependent on SARNP for their nuclear export. Violin plots show the distribution of RNA features between exports not affected (left) and export-inhibited (right) transcripts upon SARNP knockdown. The red line indicates the median, and the green line indicates quartiles. Mann-Whitney U rank test was used to calculate statistical significance. p values are shown on the top of each plot. Number of transcripts in each group (n) is mentioned below the graphs.
We then sought to identify the mRNAs blocked in the nucleus by the low levels of SARNP. RNA-seq analysis was performed in nuclear and cytoplasmic fractions of cells treated with control siRNAs or with siRNAs that target SARNP. The high levels of MALAT1 RNA and NEAT1 RNA in the nuclear fractions and the higher levels of GAPDH or actin mRNAs in the cytoplasm than in the nucleus further validated the fractionation procedure (Table S2). We identified 203 mRNAs whose nuclear/cytoplasmic (N/C) ratios were 1.5-fold or higher in SARNP knockdown cells than in control cells. The mRNAs with relative N/C ratios between 0.66 and 1.5 were considered as not affected at the export level, and there were 12,594 mRNAs in this category. We then analyzed several mRNA features to determine whether the SARNP-dependent mRNAs shared specific signatures. We found that this subset of mRNAs that are dependent on SARNP for export had significantly higher GC content than mRNAs whose export was not altered by SARNP knockdown (Figure 6E). Additionally, the high GC content was localized in the coding region and in the 3′ UTR but not in the 5′ UTR (Figures 6E–6H). Other features shared by the SARNP-dependent mRNAs include slightly shorter mRNA and pre-mRNA lengths (Figures 6I and 6J) and lower exon numbers (Figure 6K) than mRNAs unaffected by SARNP depletion. Mean exon length was not different between the SARNP-dependent and -independent mRNAs (Figure 6L). However, SARNP-dependent mRNAs had a lower mean intron length compared with the mRNAs not affected by SARNP knockdown (Figure 6M). In sum, the most robust feature shared by the SARNP-dependent mRNAs is the high GC content, which is a feature also shared by mRNAs that are dependent on DDX39B and ALYREF for nuclear export.35 This is consistent with our structural findings that SARNP and ALYREF can act together with DDX39B.
DISCUSSION
Multivalent interaction and high-order mRNP assembly
In this study, we identified a DIM in SARNP and provide a structural basis for their recognition. The number of DIMs in SARNP correlates with the complexity of the genome, perhaps to cope with larger, multiple-exon genes in higher organisms and with the added complexity of mRNP assembly that these genes may require. S. cerevisiae Tho1 has only two DIMs, and most transcripts in that organism lack introns and are relatively short. The human SARNP contains five DIMs, and the multiple DDX39B interactions that they mediate may help to package longer transcripts more efficiently. Interestingly, the TREX complex also evolves a higher-order assembly in higher eukaryotes. Yeast TREX acts as an mRNP assembly platform in a dimeric form,12–15,37 whereas the human TREX complex can form a tetramer.16 To accommodate the increasing complexity of the transcriptome, more mRNP assembly factors appear during evolution such as the EJC not present in budding yeast, which contains the DEAD-box protein eIF4A3. Endogenously isolated EJCs from human cells form megadalton-sized complexes composed of EJC multimers. Interestingly, the EJC complexes can also multimerize with SR proteins to facilitate compaction of long mRNAs into compacted mRNPs.1 In addition to forming high-order complexes with their binding partners, some DEAD-box proteins, such as DDX3, can form multivalent interactions with themselves and with RNA via their low-complexity regions.38 Interestingly, human DEAD-box protein DDX4 has been shown to multimerize both through interaction with its LOTUS-domain-containing binding partners as well as through phase separation mediated by its N-terminal low-complexity region.39,40 Together, our work and recent studies by others have revealed an emerging role of DEAD-box proteins in regulating mRNP dynamics and assembly through multivalent interactions and their formation of high-order structures.
SARNP, ALYREF, DDX39B, and TREX in high-order mRNP assembly
The SARNP binding site on DDX39B significantly overlaps with the THO binding site (Figures 5B and 5C). For the multifunctional DEAD-box proteins, it is not uncommon for mutually exclusive interactions to mediate sequential processes. For example, eIF4A3 interacts with MLN51 and the spliceosome component Snu114 in a mutually exclusive manner.41 The observation in yeast that Tho1 binds to actively transcribed chromatin in a THO- and RNA-dependent manner suggests that Tho1/SARNP function downstream of THO.33
Our structural studies indicate that simultaneous SARNP and ALYREF binding to DDX39B are compatible, which is consistent with the observation that SARNP, ALYREF, and DDX39B together form a complex.9 We suggest that SARNP and ALYREF could work together to facilitate high-order mRNP assembly. Our studies also showed that the mRNAs whose nuclear export is mostly affected by SARNP share the common feature of high GC content with those affected by ALYREF and DDX39B. Such specificity could arise for multiple possible reasons. GC-rich sequences are prone to form RNA-DNA hybrid R-loop structures that interfere with mRNA biogenesis.42 DDX39B has been shown to be a major R-loop-resolving helicase.43 Depletion of SARNP could impair the ability of DDX39B in resolving R-loop structures and thus has a stronger effect on the assembly and export of mRNPs containing GC-rich transcripts. In addition, high GC content has been shown to facilitate mRNA nuclear export.44 This could arise from direct recognition of GC-rich sequences by RNA-binding proteins, such as RBM33.45 DDX39B, which shows no preference for RNA sequences, is the only known factor that directly interacts with SARNP. The observed specificity for GC content and other features of SARNP-sensitive transcripts could be contributed by thus far unidentified SARNP partners.
