Abstract
N-glycanase defiency (NGLY1 deficiency, NGLY1-CDDG), the first autosomal recessive congenital disorder of N-linked deglycosylation (CDDG), is caused by pathogenic variants in NGLY1. The majority of affected individuals have been identified using exome or genome sequencing. To date, no reliable, clinically available biomarkers have been identified. Urine oligosaccharide analysis was included as part of a routine evaluation for possible biomarkers in patients with confirmed NGLY1-CDDG. During the qualitative review of oligosaccharide profiles by an experienced laboratory director an abnormal analyte with a proposed structure of Neu5Ac1Hex1GlcNAc1-Asn was identified in NGLY1-CDDG patient urine samples. The same species has been observed in profiles from individuals affected with aspartylglucosaminuria, although the complete spectra are not identical. Additional studies using tandem mass spectrometry confirmed the analyte’s structure. In addition to the known NGLY1-CDDG patients identified by this analysis, a single case was identified in a population referred for clinical testing who subsequently had a diagnosis of NGLY1-CDDG confirmed by molecular testing. Urine oligosaccharide screening by MALDI-TOF MS can identify individuals with NGLY1-CDDG. In addition, this potential biomarker might also be used to monitor the effectiveness of therapeutic options as they become available.
Keywords: N-glycanase, glycosylation, N-glycanase deficiency, NGLY1, oligosaccharide screening, biochemical screening, biomarker identification
1.0. Introduction:
N-glycanase (NGLY1) deficiency (OMIM: 615273) is a recently described, autosomal recessive congenital disorder of deglycosylation (CDDG), also known as NGLY1-CDDG. Pathogenic variants in NGLY1 cause NGLY1-CDDG.(Need et al 2012; Enns et al 2014; Caglayan et al 2015; He et al 2015) Clinically, affected individuals have developmental delay, hypotonia, abnormal liver function, small feet, epilepsy, peripheral neuropathy and absent tears.(Enns et al 2014; Caglayan et al 2015; Heeley and Shinawi 2015) There is currently no therapy for NGLY1-CDDG, although investigations are ongoing.(Bi et al 2017) Most reported individuals and families have been identified by large scale sequence analysis, such as exome or genome analysis, because there was no clinically available biomarker. We have identified a urine biomarker for the identification of individuals with NGLY1-CDDG. As therapeutic options become available, the utility of this biomarker may expand into monitoring the effectiveness of these treatments.
Urine oligosaccharide profiling by MALDI-TOF MS (Xia et al 2013; Bonesso et al 2014) has recently been described, and is quickly moving to replace more traditional methods of oligosaccharide screening such as thin layer chromatography.(Raymond and Rinaldo 2013) In addition to identifying classic oligosaccharidoses, screening by MALDI-TOF MS can help diagnose additional lysosomal storage disorders such as Gaucher disease and Sandhoff disease. In this paper, we describe an abnormal urine oligosaccharide profile, detectable by MALDI-TOF MS that may greatly shorten the diagnostic odyssey for NGLY1-CDDG, and may serve as a candidate biomarker for future therapeutic monitoring.
2.0. Materials and Methods:
Families were enrolled in NIH protocol #76-HG-0238 “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders” (http://clinicaltrials.gov, trial NCT00369421) and #14-HG-0071 “Clinical and Basic Investigations into Known and Suspected Congenital Disorders of Glycosylation” (http://clinicaltrials.gov, trial NCT02089789), both approved by the National Human Genome Research Institute’s Institutional Review Board. Random urine samples from patients with confirmed NGLY1-CDDG were submitted for clinical oligosaccharide screening as part of an ongoing project to identify and characterize clinical and biochemical testing parameters of individuals with congenital disorders of glycosylation.
The method for urine oligosaccharide screening by MALDI-TOF MS has been previously published. Briefly, urine samples (volume dependent on creatinine) are extracted using Sep-Pak C18 and carbograph columns, before being lyophilized and permethylated. After permethylation, samples are subject to further cleanup with a C18 column, after which the eluant is collected and evaporated to dryness. This sample is then reconstituted in 2,5-dihydroxybenzoic acid matrix and loaded on the MALDI plate for analysis.(Xia et al 2013) Instrument analysis identifies all species between m/z of 585 and 4000. Peaks in this mass range are identified by m/z and proposed structure. These results were reviewed qualitatively by board certified, (American Board of Medical Genetics and Genomics in Clinical Biochemical Genetics) experienced laboratory directors for comparison to previously confirmed disease profiles, and also to identify any additional abnormal species.
