Abstract
VP1 sequences were determined for poliovirus type 1 isolates obtained over a 189-day period from a poliomyelitis patient with common variable immunodeficiency syndrome (a defect in antibody formation). The isolate from the first sample, taken 11 days after onset of paralysis, contained two poliovirus populations, differing from the Sabin 1 vaccine strain by ∼10%, differing from diverse type 1 wild polioviruses by 19 to 24%, and differing from each other by 5.5% of nucleotides. Specimens taken after day 11 appeared to contain only one major poliovirus population. Evolution of VP1 sequences at synonymous third-codon positions occurred at an overall rate of ∼3.4% per year over the 189-day period. Assuming this rate to be constant throughout the period of infection, the infection was calculated to have started ∼9.3 years earlier. This estimate is about the time (6.9 years earlier) the patient received his last oral poliovirus vaccine dose, approximately 2 years before the diagnosis of immunodeficiency. These findings may have important implications for the strategy to eliminate poliovirus immunization after global polio eradication.
Immunity to infections by polioviruses and other human enteroviruses is mediated primarily by neutralizing antibody (24, 31). In immunocompetent persons, the duration of intestinal infection by polioviruses is typically limited to 4 to 8 weeks, as indicated by the period of excretion of poliovirus in the stool (2). In contrast, in immunodeficient persons, particularly those with B-cell deficiencies associated with hypogammaglobulinemia (31), poliovirus excretion times as long as 3.5 years have been reported (10, 38). Prolonged enteric infections with nonpolio enteroviruses of up to 6.5 years have been described for persons with hypogammaglobulinemia (24). Immunodeficient persons are at far higher risk (>3,000-fold) than immunocompetent persons for vaccine-associated paralytic poliomyelitis (VAPP) (33). For this reason, exposure of immunocompromised persons to oral poliovirus vaccine (OPV), either by receipt of the vaccine or by contact with vaccine recipients, should be avoided (4). However, because the diagnosis of immunodeficiency is often not established in the first year of life (22) and immunization with OPV may begin as early as 2 months of age, some children receive OPV before their immunodeficiency condition is recognized (33).
In this report, we describe genetic analyses of type 1 vaccine-derived polioviruses isolated from a patient with VAPP in whom common variable immunodeficiency syndrome (CVID) (31) had been diagnosed nearly 5 years earlier. The extent and rate of nucleotide divergence of the isolate sequences from the Sabin type 1 vaccine strain sequence were consistent with prolonged replication of the vaccine-derived polioviruses in the patient. The combined clinical and genetic data suggested that the infection was probably initiated by receipt of an OPV dose given at least 6.9 years before onset of paralysis and at least 2 years before diagnosis of CVID.
MATERIALS AND METHODS
Patient.
The case patient, a man born in 1964, had a history of repeated upper respiratory infections, otitis media, recurrent fever, chronic cough, sinusitis, and skin infections. At age 9 years, he was diagnosed with allergies to dogs, cats, food items, grass, and trees. At age 12 years, he was hospitalized with lung infiltrates and maxillary sinusitis and diagnosed with CVID on the basis of quantitative serum immunoglobulin (Ig) readings of 42 mg/dl for IgG (normal values, 639 to 1,349 mg/dl), 4.5 mg/dl for IgM (normal values, 56 to 352 mg/dl), and 3.8 mg/dl for IgA (normal values, 70 to 132 mg/dl). He was placed on monthly fresh frozen plasma therapy and maintained IgG levels of between 62 and 330 mg/dl from 1976 until 1981. In July 1981, at age 16 years, he developed fever and generalized weakness after a diarrheal illness. Over a period of 4 days, he developed quadriparesis requiring mechanical ventilation. Paralytic poliomyelitis was diagnosed. His subsequent clinical course included multiple hospital admissions for pneumonia and urinary tract infections. He remained ventilator dependent from 1981 until his death in October 1990. The patient had no known contact with a polio patient or vaccine recipient and had not traveled from his state of residence (Missouri) to an area where polio is endemic. He had received three doses (at ages 3, 4, and 5 months) of inactivated poliovirus vaccine, followed by four doses (at ages 3 years, 3 years 2 months, 5 years, and 10 years) of trivalent OPV (5).
Virus isolation and typing.
Serial stool specimens were obtained from the patient at 11, 23, 48, 126, 158, and 200 days after onset of paralysis. Stools were processed for virus isolation by standard methods (35) and passaged twice on monolayers of primary rhesus monkey kidney cells or RD cells (human rhabdomyosarcoma cell line; ATCC CCL136). Isolates were typed in neutralization tests with pooled hyperimmune equine sera. Isolates were passaged a third time in MA104 cells (African green monkey kidney cell line) for oligonucleotide fingerprinting and in RD cells to produce poliovirus RNA templates for nucleic acid sequencing.
Antigenic characterization of poliovirus isolates.
Poliovirus type 1 isolates were analyzed for their antigenic properties in neutralization tests with cross-absorbed antisera by the method of van Wezel and Hazendonk (34).
Oligonucleotide fingerprinting.
Viral RNAs were analyzed by the RNase T1 oligonucleotide fingerprinting method described earlier (29), as modified for smaller (16 by 16 by 0.15 cm) gels.
Dot blot hybridization.
Preparation of digoxigenin-labeled RNA transcripts, conditions for dot blot hybridization, and detection of hybrids by enzyme-linked immunosorbent assay were as described previously (8), except that the chemiluminescent substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}4-yl)phenyl phosphate (CSPD) (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) was used in the assays.