Interestingly, Yra1/ALYREF has two Sub2/DDX39B binding sites and thus can engage with two Sub2/DDX39B molecules. In yeast, the smallest assembly unit is two Sub2 molecules and one molecule each of Yra1 and Tho1 on RNA (Figure 7). Of note, Yra1 contains an RNA recognition motif (RRM) as well as arginine-rich regions that can independently bind RNA. Tho1 also contains an RNA-binding region at its C terminus.33 Given these multiple separate RNA-binding domains, there could be extensive interactions between the protein constituents and the RNA in this assembly. In humans, one SARNP molecule is capable of engaging with five DDX39B molecules as well as up to three ALYREF molecules on RNA. This unit could also repeat in order to drive assembly of high-order mRNP complexes and promote more efficient nuclear export.
Figure 7. Hypothetical model of high-order mRNP assembly mediated by SARNP, ALYREF, and DDX39B.
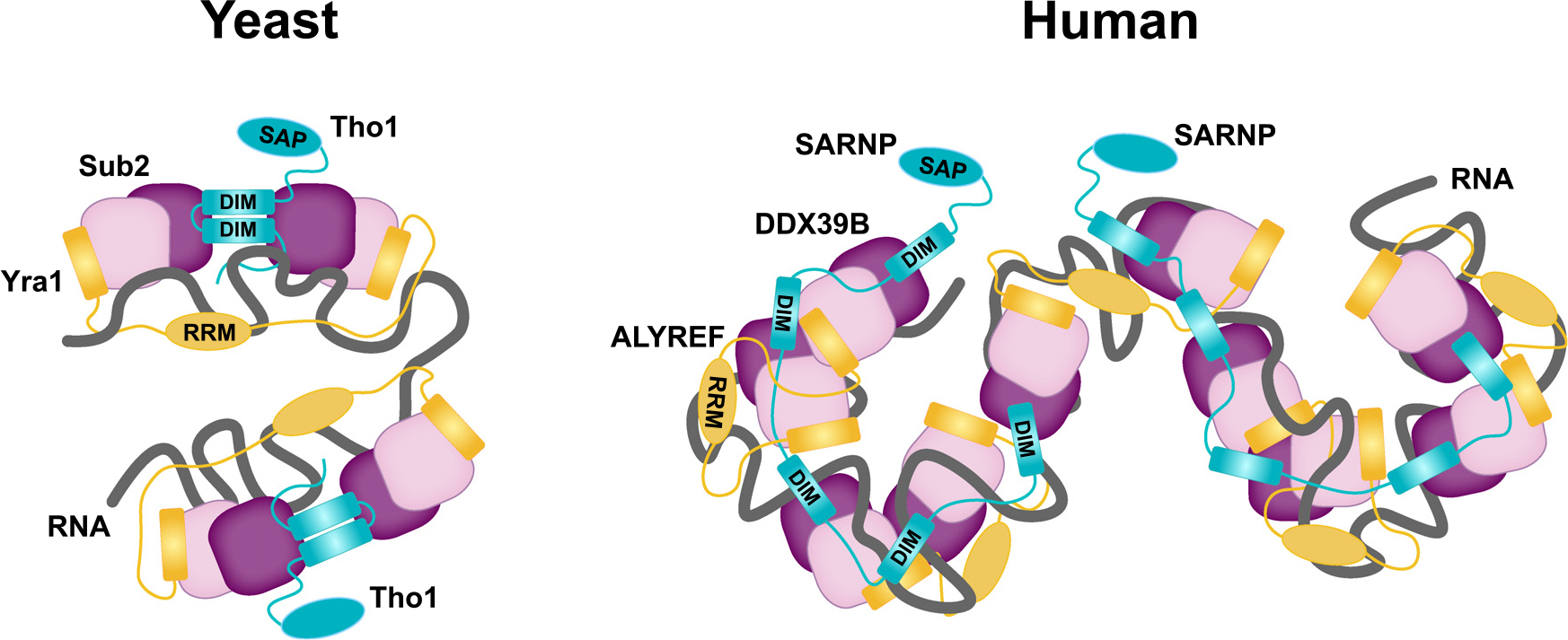
Multivalent interactions between Tho1/SARNP and the DEAD-box protein Sub2/DDX39B facilitate mRNP assembly and export. In yeast, one Tho1 molecule and one Yra1 molecule can assemble with two Sub2 molecules on RNA. In humans, one SARNP molecule can engage with five DDX39B molecules and up to three ALYREF molecules on RNA.
Increasing evidence suggests that TREX acts coordinately with the EJC and SR proteins. For example, ALYREF interacts with the EJC.22 UV crosslinking and immunoprecipitation studies indicate that they are localized in proximity on mRNPs.23 In addition, our recent studies have revealed molecular links between TREX, the SR-like protein Gbp2, and the RNA polymerase II (RNA Pol II) CTD-kinase CTDK-1 complex in yeast.13,46 SR proteins are enriched with isolated EJC from human cells and have been hypothesized to facilitate mRNP compaction.1 It is of particular interest in future studies to determine how SARNP and TREX cooperate with EJC and SR proteins to drive the highly orchestrated processes involved in nuclear mRNP maturation and assembly.
