Abstract
Currently, the incidence of diabetes mellitus is increasing rapidly, particularly in China, and its pathogenesis is still unclear. The goal of this study was to find meaningful biomarkers of metastasis in patients with diabetes and cancer using bioinformatic analysis in order to predict gene expression and prognostic importance for survival. We used the Differentially Expressed Gene, Database for Annotation Visualization and Integrated Discovery, and Gene Set Enrichment Analyses databases, as well as several bioinformatics tools, to explore the key genes in diabetes. Based on the above database, we ended up with 10 hub genes (FOS, ATF3, JUN, EGR1, FOSB, JUNB, BTG2, EGR2, ZFP36, and NR4A2). A discussion of the 10 critical genes, with extensive literature mentioned to validate the association between the 10 key genes and patients with diabetes and cancer, to demonstrate the importance of gene expression and survival prognosis. This study identifies several biomarkers associated with diabetes and cancer development and metastasis that may provide novel therapeutic targets for diabetes combined with cancer patients.
Keywords: bioinformatics analysis, biomarkers, cancer, diabetes mellitus, survival analyses
1. Introduction
Diabetes mellitus (DM) is a metabolic disease characterized by hyperglycemia caused by insulin insufficiency and insulin resistance.[1] It is more common in people over 40 years old and it has a higher prevalence in people with obesity, hypertension, dyslipidemia, a sedentary lifestyle, a family history of diabetes, and gestational diabetes. Clinically, DM manifests as “three more and one less” symptoms, which are frequently accompanied by anomalies in lipid and protein metabolism. Long-term blood sugar imbalances can result in a stroke, coronary heart disease, angina pectoris, diabetic ketoacidosis, diabetic retinopathy, diabetic nephropathy, and other complications. The global incidence of DM has increased, and according to the most recent epidemiological statistics, the number of DM fatalities in China is predicted to increase markedly from 2005 to 2020, with the social burden of the disease increasing as China’s population ages.[2–4] DM is currently incurable and the primary treatment options include glucose-lowering medications, diet, and exercise. Although these treatments have some effects on glucose metabolism, they are constrained by the limitations of long-term medication, which has severe side effects and low control rates. Therefore, there is an urgent need to develop sensitive metastasis-related biomarkers that regulate diabetes and provide new therapeutic targets for the treatment of DM.
The pathophysiology of DM is uncertain and complex and it involves considerable familial aggregation. The PI3K pathway has been reported to be an important intracellular signaling pathway in the regulation of glucose metabolism. miRNAs are important for understanding the pathogenesis of diabetes as they affect insulin release and β-cell proliferation and function.[5–7] Only a few pathogenic genes, such as MIR-21, Rfx2, HNF1A, Nus1, GP2, HBA 1c, MR-148B-3P, and MR-27A-3P, are thought to play significant roles in the development of metastasis in cancer patients with DM and associated disorders.[8–13] However, the relationship between these biomarkers and DM has not been adequately studied. Moreover, the differential expression of metastasis-related genes in patients with DM, with and without metastases, has not been studied.
Based on bioinformatics analysis, biomarkers related to DM and cancer were analyzed for their gene expression levels and value in predicting survival. To analyze the differentially expressed genes (DEGs) of the samples, 2 mRNA datasets (GSE29226 and GSE29231) were selected from the Gene Expression Omnibus database, and functional and pathway enrichment analyses of the DEGs were then performed using the database for annotation visualization and integrated discovery (DAVID) and GSEA databases to better understand the likely molecular basis of DM. Protein-protein interaction (PPI) networks were constructed using Cytoscape software, after which modules were screened for hub gene validation. WebGestalt was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the hub genes. The Gene Expression Profile Interaction Analysis (GEPIA) and KM databases were utilized to further investigate the expression levels and survival predictive ability the of hub genes. We identified 10 critical genes involved in the progression and metastasis of cancer in patients with DM, through a series of database searches, and subsequently, we investigated and validated these genes as potential novel therapeutic targets for individuals with diabetes and cancer.
2. Methods
2.1. Microarray data
We retrieved the GSE29226 and GSE29231 microarray datasets from the NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) database and transformed and normalized the probe matrix to the gene matrix. Both datasets were built on the GPL6947 platform (Illumina HumanHT-12 V3.0, expression bead chip) and included data from 12 patients with diabetes and 12 healthy individuals.