Sequencing of poliovirus RNAs.
Complete VP1 (nucleotides 2480 to 3385) and partial VP1/2A (nucleotides 3296 to 3445) sequences of the serial poliovirus isolates from the immunodeficient patient were determined in cycle sequencing reaction mixtures (20) containing fluorescent dye-labeled dideoxynucleotides (Applied Biosystems, Foster City, Calif.). The templates were 1,014-bp PCR products amplified from poliovirus RNAs by using the primer pair 3038/PCR2 (antisense [A] polarity, positions 3431 to 3452, 5′-GGGTGGCCAAGTGATAGTTGCAAAT-3′) and seroPV1,2S (sense [S] polarity, positions 2439 to 2457, 5′-TGCGIGA[C/T]ACIACICA[C/T]AT-3′) (19); deoxyinosine residues are indicated by the letter I, and primer positions having equimolar amounts of two different nucleotides are enclosed in brackets. Nucleotide sequences were determined with the aid of an automated Sequenator (Applied Biosystems). VP1/2A sequences of other poliovirus isolates were determined either by automated cycle sequencing or by manual techniques (30). RNA extracted from purified virions served as templates for extension of a synthetic DNA primer (A, 3508 to 3527, 5′-AAGAGGTCTCTATTCCACAT-3′) by avian myeloblastosis reverse transcriptase in the presence of dideoxynucleotide chain terminators (30). All primers for PCR and sequencing reactions (also including VP1/245A, A, 3212 to 3231, 5′-GTIGG[A/G]TT[A/G]TGITC[A/G]TTIAC-3′; panPV/PCR-1, A, 2915 to 2934, 5′-TTIAIIGC[A/G]TGICC[A/G]TT[A/G]TT-3′ [22]; 3038/PCR4, S, 2699 to 2719, 5′-GAGTCTAGTATAGAGTCCTTC-3′; and 3038/PCR3, S, 2927 to 2951, 5′-GCCTTAAATCAAGTATACCAAATTA-3′) were prepared and purified as described previously (36).
Separation of poliovirus populations by plaque purification and sequence analysis.
Two sequence variants (major [M] and minor [m]) in the day 11 isolate were separated by plaque purification of HEp-2 (human larynx epidermoid carcinoma cell line; ATCC CCL23) cell monolayers (27). The virus of 10 well-separated plaques was grown on RD monolayers in tubes. RNA was extracted directly from cell culture lysates for amplification of poliovirus sequences by PCR (36). Amplicons were sequenced over the interval 2954 to 3385 by automated cycle sequencing methods.
Selective amplification of m variant poliovirus sequences by PCR.
A nested, two-step PCR assay was developed for selective amplification of m variant sequences. In the first step, nonselective primers VP1/245A and 3038/PCR4, equivalently matching M and m templates, were used in limiting quantities (2 pmol each) in 50-μl amplification reaction mixtures (denaturation, 94°C, 45 s; annealing, 42°C, 45 s; extension, 60°C, 90 s; 30 cycles) otherwise performed as previously described (36), producing a 533-bp amplicon. Ten microliters of the first-step reaction mixture was used as template in the second-step reaction mixture containing 10 pmol each of selective primers 2677/m− (A, 3127 to 3144, 5′-GCCGACTGGTCTTTCAGC-3′) and 2677/m+ (S, 2759 to 2776, 5′-AACTCAGTTTCCACCGGG-3′) designed to perfectly match VP1 sequences of the m variant but to mismatch the M variant in the two terminal nucleotide positions at the 3′-donor ends of each primer. Conditions for programmed amplification (25 cycles) were as described above, except that extensions were performed at 62°C. Identifications of the 386-bp amplicons were confirmed by digestion with the restriction endonucleases XbaI (recognition site at position 2861 [m variant only], yielding fragments of 281 and 105 bp) and EcoRI (recognition sites at position 2846 [m variant only] and 2876 [both variants], yielding fragments of 266 and 120 bp [M variant] or 266, 90, and 30 bp [m variant]) and electrophoretic separation of the digestion products in 12.5% polyacrylamide gels (36).
Analysis of VP1/2A nucleotide sequences.
Evolutionary distances between poliovirus genomes were estimated from VP1/2A sequences (all codon positions), using the two-parameter method of Kimura (21) to correct for multiple substitutions at a site. Calculations were performed by the program DNADIST of the PHYLIP 3.5c program package (11), with a value of 10 used for the transition/transversion ratio. VP1/2A evolutionary distances among poliovirus isolates were summarized in a tree constructed by the neighbor-joining method with the program NEIGHBOR (11).
Estimation of the time of initial infection from the VP1 evolution rate.
The evolution rate of VP1 at third codon positions was estimated for the M lineage from day 11 to day 200. For each time point, all positions with a new substitution were given a score of 1; all others were scored as 0 (no change from preceding sequence). At ambiguous sites, only the base found in a higher molar proportion was scored. The two-parameter correction method (21) was used to convert base substitution differences to evolutionary distances (expressed as VP1 third-position substitutions/100 nucleotides). The rate of evolution over the 189-day period was estimated by drawing a linear regression line through the evolutionary distance points. The age of initial poliovirus infection was estimated by increasing all evolutionary distances by 31.2 (the evolutionary distance between the Sabin 1 VP1 and the day 11 M variant VP1) and plotting the data points on rescaled axes (abscissa, patient age in years; ordinate, evolutionary distance from the Sabin 1 sequence). The estimated age of initial poliovirus infection was the intercept obtained by extrapolating the evolution rate line back to zero substitutions in the Sabin 1 VP1.