3.0. Results
Summary information about the patients and the results of urine oligosaccharide screening are shown in Table 1. Further clinical information about a subset of these patients has been previously published.(Lam et al 2017) After the initial identification of an abnormal profile (n = 14) in confirmed patients (PLH and JDS at EGL Genetics, formerly known as Emory Genetics Laboratory), a verification set of samples (n = 11) was submitted to a second laboratory employing a similar urine oligosaccharide method to confirm that an abnormal profile could be identified by qualitative review (KMR at Biochemical Genetics Laboratory at Mayo Clinic). A set of samples submitted for clinical testing was reviewed (n = 250) to determine if this profile had been observed in the clinical population referred for urine oligosaccharide screening by a health care provider.
Table 1:
Patient population and qualitative assessment of excretion of Asn-N in individuals with NGLY1-CDDG
| Patient ID |
Age at Collection (Yrs) |
Gender | Lab 1 Asn-N Excretion |
Lab 2 Asn-N Excretion |
|---|---|---|---|---|
| 1 | 2.4 | M | Moderate | Minimal |
| 2 | 4.3 | F | Minimal | Minimal |
| 3 | 4.9 | M | Prominent | Moderate |
| 4 | 6.5 | M | Absent* | Moderate |
| 5 | 7.7 | F | Minimal | Moderate |
| 6 | 10.8 | M | Moderate | Prominent |
| 7 | 18.4 | F | Prominent | Moderate |
| 8 | 21.4 | F | Minimal | Moderate |
| 9 | 3.9 | M | Moderate | Absent |
| 10 | 16.9 | F | Prominent | Moderate |
| 11 | 15.6 | M | Moderate | Moderate |
| 12 | 6.7 | M | Minimal | NP |
| 13 | 3.1 | F | Minimal | NP |
| 14 | 0.4 | M | Minimal | NP |
No Asn-N was detected in multiple samples, however an abnormal species with a small mass shift (~ 0.5) was detected. This may represent a mass calibration issue for this sample. Reanalysis was not possible.
NP: Not Performed, Yrs: Years, M: Male, F: Female, Asn-N: oligosaccharide species identified in NGLY1-CDDG patients
Qualitative review criteria:
Prominent: > 50 % relative abundance based on overall peak height in the complete profile
Moderate: 20 – 50 % relative abundance based on overall peak height in the complete profile
Minimal: 5 – 20 % relative abundance based on overall peak height in the complete profile
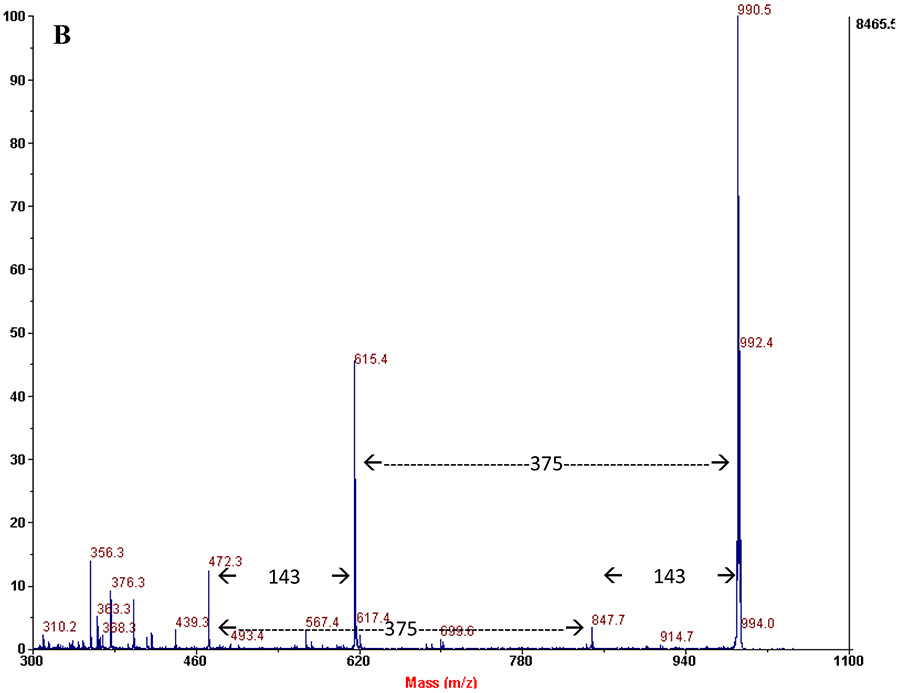
Qualitative review of urine oligosaccharide profiles of individuals with confirmed NGLY1-CDDG identified a single abnormal species with an approximate m/z = 990. The predicted structure of this species is the permethylated sodium adduct of Neu5Ac1Hex1GlcNAc1-Asn (Asn-N). MALDI-TOF/TOF MS analysis confirmed this structure is the same as has been previously identified in aspartylglucosaminuria (OMIM: 208400) patients (Figure 1). The additional abnormal species found in aspartylglucosaminuria, particularly the characteristic high mass species (m/z = 1078, 1439, 1824, 2186), were not detected in the profiles of the patients with NGLY1-CDDG.(Xia et al 2013) To confirm the proposed structure, samples were analyzed on a liquid chromatography – tandem mass spectrometry system with a quadrupole and time of flight detector (SCIEX Triple TOF 5600+). This allowed fragmentation of the permethylated glycan structure without the sodium adduct, providing an exact mass of 968.7203 amu after the loss of methanol (32 amu). The product ion spectrum for this peak is shown in Figure 2. All significant fragments in the spectrum are identified, and confirm the proposed structure from the original method paper.(Xia et al 2013) Additional information about MS/MS parameters for this experiment and fragment generation is shown in Supplement 1.