2.2. DEG analysis
Fold-change (FC) is a metric used to compare the change in absolute gene expression between 2 groups, and is usually converted to log2 FC for a more intuitive presentation. GEO2R is an online tool (http://www.ncbi.nlm.nih.gov/geo/geo2r) for comparing 2 or more sets of gene expression data. We used GEO2R to identify DEGs between samples with and without diabetes, with selection criteria of P < .05 and |log (FC)| acuity of 1.0, and visualization analysis. We classified DEGs with logFC > 0 as upregulated genes and those with logFC < 0 as downregulated genes. To identify the cross-genes between GSE29226 and GSE29231, we used the online tool Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/), and generated DEG heatmaps using the OmicStudio platform (v5.3.0).
2.3. GO functional and pathway enrichment analyses
To determine the function of the DEGs, we used the DAVID Database (version 6.8; https://david.ncifcrf.gov/) to enrich for GO and KEGG functions and pathways. The GO terms used in the enrichment analysis were biological process (BP), cellular component (CC), and molecular function (MF). We used a Bengemini-Hochberg false discovery rate (FDR) < 0.05 to indicate a statistically significant difference.
2.4. GSEA analysis
GSEA 4.3.2 software was downloaded from the GSEA website (http://software.broadinstitute.org/gsea/downloads.jsp) and operated in a Java environment to perform GO and KEGG pathway enrichment analyses. The reference gene sets, c5.go.bp.v2022.1. Hs.symbols.gmt, c5.go.cc.v2022.1.Hs.symbols.gmt, c5.go.mf.v2022.1.Hs.symbols.gmt, and c2.cp.v2022.1 were chosen from MSigDB for GSEA analysis. An enrichment score > 0.6, P < .05, and an FDR q-value < 0.25 were chosen as the crucial thresholds.
2.5. PPI network construction and module analysis
We constructed a PPI network using the STRING database (https://cn.string-db.org/) for data filtering and analysis. We first set the cutoff criterion in the STRING database as a combined score > 0.4 and we then retrieved the data. The PPI network was mapped using Cytoscape (version 3.9.1). The Molecular Complex of the Cell Landscape plugin (version 2.0.2) was used to detect the hub genes of the key modules of MCODE. The following criteria were used: MCODE score > 5, degree cutoff = 2, node score cutoff = 0.2, maximum depth = 100, and k-core = 2. The most significant DEGs were then analyzed using the ClueGO (version 2.5.9) and CluePedia (version 1.5.9) plugins of the Cytoscape platform, with an FDR cutoff of 0.05 and a coefficient cutoff of 0.4.
2.6. Hub gene selection and analysis
Based on the hub genes selected in MCODE, we used the Cytoscape CytoHubba plugin (version 0.1) to identify the 10 most significant genes, with P < .05 considered statistically significant. Then, using FDR < 0.05 as the criterion, WebGestalt (http://www.webgestalt.org/) was used to perform GO and KEGG enrichment analyses of these 10 key genes.
2.7. Differential expression analysis
GEPIA (http://gepia2.cancer-pku.cn/#index) is a new interactive website that provides patient survival data, tumor/normal tissue differential expression data, and other relevant information. We examined the expression of the 10 hub genes in diabetic and non-diabetic tissues. The following criteria were used to indicate statistical significance: P < .05, and |Log2FC| > 1.
2.8. Survival analysis
We confirmed the prognostic relevance of the 10 hub genes in patients with DM using KM Plotter (http://kmplot.com/). Based on the median expression level of each hub gene, we divided the patients with DM into high- and low-expression groups, confirmed the overall survival and recurrence-free survival associated with each of the 10 hub genes, and generated KM survival maps. The hazard ratios and associated 95% confidence intervals and log-rank tests were used to generate P values. A log-rank test result of P < .05 was considered statistically significant.
3. Results
3.1. Identification of DEGs
We retrieved 487 DEGs from GSE29226 (312 upregulated and 175 downregulated) and 916 DESs from GSE29231 (126 upregulated and 790 downregulated) using the online resource, GEO2R. First, a volcano plot was created by analyzing the genes that were differentially expressed in the DM and non-DM samples (Fig. 1). A Venn diagram was used to conduct additional analyses, which revealed 63 overlapping DEGs in the 2 datasets, including 10 upregulated and 53 downregulated genes (Fig. 2A; Table 1). Two heat maps (Fig. 2B) were generated using the 63 DEGs that overlapped in the GSE29226 and GSE29231 datasets.