SARNP and DDX39B in cancer and viral infection
SARNP was initially identified as a cytokine-induced protein involved in cell proliferation.27 It has been shown to be expressed at elevated levels in a variety of cancer cell types.28–30 DDX39B is also upregulated in various cancer cells, and its overexpression promotes cancer cell growth and metastasis.47–51 In addition, ALYREF and THO are overexpressed in several different types of human cancers.52,53 The upregulation of these different mRNP assembly factors in human cancers and their connection to cell proliferation indicate a crucial role for more efficient mRNP assembly and their increased export in giving a growth advantage to cancer cells. Indeed, it has been shown that depletion of DDX39B sensitizes HeLa cells to DNA-damaging chemotherapeutic agents.54
SARNP and DDX39B are also highly relevant in the context of viral infections. For example, SARNP was identified as one of the top host proteins with antiviral activity, specifically against Middle East respiratory syndrome (MERS) virus in a CRISPR screen targeting host RNA-binding proteins that bind viral RNA.55 Additionally, DDX39B is exploited by influenza A virus to facilitate viral RNP (vRNP) assembly through interaction with influenza NP protein.56,57 Furthermore, DDX39B knockdown inhibits influenza virus mRNA export.58–61 DDX39B was also shown to be an antiviral target for Kaposi’s sarcoma-associated herpesvirus (KSHV).62
Given its many roles at various steps of mRNP maturation and export, as well as its known importance in both cancer and viral infection, DDX39B is an attractive target for the development of therapeutic agents. So far, molecules utilized to inhibit DDX39B target its ATP-binding pocket.62 Since DDX39B also functions in other aspects of RNA metabolism, such as splicing and R-loop resolution,43,63 inhibition of its ATPase activity would affect all these processes and could have undesired off-target effects. Our studies here provide crucial mechanistic insights into SARNP/DDX39B/RNA assembly, and the highly conserved and specific interaction between SARNP-DIM and DDX39B is thus an attractive drug target that warrants further investigation.
Limitations of the study
Although our structural and biochemical studies provide strong evidence for a high-order assembly of the SARNP-DDX39B complex, we have not been able to reconstitute a defined complex in vitro due to the modest stability of the complex. The configuration and the functional importance of the high-order SARNP-DDX39B complex in the cellular context remain to be elucidated. In addition, our RNA-seq analysis reveals that mRNAs that are dependent on SARNP for nuclear export share the feature of high GC content with mRNAs that are dependent on DDX39B and ALYREF. How the TREX complex and related factors such as ALYREF and SARNP coordinate and/or divide the labor in mRNP assembly is an interesting future direction. Last, our data cannot differentiate whether multiple DIMs in SARNP bridge long-range interactions within one mRNA or between different mRNAs. Given the latter possibility, it would be interesting to examine whether SARNP plays a role in the formation of RNA granules or membraneless compartments in future studies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yi Ren (yi.ren@vanderbilt.edu).
Materials availability
Plasmids generated in this study will be available without restrictions upon request.
Data and code availability
The atomic coordinates of the crystal structure have been deposited in the RCSB Protein DataBank with the ID 8ENK. The RNA-seq data have been deposited in the BioProject database with the accession number PRJNA893026. All data are publicly available as of the date of the publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell lines
A549 human lung adenocarcinoma cells were obtained from the American Type Culture Collection (ATCC, Cat. CCL-185). Cells were maintained in DMEM media (Thermo Fisher, Cat. 11965092) supplemented with 10% fetal bovine serum (Sigma-Aldrich, Cat. F4135) and 100 units/mL Pen Strep antibiotics (Thermo Fisher Cat. 15140–122) at 37°C with 5% CO2. Cells were tested once a month and proven negative for mycoplasma contamination.
METHOD DETAILS
Antibodies
SARNP polyclonal antibody (Invitrogen, PA5–56586) was used at a 1:250 dilution in immunofluorescence assay (IFA) and 1:1000 dilution for Western blot assay. β-actin monoclonal antibody (Invitrogen AM4302) was used at 1:5000 dilution in Western blot. SC35 monoclonal antibody (Sigma Aldrich SAB4200725) was used at 1:500 dilution for IFA. SON polyclonal antibody (Genetex GTX129778) was used at 1:500 dilution for IFA.
Plasmids
SARNP (residues 1–210), SARNP-N (residues 1–106), SARNP-C (residues 107–210), SARNP-mut1 (R106A/F110A/R123A/F127A/R153A/F157A/R177A/F181A/R203A/F207A), and SARNP-mut2 (R153A/F157A/R177A/F181A/R203A/F207A), each followed by a C-terminal hexahistidine tag, were cloned into a pGEX-4T-1 vector modified to contain a TEV protease cleavable N-terminal GST tag. DDX39B (residues 44–428), DDX39B-CTD (residues 259–428), DDX39B-CTD-D283R, Tho1 (residues 1–218), Tho1 (residues 123–178), and Tho1-mut (R139A/F143A/R159A/F163A) were cloned into the same modified pGEX-4T-1 vector. DDX39B (residues 1–428) was cloned into the pProEx-HTb vector (Thermo Fisher Scientific).
Protein expression and purification
All proteins were expressed in E. coli Rosetta cells (Sigma-Aldrich). Protein expression was induced by 0.5 mM IPTG at 20°C overnight. Cells were lysed in a lysis buffer containing 50 mM Tris, pH 8.0, 300 mM NaCl, 0.5 mM TCEP, 1 mM PMSF, and 5 mg/L aprotinin. The GST-tagged proteins were pulled down using Glutathione Sepharose 4B resin and the His-tagged DDX39B was pulled down using Ni Sepharose resin (Cytiva). GST-tagged DDX39B-CTD variants were digested with GST-TEV overnight and purified on a Source 15Q column (Cytiva). The proteins were then passed over Glutathione Sepharose 4B resin to remove remaining GST and uncleaved protein. Purified DDX39B-CTD variants were concentrated, aliquoted, and stored at −80°C in 10 mM Tris, pH 8.0, 300 mM NaCl, 0.5 mM TCEP, and 10% glycerol. The other affinity-purified proteins were first purified on a mono Q column and were subjected to overnight digestion with GST-TEV (for GST-tagged proteins) or His-TEV (for His-tagged DDX39B) to remove the affinity tag. The digested proteins were passed over Glutathione Sepharose 4B resin or Ni Sepharose resin to remove uncleaved protein and TEV. The proteins were further purified on a Superdex 200 column equilibrated with 10 mM Tris, pH 8.0, 150 mM NaCl, and 0.5 mM TCEP. All purified proteins were concentrated, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C.