Estimation of time of divergence of the M and m lineages.
The estimated time of divergence (timediv M,m) of the M and m lineages was calculated from the following relationship:
 |
where ageday 11 is the patient age at day 11, ageinit is the patient age at the time of initiation of infection (estimated as described above), evoldistday 11 M,m is the evolutionary distance (calculated from VP1 third-position substitutions) between the M and m variants, and evoldistSab1, day 11 M is the evolutionary distance between Sabin 1 and the M variant.
Nucleotide sequence accession number.
VP1 sequences of poliovirus isolates from the immunodeficient patient described in this article have been deposited in the GenBank data library and assigned accession no. AF083931 to AF083944.
RESULTS
Initial characterization of isolates.
Clinical specimens from the patient were obtained as part of the routine surveillance of poliomyelitis cases in the United States. Poliovirus type 1 was isolated from each specimen. The initial (day 11) isolate was characterized in neutralization assays by using cross-absorbed antisera (34) and was found to have “non-vaccine-like” antigenic properties (13). However, it is known that the Sabin vaccine strains, especially the type 1 strain, frequently revert to non-vaccine-like antigenicities upon replication in the human intestine (3, 27). Therefore, the isolates were further tested by the independent method of RNase T1 oligonucleotide fingerprinting.
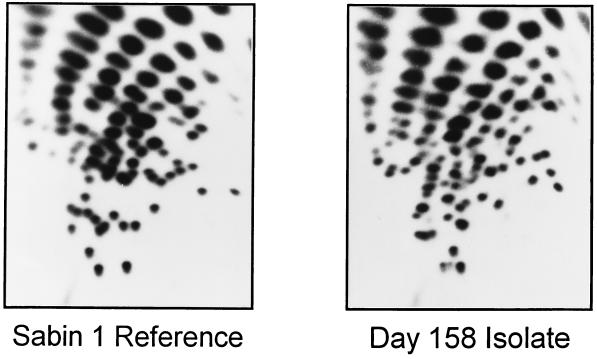
The fingerprint of the day 11 isolate (not shown) had a high background of secondary oligonucleotide spots. The fingerprint of a later (day 158) isolate had a low background and a complexity typical of a poliovirus genome (Fig. 1) (18, 29). The fingerprint of the day 158 isolate was similar to the primary spots in the fingerprint of the day 11 isolate. However, the pattern was very different from that of the Sabin type 1 vaccine strain, LSc 2ab (Fig. 1). Sabin 1 vaccine-related isolates generally have fingerprints that are very similar (>75% comigrating oligonucleotides) to that of the reference vaccine strain (18, 29). The fingerprint of the day 158 isolate also differed from those of the last wild type 1 isolates from the United States, from cases that occurred in 1979 in the states of Missouri (the last case of the Turkey→Netherlands→Canada→USA epidemic) (29) and Illinois (importation from Mexico) (18). Moreover, no other wild poliovirus isolate was found to have a fingerprint similar to that of the day 158 isolate (18, 29). Thus, the initial characterizations did not identify the isolates from the immunodeficient patient as vaccine derived.
FIG. 1.
RNase T1 oligonucleotide fingerprints of the Sabin type 1 OPV reference strain (LSc 2ab) (left) and the day 158 isolate from the immunodeficient patient (right). Electrophoretic separation of oligonucleotides in the first dimension (separation by base composition) is from left to right and in the second dimension (separation by chain length) is from bottom to top. The longer oligonucleotides (chain lengths of ≥12 nucleotides) (18), which having unique sequences representing ∼15% of the genome, are resolved into patterns (fingerprints) that are highly characteristic of the RNA sequence.
Sequence properties of the day 11 isolate.
At the time the case of infection was identified, oligonucleotide fingerprinting was the most accurate routine method for characterizing poliovirus isolates. However, fingerprinting is highly sensitive to changes in RNA sequence, such that quantitative estimates of relatedness are most reliable when RNA molecules share >95% base sequence similarity (1). Therefore, if a vaccine-derived poliovirus had evolved extensively, it might not be correctly identified by fingerprinting.
The most definitive method for characterizing poliovirus isolates is by nucleotide sequencing. When we determined the VP1 sequences of the day 11 isolate, they differed from the Sabin 1 sequence at >100 sites. However, 50 of the 906 codon positions were ambiguous, usually containing either A+G or C+T. The sequence ambiguity suggested that the day 11 isolate contained a mixture of related type 1 polioviruses. To resolve the mixture, the day 11 isolate was plaque purified, and the progeny of 10 plaques were sequenced over a 432-base VP1 interval (2954 to 3385) having 24 ambiguous sites. Seven of the plaque isolates had one unambiguous sequence; the other three plaque isolates had another unambiguous sequence that differed from the first only at the ambiguous sites. The sequence variant represented by the seven plaques did indeed appear to correspond to the major population in the original day 11 isolate, because its nucleotides had the stronger bands in the sequencing gels at the positions of ambiguity. The day 11 isolate apparently contained only two principal populations, because the combination of the M and m variant sequences fully accounted for the observed sequence ambiguity. The presence of two distinct sequence variants probably also accounted for the high background in the oligonucleotide fingerprint of the day 11 isolate.
