Abstract
COVID-19 has represented an issue for global health since its outbreak in March 2020. It is now evident that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection results in a wide range of long-term neurological symptoms and is worryingly associated with the aggravation of Alzheimer’s disease. Little is known about the molecular basis of these manifestations. Here, several strain variants were used to infect SH-SY5Y neuroblastoma cells and K18-hACE C57BL/6J mice. The Tau phosphorylation profile and aggregation propensity upon infection were investigated on cellular extracts, subcellular fractions, and brain tissue. The viral proteins spike, nucleocapsid, and membrane were overexpressed in SH-SY5Y cells, and the direct interaction and effect on Tau phosphorylation were checked using immunoblot experiments. Upon infection, Tau is phosphorylated at several pathological epitopes associated with Alzheimer’s disease and other tauopathies. Moreover, this event increases Tau’s propensity to form insoluble aggregates and alters its subcellular localization. Our data support the hypothesis that SARS-CoV-2 infection in the central nervous system triggers downstream effects altering Tau function, eventually leading to the impairment of neuronal function.
Keywords: SARS-CoV-2, COVID-19, neuro-COVID, Tau, phospho-Tau
Significance Statement.
The significance of this study lies in its investigation of the molecular basis of the long-term neurological symptoms associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and its potential link to the aggravation of Alzheimer's disease. By demonstrating that SARS-CoV-2 infection in neuronal cells triggers aberrant Tau phosphorylation at several pathological epitopes, this study provides valuable insights into the downstream pathophysiological aberrations that may occur in neuro-coronavirus disease (COVID) cases. The results support the evidence that SARS-CoV-2 infection in the central nervous system alters Tau properties, eventually leading to its aggregation and the impairment of neuronal function. This study contributes to our understanding of the long-term neurological consequences of COVID-19 and may have important implications for the development of therapeutic interventions to limit long-term consequences.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for coronavirus disease 19 (COVID-19), has been causing a major pandemic with considerable morbidity and mortality since its outbreak in March 2020 (1–3).
Despite mainly leading to respiratory disease, increasing studies have reported that this virus can spread to other organs, including the central nervous system (CNS). COVID-19 patients present indeed with acute neurological manifestations, such as altered smell and taste, as well as with long-term cognitive symptoms, namely fatigue, headache, attention disorders, dyspnea, and cognitive impairment (i.e. brain fog) (4, 5). This latter condition is defined as “neuro-COVID” and represents a serious concern for global health as it has also been observed in individuals who experienced mild COVID-19 symptoms (6, 7). The pathophysiology of this postviral syndrome is complex as it might involve hypoxia, blood–brain barrier breakage, and, notably, a direct SARS-CoV-2-mediated neuronal tissue invasion and damage. The latter statement is based on the recent confirmation of SARS-CoV-2 neurotropism and the observed loss of gray matter and brain size upon viral infection (8). This hypothesis is further corroborated by the brain anatomical and metabolic alterations observed in COVID-19 survivors (2, 9–11). Current follow-up studies are beginning to document the feared connection between COVID-19 and neurodegenerative diseases (11). COVID-19 can exacerbate and accelerate preclinical dementia in people with preexisting Alzheimer’s disease (12, 13). At the same time, it leads to a greater propensity for Alzheimer’s disease in patients without preexisting dementia compared to noninfected individuals, suggesting the association between SARS-CoV-2 and new-onset mental decay (14, 15).
Despite such reports, to what extent the neuronal effects in patients are a direct consequence of neuronal infection by SARS-CoV-2 or are an indirect effect of the infection of nonneuronal cells is still debated (16), the molecular basis for SARS-CoV-2 neuro-infection is yet to be elucidated in healthy neurons or neurodegeneration-primed cells. Within this context, it has been described that COVID-19 patients experiencing neurological symptoms display increased serum levels for brain injury biomarkers, including total Tau and phospho-Tau 181, two hallmarks of Alzheimer’s pathology and other neurodegenerative disorders (15, 17).
Here, we investigated the interplay between SARS-CoV-2 and the microtubule-associated protein Tau in neurons. Our results indicate that Tau undergoes hyperphosphorylation at pathological epitopes upon infection both in vitro and in vivo. Moreover, the infection increases Tau’s propensity to accumulate in the insoluble cellular fraction. Both these features are associated with Alzheimer’s disease and other tauopathies, suggesting that SARS-CoV-2 could trigger downstream mechanisms that alter Tau functions.
Results
SARS-CoV-2 is associated with Tau in human neuron–like cells
To investigate SARS-CoV-2 dynamics and its downstream effects in neurons, we infected SH-SY5Y cells, a neuroblastoma line widely used as an in vitro model of neurodegenerative disorders and competent for productive infection with SARS-CoV-2 (18–26).
Three clinical strains belonging to the following variants: B.1, B.1.1.7, and B.1.617.2 have been used and the infection was verified by immunoblot assays and confocal microscopy imaging 48-h postinfection. We detected spike (S) and nucleocapsid (N) viral proteins in both infected neuroblastoma and control VeroE6 and HuH7 cells, which are routinely used for viral amplification and isolation (Fig. 1A).
Fig. 1.
SARS-Cov2 infects human neuron–like cells. Viruses detected in infected cells. A) Viral proteins detected in cellular extracts of infected and control cells by immunoblot. B) Superresolution imaging of viral particles detected in infected SH-SY5Y cells 48-h postinfection (hpi). SARS-CoV-2 Spike (S), tubulin (Tub), and DAPI (nuclei). C) Immunoblot of virions released 24- and 48 hpi in the supernatant of SH-SY5Y cells infected with B.1 or B.1.1.7 strains. Total Tau has been detected by anti Tau-13 antibodies. D) Superresolution imaging of infected cells. SARS-CoV-2 (S), Tau, and DAPI (nuclei). Scale bar: 10 µm.
As shown by the tubulin and S staining in Fig. 1B, infected cells displayed an intact microtubule network; although only partially, a few viral particles appeared aligned along the microtubule tracks. Immunoblot assays on collected cell media further showed that SH-SY5Y cells underwent productive infection, as new viral particles were generated and released into the medium. Unexpectedly, the viral progeny contained, or was associated with, the microtubule-associated protein Tau (Fig. 1C).
We further investigated the interplay between viral particles and Tau by superresolution imaging of infected cells. As shown in Fig. 1D, viral puncta colocalized with the Tau signal. Remarkably, Tau formed spots facing viral signals.