Crystallization and structure determination
DDX39B (residues 44–428), Tho1 (residues 123–178), and poly(U) 15-mer RNA were mixed in the presence of 2 mM MgCl2 and ADP-BeF3 (prepared in a 1:4:20 ratio of ADP:Be:F). Crystals of DDX39B/Tho1/RNA were obtained at 20°C by vapor diffusion in sitting drops using 0.5 μL protein complex and 0.5 μL reservoir solution containing 0.025 M HEPES, pH 7.4, 0.2 M sodium chloride, and 18% PEG4000. The crystals were transferred in three steps of increasing glycerol concentration to a cryoprotectant solution containing 0.025 M HEPES, pH 7.4, 0.2 M sodium chloride, 18% PEG4000, and 20% glycerol. X-ray diffraction data were collected at the 21-ID-G beamline at the Advanced Photon Source, Argonne National Laboratory.
X-ray intensities were processed using the HKL2000 package.66 An initial electron density map was determined by molecular replacement using Phenix.68 The DDX39B (PDB ID 1XTJ) structure was used as a molecular replacement model. Tho1 and RNA were manually built in Coot.64 Refinement with Phenix68 led to a DDX39B/Tho1/RNA model at 2.50 Å resolution. The model consists of two DDX39B molecules (each bound to an ADP-BeF3 and an RNA oligo) and one Tho1 molecule. Details of the data collection and refinement statistics are in Table S1. Figures were prepared using PyMOL (Molecular Graphics System, Schrodinger, LLC).
Electrophoretic mobility shift assay
Poly(U) 15-mer RNA (100 nM) labeled with Alexa Fluor 488 at the 5′ end was mixed with indicated proteins in a buffer containing 20 mM HEPES (pH 7.0), 50 mM NaCl, 0.5 mM TCEP, 50 μg/mL BSA, 5% glycerol, and 0.5 U/μl SUPERase•In RNase Inhibitor (Thermo Fisher Scientific). Reactions in the presence of Sub2 or DDX39B also contained 1 mM MgCl2 and 1 mM ATP-γ-S. The mixtures were incubated at room temperature for 30 min. Samples were separated on a 5% native PAGE gel. RNA was visualized with a ChemiDoc MP Imaging System (Bio-Rad). The experiments were repeated three times independently.
Microscale thermophoresis (MST)
MST experiments were performed with Monolith NT.115 (NanoTemper Technologies). His-tagged SARNP proteins were labeled with the RED-tris-NTA 2nd Generation dye. Binding reactions contained 30 nM RED-tris-NTA, 120 nM His-tagged SARNP, and DDX39B-CTD at various concentrations in 20 mM HEPES pH 7.5, 150 mM NaCl, 1% glycerol, 0.01% Tween 20, and 3 mM DTT. The reaction samples were loaded into Monolith Premium Capillaries and measured at room temperature with 80% excitation power and medium MST power.
RNA interference and transfection
A pool of 4 siRNAs that target SARNP (SMARTpool siRNA, Dharmacon) was used for RNA knockdown of SARNP. Nontargeting siRNAs (a pool of 4 siRNAs, Dharmacon) were used as control. siRNAs were used at a final concentration of 50 nM. A549 cells were reverse transfected with siRNAs using RNAiMAX (Invitrogen), according to the manufacturer’s instruction. After 48 h, cells were fixed for imaging analysis or lysed for biochemical approaches.
Western blot
Western blot was performed as we previously described.70 Cells were lysed in sample buffer [63 mM tris (pH 7.0), 10% glycerol, and 2% SDS in nuclease free water]. Cell lysates were heated at 95°C for 10 min and sonicated briefly. Cell debris were removed by centrifugation at 12000 rpm for 5 min at 4°C. The supernatant was subjected to 8% SDS-PAGE in running buffer (Invitrogen Cat.B0002–02), followed by Western blot. Proteins were transferred to PVDF membrane with 0.45 μm pore size (Millipore, Cat. IPVH00010) in transfer buffer (Bio-Rad, Cat. 161–0771). The membrane was blocked with 5% nonfat milk in TBS-T buffer (Thermo Fisher, Cat. 28360) supplemented with 1% bovine serum albumin (HyClone, Cat. SH30574.02) at room temperature for 1 h. Primary and secondary antibodies were diluted with TBS-T supplemented with 1% BSA. Incubation of antibodies was performed for 1 h at room temperature. Membrane was washed 5 times for 5 min each time with TBS-T. LI-COR Odyssey imaging system was used to detect the chemiluminescence signal. Quantification of protein band intensity was determined by generating a ratio of mean intensity over median intensity using Adobe Photoshop (CS4). Results were normalized to β-actin.
Immunofluorescence assay
Immunofluorescence assay was performed as we previously described.60 Cells grown on coverslips were fixed for 15 min in 4% paraformaldehyde (PFA, Electron Microscopy Sciences, Cat. 15710-S) and then permeabilized for 5 min in PBS with 0.5% Triton X-100. After blocking (30 min, 5% BSA in PBS), cells were immunostained for 1 h at 37°C with the indicated primary antibodies. The coverslips were washed three times with PBS and labeled with the secondary antibody for 1 h at 37°C. Coverslips were then washed three times with PBS, stained with 1 μg/mL Hoechst 33258 (Molecular Probes Life Technologies) for 5 min, and briefly washed in PBS. Coverslips were mounted in ProLong Gold antifade reagent (Life Technologies).