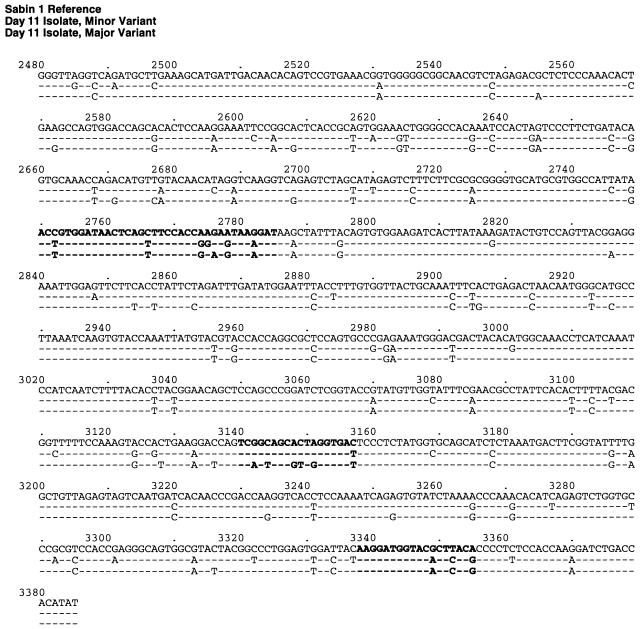
The complete VP1 nucleotide sequences were determined for a plaque isolate of each variant. The M and m variants were related to, but nearly equally divergent from, Sabin 1, differing at 9.6% (87 of 906 [M variant]) and 10.0% (91 of 906 [m variant]) of the VP1 nucleotide positions (Fig. 2). They were most closely related to each other, differing at 5.5% (50 of 906) of the bases. The M and m variants probably diverged from a single lineage derived from Sabin 1, because they shared ∼72% of their VP1 base differences from the vaccine strain sequence. Most (85% [151 of 178]) of the base substitutions occurred in the third codon position, 97% (147 of 151) of which generated synonymous codons.
FIG. 2.
VP1 nucleotide sequence alignment of the Sabin 1 reference strain (line 1), the day 11 isolate m variant (line 2), and the day 11 isolate M variant (line 3). Sabin 1 nucleotide positions are numbered according to the system of Nomoto et al. (28); those of the day 11 isolates are numbered similarly for comparability. Boldface letters identify codons of amino acid residues that form NAg I (2750 to 2785), NAg II (3140 to 3157), and NAg III (3338 to 3355) (25).
A small proportion (18% [16 of 91], M variant; 16% [14 of 87], m variant) of base changes encoded amino acid substitutions (Fig. 2 and 3). In both variants, five amino acid substitutions clustered in the interval 1090 to 1106, spanning neutralizing antigenic site I (NAg I) (Fig. 3). Two of these mutations (I1090M and T1106A) were reversions to the parental Mahoney sequence, two others (K1099E [M variant] and K1099G [m variant]) eliminated a trypsin cleavage site characteristic of Sabin 1 (12), and another (N1100S) restored the consensus residue for type 1 polioviruses (26, 37). Another substitution (A1222V) mapped to NAg II of the M variant (Fig. 3). The remaining substitutions were conservative (16) and occurred in domains internalized in the intact virion (14).
FIG. 3.
VP1 amino acid sequence alignment of the Sabin 1 reference strain (line 1), the day 11 isolate m variant (line 2), and the day 11 isolate M variant (line 3). Capsid amino acid positions are indicated by a four-digit number: the first digit identifies the virion protein, and the next three digits specify the residue position (e.g., 1001 indicates residue 1 of VP1). Boldface letters identify virion surface residues that form NAg I (1101 to 1112), NAg II (1221 to 1226), and NAg III (1287 to 1292) (25).
Relationships of case isolates to other type 1 polioviruses.
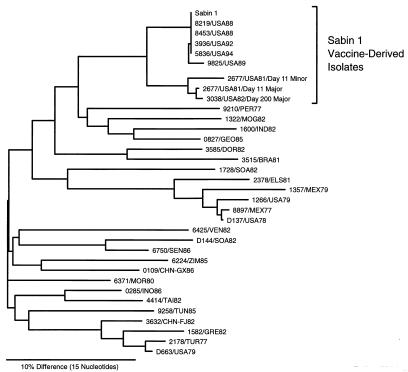
Nucleotide sequencing can detect much more distant genetic relationships than can oligonucleotide fingerprinting (1, 30). To survey the genetic relationships among poliovirus isolates, we compared sequences in the VP1/2A junction (nucleotides 3296 to 3445) (17, 30). VP1/2A sequences were determined for the M and m variants from the day 11 isolate, the day 200 isolate, the Sabin 1 vaccine strain, 5 Sabin 1 vaccine-derived isolates from immunodeficient VAPP patients in the United States, and 25 type 1 wild polioviruses from cases that occurred in different parts of the world during 1977 to 1986. The sequence relationships, summarized in a tree (Fig. 4), confirmed that the isolates from the immunodeficient patient were most closely related to vaccine-derived polioviruses (∼8% VP1/2A sequence difference). In contrast, wild poliovirus isolates from the same period were more divergent (19 to 24% sequence differences) (Fig. 4). For example, the day 11 isolates differed from the last wild isolates from the United States at ∼23% (1979 Missouri case) and ∼18% (1979 Illinois case) of nucleotide positions.