Figure 1.
Volcano plots of DEGs detected from the datasets of GSE29226 and GSE29231. (A) GSE29226, (B) GSE29231. The red dots represent upregulated DEGs; the green dots mean downregulated DEGs; the black dots denote no differentially expressed genes. DEG = differentially expressed gene.
Figure 2.
Identification of DEGs associated with DM metastasis. (A) The Venn diagram demonstrates the crossed genes shared by GSE29226 and GSE29231 datasets. The left panel represents the downregulated intersectional expressed genes between GSE29226 and GSE29231 datasets, whereas the right panel represents the upregulated co-differentially genes between the 2 datasets. (B) Heatmap of the top 63 DEGs associated with DM metastasis in the 2 datasets. Left, GSE29226; right, GSE29231. DEG = differentially expressed gene, DM = diabetes mellitus.
Table 1.
Identification of DEGs associated with DM.
| Regulation | Genes |
|---|---|
| Upregulated (n = 10) | CLOCK, BRWD1, MCOLN3, SUCNR1, CLC, PRG2, CNGA1, THNSL2, ALDOC, PLK4 |
| Downregulated (n = 53) |
ERAP2, EGR2, EGFLAM, EGR1, BTG2, FOS, FOSB, PTGS2, ZFP36, GOLGA6B, NFKBIZ, MCL1, ATF3, JUN, LOC401357, JUNB, ADCY1, TSC22D3, CCL3L3, CYR61, BRSK1, CXCL8, NFIL3, IER3, CSF3R, SLC2A3, NR4A2, SERPINA1, SLCO4A1, FCN3, SERPINA1, HBEGF, HLA-DRB6, PLAUR, VNN2, NAMPT, MMP25, RGS2, ARRDC4, HSPA1A, NFE2, AQP9, CSRNP1, IL18RAP, CXCR4, MNDA, SLC11A1, FAM65B, NKD2, SORL1, LILRA5, RGS1, HSPA1B |
DEG = differentially expressed gene, DM = diabetes mellitus.
3.2. Functional enrichment analysis of DEGs
Using the DAVID database, we further explored the 63 overlapping DEGs and examined the functions of the enriched GO and KEGG pathways. In the BP category, 63 overlapping DEGs were primarily enriched for inflammatory responses, with differences in skeletal muscle cells being enriched to the same extent as the cellular response to calcium (Fig. 3A; Table 2). We found that the DEGs were primarily enriched in the plasma membrane, extracellular regions, and chromatin, with the transcription factor activator protein-1 (AP-1) being the least enriched, and the ficolin-1-rich granule lumen being enriched to the same extent as RNA polymerase II transcriptase (Fig. 3A; Table 2). In terms of MF, transcription factor activators, sequence-specific double-stranded RNA polymerase II transcripts, and DNA binding were all enriched in DEGs to the same extent, with protein binding being the main molecular activity (Fig. 3A; Table 2). The P-values ranged from lowest to highest for the following genes: hsa04928, parathyroid hormone synthesis, secretion, and action; hsa04915, estrogen signaling pathway; hsa04657, IL-17 signaling pathway; and hsa05142, Chagas disease (Fig. 3B; Table 3).
Figure 3.
GO and KEGG pathway enrichment analyses of DEGs using the DAVID database. (A) Biological processes, cellular components, molecular functions, and (B) the signaling pathways from the GSE29226 and GSE29231 datasets. DAVID = database for annotation visualization and integrated discovery, DEG = differentially expressed gene, GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
Table 2.