RNA-FISH, imaging quantification, and statistics
RNA-FISH was performed as described in our previous publication.60 Cells grown on coverslips were fixed in 4% PFA solution for 15 min, and then permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. Cells were briefly washed with PBS and immunostained with primary antibodies diluted in inhibition buffer (0.2% Triton X-100, 200 units/mL RNasin rnase inhibitor, 1mM DTT in PBS) at 37°C for 1 h. After incubation, coverslips were washed with 0.2% Triton X-100 in PBS once and with PBS twice. Wash buffer [2X SSC buffer (Invitrogen, Cat. 15557–036), 10% formamide (Sigma-Aldrich, Cat. F9037) in nuclease free water] was added for 5 min. After removing the wash buffer, cells were incubated with poly(A) probe60 diluted in hybridization buffer (0.1 g/mL dextran sulfate in wash buffer) at 37°C for 4 h. Then, the probe was removed, and cells were washed with wash buffer at 37°C for 30 min and subsequently washed twice with PBS at room temperature. Secondary antibodies were diluted in inhibition buffer and coverslips were incubated with it for 1 h at 37°C. Coverslips were washed 3 times with PBS, stained with Hoechst 33258 for 10 min, washed briefly with PBS and mounted in ProLong Gold antifade reagent. ImageJ was used to calculate the plot profile of the selected images.
Cell fractionation and RNA purification
Cell fractionation was performed as previously described.71 Cells were harvested by trypsinization and centrifugation. The cell pellets were washed once with cold PBS and pelleted at 3000 rpm for 5 min at 4°C. The cell pellets were resuspended with 1mL RSB buffer (10 mM tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2), incubated for 3 min on ice and then centrifuged at 3000 rpm for 5 min at 4°C. The volume of the pelleted cells was estimated and four times volume of lysis buffer RSBG40 (10% glycerol, 0.5% IGEPAL CA-630, 0.5 mM DTT and 100 U/mL RNasin RNase inhibitor added in RSB buffer) was used to resuspend the pellet by slow pipetting. Nuclei were pelleted by centrifugation at 7000 rpm for 3 min, and the supernatant was recovered and saved as the cytoplasmic fraction. Nuclear pellets were resuspended in RSBG40, and one-tenth volume of detergent [3.3% (w/w) sodium deoxycholate and 6.6% (v/v) Tween 40] was added with slow vortexing, followed by incubation on ice for 5 min. Nuclei were again pelleted and the supernatant was pooled with the previous cytoplasmic fraction. Nuclear pellets were washed once in RSBG40, collected at 10,000 rpm for 5 min, and the resulting pellet was used for nuclear RNA extraction. RNA was isolated with RNeasy Plus Mini Kit (Qiagen).
mRNA-seq
Samples were run on the Agilent Tapestation 4200 to determine the level of degradation thus ensuring that only high quality RNA is used (RIN Score 8 or higher). The Qubit fluorimeter was used to determine the concentration prior to starting library preparation. One microgram of total DNAse treated RNA was then prepared with the TruSeq Stranded mRNA Library Prep Kit from Illumina. Poly(A) RNA was purified and fragmented before strand specific cDNA synthesis. cDNA were then a-tailed and indexed adapters were ligated. After adapter ligation, samples were PCR amplified and purified with AmpureXP beads, then validated again on the Agilent Tapestation 4200. Before being normalized and pooled, samples were quantified by Qubit then run on the Illumina NextSeq 500 using V2.5 reagents.
RNA-seq data analysis
Raw sequence data was trimmed using Trimmomatic.72 QC filtered trimmed sequences were aligned to hg19 using STAR.73 All sub-sequent analysis was performed using R version 4.0.2 and Bioconductor 3.11 in RStudio.74 Raw counts were obtained from BAM files using FeatureCounts from the Rsubread package,75 and TPM was calculated from raw counts. TPM values were used for calculating N/C ratio of mRNAs for control and SARNP knockdown samples. Relative change in N/C ratio was calculated comparing SARNP knockdown to control sample. Transcripts with relative change in N/C ratio ≥ 1.5 were considered as export inhibited and mRNAs with relative change in N/C ratio ≥ 0.66 and <1.5 in two independent experiments were considered as export not affected. RNA Features (mRNA length, pre-mRNA length, exon count, mean exon length, mean intron length, %GC content of full-length mRNA, 5′UTR, CDS, and 3′UTR) of export inhibited and not affected transcripts were extracted based on Ensembl GTF annotations (GRCh38, release version 104) for each Ensembl Canonical transcript. RNA features of transcripts belonging to protein coding and lncRNA biotypes were used for further analysis. TPM values, relative N/C ratios, and RNA features for different transcripts are provided in Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
MST data were analyzed by MO. Affinity Analysis v2.3 (NanoTemper Technologies) and are represented as mean ± SD of three independent measurements (Figure 4C). RNA-Seq was performed using samples from two independent experiments. The statistical significance of different RNA features between the export inhibited and not affected groups was determined by two-tailed Mann-Whitney U rank test using R. p values ≤0.05 are considered significant. Exact p values are indicated in the figures. Violin plots were generated to compare the distribution of RNA features between the two groups using GraphPad Prism 9. The number of genes present in each group are indicated in the figures. For each RNA feature, distribution of values with the same shape in both the export inhibited and not affected groups was confirmed using the frequency distribution analysis tool available in GraphPad Prism 9.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| β-actin monoclonal antibody | Invitrogen | Cat# AM4302; RRID: AB_437394 |
| SARNP antibody | Invitrogen | Cat# PA5–56586; RRID:AB_2646955 |
| SC35 monoclonal antibody | Sigma Aldrich | Cat# SAB4200725 |
| SON polyclonal antibody | Genetex | Cat# GTX129778; RRID:AB_2886086 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli strain Rosetta (DE3) | Sigma-Aldrich | Cat# 70954 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Aprotinin | Santa Cruz | Cat# sc-3595A |