FIG. 4.
Tree summarizing sequence relatedness across the interval of nucleotides 3296 to 3445 (VP1/2A region) among the Sabin 1 vaccine strain, 3 isolates from the immunodeficient patient, 5 type 1 vaccine-related isolates from other immunodeficient VAPP patients in the United States, and 25 type 1 wild-type polioviruses isolated in different regions of the world from cases occurring within 5 years of onset of paralysis in the immunodeficient patient. BRA, Brazil; CHN, China (FJ, Fujian; GX, Guangxi); DOR, Dominican Republic; ELS, El Salvador; GEO, Georgia; GRE, Greece; IND, India; INO, Indonesia; MEX, Mexico; MOG, Mongolia; MOR, Morocco; PER, Peru; SEN, Senegal; SOA, South Africa; TAI, Taiwan; TUN, Tunisia; TUR, Turkey; USA, United States; VEN, Venezuela; and ZIM, Zimbabwe (35).
The M and m variants were also recognized as Sabin 1 related in hybridization reactions (not shown) with the specific RNA probe Sab1/VP1, currently used for routine identification of type 1 poliovirus isolates (8). Hybridization signal intensities were reduced relative to that of the control Sabin 1 RNA, probably because mismatches with the probe (5.3%, M variant; 7.5%, m variant) reduced the stabilities of the hybrids.
Disappearance of the m variant after day 11.
The sequences of all isolates from specimens taken after day 11 had very low ambiguity, and all appeared to be derived from the M variant. However, if the m variant was present in proportions of <5%, the resulting sequence ambiguity would be difficult to detect in sequencing gels. To test for the presence of m variant sequences in the later isolates, we performed PCR assays using primers that can selectively amplify m variant sequences present in trace amounts (as low as 0.01%) in preparations of M variant RNA templates. Using this sensitive PCR assay, we detected m variant sequences only in the day 11 isolate (data not shown), suggesting that the m variant lineage had died out between days 11 and 23.
Continued evolution of the M variant lineage.
Additional nucleotide substitutions accumulated in the VP1 of the M variant lineage from day 11 to day 200 (Table 1). Most (10 of 13) of the observed substitutions generated synonymous codons, and 80% (8 of 10) of these occurred in the third codon position. Of the three observed amino acid substitutions, one (A1223V) occurred within NAg II, another (L1104I) mapped near NAg I, and the third (I1056V) restored the residue found in the Sabin 1 VP1 (Table 1 and Fig. 3). The overall direction of genetic change during the 189-day period was further divergence away from the day 11 isolate (by 0.8%, uncorrected) and from the Sabin 1 vaccine strain (from 9.6% [day 11] to 9.9% [day 200], uncorrected). However, the pattern of VP1 variability was much more dynamic than could be seen from the net genetic change alone. For example, nearly half (6 of 13) of the observed substitutions (at positions 2659, 2693, and 2878) were paired, such that a substitution found in an isolate from one time point appeared to have reverted by the time of the next isolate. Five of the 13 substitutions (A2645G, G2659A, C2693A, G2878A, and G3124A) restored the Sabin 1 base. At least one isolate (day 158) contained two significant populations, indicated by sequence ambiguity at position 3316 (Table 1). The dynamics of variability within the virus quasispecies population was probably far higher than what was actually observed (9).
TABLE 1.
VP1 sequence evolution of day 23 to day 200 isolates
| Day of specimen collection | Sequence evolutiona |
|---|---|
| 23 | A2693C (1) |
| 48 | C2693A (1) |
| 126 | C2722T (3) C2789A (1) C2968T (3) G3124A (3) C3147T (2) |
| 158 | A2645G (1) G2659A (3) A2878G (3) |
| 200 | A2659G (3) G2878A (3) T2887C (3) |
Base substitutions are indicated as original base/nucleotide number/mutant base (codon position in parentheses). Nucleotides are numbered as in Fig. 3. Base changes encoding amino acid substitutions are underlined. The amino acid substitutions (original residue/amino acid number/mutant residue) were L1104I for C2789A, A1223V for C3147T, and I1056V for A2645G. Amino acids are numbered as in Fig. 4. Except where indicated, substitutions found in a preceding isolate were present in all subsequent isolates. VP1 position 3316 of the day 158 isolate contained a base mixture (T>C).
Estimation of the duration of the chronic infection from the evolution rate of the M variant lineage.
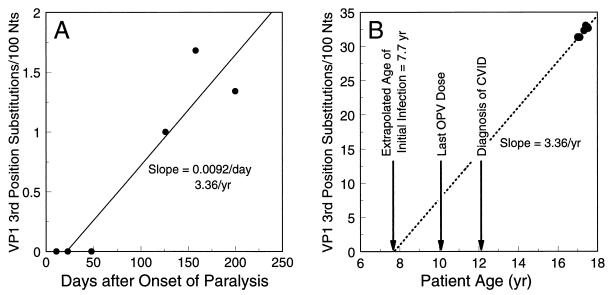
We estimated the rate of fixation of third-codon-position substitutions into the VP1 of the main M variant lineage from day 11 to day 200. Third-position substitutions were used in the analysis because all were associated with synonymous mutations, which are most likely to arise through genetic drift rather than by positive selection, and thus would be expected to accumulate at a fairly uniform rate. Evolutionary distances were calculated from the observed VP1 third-position substitutions by the two-parameter method of Kimura (21) to correct for multiple substitutions at a site. When the evolutionary distances were plotted from day 11 to day 200, the rate of change was estimated to be 0.0092% third-position substitutions/day, or 3.36% third-position substitutions/year (Fig. 5A). The evolutionary distance data were replotted on rescaled axes (Fig. 5B), with all points shown in Fig. 5A increased by 31.2 third-position substitutions/100 nucleotides (the evolutionary distance between Sabin 1 and the day 11 M variant) and the time coordinate expressed as the patient age in years. When the line for the evolution rate of 3.36%/year was extrapolated back to zero substitutions in the Sabin 1 VP1, the intercept was at the patient age of ∼7.7 years, about 2 years prior to receipt of his last OPV dose (Fig. 5B). The time from the initial infection to the onset of paralysis was estimated to be 9.3 years.