These data suggest that SARS-CoV-2, like other CoVs (27), might hijack the neuronal cytoskeleton during the viral life cycle in neurons. In particular, the involvement of Tau in the viral life cycle in both infected cells and newly formed virions suggests possible downstream implications for neuronal homeostasis and highlights a novel aspect of SARS-CoV-2 molecular pathology possibly based on cell-to-cell Tau spreading.
SARS-CoV-2 leads to Tau hyperphosphorylation and aggregation
It has been reported that SARS-CoV-2 induces a global phosphorylation signaling as a primary host response to the infection (28–34). Surjit et al. (35) previously demonstrated that SARS-CoV N protein significantly up-regulates p38 mitogen-activated protein kinase (MAPK) cascade, whose activation is involved in actin remodeling, neuroinflammation, and, notably, Tau hyperphosphorylation (36). Recent studies have confirmed that SARS-CoV-2 activates p38 activity in infected cells likewise (37, 38).
Therefore, we assessed the Tau phosphorylation profile at epitopes considered pathological hallmarks of numerous neurodegenerative disorders (39, 40). By immunoblot experiments, we found a significant increase in Tau phosphorylation at Ser262 (p262) and Ser396 (p396); however, the signal from Tau p231 was unaltered (Fig. 2A; Fig. S1A). Of note, all tested viral strains lead to the same phosphorylation profile, suggesting that their differences in transmissibility have no impact on downstream Tau modifications.
Fig. 2.
Tau is phosphorylated and accumulates in the insoluble fraction upon SARS-CoV-2 infection. A) Immunoblot analysis of SH-SY5Y cells not infected (Ctrl) and infected with SARS-CoV-2 strains and relative quantification (n = 5/9) Kruskal–Wallis nonparametric ANOVA followed by Mann–Whitney post hoc comparison; *P < 0.05. FC, fold changes. B) Immunoblot of soluble and insoluble cellular fractions after detergent fractionation of cells infected with B.1 and B.1.617.2 strains and control cells. Quantification of total Tau in the insoluble fraction. Kruskal–Wallis ANOVA and Mann–Whitney test. *P < 0.05.
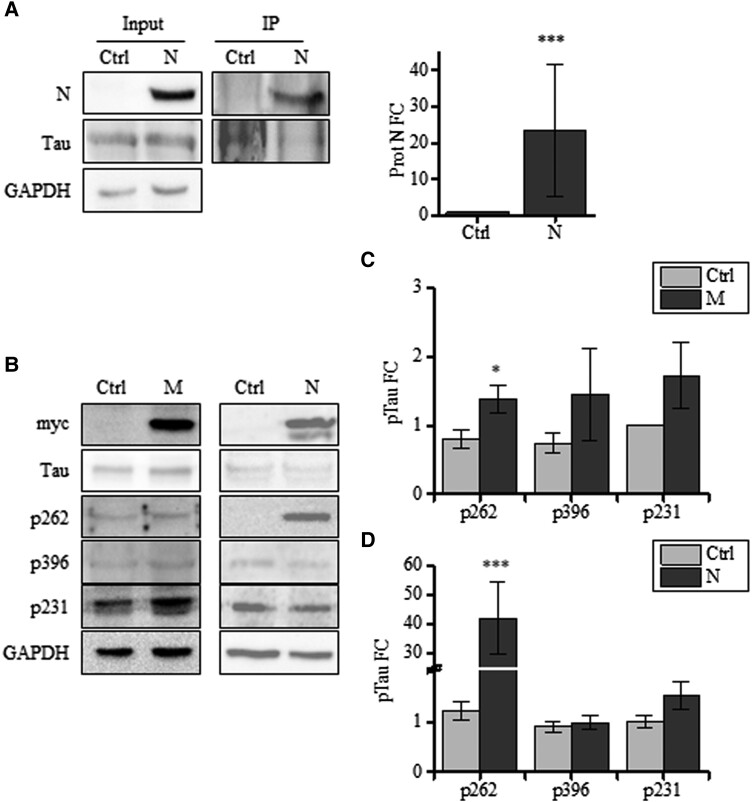
It is well known that Tau hyperphosphorylation disrupts its interaction with microtubules, thereby leading to its mislocalization and increasing its propensity to form insoluble aggregates (41, 42). To investigate the solubility of Tau protein, we checked the presence of total Tau in the soluble and insoluble fractions of infected cells, and we found a robust accumulation of Tau in the insoluble fraction (Fig. 2B; Fig. S1B). Again, the formation of insoluble species was significantly induced by all the tested viral strains similarly. Remarkably, the S protein could be observed in the soluble fraction, whereas protein N was enriched in the insoluble fraction with Tau, suggesting a putative interplay between these proteins, which might account for Tau pathological alterations. Due to the presence of both N and Tau in the insoluble fraction of infected cells, we first checked their protein–protein interaction. As shown in Fig. 3A, in cells transfected with the plasmid encoding SARS-CoV-2 N, the coimmunoprecipitation (co-IP) with anti-Tau antibodies revealed a direct interaction between Tau and N.
Fig. 3.
N and M lead to Tau phosphorylation at S262. A) Co-IP with anti-Tau antibodies in cellular extracts from control and N expressing cells (N) and relative quantification (total extract = input; immunoprecipitate = IP). B) Immunoblot analyses of cells expressing N, M, and control cells. Detection of total Tau (anti-Tau13 antibodies) and p231, p396, AT8, and p262. C) Quantification of pTau in M expressing cells in B. D) Quantification of pTau in N expressing cells in B. Mann–Whitney: *P < 0.05; ***P < 0.001.
To further investigate this aspect and gain insights into the molecular mechanism underlying SARS-CoV-2-dependent Tau phosphorylation, we examined whether the virus itself triggered these alterations. Hence, we dissected the viral components and tested their separate ability to impact Tau phosphorylation.
To this aim, we transfected neuroblastoma cells with plasmids expressing SARS-CoV-2 viral protein. Protein N expression induced a significant increase in Tau p262, while p231 and p396 were not modulated. The expression of membrane (M) induced the phosphorylation at Serine 262, as well (Fig. 3B and C). On the contrary, protein S did not alter Tau phosphorylation at these analyzed residues (Fig. S2).