| DMEM (Dulbecco’s Modified Eagle Medium) | Gibco | Cat# 11965092 |
| FBS | Sigma-Aldrich | Cat# F4135 |
| Glutathione Sepharose 4B resin | Cytiva | Cat# 17075605 |
| Ni Sepharose resin 6FF | Cytiva | Cat# 17531802 |
| PEG4000 | Hampton Research | Cat# HR2–605 |
| SUPERase•In RNase Inhibitor | Thermo Fisher | Cat# AM2694 |
| TCEP | Hampton Research | Cat# HR2–801 |
|
| ||
| Critical commercial assays | ||
|
| ||
| NEBuilder HiFi DNA Assembly | NEB | Cat# E2621S |
| RED-tris-NTA 2ND generation | Nanotemper | Cat# MOL018 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74034 |
|
| ||
| Deposited data | ||
|
| ||
| Structure of the DDX39B-Tho1-RNA complex | This paper | PDB: 8ENK |
| RNA sequencing data | This paper | BioProject: PRJNA893026 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human lung adenocarcinoma epithelial cells (A549) | ATCC | CCL-185 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Human SARNP siRNA | Dharmacon | Cat# L-015061–02-0005 |
| Poly(U) 15-mer RNA with Alexa 488 on the 5' end | IDT | N/A |
| Poly(U) 15-mer RNA | IDT | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pGEX-SARNP(1–210)-His | This paper | N/A |
| pGEX-SARNP(1–106)-His | This paper | N/A |
| pGEX-SARNP(107–210)-His | This paper | N/A |
| pGEX-SARNP(1–210)-His-(R153A/F157A/R177A/F181A/R203A/F207A) | This paper | N/A |
| pGEX-SARNP(1–210)-His-(R106A/F110A/R123A/F127A/R153A/F157A/R177A/F181A/R203A/F207A) | This paper | N/A |
| pGEX-DDX39B(44–428) | This paper | N/A |
| pGEX-Sub2(1–446) | This paper | N/A |
| pGEX-Tho1(1–218) | This paper | N/A |
| pGEX-Tho1(123–178) | This paper | N/A |
| pGEX-Tho1-(R139A/F143A/R159A/F163A) | This paper | N/A |
| pGEX-DDX39B(259–428) | This paper | N/A |
| pGEX-DDX39B(259–428)-D283R | This paper | N/A |
| pProEX-DDX39B(1–428) | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Coot | Emsley et al.64 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| ESPript 3.0 web server | Robert and Gouet65 | https://espript.ibcp.fr |
| HKL2000 | Otwinowski and Minor66 | https://hkl-xray.com/hkl-2000 |
| ImageJ | Schneider et al.67 | https://imagej.nih.gov/ij |
| MO. affinity analysis v2.3 | NanoTemper Technologies | https://www.nanotempertech.com |
| Phenix | Afonine et al.68 | https://www.phenix-online.org |
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism |
| PyMOL | Schrödinger, LLC | https://pymol.org/2/ |
| R 4.0.2 | The R Fouondation | https://www.R-project.org |
| WebLogo | Crooks et al.69 | https://weblogo.berkeley.edu/ |
Highlights.
Crystal structure of a ScTho1SARNP/DDX39B/RNA complex
SARNP binds DDX39B through multivalent interaction with tandem DDX39B interacting motifs
mRNAs that are dependent on SARNP for nuclear export show high GC content
SARNP, ALYREF, and DDX39B could coordinate to facilitate mRNP assembly
ACKNOWLEDGMENTS
We thank the staff at the Life Sciences Collaborative Access Team beamline 21-ID-F and 21-ID-G at the Advanced Photon Source for help with X-ray data collection. This work was supported by NIH R35 GM133743 to Y.R., NIH R01 AI154635 to B.F., NIH R01 AI125524 to K.W.L. and B.F., and funds from Vanderbilt University School of Medicine to Y.R.. B.P.C. was supported by NIH/NCI training grant T32CA119925.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112988.
REFERENCES
- 1.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, and Moore MJ (2012). The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein Nexus. Cell 151, 915–916. 10.1016/j.cell.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 2.Metkar M, Ozadam H, Lajoie BR, Imakaev M, Mirny LA, Dekker J, and Moore MJ (2018). Higher-order organization principles of pre-translational mRNPs. Mol. Cell 72, 715–726.e3. 10.1016/j.molcel.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y, and Ren Y (2019). Mechanisms of nuclear mRNA export: a structural perspective. Traffic 20, 829–840. 10.1111/tra.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batisse J, Batisse C, Budd A, Böttcher B, and Hurt E (2009). Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem. 284, 34911–34917. 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh G, Pratt G, Yeo GW, and Moore MJ (2015). The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 84, 325–354. 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller-McNicoll M, and Neugebauer KM (2013). How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14, 275–287. 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 7.Wende W, Friedhoff P, and Strãßer K (2019). Mechanism and regulation of co-transcriptional mRNP assembly and nuclear mRNA export. Adv. Exp. Med. Biol. 1203, 1–31. 10.1007/978-3-030-31434-7_1. [DOI] [PubMed] [Google Scholar]
- 8.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, and Reed R (2001). Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413, 644–647. 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 9.Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, and Reed R (2010). ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 24, 2043–2053. 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strässer K, and Hurt E (2001). Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413, 648–652. 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 11.Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, and Hurt E (2002). TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308. 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 12.Ren Y, Schmiege P, and Blobel G (2017). Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1. Elife 6, e20070. 10.7554/eLife.20070.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Y, Clarke BP, Kim YJ, Ivey AL, Hill PS, Shi Y, and Ren Y (2021). Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2. Elife 10, e65699. 10.7554/eLife.65699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuller SK, Schuller JM, Prabu JR, Baumgärtner M, Bonneau F, Basquin J, and Conti E (2020). Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex. Elife 9, e61467. 10.7554/eLife.61467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Tan M, Wu Z, Zhang Y, He F, Lu Y, Li S, Cao M, Li G, Wu J, et al. (2021). Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex. Sci. Bull 66, 2347–2352. 