FIG. 5.
Estimation of the duration of chronic type 1 poliovirus vaccine infection from the rate of evolution of VP1 nucleotide sequences. (A) Rate of fixation of third-codon-position substitutions into VP1 from day 11 (M variant) to day 200, estimated from evolutionary distance calculations. On the abscissa, time zero is the date of onset of paralysis. The ordinate shows evolutionary distances from the sequence of the day 11 M variant. (B) Evolution rate calculated in panel A extrapolated back (dashed line) to zero substitutions in the Sabin 1 VP1. On the abscissa, time zero is the patient’s date of birth. The ordinate shows evolutionary distances from the Sabin 1 sequence (value for day 11 M variant = 31.2 VP1 third-position substitutions/100 nucleotides [Nts]).
Approximate time of divergence of the M and m lineages.
The nearly equivalent evolutionary distances of the M and m variants from Sabin 1 suggested that the two lineages had diverged from a single Sabin 1-derived lineage. We assumed that the evolutionary distance of each variant to the point of divergence was equal to half the total evolutionary distance separating the two (16.6/2 = 8.3). This represented 27% (8.3/31.2 = 0.27) of the total distance from Sabin 1 to the M variant. By further assuming that the rate of evolution was constant over the period of the infection, the time of divergence was calculated from the estimated duration of infection (9.3 years × 0.27) to be 2.5 years (Fig. 6).
FIG. 6.
Time course of chronic type 1 poliovirus vaccine infection estimated from the VP1 evolution rate shown in Fig. 5. If the infection was initiated by the last OPV dose received by the immunodeficient patient, then the actual duration of infection to day 200 would be reduced by 24%, from 9.8 years to 7.4 years.
DISCUSSION
The type 1 polioviruses isolated from the immunodeficient patient described here are, to our knowledge, the most extensively evolved derivatives of the Sabin 1 vaccine strain reported so far. The estimated duration of the prolonged poliovirus infection of ∼9.8 years, calculated from the rate of VP1 sequence evolution, is over twice as long as has been described in any previous report (10, 38). Furthermore, we do not know whether poliovirus excretion continued beyond the period of sampling.
Because stool specimens were obtained only after onset of paralysis, we could not determine from direct measurement precisely when the patient was first infected with the vaccine-derived polioviruses. Nonetheless, the estimate obtained from the indirect genetic data fits reasonably well with the clinical history of the patient. The finding that the patient was infected with two virus populations that were nearly equally divergent from the Sabin 1 vaccine strain strongly suggests that the infection started with a single OPV dose. Although this dose could have been administered to another person and transmitted by contact to the immunodeficient child at any time before the divergence of the two lineages, it seems more likely that the exposure was by direct immunization. The patient received his last OPV dose at 10 years of age, 2 years before diagnosis of CVID and 6.9 years before onset of paralysis. We suggest that this last OPV dose was the initiating dose and that the patient’s still undiagnosed CVID condition had prevented clearance of infection with the Sabin 1 component of the vaccine. Although the interval between infection and onset of paralysis estimated from the evolution rate data (∼9.3 years) does not preclude that the infection may have started with an earlier OPV dose, it appears unlikely that the infection began much later than the time of the last OPV dose.
The evolution rate measurements used to estimate the duration of infection have several limitations to their precision. First, the number of specimens available for analysis was small (six stools), and the time interval for monitoring the sequence evolution of the vaccine-derived virus was short (189 days) relative to the estimated duration of the infection. Second, only a portion of the sequence information potentially available was used for our evolution rate estimates. Beyond limiting the sequence comparisons to VP1, we analyzed only synonymous third-position substitutions in order to minimize the effects of positive selection on the evolution rates. Finally, the underlying assumption that the rate of VP1 sequence evolution in immunodeficient persons is effectively constant over several years is unproven. Despite these limitations, the estimated rate of VP1 evolution (∼3.36% third-position substitutions/year) is close to the rate (∼3.07% third-position substitutions/year) obtained for a wild poliovirus lineage circulating over 10-year period (7, 17). The apparent similarities in the evolution rates of synonymous nucleotide sites during replication in immunodeficient and normal persons may indicate that the frequencies of genetic bottleneck events driving the evolution of these presumably neutral sites are similar under both conditions of infection.
Two aspects of the chronic infection of the immunodeficient patient were difficult to explain. The first surprising observation was that two variant populations had apparently coevolved (and therefore coreplicated) in the patient for years. Replication of the two variants may have been localized to different sites in the gastrointestinal tract, and extensive colonization of each site by virus from the other site may have been blocked by intracellular interference. The second observation was the apparent disappearance of the m variant soon after onset of paralysis. The appearance of paralysis was probably an indication that circulating antibodies could no longer suppress dissemination of virus to other cells and tissues. Under these new physiologic conditions, the M variant may have had a higher fitness for replication and spread (9).