The top 3 enriched GO terms of DEGs associated with SS metastasis.
| Category | Term | Count | FDR |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0035914–skeletal muscle cell differentiation | 5 | 0.00838327492717679 |
| GOTERM_BP_DIRECT | GO:0006954–inflammatory response | 9 | 0.00936662298578149 |
| GOTERM_BP_DIRECT | GO:0071277–cellular response to calcium ion | 5 | 0.0379982817297754 |
| GOTERM_CC_DIRECT | GO:0090575–RNA polymerase II transcription factor complex | 6 | 0.00152721209864361 |
| GOTERM_CC_DIRECT | GO:1904813–ficolin-1-rich granule lumen | 6 | 0.00152721209864361 |
| GOTERM_CC_DIRECT | GO:0035976–transcription factor AP-1 complex | 3 | 0.00319859162710518 |
| GOTERM_MF_DIRECT | GO:0001228–transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | 10 | 0.00163773946264629 |
| GOTERM_MF_DIRECT | GO:0003700–transcription factor activity, sequence-specific DNA binding | 10 | 0.00183452190316464 |
| GOTERM_MF_DIRECT | GO:1990837–sequence-specific double-stranded DNA binding | 10 | 0.00183452190316464 |
DEG = differentially expressed gene, GO = gene ontology.
Table 3.
The top 2 enriched KEGG pathways of DEGs associated with DM metastasis.
| Category | Term | Count | FDR |
|---|---|---|---|
| KEGG_PATHWAY | hsa04928: Parathyroid hormone synthesis, secretion, and action | 6 | 0.016035691 |
| KEGG_PATHWAY | hsa04915: Estrogen signaling pathway | 6 | 0.027540363 |
DEG = differentially expressed gene, DM = diabetes mellitus, KEGG = Kyoto Encyclopedia of Genes and Genomes.
3.3. PPI network and module analysis
To identify the associations between the 63 DEGs, we built a PPI network from a text database with 60 nodes and 145 edges (Fig. 4A). The top 5 genes highly associated with the 63 genes used to generate the Sankey map of DM using online tools were ADCY1, ALDOC, AQP9, ARRDC4, and ATF3. The BCL2 family of apoptosis regulators, BR serine/threonine kinase 1, BTG anti-proliferative factor 2, C-C motif chemokine ligand 3-like 3, and C-X-C motif chemokine ligand 8 were the top 5 pathways highly associated with the 63 pathways in DM (Fig. 4B). The highest-scoring key genes made up 12 nodes and 53 edges based on the screening results. Next, the ClueGO + CluePedia plugin for Cytoscape was used to perform GO and KEGG pathway enrichment analyses. The results showed that skeletal muscle cell differentiation in the BP group accounted for 42. 31% (Fig. 5A); ficolin-1-rich granule lumen in the CC group accounted for 50% and the AP-1 transcription factor complex accounted for 50% (Fig. 5B); and tau-protein kinase activity in the MF group accounted for 100% (Fig. 5C). In the KEGG pathway analysis, parathyroid hormone synthesis secretion and action accounted for 53.85%, viral protein interaction with cytokine and cytokine receptor interactions accounted for 7.69%, and the IL-17 signaling pathway accounted for 38.46% (Fig. 5D).
Figure 4.
PPI network analysis and identification of hub genes. (A) Protein-protein interactions of DEGs were analyzed using the STRING database. (B) Sankey diagram of DM: The picture demonstrates 63 genes and pathways associated with DM. DEG = differentially expressed gene, DM = diabetes mellitus, PPI = protein-protein interaction, STRING = search tool for the retrieval of interacting genes.
Figure 5.
GO analyses of DEGs in the most significant module using Cytoscape plugins ClueGO and CluePedia. (A) Percentages of biological processes terms per group. (B) Percentages of cellular components terms per group. (C) Percentages of molecular functions terms per group. (D) Percentages of Kyoto Encyclopedia of Genes and Genomes terms per group. DEG = differentially expressed gene, GO = gene ontology.
3.4. Hub gene selection and survival analysis
The CytoHubba plugin was used to screen the top 10 hub genes, FoS, ATF3, JUN, EGR1, FoSB, JUNB, BTG2, EGR2, ZFP36, and NR4A2, from the major modules (Fig. 6A; Table 4). Using the online tool, WebGestFLT, these 10 hub genes were analyzed for GO and KEGG pathway enrichment. The 10 BP hub genes were highly enriched in the inflammatory response (Fig. 6B); the CC genes were mainly enriched in the plasma membrane, extracellular areas, and chromatin (Fig. 6C); and the MF genes were mainly enriched in protein binding (Fig. 6D). The 10 KEGG pathway hub genes were found to be particularly critical in parathyroid hormone synthesis and release and interaction with the estrogen signaling pathway (Fig. 6E).
Figure 6.