These data suggest a direct role for both SARS-CoV-2 N and M in altering Tau function, which might potentially trigger downstream events, possibly leading, or contributing to neuro-COVID.
Taken together, our findings suggest that SARS-CoV-2 infection sustains Tau hyperphosphorylation at several pathological epitopes, an effect possibly mediated by its integral proteins N and M, which eventually leads to its mislocalization and aggregation in neuroblastoma cells.
SARS-CoV-2 leads to Tau phosphorylation and mislocalization in the mouse brain
Given our findings in neuron-like cells, we assessed Tau modulation in vivo by challenging a transgenic mouse line expressing human ACE2 (K18-hACE2) model (43) with the SARS-CoV-2 B.1 viral strain. As previously shown by Winkler et al., infected mice developed severe respiratory syndrome, which eventually led to premature death within about 7 days.
Productive infection has been verified by RT-PCR (Fig. S3) and by immunoblot analysis on brain tissues from mice sacrificed 6 days after infection (Materials and methods), we observed that SARS-CoV-2 infection caused a significant increase of Tau p262 and p396 (Fig. 4A).
Fig. 4.
Tau is hyperphosphorylated and mislocalized in the infected mouse brain. A) Immunoblot assay of brain lysates from K18-hACE2 mice after SARS-CoV-2 infection and relative quantification; nonparametric Mann–Whitney: *P < 0.05; **P < 0.01. B) Confocal images of brain cross sections from K18-hACE2 mice either uninfected (Ctrl) or infected (SARS-CoV-2) with B.1. strain. Nuclei stained with TOTO3, Nucleocapsid (N), and Tau. Scale bar: 20 µm.
To understand whether this modulation altered Tau subcellular distribution and function, we assessed its localization in neurons. As shown in Fig. 4B, brain slices from control animals displayed Tau enriched at neurites decorating the neuronal cytoskeleton, as usually observed. However, Tau appeared widespread in the cytoplasm after viral infection. These data support the evidence that SARS-CoV-2 infection in the CNS might result in symptoms commonly associated with neurodegenerative disorders due to its involvement in Tau posttranslational modification and, eventually, its mislocalization.
Discussion
New SARS-CoV-2 variants are continuously emerging causing new waves of infection and major worldwide burden. Although most COVID-19 patients primarily develop respiratory signs, they may experience neurological symptoms with cognitive and psychiatric impairments, which may persist after the acute phase of infection (5–7). This new illness is of great concern as it might affect mental and social well-being by interfering with daily life.
As the virus keeps mutating and spreading worldwide, it is a matter of necessity and urgency to provide a molecular mechanism underlying COVID-19-related long-term neurological complications. Filling this gap would eventually contribute to finding strategies for treatment and prevention. This work provides new insights by exploring viral downstream effects in neurons.
Given that SARS-CoV-2 neurotropism has been widely proven to be comparable to other CoVs, we exploited SH-SY5Y cells as a human neuronal–like cellular model to investigate the molecular biology of the viral–host interplay (9, 44–46).
In these cells, the viral replication level is compared to VeroE6 and HuH7 cells, which are the gold standard platforms to amplify and isolate this virus. Although their infection rate was lower than that of reference cells, SH-SY5Y cells exhibited active viral replication, and the viral progeny they released was associated with the neuronal protein Tau. This event was not surprising since several enveloped virions from different taxa have been described to package host cell proteins into or onto their surface (47). However, this finding needs further investigation due to its potential long-term complications. A common mechanism for the progression of several neurodegenerative disorders is indeed the transcellular transmission of pathological proteins and the subsequent misfolding of their wild-type counterparts in recipient neurons (48). Within this context, a well-studied pathway is the spreading of hyperphosphorylated Tau aggregates in the brain of Alzheimer's disease patients (49, 50).
It is widely recognized that most neurotropic viruses, including betacoronaviruses like SARS and Middle East Respiratory Sindrome (MERS), as well as human immunodeficiency virus (HIV-1), herpes simplex virus (HSV), and zika virus (ZIKV), hijack the cytoskeleton network to fulfill a successful infection. The fine interplay between viral and host cytoskeletal elements is relevant for the pathogen spreading across the cytoplasm toward the ER-golgi-intermediate compartment (ERGIC) compartment or the nucleus (27, 51, 52). In this work, we observed that only a few viral particles colocalize with tubulin on the microtubules network, but there is a strong association with Tau, suggesting that the SARS-CoV-2 neurotropic mechanism employs the neuronal cytoskeleton. In addition, we found that the infection results in increased phosphorylation at Ser262 and Ser396 residues of Tau, both in vitro and in vivo. These epitopes are widely described as hallmarks of Alzheimer's disease and other tauopathies (39, 40). It is known that hyperphosphorylated Tau displays decreased interaction with microtubules, and certain residues such as Ser262 are the major sites inhibiting this binding efficiency (53). This event could cause the formation of insoluble aggregates, which are linked with neuron toxicity and are considered the main trigger of tauopathies (54). In line with these data, we demonstrated that SARS-CoV-2 infection is associated with Tau overspreading and mislocalization in the mouse brain. This observation is also in agreement with high-resolution imaging data from SARS-CoV-2-infected human brain organoids (9), showing that it is associated with altered distribution of Tau from axons to soma. Moreover, we detected a significant increase of Tau in the insoluble fraction of infected cells, which indicates that SARS-CoV-2 affects not only the phospho-Tau profile but also its biochemical properties. Remarkably, we found N in the insoluble fraction, suggesting a possible interplay between N and Tau. Indeed, we found a direct interaction between Tau and N by co-IP experiments, suggesting that the pathological alteration of Tau protein might be mediated by N. It is also possible that the interaction between SARS-CoV-2 and Tau is indirect, via a nonidentified intermediate protein. While future mass spectrometry or yeast two-hybrid experiments will allow dissecting between a direct and an indirect interaction between SARS-CoV-2 protein N and Tau, a conservative conclusion is that the two proteins are part of a protein–protein interaction multicomplex.
These findings suggest that SARS-CoV-2 infection in neurons causes Tau pathological changes, which could eventually contribute to neuro-COVID symptoms by impairing neuronal homeostasis.