The case described in this report is exceptional because it is the only known VAPP case in an immunodeficient person in which immunodeficiency had been diagnosed before onset of paralysis. In all other cases of VAPP among immunodeficient persons, the appearance of the paralysis was the event that prompted consideration of the diagnosis of immunodeficiency. While there is no evidence that this vaccine-derived poliovirus caused other cases of VAPP, we do not know whether the virus caused any subclinical infections of contacts.
Apart from hypogammaglobulinemic persons, poliovirus infections of other immunodeficient persons do not appear to be prolonged (32). However, we currently do not know the prevalence or typical duration of chronic poliovirus excretion among hypogammaglobulinemic persons. Stool specimens from immunodeficient vaccine recipients or contacts are normally taken only after appearance of paralysis. If poliovirus infection is confirmed from the initial specimens, subsequent specimens are not routinely taken for virologic monitoring.
The findings reported here may have implications for the global polio eradication initiative of the World Health Organization (15), in particular how and when to discontinue immunization after wild polioviruses have been eradicated (6). The strategy of displacing circulating wild polioviruses with vaccine-derived strains through well-synchronized mass OPV immunization campaigns, combined with routine OPV immunization, has been highly effective (15). Vaccine-derived strains normally disappear rapidly from the community after cessation of immunization with OPV. This has been most clearly shown in Cuba, where all OPV is given in two rounds of mass campaigns and where extensive samplings of young children and the environment were found to be consistently negative for polioviruses at 3 months after the last round of immunization (23). Additional evidence against the long-term persistence of vaccine-derived strains in the community comes from the epidemiology of polio outbreaks among unimmunized populations in countries where poliovirus is nonendemic. If any residual circulation of vaccine-derived strains had occurred, it was apparently quite limited, because herd immunity was insufficient to prevent epidemics caused by imported wild polioviruses (17, 18, 29). However, even in unimmunized populations, chronic infection of a small number of immunodeficient persons might continue undetected for some time. Such persons may represent a potential reservoir for polioviruses after wild polioviruses have been eradicated and OPV immunization stopped. Systematic studies of poliovirus excretion among immunodeficient persons are therefore needed to assess the most appropriate strategy to eliminate all poliovirus infections of humans.
ACKNOWLEDGMENTS
We thank George Marchetti for isolating the polioviruses from the clinical specimens and Milford Hatch for performing the antigenic characterization of the initial poliovirus isolate. The contributions of Lina De, Rebeca Rico-Hesse, and Su-Ju Yang to the database of wild-type poliovirus sequences are appreciated. We thank Larry Anderson, Steve Cochi, Walter Dowdle, Howard Gary, Milford Hatch, Brian Mahy, John O’Connor, and Melinda Wharton for constructive suggestions and review of the manuscript and Andrew Ball, John Modlin, Neal Nathanson, Larry Schonberger, Gail Wertz, Catherine Wilfert, Jerry Winkelstein, and Peter Wright for helpful discussions. The cooperation and assistance of the Missouri State Health Department and the Task Force for Child Survival and Development are appreciated.
REFERENCES
- 1.Aaronson R P, Young J F, Palese P. Oligonucleotide mapping: evaluation of its sensitivity by computer simulation. Nucleic Acids Res. 1982;10:237–246. doi: 10.1093/nar/10.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, J. P., Jr., H. E. Gary, Jr., and M. A. Pallansch. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J. Infect. Dis. 175(Suppl. 1):S176–S182. [DOI] [PubMed]
- 3.Blondel B, Crainic R, Fichot O, Dufraisse G, Candrea A, Diamond D, Girard M, Horaud F. Mutations conferring resistance to neutralization with monoclonal antibodies in type 1 poliovirus can be located outside or inside the antibody-binding site. J Virol. 1986;57:81–90. doi: 10.1128/jvi.57.1.81-90.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Poliomyelitis prevention in the United States: introduction of a sequential vaccination schedule of inactivated poliovirus vaccine followed by oral poliovirus vaccine. Recommendations of the Advisory Committee on Immunization Practices (ACIP) Morbid Mortal Weekly Rep. 1997;46:1–25. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Prolonged poliovirus excretion in an immunodeficient person with vaccine-associated paralytic poliomyelitis. Morbid Mortal Weekly Rep. 1997;46:641–643. [PubMed] [Google Scholar]
- 6.Cochi S L, Sutter R W, Kew O M, Pallansch M A, Dowdle W R. A decision tree for stopping polio immunization. Technical Consultation on Global Eradication of Poliomyelitis. EPI/POLIO/TECH.97/WP18. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 7.De L, Jorba J, Boshell J, Salas R, Kew O. Abstracts of the 17th Annual Meeting of the American Society for Virology. Milwaukee, Wis: American Society for Virology; 1998. Calibration of a clock for VP1 nucleotide variation in poliovirus type 1 during circulation in humans, abstr. W36-3; p. 123. [Google Scholar]
- 8.De L, Nottay B K, Yang C-F, Holloway B P, Pallansch M, Kew O. Identification of vaccine-related polioviruses by hybridization with specific RNA probes. J Clin Microbiol. 1995;33:562–571. doi: 10.1128/jcm.33.3.562-571.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 10.Dowdle, W. R., and M. E. Birmingham. 1997. The biologic principles of poliovirus eradication. J. Infect. Dis. 175(Suppl. 1):S286–S292. [DOI] [PMC free article] [PubMed]
- 11.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 12.Fricks C E, Icenogle J P, Hogle J M. Trypsin sensitivity of the Sabin strain of type 1 poliovirus: cleavage sites in virions and related particles. J Virol. 1985;54:856–859. doi: 10.1128/jvi.54.3.856-859.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatch, M. Unpublished data.