10.1016/j.scib.2021.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Pühringer T, Hohmann U, Fin L, Pacheco-Fiallos B, Schellhaas U, Brennecke J, and Plaschka C (2020). Structure of the human core transcription-export complex reveals a hub for multivalent interactions. Elife 9, e61503. 10.7554/eLife.61503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, and Le Hir H (2005). The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12, 861–869. 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 18.Bono F, Ebert J, Lorentzen E, and Conti E (2006). The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725. 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen KH, Chamieh H, Andersen CBF, Fredslund F, Hamborg K, Le Hir H, and Andersen GR (2009). Mechanism of ATP turnover inhibition in the EJC. RNA 15, 67–75. 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, Pedersen JS, Séraphin B, Le Hir H, and Andersen GR (2006). Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972. 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 21.Gehring NH, Lamprinaki S, Kulozik AE, and Hentze MW (2009). Disassembly of exon junction complexes by PYM. Cell 137, 536–548. 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 22.Gromadzka AM, Steckelberg AL, Singh KK, Hofmann K, and Gehring NH (2016). A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 44, 2348–2361. 10.1093/nar/gkw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viphakone N, Sudbery I, Griffith L, Heath CG, Sims D, and Wilson SA (2019). Co-transcriptional loading of RNA export factors shapes the human transcriptome. Mol. Cell 75, 310–323.e8. 10.1016/j.molcel.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hautbergue GM, Hung ML, Walsh MJ, Snijders APL, Chang CT, Jones R, Ponting CP, Dickman MJ, and Wilson SA (2009). UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 19, 1918–1924. 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CT, Hautbergue GM, Walsh MJ, Viphakone N, van Dijk TB, Philipsen S, and Wilson SA (2013). Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 32, 473–486. 10.1038/emboj.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viphakone N, Cumberbatch MG, Livingstone MJ, Heath PR, Dickman MJ, Catto JW, and Wilson SA (2015). Luzp4 defines a new mRNA export pathway in cancer cells. Nucleic Acids Res. 43, 2353–2366. 10.1093/nar/gkv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda S, Wu DW, Stark K, and Pelus LM (2002). Cloning and characterization of a proliferation-associated cytokine-inducible protein, CIP29. Biochem. Biophys. Res. Commun. 292, 593–600. 10.1006/bbrc.2002.6680. [DOI] [PubMed] [Google Scholar]
- 28.Kang GJ, Park MK, Byun HJ, Kim HJ, Kim EJ, Yu L, Kim B, Shim JG, Lee H, and Lee CH (2020). SARNP, a participant in mRNA splicing and export, negatively regulates E-cadherin expression via interaction with pinin. J. Cell. Physiol. 235, 1543–1555. 10.1002/jcp.29073. [DOI] [PubMed] [Google Scholar]
- 29.Choong ML, Tan LK, Lo SL, Ren EC, Ou K, Ong SE, Liang RC, Seow TK, and Chung MC (2001). An integrated approach in the discovery and characterization of a novel nuclear protein over-expressed in liver and pancreatic tumors. FEBS Lett. 496, 109–116. 10.1016/s0014-5793(01)02409-7. [DOI] [PubMed] [Google Scholar]
- 30.Hashii Y, Kim JY, Sawada A, Tokimasa S, Hiroyuki F, Ohta H, Makiko K, Takihara Y, Ozono K, and Hara J (2004). A novel partner gene CIP29 containing a SAP domain with MLL identified in infantile myelomonocytic leukemia. Leukemia 18, 1546–1548. 10.1038/sj.leu.2403450. [DOI] [PubMed] [Google Scholar]
- 31.Germain H, Qu N, Cheng YT, Lee E, Huang Y, Dong OX, Gannon P, Huang S, Ding P, Li Y, et al. (2010). MOS11: a new component in the mRNA export pathway. PLoS Genet. 6, e1001250. 10.1371/journal.pgen.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sørensen BB, Ehrnsberger HF, Esposito S, Pfab A, Bruckmann A, Hauptmann J, Meister G, Merkl R, Schubert T, Längst G, et al. (2017). The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol. Biol. 93, 283–298. 10.1007/s11103-016-0561-9. [DOI] [PubMed] [Google Scholar]
- 33.Jimeno S, Luna R, García-Rubio M, and Aguilera A (2006). Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol. Cell Biol. 26, 4387–4398. 10.1128/Mcb.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piruat JI, and Aguilera A (1998). A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17, 4859–4872. 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuckerman B, Ron M, Mikl M, Segal E, and Ulitsky I (2020). Gene architecture and sequence composition underpin selective dependency of nuclear export of long RNAs on NXF1 and the TREX complex. Mol. Cell 79, 251–267.e6. 10.1016/j.molcel.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Linder P, and Jankowsky E (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516. 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 37.Peña A, Gewartowski K, Mroczek S, Cuéllar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. (2012). Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 31, 1605–1616. 10.1038/emboj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, and Weis K (2019). DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148. 10.1038/s41586-019-1502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu C, Cao Y, and Bao J (2021). Building RNA-protein germ granules: insights from the multifaceted functions of DEAD-box helicase Vasa/Ddx4 in germline development. Cell. Mol. Life Sci. 79, 4. 10.1007/s00018-021-04069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubíková J, Reinig R, Salgania HK, and Jeske M (2021). LOTUS-domain proteins - developmental effectors from a molecular perspective. Biol. Chem. 402, 7–23. 10.1515/hsz-2020-0270. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Yan C, Hang J, Finci LI, Lei J, and Shi Y (2017). An atomic structure of the human spliceosome. Cell 169, 918–929.e14. 10.1016/j.cell.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 42.Niehrs C, and Luke B (2020). Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 21, 167–178. 