Previous work has shown the association between SARS-CoV and SARS-CoV-2 infections and the increased activity of the p38 MAPK pathway, which is involved in actin remodeling and Tau phosphorylation (35, 36, 38). MAPKs such as the extracellular signal-regulated kinases 1 and 2, c-Jun amino-terminal kinases, protein-kinase C (PKC), and p38 γ and δ isoforms are activated by SARS-CoV-2 and are involved in neurodegeneration as well (29, 55). Of note, the Ser396 and Ser262 residues are more susceptible to proline-rich kinases such as glycogen synthase kinase 3-beta, MAPKs, and PKC (56–59). Future research should address which kinases responsible for Tau phosphorylation are overactivated upon viral entry in neurons (Fig. 5). The increased phosphorylation of Tau is also mediated by the inhibition of phosphatases activity (59). Goel et al. recently reported that SARS-CoV-2 induced reduction of the phosphatases DUSP1 and DUSP5 gene expression. Remarkably, DUSP reduction has been found in AD brains concomitantly with kinase activation and correlates with Tau pathology (37).
Fig. 5.
Schematic representation of the residues in the longest isoform of Tau that can be phosphorylated. Residues phosphorylated upon SARS-CoV-2 infection are represented. Kinases involved in the phosphorylation of residues during neurodegeneration are reported; kinases involved also in SARS-CoV-2 infection are highlighted. Created with BioRender.com.
It is still unclear whether Tau aberrant phosphorylation profile and its consequent aggregation are caused by the virus itself upon infection or/and by an indirect cellular response. In this regard, we observed that the expression of SARS-CoV-2 M and N proteins is sufficient to increase Tau phosphorylation, indicating that Tau posttranslational modulation could be at least partially modulated by a direct function of these viral proteins, as already described for their SARS-CoV orthologs (35, 36). By making microtubules less stable, Tau phosphorylation could represent a cell defensive strategy to detach the virus from microtubules, preventing its transport to the ERGIC compartment, i.e. the assembly and budding site of newly synthesized virions. Of note, pTau profile is not altered by the expression of S, which is subject to a higher mutation rate than N and M (60), which potentially highlights these two viral proteins as a druggable target to counteract COVID-dependent neuropathology.
We cannot exclude a contribution of the inflammatory pathways activated by the virus. Indeed, neuroinflammation has been reported in COVID-19 patients, especially in those requiring hospital or intensive care unit treatment. Interestingly, several inflammatory mediators such as TNF-α, IL-1β, NF-κB, IL-6, and MHCII, along with Iba-1 and GFAP-positive cells, are AD biomarkers and are activated in COVID-19 (61–64). This suggests a possible common mechanism leading to the increasing risk of neurological diseases.
In conclusion, we demonstrated that SARS-CoV-2 infection in neuronal cells triggers aberrant Tau phosphorylation at several pathological epitopes that are associated with Alzheimer's disease and other tauopathies (Fig. 5). By altering Tau properties, this event eventually leads to its aggregation and the impairment of neuronal function as a consequence of the displacement of host cytoskeletal components.
Despite the study was conducted in mice during the acute phase of infection, our data open up a new molecular mechanism underlying post-COVID neurological manifestations and acknowledge the potential scale of the disease’s long-term course. Although vaccination is currently the primary strategy for the management of the SARS-CoV-2 pandemic, patients who already experienced COVID-19 must not be side-lined. To limit long-term consequences, it is relevant for clinicians to be aware of the downstream pathophysiological aberrations when dealing with neuro-COVID cases.
Materials and methods
Cell lines and culture
Human neuroblastoma SH-SY5Y cells (CRL-2266), African green monkey kidney epithelial VeroE6 cells (CRL-1586), and human hepatocyte carcinoma HuH7 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and were already available in the lab. VeroE6-TMPRSS2 cells were kindly provided by Dr. Nicola Clementi, Vita-Salute San Raffaele University Hospital, Milan, Italy. SH-SY5Y cells were routinely cultured in Dulbecco's Modified Eagle Medium/nutrient mixture F12 (DMEM/F12; GIBCO) supplemented with 10% heat-inactivated fetal bovine serum (FBS, EuroClone), 2 mM l-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin; VeroE6, HuH7, and VeroE6-TMPRSS2 cells were maintained in DMEM high glucose medium (Sigma-Aldrich) supplemented with 10% FBS, 2 mM l-glutamine, 10 U/mL penicillin, and 10 mg/mL streptomycin. All lines were grown at 37°C with 5% CO2.
Virus isolation and amplification
SARS-CoV-2 manipulation was performed in the biohazard safety level 3 (BLS3) facility of the Virology Unit, Pisa University Hospital, Pisa, Italy, and in compliance with the European Committee and the World Health Organization laboratory biosafety guidelines. The SARS-CoV-2 strains used belonged to B.1 (hCoV-19/Italy/LOM-UniSR10/2021; GISAID accession ID: EPI_ISL_2544194), B.1.1.7 (hCoV-19/Italy/LOM-UniSR7/2021; GISAID accession ID: EPI_ISL_1924880), and B.1.617.2 (hCoV-19/Italy/TUS-AOUP-FOAN-004/2021; GISAID accession ID: EPI_ISL_3184308.1). All strains were isolated from nasopharyngeal swab specimens of infected individuals. Briefly, VeroE6 and Vero-TMPRSS2 cells were plated at approximately 80% confluence and infected with 500 µL of the nasal swab buffer diluted in 5 mL of medium. Culture plates were incubated at 37°C, 5% CO2, and shaken every 15 min for 2 h. At the end, medium supplemented with 5% serum was added and cells were cultured until a full cytopathic effect was achieved. Cells were then resuspended in lysis buffer and centrifuged at 900g for 10 min. The supernatants containing virus particles were filtered with a 0.45 µM strainer and aliquots were stored at −80°C. Viral stocks were titrated by limited dilution according to the Reed and Muench method (65) and expressed as Tissue Culture Infectious Dose 50%/mL (TCID50/mL). The titers achieved were B.1 7 × 108 TCID50/mL; B.1.1.7 7 × 106 TCID50/mL; B.1.617.2 5.2 × 106 TCID50/mL; and B.A.1 1.5 × 106 TCID50/mL.