Figure 1:
MS/MS product ion scans of sodium adduct of Asn-N in a patient with NGLY1-CDDG (A) and aspartylglucosaminuria (B), showing identical fragmentation and suggesting a common pathway for origin.
Figure 2:
Product ion scan of unadducted Asn-N by LC-MS/MS. Fragmentation confirms the proposed structure of the species. All fragments can be assigned to portions of the glycan. No discrepant bonds were identified from the original proposed structure.
This biomarker was present in samples analyzed for NGLY1-CDDG patients and is not a part of the normal background control samples at significant levels. Review of 250 clinical samples submitted for testing identified only two additional samples with this abnormal analyte. In one of these individuals, a 7-year-old female, subsequent sequence analysis showed homozygosity for a previously described pathogenic variant in NGLY1 (c.1891delC; biparental inheritance confirmed). Clinically, this patient had a history of developmental delay, decreased thyroid hormone (low T4, free T4), visual disturbance, absent tears and cirrhosis. Developmental delay and abdominal distension were noted by 9 months of age, with a liver biopsy at 10 months indicating bridging fibrosis. To our knowledge, this is the first individual with NGLY1-CDDG identified by biochemical screening methods. The second individual with this profile was lost to follow-up by the testing laboratory and no further information is available. Urine oligosaccharide screening has a broad range of clinical indications, including developmental delay, dysmorphic features, seizures and hypotonia. This number of additional cases of NGLY1-CDDG would not be unexpected in the clinical population of individuals screened for oligosaccharidoses, due to common clinical features.
Results of urine oligosaccharide analysis for previously identified NGLY1-CDDG patients are shown in Table 1. Thirteen of 14 patients (93 %) screened initially had detectable amounts of Asn-N. Ten of 11 patients (91 %) who had samples submitted to the verification laboratory for testing had detectable amounts of Asn-N and data are also shown in Table 1. Taken together, and assuming the single unconfirmed positive clinical sample is a false positive, the sensitivity of this finding is 92.3 % (73.4 % - 98.6 % = 95 % confidence interval), and the specificity is 99.6 % (97.4 % - 100.0 % = 95 % confidence interval). In this particular population, the positive predictive value was 96.0% and the negative predictive value 99.2 %. The relative amounts of Asn-N per patient varied, but upon qualitative review, it was reliably detected. Variability among specimens from individual patients suggests that in some individuals the excretion of this metabolite may be intermittent. The overlap between the two groups showed that every affected individual had this abnormal metabolite detected in at least one specimen.