Significantly enriched GO and KEGG pathways for the 10 hub genes using the WebGestalt database. (A) 10 hub genes were identified from the most significant module. (B) Biological process, (C) Cellular component, (D) Molecular function, (E) KEGG pathways for the 10 hub genes. GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
Table 4.
Hub genes are identified in the key module.
| Genes | Description | Degree | P value | Log FC |
|---|---|---|---|---|
| FOS | FOS proto-oncogene, AP-1 transcription factor subunit | 11 | .00000872 | −1.2275013 |
| ATF3 | Activating transcription factor 3 | 11 | .000058 | −2.7859614 |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit | 11 | .000132 | −2.1625615 |
| EGR1 | Early growth response 1 | 11 | .00000268 | −1.9486935 |
| FOSB | FOSB proto-oncogene, AP-1 transcription factor subunit | 10 | .0000187 | −1.9342314 |
| JUNB | JunB proto-oncogene, AP-1 transcription factor subunit | 9 | .000298 | −2.2360312 |
| BTG2 | BTG anti-proliferation factor 2 | 9 | .00000276 | −1.0029259 |
| EGR2 | Early growth response 2 | 8 | .000000636 | −1.4740463 |
| ZFP36 | ZFP36 ring finger protein | 8 | ||
| NR4A2 | Nuclear receptor subfamily 4 group A member 2 | 7 | .0000393 | −1.0055742 |
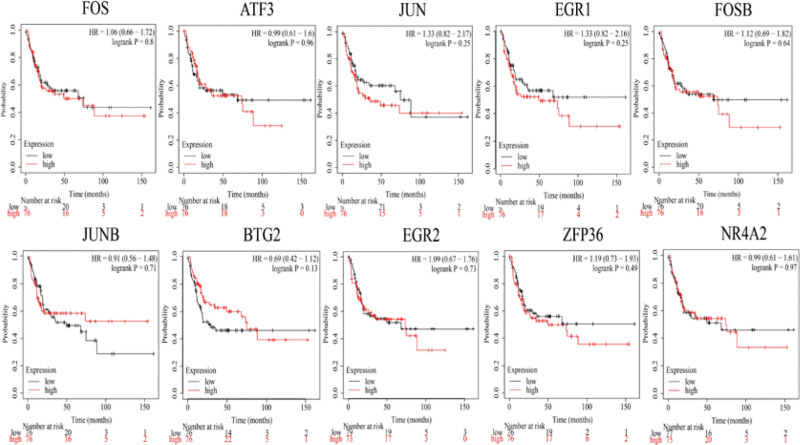
The GEPIA database was used to compare the differential expression of the 10 hub genes. The expression levels of the 10 hub genes were noticeably higher in DM patients than in non-DM patients (Fig. 7), and KM plots were used to investigate the impact of the 10 hub genes on overall survival (Fig. 8). As shown in Figure 8, diabetic patients with high JUN gene expression levels had lower overall survival. In addition, a shorter relapse-free survival time was associated with higher JUN expression levels in patients with DM (Fig. 9).
Figure 7.
Validation of the 10 hub genes overexpression in DM metastasis using the GEPIA database. *P < .05, unpaired Student t test. DM = diabetes mellitus, GEPIA = Gene Expression Profile Interaction Analysis.
Figure 8.
Kaplan–Meier curves for overall survival analysis of the 10 hub genes in DM patients. The red line represented the high expression group, whereas the black line denoted the low expression group. DM = diabetes mellitus.
Figure 9.
Kaplan–Meier curves for recurrence-free survival analysis of the 10 hub genes in DM patients. The red line represented the high expression group, whereas the black line denoted the low expression group. DM = diabetes mellitus.
4. Discussion
We identified 63 DEGs linked with metastases in cancer patients with DM from the GSE29226 and GSE29231 datasets. These DEGs comprised 10 upregulated and 53 downregulated genes. GO and KEGG pathway enrichment analyses of the 63 DEGs revealed a high level of enrichment in the inflammatory response. After constructing a PPI network, we identified a crucial module with 60 nodes and 145 edges. Based on this critical module, we screened 10 hub genes were found to be closely connected with the progression and prognosis of diabetes.