Animals and experimental infection
In vivo procedures were approved by the Ethics Committee for Animal Experimentation of University of Pisa and Italian Ministry of Health (authorization n. 834/2021-PR). Heterozygous K18-hACE c57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were purchased from Charles River Laboratories (Calco, Italy). Animals had ad libitum access to standard rodent chow (Mucedola, Milano, Italy) and water under a 12-h light/dark cycle. Briefly, 8-week-old male and female mice were anesthetized with ketamine 100 mg/kg and infected by intranasal route with 30 µL of virus SARS-COV-2, B.1 strain, 6 × 103 TCID50/mice. Animals were monitored daily for the appearance of symptoms and 6-day postinfection were sacrificed by cervical dislocation to harvest brains for qRT-PCR, western blot, and histology.
Plasmids and transfection
SARS-CoV-2 N and S plasmids in a pCMV14-3X-Flag backbone were a kind gift from S. Lisi from Bio@SNS laboratory, Scuola Normale Superiore, Italy. The construct encoding SARS-CoV-2 matrix (M) protein was cloned by PCR amplification of M cDNA from the pDONR 207 SARS-CoV-2 M plasmid (Addgene #141274) using a Forward primer inserting a restriction site for EcoRI and a reverse primer bearing a restriction site for BamHI and a Myc tag: proteinM-fwd, 5′-GCT GAA TTC GCA TGG CTG ACT CTA ACG GT-3′; proteinM-rev, 5′-AGC GGA TCC TGC AGA TCC TCT TCA GAG ATG AGT TTC TGC TCC CCC TGC ACC AGC AGG GCG AT-3′. SH-SY5Y was seeded onto 6-well plates (200.000 cells/well) and transfected using the Lipofectamine 2000 reagent (Thermo Fisher Scientific, Milan, Italy) according to the manufacturer's instructions. Cells were routinely harvested after 48 h of transient expression.
Immunoprecipitation
Cells were lysed in lysis buffer (Tris-HCl 20 mM pH 7.5, NaCl 20 mM, glycerol 10%, NP-40 1%, and EDTA 10 mM, supplemented with protease and phosphatase inhibitors) followed by incubation for 30 min on ice. Cell lysates were centrifuged at 16,000 rcf for 15 min at 4°C. The supernatant was collected and quantified by the Bradford method. For Co-IP experiment, 2 mg of total extracts have been incubated with 4 µg of anti-Tau antibodies (Tau13) for 3 h at 4°C. Thirty microliters of prewashed protein A/G have been added to protein extract O/N at 4°C. The following day, IP has been washed five times with TBS 1× and centrifuged (12,000 rpm, 30 s, 4°C). Pellet has been resuspended in Laemmli buffer and boiled at 95°C for 10 min. Input and IP have been analyzed by western blot. Antibodies used were mouse anti-Tau (Tau13) (sc-21796 Santa Cruz Biotechnology, Dallas, TX, USA).
Cell infection and western blot analysis
VeroE6, HuH7, and SH-SY5Y cells (2 × 105 cells/well) were infected with B.1 and B.1.1.7 strains (MOI 0.1). Neuroblastoma cells were additionally challenged with the B.1.617.2 and with B.A.1 variants (MOI 0.01). Following a 2-h adsorption period, cells were maintained in complete culture medium; 48-h postinfection, the medium was harvested and stored at −80°C while cells were detached with double phosphate-buffered saline (PBS) washes and centrifuged at 900g for 10 min. Total protein extraction was carried out in RIPA buffer (Millipore, USA) for infected cells and in aTris-HCl 20 mM pH 8.1, NaCl 20 mM, glycerol 10%, NP-40 1%, and EDTA 10 mM buffer for transfected cells. Both reagents were supplemented with protease and phosphatase inhibitors (Roche). The extracts were centrifuged at 16,000 rcf for 15 min at 4°C to collect the supernatants, which were quantified with the Bradford method before loading buffer resuspension. Western blotting experiments on viral particles were carried out on the collected culture media concentrated with Vivaspin 6 MWCO 30-kDa membranes (Sigma-Aldrich). Samples were then reduced and denatured by boiling in 4× Laemmli sample buffer at 95°C for 5 min. Triton-X 100 fractionation was performed as previously reported in Siano et al. (25). Briefly, infected cell pellets were resuspended in 1% Triton-X 100 in PBS lysis buffer supplemented with protease and phosphatase inhibitors. Lysates were subsequently centrifuged at 16,000g for 15 min at 4°C. Pellets (Triton X-100 insoluble fractions) were further dissolved in 1% SDS and 1% Triton-X in PBS lysis buffer, whereas supernatants (Triton X-100 soluble fraction) were directly processed. Brains excised from infected and control K18-hACE mice were homogenized in RIPA buffer added with proteinase and phosphatase inhibitors and incubated overnight at 4°C. The following day, extracted proteins were quantified by the Bradford method and processed for western blot analysis as follows. Equal amounts of protein preparations were separated by 10% Tris-glycine SDS–PAGE (Bio-Rad) and then transferred onto a nitrocellulose M (Amersham Biosciences). Ms were blocked with EveryBlot Blocking buffer solution (Bio-Rad) for 5 min at room temperature according to the manufacturer's instruction and hybridized with the primary antibody in the same blocking buffer overnight at 4°C. The following day, incubation with Horseradish Peroxidase (HRP)-conjugated antibodies diluted in blocking solution was carried out at room temperature for 1 h. M washing was performed three times for 5 min in Tris-buffered saline with 0.1% Tween20 (TBST). Blots were developed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and acquired by ChemiDoc Imaging Systems (Bio-Rad). Primary antibodies are as follows: rabbit SARS-CoV-2 Spike S1 antibody (HL6), 1:1,000 (GTX635654, GeneTex, Irvine, CA, USA); mouse SARS-CoV/SARS-CoV-2 Spike S2 antibody (1A9), 1:1,000 (GTX632604, GeneTex, Irvine, CA, USA); rabbit SARS-CoV-2 Nucleocapsid antibody, 1:1,000 (GTX135357, GeneTexm Irvine, CA, USA); mouse anti-Tau (Tau5), 1:1,000 (ab80579, Abcam); mouse anti-Tau (Tau13), 1:1,000 (sc-21796 Santa Cruz Biotechnology, Dallas, TX, USA); rabbit anti-pTau (Ser262), 1:500 (OPA1-03142, Thermo Fisher Scientific); mouse anti p-Tau (Ser396), 1:500 (#9632, Cell Signaling technology, Danvers, MA, USA); mouse anti p-Tau Thr231, 1:500 (# MN1040, Thermo Fisher Scientific); mouse anti-GAPDH, 1:10,000 (10R-G109a, Fitzgerald Industries international, Acton, MA, USA); rabbit anti-β-tubulin, 1.1000 (#2146, Cell Signaling Technology); rabbit anti-histone H2B, 1:1,000 (sc-515808, Santa Cruz Biotechnology, Dallas, TX, USA); rabbit anti-actin, 1:5,000 (A300-485A, Bethyl Laboratories); and HRP-conjugated anti-Myc-Tag (9B11) (#2040 Cell Signaling Technology). Secondary antibodies are as follows: goat antimouse IgG-HRP, 1:10,000 (sc-516102, Santa Cruz Biotechnology, Dallas, TX, USA); and mouse antirabbit IgG-HRP, 1:10,000 (sc-2357, Santa Cruz Biotechnology, Dallas, TX, USA).