- 14.Hogle J M, Chow M, Filman D J. The three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 15.Hull, H. F., M. E. Birmingham, B. Melgaard, and J. W. Lee. 1997. Progress towards global polio eradication. J. Infect. Dis. 175(Suppl. 1):S4–S9. [DOI] [PubMed]
- 16.Jones D T, Taylor W R, Thornton J M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 17.Kew O M, Mulders M N, Lipskaya G Y, da Silva E E, Pallansch M A. Molecular epidemiology of polioviruses. Semin Virol. 1995;6:401–414. [Google Scholar]
- 18.Kew, O. M., and B. K. Nottay. 1984. Molecular epidemiology of polioviruses. Rev. Infect. Dis. 6(Suppl. 2):S499–S504. [DOI] [PubMed]
- 19.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, da Silva E, Peñaranda S, Pallansch M, Kew O. Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1998;36:352–357. doi: 10.1128/jcm.36.2.352-357.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, Mulders M N, Holloway B P, Pallansch M A, Kew O M. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1996;34:2990–2996. doi: 10.1128/jcm.34.12.2990-2996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 22.Lederman H M, Winkelstein J A. X-linked agammaglobulinemia: an analysis of 96 patients. Medicine. 1985;64:145–156. [PubMed] [Google Scholar]
- 23.Más Lago P, Louzará C, Beltrán J, Jacobo M, Palomera R. Circulación de poliovirus en la población infantil de Cuba. Bol Oficina Sanit Panam. 1975;87:443–449. [PubMed] [Google Scholar]
- 24.McKinney R E, Jr, Katz S L, Wilfert C M. Chronic enteroviral meningoencephalitis in agammaglobulinemic patients. Rev Infect Dis. 1987;9:334–356. doi: 10.1093/clinids/9.2.334. [DOI] [PubMed] [Google Scholar]
- 25.Minor P D. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol. 1990;161:121–154. doi: 10.1007/978-3-642-75602-3_5. [DOI] [PubMed] [Google Scholar]
- 26.Minor P D, Ferguson M, Phillips A, Magrath D I, Huovilainen A, Hovi T. Conservation in vivo of protease cleavage sites in antigenic sites of poliovirus. J Gen Virol. 1987;68:1857–1865. doi: 10.1099/0022-1317-68-7-1857. [DOI] [PubMed] [Google Scholar]
- 27.Nakano J H, Gelfand H M, Cole J T. The use of a modified Wecker technique for the serodifferentiation of type 1 polioviruses related and unrelated to Sabin’s vaccine strain. II. Antigenic segregation of isolates from specimens collected in field studies. Am J Hyg. 1963;78:214–226. doi: 10.1093/oxfordjournals.aje.a120339. [DOI] [PubMed] [Google Scholar]
- 28.Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci USA. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nottay B K, Kew O M, Hatch M H, Heyward J T, Obijeski J F. Molecular variation of type 1 vaccine-related and wild polioviruses during replication in humans. Virology. 1981;108:405–423. doi: 10.1016/0042-6822(81)90448-7. [DOI] [PubMed] [Google Scholar]
- 30.Rico-Hesse R, Pallansch M A, Nottay B K, Kew O M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987;160:311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- 31.Rosen F S, Cooper M D, Wedgwood R J P. The primary immunodeficiencies. N Engl J Med. 1995;333:431–440. doi: 10.1056/NEJM199508173330707. [DOI] [PubMed] [Google Scholar]
- 32.Savilahti E, Klemola T, Carlsson B, Mellander L, Stenvik M, Hovi T. Inadequacy of mucosal IgM antibodies in selective IgA deficiency: excretion of attenuated polioviruses is prolonged. J Clin Immunol. 1988;8:89–94. doi: 10.1007/BF00917895. [DOI] [PubMed] [Google Scholar]
- 33.Sutter R W, Prevots R. Vaccine-associated paralytic poliomyelitis among immunodeficient persons. Infect Med. 1994;11:426–438. doi: 10.1086/379791. [DOI] [PubMed] [Google Scholar]
- 34.van Wezel A L, Hazendonk A G. Intratypic serodifferentiation of poliomyelitis virus by strain-specific antisera. Intervirology. 1979;11:2–8. doi: 10.1159/000149005. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. Manual for the virologic investigation of poliomyelitis. WHO/EPI/GEN/97.1. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 36.Yang C-F, De L, Holloway B P, Pallansch M A, Kew O M. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 1991;20:159–179. doi: 10.1016/0168-1702(91)90107-7. [DOI] [PubMed] [Google Scholar]
- 37.Yang, C.-F., and S.-J. Yang. Unpublished data.
- 38.Yoneyama T, Hagiwara A, Hara M, Shimojo H. Alteration in oligonucleotide fingerprint patterns of the viral genome in poliovirus type 2 isolated from paralytic patients. Infect Immun. 1982;37:46–53. doi: 10.1128/iai.37.1.46-53.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]