4.1. Screening of the DEGs using a PPI network
Ten hub genes, namely FoS, ATF3, JUN, EGR1, FoSB, JUNB, BTG2, EGR2, ZFP36, and NR4A2 may have crucial roles in DM metastasis. The FoS proto-oncogene, an AP-1 transcription factor subunit, which is linked with the inflammatory response, is dysregulated in numerous clinical situations and can suppress cervical cancer proliferation by targeting and modulating FoS gene expression.[14,15] Previous studies have demonstrated that AP-1 inhibitors can slow cancer progression.[16] Patients with diabetes and cancer who carry the causal FoS gene variant can thus be rapidly identified and treated with targeted medicines. Defective AP-1 activity has been detected in a number of disorders, most notably cancer and inflammatory diseases.[17] It is widely established that the FoSB gene is critical for regulating drug sensitivity and invasive activity, and that FoS functions as a tumor suppressor during prostate cancer growth and invasion, while FoSB also plays a key role in tumor proliferation, migration, and invasion.[18–20] Bioinformatic investigation of the FoS and FoSB genes in diabetic individuals revealed 2 hub genes that can be exploited as diagnostic biomarkers for liver cancer.[21]
Early growth response factor 1 (Egr1) is a zinc finger transcription factor that regulates several physiological processes, including the immune response.[22] Increased expression levels of renal NOX5 were found to be related to the levels of EGR1, a renal reactive-oxygen-species factor, in diabetic patients, and previous studies have demonstrated that the early stress-coupling agent, EGR1, is a critical factor in the development of islet failure.[23,24] There are also many studies on EGR1 as a therapeutic and prognostic biomarker in diabetic nephropathy, cervical cancer, and glioblastoma, which provide a deeper understanding of the targeting mechanisms and therapeutic targets for diabetic patients with cancer.[25–27] A previous study found that miR-25 regulates the proliferation and death of gastric cancer cells by targeting EGR2 and that knocking down EGR2 markedly decreases the anti-cancer activity of miR-17-5p inhibitors.[28,29] According to survival studies, EGR1 and EGR2 levels are associated with clinical outcomes in patients with liver cancer.[30] Activating transcription factor 3 (ATF3) causes inflammation associated with diabetic neuropathy and can be employed as a neuronal marker of injury and a marker for early diabetes diagnosis.[31–33] ATF3 has been shown to play a complex and multifaceted role in pancreatic cancer pathology, but the processes underlying its role in the diagnosis and treatment of gastric cancer, clear cell renal cell carcinoma, and ovarian tumors are quite evident.[34–37]
The product of the proto-oncogene JUNB is also a component of AP-1. JUNB is an ER-dependent gene found only in premenopausal women.[38,39] The JUNB gene has been shown to play an important role in the advancement of hepatocellular carcinoma and lung cancer and it is strongly associated with patient survival, suggesting a novel function of JUNB in cell proliferation and tumor aggressiveness.[40,41] We discovered little convincing evidence for the JUNB gene in our search of the literature, and therefore, the bioinformatics analysis performed in this study increases our understanding of this gene. Based on the results of our analysis, it is clear that the JUN gene is highly expressed in diabetic patients and this is associated with poorer overall survival and relapse-free survival.
NR4A2 is an immediate-early-response gene that functions as a transcription factor and has a pro-carcinogenic effect on glioblastoma.[42–44] BTG2 is linked to immune cell infiltration, has an inhibitory effect on ovarian cancer, and can be used as a biomarker in patients with lung adenocarcinoma.[45,46]A recent study found a link between ZFP36 and postoperative osteosarcoma, implying that it could be a potential prognostic biomarker as well as a new therapeutic target for osteosarcoma.[47]
Although we identified 10 hub genes in diabetic patient samples, the direct participation of these genes in the progression of diabetes has not been established. These 10 hub genes are known to be involved in the development of different malignancies. We anticipate that these hub genes will have prognostic value for the survival of patients with diabetes and cancer, and will provide new targets for the treatment of diabetes and cancer.
Diabetes research has been a hot topic worldwide, but there is still a scarcity of supporting literature on the 10 hub genes identified in this study, which limits the interpretation of our findings. However, we were able to extract relevant data from the literature to support the hypothesis that these 10 hub genes provide potential direction for the identification of treatments for diabetes; thus, our findings address current gaps in the field. However, our findings are yet to be confirmed by additional experimental analyses, leaving them open for further investigation in the future.