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez-Calero C, Bayona-Feliu A, Xue X, Barroso SI, Muñoz S, González-Basallote VM, Sung P, and Aguilera A (2020). UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 34, 898–912. 10.1101/gad.336024.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mordstein C, Savisaar R, Young RS, Bazile J, Talmane L, Luft J, Liss M, Taylor MS, Hurst LD, and Kudla G (2020). Codon usage and splicing jointly influence mRNA localization. Cell Syst. 10, 351–362.e8. 10.1016/j.cels.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas A, Rehfeld F, Zhang H, Chang TC, Goodarzi M, Gillet F, and Mendell JT (2022). RBM33 directs the nuclear export of transcripts containing GC-rich elements. Genes Dev. 36, 550–565. 10.1101/gad.349456.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Y, Lord CL, Clarke BP, Ivey AL, Hill PS, McDonald WH, Wente SR, and Ren Y (2021). Structure and activation mechanism of the yeast RNA Pol II CTD kinase CTDK-1 complex. Proc. Natl. Acad. Sci. USA 118, e2019163118. 10.1073/pnas.2019163118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiura T, Nagano Y, and Noguchi Y (2007). DDX39, upregulated in lung squamous cell cancer, displays RNA helicase activities and promotes cancer cell growth. Cancer Biol. Ther. 6, 957–964. 10.4161/cbt.6.6.4192. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Li P, Wang C, Zhang D, Zeng L, Liu X, and Lin J (2020). DEAD-box RNA helicase 39 promotes invasiveness and chemoresistance of ER-positive breast cancer. J. Cancer 11, 1846–1858. 10.7150/jca.37247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T, Ma Z, Liu L, Sun J, Tang H, Zhang B, Zou Y, and Li H (2018). DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/b-catenin pathway. Cell Death Dis. 9, 675. 10.1038/s41419-018-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing C, Tian H, Zhang Y, Guo K, Tang Y, Wang Q, Lv L, and Wang L (2020). DDX39 overexpression predicts a poor prognosis and promotes aggressiveness of melanoma by cooperating with SNAIL. Front. Oncol. 10, 1261. 10.3389/fonc.2020.01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He C, Li A, Lai Q, Ding J, Yan Q, Liu S, and Li Q (2021). The DDX39B/FUT3/TGFbR-I axis promotes tumor metastasis and EMT in colorectal cancer. Cell Death Dis. 12, 74. 10.1038/s41419-020-03360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domínguez-Sánchez MS, Sáez C, Japón MA, Aguilera A, and Luna R (2011). Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer 11, 77. 10.1186/1471-2407-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue C, Zhao Y, Li G, and Li L (2021). Multi-omic analyses of the m(5)C regulator ALYREF reveal its essential roles in hepatocellular carcinoma. Front. Oncol. 11, 633415. 10.3389/fonc.2021.633415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z, Li X, Li H, Nie C, Liu W, Li S, Liu Z, Wang W, and Wang J (2020). Suppression of DDX39B sensitizes ovarian cancer cells to DNA-damaging chemotherapeutic agents via destabilizing BRCA1 mRNA. Oncogene 39, 7051–7062. 10.1038/s41388-020-01482-x. [DOI] [PubMed] [Google Scholar]
- 55.Flynn RA, Belk JA, Qi Y, Yasumoto Y, Wei J, Alfajaro MM, Shi Q, Mumbach MR, Limaye A, DeWeirdt PC, et al. (2021). Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell 184, 2394–2411.e16. 10.1016/j.cell.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Momose F, Basler CF, O’Neill RE, Iwamatsu A, Palese P, and Nagata K (2001). Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75, 1899–1908. 10.1128/jvi.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris AK, Wang Z, Ivey AL, Xie Y, Hill PS, Schey KL, and Ren Y (2020). Cellular mRNA export factor UAP56 recognizes nucleic acid binding site of influenza virus NP protein. Biochem. Biophys. Res. Commun. 525, 259–264. 10.1016/j.bbrc.2020.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Read EKC, and Digard P (2010). Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J. Gen. Virol. 91, 1290–1301. 10.1099/vir.0.018564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wisskirchen C, Ludersdorfer TH, Müller DA, Moritz E, and Pavlovic J (2011). The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 85, 8646–8655. 10.1128/jvi.02559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mor A, White A, Zhang K, Thompson M, Esparza M, Muñoz-Moreno R, Koide K, Lynch KW, García-Sastre A, and Fontoura BMA (2016). Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 1, 16069. 10.1038/nmicrobiol.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esparza M, Mor A, Niederstrasser H, White K, White A, Zhang K, Gao S, Wang J, Liang J, Sho S, et al. (2020). Chemical intervention of influenza virus mRNA nuclear export. PLoS Pathog. 16, e1008407. 10.1371/journal.ppat.1008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schumann S, Jackson BR, Yule I, Whitehead SK, Revill C, Foster R, and Whitehouse A (2016). Targeting the ATP-dependent formation of herpesvirus ribonucleoprotein particle assembly as an antiviral approach. Nat. Microbiol. 2, 16201. 10.1038/nmicrobiol.2016.201. [DOI] [PubMed] [Google Scholar]
- 63.Fleckner J, Zhang M, Valcárcel J, and Green MR (1997). U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11, 1864–1872. 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 67.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang K, Shang G, Padavannil A, Wang J, Sakthivel R, Chen X, Kim M, Thompson MG, García-Sastre A, Lynch KW, et al. (2018). Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression. Proc. Natl. Acad. Sci. USA 115, E12218–E12227. 10.1073/pnas.1818012115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Zhu W, and Levy DE (2006). Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods (San Diego, Calif 39, 356–362. [DOI] [PubMed] [Google Scholar]
- 72.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao Y, Smyth GK, and Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. 10.1093/nar/gkz114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of the crystal structure have been deposited in the RCSB Protein DataBank with the ID 8ENK. The RNA-seq data have been deposited in the BioProject database with the accession number PRJNA893026. All data are publicly available as of the date of the publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