Immunofluorescence
SH-SY5Y (10,000 cells/well) was seeded onto eight well-chamber microscope slides (Nunc Lab Tech, Thermo Fisher Scientific) and infected with the SARS-CoV-2 B.1 strain (MOI 0.1) as described above. After a 24-h adsorption period, cells were washed from the viral inoculum three times in PBS and fixed with 100% ice-cold methanol for 15 min.
Samples were then permeabilized in Triton 0.1% in PBS for 5 min and blocked in a 1% BSA-Tween 0.1% in PBS solution for 30 min at room temperature. Hybridization with primary antibodies diluted in blocking solution was performed overnight at 4°C. The following day, cells were permeabilized and blocked again for 5 min and incubated with Alexa Fluor-conjugated secondary antibodies diluted in blocking solution for 1 h at room temperature. Nuclei were stained with DAPI (Sigma-Aldrich). Brain tissues were fixed in buffered formalin solution and stored at 4°C overnight. The day after, they were included in OCT solution (Bio-Optica, Milan, Italy) and cut into 50-µm coronal slices with a freezing microtome. Free-floating sections were then blocked in 10% normal goat serum (NGS)-0.3% Triton in PBS 1× for 2 h at room temperature and then hybridized with primary antibodies overnight at 4°C. On the following day, slices were incubated with the secondary antibody for 2 h at room temperature. Both primary and secondary antibodies were diluted in a 1% NGS-0.1% Triton in PBS solution. Primary antibodies are as follows: anti-S IgG rabbit monoclonal antibody (40592-V05H, Sino Biological); rabbit SARS-CoV-2 N antibody, 1:1,000 (GTX135357, GeneTex Irvine, CA, USA); anti-α-tubulin IgG mouse monoclonal antibody (T5168, Merck); and mouse anti-Tau (Tau13), 1:500 (sc-21796 Santa Cruz Biotechnology, Dallas, TX, USA). Secondary antibodies (1.200 dilution) are as follows: goat antirabbit IgG Alexa488-labeled monoclonal antibody; donkey antimouse IgG Alexa568-labeled monoclonal antibody; goat antimouse IgG Alexa546-labeled monoclonal antibody goat antimouse IgG Alexa633-labeled monoclonal antibody (Thermo Fisher Scientific); DAPI, 1:10,000 (28718-90-3, Sigma-Aldrich); and TOTO-3 iodide, 1:5,000 (T3604, Thermo Fisher Scientific). Images were acquired on a Zeiss laser scanning (LSM) 880 confocal microscope (Carl Zeiss, Jena, Germany) supplied with GaAsP (Gallium:Arsenide:Phosphide) detectors. Samples were viewed with a 63× Apochromat oil immersion (1.4 NA) DIC objective. Whole-cell images were acquired with a z-stack series of usually ten slices with 0.5-µm intervals and summed up with the z-projection tool from Fiji.
SARS-CoV-2 real-time PCR
Weighted brains were stored in trizol reagent and homogenized at 4°C, the RNA was extracted by Chomczynski protocol and quantified and assessed for purity by nanodrop. qRT-PCR was performed to quantify the SARS-COV2 viral load present in the tissue. SARS-CoV-2 RNA relative amounts detected for each experimental condition as a cycle threshold (Ct) value were compared, with a mean Ct value determined for the positive infection control. The purified RNA was then used to perform the synthesis of first-strand complementary DNA, using the One-Step TB Green PrimeScript RT-PCR Kit II (Takara) for RT-PCR. Real-time PCR, using the SYBR green dye–based PCR amplification and detection method, was performed to detect the complementary DNA. We used the forward primer SF(CTCATCACGTAGTCGCAACAGTTC) the reverse primer SR (CAAGCTGGTTCAATCCTGTCAAGCA) for S protein and the forward primer AF (CTCCATCCTGGCCTCACTGT) and the reverse primer AR (GAGGGGCCGGACTCATCGT) for actin. The PCR conditions were as follows: 42°C for 2 min, 45 cycles of 95°C for 10 s, annealing at 95°C for 5 s, and elongation at 62°C for 30 s, followed by a final elongation at 72°C for 10 min. RT-PCR was performed using the ABI-PRISM 7900HT Fast Real-Time instrument (Applied Biosystems) and optical-grade 96-well plates. Samples were run in duplicate, with a total volume of 20 µL.
Statistical analysis
In western blot assays, differences between means were analyzed by the nonparametric Mann–Whitney test (for n = 2 independent experimental groups) or Kruskal–Wallis test followed by post hoc Mann–Whitney (for n > 2 independent experimental groups). Fold changes were calculated with respect to control groups, to which a theoretical mean of 1 was attributed. All results are shown as mean (bars) ± SEM (whiskers) from at least three independent experiments. Levels of significance are depicted as * for P < 0.05, ** for P < 0.01, *** for P < 0.001, and n.s. for nonsignificant. Origin 9.0 (OriginLab, Northampton, MA) software was used to perform statistical analysis and plotting.
Supplementary Material
Acknowledgments
The authors are grateful to Alessandro Viegi, Simonetta Lisi, and Elena Novelli for technical support.
Contributor Information
Cristina Di Primio, Institute of Neuroscience, Italian National Research Council (CNR), Pisa 56124, Italy.
Paola Quaranta, Institute of Neuroscience, Italian National Research Council (CNR), Pisa 56124, Italy; Retrovirus Center, Virology Section, Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa 56100, Italy.
Marianna Mignanelli, Laboratorio di Biologia Bio@SNS, Scuola Normale Superiore di Pisa, Pisa 56126, Italy.