4.0. Discussion
The presence of an identical species in NGLY1-CDDG and aspartylglucosaminuria can be explained by similar functionalities of the deficient enzymes in each disorder. N-glycanase and aspartylglucosaminidase both cleave the bond between GlcNAc and Asn. N-glycanase acts in the cytoplasm and aspartylglucosaminidase in the lysosome; however in the case of either enzyme deficiency, undegraded substrate can eventually be excreted in the urine, regardless of its subcellular origin.(Sugahara et al 1977; Huang et al 2015) This study describes an important marker for identifying NGLY1-CDDG patients, and it could serve as a biomarker when treatment options are being evaluated. Based on oligosaccharide profiles and molecular testing, aspartylglucosaminuria can be distinguished from NGLY1-CDDG.
Regarding the origin of the Asn-N species, it is likely derived from N-linked glycans. Thus, GlcNAc must first be cleaved from the chitobiose core (GlcNAc-GlcNAc-Asn) followed by addition of Hex and Neu5Ac. In the absence of NGLY1 activity, ENGase generates GlcNAc-Asn(Huang et al 2015), which could then potentially serve as a primer for the addition of Hex and Neu5Ac, respectively.(Bengtson et al 2016) The mechanism is unknown, but it could occur by transglycosylation within the cell, possibly in lysosomes. Autophagy-mediated delivery of GlcNAc-Asn from the cytoplasm to the lysosome would co-localize substrates and enzymes, which may lend credence to this pathway. Alternatively, synthesis could occur in the plasma.
Recent studies identified both functional extracellular glycosyltransferases(Lee-Sundlov et al 2016) and nucleotide sugar donors(Lee et al 2014) in plasma. Finally, it is possible that GlcNAc-Asn enters the Golgi where it serves as a primer for the addition of galactose and sialic acid using the resident transferases and the imported nucleotide sugars, however, there is currently no known mechanism for the uptake or transport of GlcNAc-Asn into the Golgi. Further work to elucidate the mechanism of formation for this abnormal glycan is needed, which will also be important to evaluate its potential as a biomarker.
Most of the patients described in this study were identified through exome sequencing or targeted testing after an affected sibling had been diagnosed. Molecular techniques, such as sequence analysis, can provide important information, but they are expensive, and can take a significant amount of time to receive results, particularly when looking at a large number of genes. Traditionally available screening tests for congenital disorders of glycosylation, such as carbohydrate deficient transferrin analysis, N-glycan profiling, and O-glycan profiling are uninformative in NGLY1-CDDG. The availability of a clinically obtainable biochemical screening assay, such as urine oligosaccharide profiling, will allow quick and efficient screening, and can provide important clarification of uncertain molecular results, either in the case of variants that cannot be classified as pathogenic or when a second variant is not identified, but the disorder is still strongly suspected. A biochemical phenotype can be helpful supporting evidence that the correct causative gene is being analyzed, particularly with overlapping clinical phenotypes or large amounts of sequence data.(Narravula et al 2017)
5.0. Conclusions:
Urine oligosaccharide screening by MALDI-TOF MS is a fast, and relatively inexpensive means to screen patients suspected of having NGLY1-CDDG. The availability of a clinical assay that can identify patients will shorten the diagnostic odyssey in the future, while reducing costs and providing additional information that can be used to help interpret variants of uncertain significance. Urine oligosaccharide screening can be ordered by any physician, does not require any special patient preparation, and is non-invasive. Results are available quickly, often less than 10 days after ordering the test. Several clinical indications for ordering routine urine oligosaccharide screening overlap with the clinical presentation of NGLY1-CDDG, including developmental delay, epilepsy and hypotonia, and the assay can be used to screen for a large number of conditions early in a diagnostic workup. Qualitative examination of profiles by an experienced laboratory director is still a valuable part of the review of certain laboratory tests and can be used to expand the clinical utility of an already available test. Once known abnormal profiles have been identified, and a sufficient number of cases have been identified, post-analytical tools may be beneficial to ensure abnormal profiles are flagged by a quantitative review system in addition to the more traditional qualitative review.(Hall et al 2014) Significant work remains to fully characterize this potential biomarker, and evaluate its utility for the monitoring of therapeutic options, but the discovery of a biochemical phenotype with an appropriate screening test for the identification of patients is an important milestone.