Giacomo Siano, Laboratorio di Biologia Bio@SNS, Scuola Normale Superiore di Pisa, Pisa 56126, Italy.
Matteo Bimbati, Laboratorio di Biologia Bio@SNS, Scuola Normale Superiore di Pisa, Pisa 56126, Italy; Department of Biotechnology, University of Verona, Verona 37134, Italy.
Arianna Scarlatti, Laboratorio di Biologia Bio@SNS, Scuola Normale Superiore di Pisa, Pisa 56126, Italy.
Carmen Rita Piazza, Retrovirus Center, Virology Section, Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa 56100, Italy; Department of Medical Biotechnologies, University of Siena, Siena 53100, Italy.
Piero Giorgio Spezia, Retrovirus Center, Virology Section, Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa 56100, Italy.
Paola Perrera, Retrovirus Center, Virology Section, Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa 56100, Italy.
Fulvio Basolo, Department of Surgical, Medical and Molecular Pathology, University Hospital of Pisa, Pisa 56124, Italy.
Anello Marcello Poma, Department of Surgical, Medical and Molecular Pathology, University Hospital of Pisa, Pisa 56124, Italy.
Mario Costa, Institute of Neuroscience, Italian National Research Council (CNR), Pisa 56124, Italy.
Mauro Pistello, Retrovirus Center, Virology Section, Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa 56100, Italy; Virology Unit, Pisa University Hospital, Pisa 56100, Italy.
Antonino Cattaneo, Laboratorio di Biologia Bio@SNS, Scuola Normale Superiore di Pisa, Pisa 56126, Italy.
Supplementary material
Supplementary material is available at PNAS Nexus online.
Funding
This work was supported by the following funding agencies: Immunohub (Ministero della Salute) to M.P.; Piano Nazionale di Ripresa e Resilienza (PNRR) Missione 4, Componente 2, Investimento 1.4 “Centro Nazionale per lo sviluppo di terapia genica e farmaci con tecnologia a RNA”, and Spoke 3– “Neurodegeneration” to M.P.; EU funding within the NextGeneration EU-MUR PNRR TUSCANY HEALTH Ecosystem (THE) (project no. ECS_00000017) spoke 1 to M.C. and 8 to A.C. and C.D.P.; CARIPLO (PANANTICOVID19 R1.2020.0002411) to M.P.; EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (project no. PE00000007, INF-ACT) to M.C. and C.D.P.; Ricerca Salute 2018 “Tuscany Antiviral Research Network (TUSCAVIR.NET)” to M.P.; and COVID-19 Toscana 2020 “Suppression of Airborne Viral Epidemic Spread by Ultraviolet light barriers (SAVES-US)” to M.P.
Author contributions
Conceptualization: C.D.P. and P.Q. Methodology: C.D.P., P.Q., M.M., M.P., F.B., and M.C. Investigation: C.D.P., P.Q., M.M., G.S., M.B., C.R.P., A.S., A.M.P., and P.P. Supervision: C.D.P., M.P, A.C., and M.C. Writing: C.D.P., M.M., G.S., P.Q., and A.C. All authors commented and approved the final version.
Preprint
This manuscript was posted on a preprint: bioRxiv 2023.05.17.541098.
Data availability
All data are included in the manuscript and/or supporting information.
References
- 1. Hu B, Guo H, Zhou P, Shi Z-L. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 19:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yong SJ. 2021. Long COVID or post-COVID-19 syndrome: putative pathophysiology, risk factors, and treatments. Infect Dis (London, England). 53:737–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yüce M, Filiztekin E, Özkaya KG. 2021. COVID-19 diagnosis—a review of current methods. Biosens Bioelectron. 172:112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellul MA, et al. 2020. Neurological associations of COVID-19. Lancet Neurol. 19:767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leon SL, et al. 2021. More than 50 long–term effects of COVID–19: a systematic review and meta–analysis Middle East respiratory syndrome. Sci Rep. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paterson RW, et al. 2020. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain 143:3104–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raveendran AV, Jayadevan R, Sashidharan S. 2021. Long COVID: an overview. Diabetes Metab Syndr. 15:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Douaud G, et al. 2022. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature 604:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramani A, et al. 2020. SARS-CoV-2 targets neurons of 3 D human brain organoids. EMBO J. 39:e106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun S-H, et al. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 28:124–133.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. von Weyhern CH, Kaufmann I, Neff F, Kremer M. 2020. Early evidence of pronounced brain involvement in fatal COVID-19 outcomes. Lancet (London, England) 395:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emmi A, Sandre M, Porzionato A, Antonini A. 2022. Smell deficits in COVID-19 and possible links with Parkinson's disease. Int Rev Neurobiol. 165:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciaccio M, et al. 2021. COVID-19 and Alzheimer's disease. Brain Sci. 11(3):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naughton SX, Raval U, Pasinetti GM. 2020. Potential novel role of COVID-19 in Alzheimer's disease and preventative mitigation strategies. J Alzheimers Dis. 76:21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frontera JA, et al. 2022. Comparison of serum neurodegenerative biomarkers among hospitalized COVID-19 patients versus non-COVID subjects with normal cognition, mild cognitive impairment, or Alzheimer's dementia. Alzheimers Dement. 18:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clijsters M, Wauters J, Mombaerts P, Van Gerven L. 2021. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 184:5932–5949.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kokkoris S, et al. 2022. Serum inflammatory and brain injury biomarkers in COVID-19 patients admitted to intensive care unit: a pilot study. eNeurologicalSci. 29:100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torices S, et al. 2021. Expression of SARS-CoV-2-related receptors in cells of the neurovascular unit: implications for HIV-1 infection. Res Sq. 2:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bielarz V, et al. 2021. Susceptibility of neuroblastoma and glioblastoma cell lines to SARS-CoV-2 infection. Brain Res. 1758:147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benedetti F, et al. 2021. Comparison of SARS-CoV-2 receptors expression in primary endothelial cells and retinoic acid-differentiated human neuronal cells. Viruses 13(11):2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartolomeo CS, et al. 2022. SARS-CoV-2 infection and replication kinetics in different human cell types: the role of autophagy, cellular metabolism and ACE2 expression. Life Sci. 308:120930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordon J, Amini S, White MK. 2013. General overview of neuronal cell culture. Methods Mol Biol. 1078:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slanzi A, Iannoto G, Rossi B, Zenaro E, Constantin G. 2020. In vitro models of neurodegenerative diseases. Front Cell Dev Biol. 8:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin K, Baillie GJ, Vetter I. 2016. Neuronal cell lines as model dorsal root ganglion neurons: a transcriptomic comparison. Mol Pain. 12:1744806916646111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Siano G, et al. 2019. Identification of an ERK inhibitor as a therapeutic drug against Tau aggregation in a new cell-based assay. Front Cell Neurosci. 13:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siano G, et al. 2019. Modulation of Tau subcellular localization as a tool to investigate the expression of disease-related genes. J Vis Exp. 10.3791/59988. [DOI] [PubMed] [Google Scholar]