Supplementary Material
Acknowledgements
This study was supported by Intramural Research Program of the National Human Genome Research Institute/National Institutes of Health, Bethesda, Maryland, USA. We sincerely thank all the individuals and their families for participating in this study, and the advocacy groups NGLY1.org and the Grace Science Foundation, which both provided patient recruitment, as well as family support. HHF would also like to acknowledge support from the Bertrand Might Foundation.
Abbreviations
- NGLY1
N-glycanase
- MALDI-TOF
matrix assisted laser desorption time of flight
- CDG
congenital disorder of glycosylation
- CDDG
congenital disorder of deglycosylation
- Hex
hexose
- Glc
glucose
- Neu5Ac
sialic acid
- HexNac
N-acetylhexosamine
- Asn
asparagine
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bengtson P, Ng BG, Jaeken J, Matthijs G, Freeze HH, Eklund EA (2016) Serum transferrin carrying the xeno-tetrasaccharide NeuAc-Gal-GlcNAc2 is a biomarker of ALG1-CDG. Journal of inherited metabolic disease 39: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Might M, Vankayalapati H, Kuberan B (2017) Repurposing of Proton Pump Inhibitors as first identified small molecule inhibitors of endo-beta-N-acetylglucosaminidase (ENGase) for the treatment of NGLY1 deficiency, a rare genetic disease. Bioorganic & medicinal chemistry letters 27: 2962–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonesso L, Piraud M, Caruba C, Van Obberghen E, Mengual R, Hinault C (2014) Fast urinary screening of oligosaccharidoses by MALDI-TOF/TOF mass spectrometry. Orphanet journal of rare diseases 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan AO, Comu S, Baranoski JF, et al. (2015) NGLY1 mutation causes neuromotor impairment, intellectual disability, and neuropathy. European journal of medical genetics 58: 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns GM, Shashi V, Bainbridge M, et al. (2014) Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genetics in medicine : official journal of the American College of Medical Genetics 16: 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall PL, Marquardt G, McHugh DM, et al. (2014) Postanalytical tools improve performance of newborn screening by tandem mass spectrometry. Genetics in medicine : official journal of the American College of Medical Genetics 16: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Grotzke JE, Ng BG, et al. (2015) A congenital disorder of deglycosylation: Biochemical characterization of N-glycanase 1 deficiency in patient fibroblasts. Glycobiology 25: 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeley J, Shinawi M (2015) Multi-systemic involvement in NGLY1-related disorder caused by two novel mutations. American journal of medical genetics Part A 167a: 816–820. [DOI] [PubMed] [Google Scholar]
- Huang C, Harada Y, Hosomi A, et al. (2015) Endo-beta-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells. Proceedings of the National Academy of Sciences of the United States of America 112: 1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C, Ferreira C, Krasnewich D, et al. (2017) Prospective phenotyping of NGLY1-CDDG, the first congenital disorder of deglycosylation. Genetics in medicine : official journal of the American College of Medical Genetics 19: 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Sundlov MM, Ashline DJ, Hanneman AJ, et al. (2016) Circulating blood and platelets supply glycosyltransferases that enable extrinsic extracellular glycosylation. Glycobiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Nasirikenari M, Manhardt CT, et al. (2014) Platelets support extracellular sialylation by supplying the sugar donor substrate. The Journal of biological chemistry 289: 8742–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narravula A, Garber KB, Askree SH, Hegde M, Hall PL (2017) Variants of uncertain significance in newborn screening disorders: implications for large-scale genomic sequencing. Genetics in medicine : official journal of the American College of Medical Genetics 19: 77–82. [DOI] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, et al. (2012) Clinical application of exome sequencing in undiagnosed genetic conditions. Journal of medical genetics 49: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K, Rinaldo P (2013) From art to science: oligosaccharide analysis by maldi-tof mass spectrometry finally replaces 1-dimensional thin-layer chromatography. Clinical chemistry 59: 1297–1298. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Akasaki M, Funakoshi I, Aula P, Yamashina I (1977) Structural studies of glycoasparagines from urine of a patient with aspartylglycosylaminuria (AGU). FEBS letters 78: 284–286. [DOI] [PubMed] [Google Scholar]
- Xia B, Asif G, Arthur L, et al. (2013) Oligosaccharide analysis in urine by maldi-tof mass spectrometry for the diagnosis of lysosomal storage diseases. Clinical chemistry 59: 1357–1368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.