- 27. Wen Z, Zhang Y, Lin Z, Shi K, Jiu Y. 2020. Cytoskeleton—a crucial key in host cell for coronavirus infection. J Mol Cell Biol. 12:968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xia X, Wang Y, Zheng J. 2021. COVID-19 and Alzheimer's disease: how one crisis worsens the other. Transl Neurodegener. 10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bouhaddou M, et al. 2020. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 182:685–712.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maginnis MS. 2023. β-Arrestins and G protein-coupled receptor kinases in viral entry: a graphical review. Cell Signal 102:110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boytz R, et al. 2023. Anti-SARS-CoV-2 activity of targeted kinase inhibitors: repurposing clinically available drugs for COVID-19 therapy. J Med Virol. 95:e28157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stukalov A, et al. 2021. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 594:246–252. [DOI] [PubMed] [Google Scholar]
- 33. Liu X, et al. 2022. SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response. Sci Signal. 15:eabg8744. [DOI] [PubMed] [Google Scholar]
- 34. Chatterjee B, Thakur SS. 2022. SARS-CoV-2 infection triggers phosphorylation: potential target for anti-COVID-19 therapeutics. Front Immunol. 13:829474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Surjit M, Liu B, Jameel S, Chow VTK, Lal SK. 2004. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem J. 383:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. 2004. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun. 319:1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goel S, et al. 2021. SARS-CoV-2 switches ‘on ‘MAPK and NF κ B signaling via the reduction of nuclear DUSP1 and DUSP5 expression. Front Pharmacol. 12:631879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grimes JM, Grimes KV. 2020. P38 MAPK inhibition: a promising therapeutic approach for COVID-19. J Mol Cell Cardiol. 144:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Avila J. 2009. The Tau code. Front Aging Neurosci. 1:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alonso AD, et al. 2010. Phosphorylation of Tau at Thr212, Thr231, and Ser262 combined causes neurodegeneration. J Biol Chem. 285:30851–30860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Avila S, Lucas JJ, Pérez M, Hernández F. 2022. Role of Tau protein in both physiological and pathological conditions. Physiol Rev. 84:361–384. [DOI] [PubMed] [Google Scholar]
- 42. Johnson GVW, Stoothoff WH. 2004. Tau phosphorylation in neuronal cell function and dysfunction. J Cell Sci. 117:5721–5729. [DOI] [PubMed] [Google Scholar]
- 43. Winkler ES, et al. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 21:1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen R, et al. 2020. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in the human and mouse brains. Front Neurol. 11:573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang C, et al. 2021. ApoE-isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell 28:331–342.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mesci P, et al. 2022. SARS-CoV-2 infects human brain organoids causing cell death and loss of synapses that can be rescued by treatment with sofosbuvir. PLoS Biol. 20:e3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Me S, Tremblay MJ. 2005. Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J Virol. 79:6577–6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng C, Trojanowski JQ, Lee VM-Y. 2020. Protein transmission in neurodegenerative disease. Nat Rev Neurol. 16:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saman S, et al. 2012. Exosome-associated Tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 287:3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Y, et al. 2017. The release and trans-synaptic transmission of Tau via exosomes. Mol Neurodegener. 12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lehmann M, Nikolic DS, Piguet V. 2011. How HIV-1 takes advantage of the cytoskeleton during replication and cell-to-cell transmission. Viruses 3:1757–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nie Y, et al. 2021. Rearrangement of actin cytoskeleton by Zika virus infection facilitates blood—testis barrier hyperpermeability. Virol Sin. 36:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Drewes G, et al. 1995. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates Tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem. 270:7679–7688. [DOI] [PubMed] [Google Scholar]
- 54. Alonso A, Zaidi T, Novak M, Grundke-Iqbal I, Iqbal K. 2001. Hyperphosphorylation induces self-assembly of Tau into tangles of paired helical filaments/straight filaments. Proc Natl Acad Sci U S A. 98:6923–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Falcicchia C, Tozzi F, Arancio O, Watterson DM, Origlia N. 2020. Involvement of p38 MAPK in synaptic function and dysfunction. Int J Mol Sci. 21(16):5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li T, Paudel HK. 2006. Glycogen synthase kinase 3beta phosphorylates Alzheimer's disease-specific Ser396 of microtubule-associated protein Tau by a sequential mechanism. Biochemistry 45:3125–3133. [DOI] [PubMed] [Google Scholar]
- 57. Richards A, Jaenisch R, Leopold P. 2022. Starting signal: aberrant kinase activation as a trigger for SARS- CoV-2 induced axonal damage. Front Virol. 2:1–6. [Google Scholar]
- 58. Solution P, Holms RD. 2022. The COVID-19 cell signalling problem: spike, RAGE, PKC, p38, NF κ B & IL-6 hyper-expression and the human ezrin. Immuno. 2:260–282. [Google Scholar]
- 59. Tenreiro S, Eckermann K, Outeiro TF. 2014. Protein phosphorylation in neurodegeneration: friend or foe? Front Mol Neurosci. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abavisani M, Rahimian K, Mahdavi B, Tokhanbigli S. 2022. Mutations in SARS–CoV–2 structural proteins: a global analysis. Virol J. 19:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roczkowsky A, et al. 2023. COVID-19 induces neuroinflammation and suppresses peroxisomes in the brain. Ann Neurol. 94:531–546. [DOI] [PubMed] [Google Scholar]
- 62. Vanderheiden A, Klein RS. 2022. Neurobiology. Curr Opin Neurobiol. 76:102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kinney JW, et al. 2018. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement (NY). 4:575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Novoa C, et al. 2022. Inflammation context in Alzheimer’s disease, a relationship intricate to define. Biol Res. 55:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 27:493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and/or supporting information.