5. Conclusions
In summary, our study successfully identified 10 hub genes (FoS, ATF3, JUN, EGR1, FoSB, JUNB, BTG2, EGR2, ZFP36, and NR4A2) by bioinformatics analysis of DEGs in metastatic and non-metastatic tissues from diabetic patients. Moreover, herein we provide an extensive review of the literature on the functions of these 10 hub genes, which will help to further identify biological markers of diabetes combined with cancer and will provide a better understanding of the molecular mechanisms.
Author contributions
Conceptualization: Yiting Li, Shinong Gu.
Data curation: Shinong Gu, Qing Huang.
Formal analysis: Shinong Gu, Qing Huang.
Funding acquisition: Shinong Gu.
Investigation: Shinong Gu, Qing Huang.
Methodology: Yiting Li, Shinong Gu, Qing Huang.
Project administration: Qing Huang.
Resources: Shinong Gu, Qing Huang.
Software: Shinong Gu, Xuanwen Li.
Supervision: Yiting Li.
Validation: Shinong Gu, Qing Huang.
Visualization: Yiting Li.
Writing – original draft: Yiting Li, Xuanwen Li.
Writing – review & editing: Xuanwen Li.
Abbreviations:
- AP-1
- activator protein-1
- BP
- biological process
- CC
- cell component
- DAVID
- database for annotation visualization and integrated discovery
- DEG
- differentially expressed gene
- DM
- diabetes mellitus
- EGR1
- early growth response 1
- FC
- fold change
- GEPIA
- Gene Expression Profile Interaction Analysis
- GO
- gene ontology
- GSEA
- Gene Set Enrichment Analyses
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- KM
- Kaplan–Meier
- MCODE
- molecular complex detection
- MF
- molecular function
- MSigDB
- molecular signatures database
- PPI
- protein-protein interaction
- STRING
- search tool for the retrieval of interacting genes
- VEGF
- vascular endothelial growth factor
- WebGestalt
- WEB-based Gene SeT AnaLysis Toolkit
This research was supported by the Natural Science Foundation of Xiamen, China (grant no: 3502Z20227318), and the funder had no role during the entire process of this study.
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
How to cite this article: Li Y, Gu S, Li X, Huang Q. To identify biomarkers associated with the transfer of diabetes combined with cancer in human genes using bioinformatics analysis. Medicine 2023;102:37(e35080).
Contributor Information
Yiting Li, Email: 87887971@qq.com.
Shinong Gu, Email: Gusn@hxxy.edu.cn.
Xuanwen Li, Email: 87887971@qq.com.
References
- [1].Zhang LW, Ruan MH, Liu JL, et al. Progress on research and development in diabetes mellitus. Yi Chuan. 2022;44:824–39. [DOI] [PubMed] [Google Scholar]
- [2].Liu R, Li L, Shao C, et al. The impact of diabetes on vascular disease: progress from the perspective of epidemics and treatments. J Diabetes Res. 2022;2022:1531289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang F, Wang W, Yin P, et al. Mortality and years of life lost in diabetes mellitus and its subcategories in China and Its Provinces, 2005-2020. J Diabetes Res. 2022;2022:1609267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Writing Committee of the Report on Cardiovascular Health and Diseases in China. Report on Cardiovascular Health and Diseases in China 2021: an updated summary. Biomed Environ Sci. 2022;35:573–603. [DOI] [PubMed] [Google Scholar]
- [5].Maddaloni E, Bolli GB, Frier BM, et al. C-peptide determination in the diagnosis of type of diabetes and its management: a clinical perspective. Diabetes Obes Metab. 2022;24:1912–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pla Peris B, Arranz Martin A, Ballesteros García A, et al. Alpelisib-Induced diabetes mellitus: case report, pharmacodynamics and management considerations. Front Endocrinol (Lausanne). 2022;13:802612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kaur P, Kotru S, Singh S, et al. Role of miRNAs in diabetic neuropathy: mechanisms and possible interventions. Mol Neurobiol. 2022;59:1836–49. [DOI] [PubMed] [Google Scholar]
- [8].Liu S, Wu W, Liao J, et al. MicroRNA-21: a critical pathogenic factor of diabetic nephropathy. Front Endocrinol (Lausanne). 2022;13:895010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stankute I, Kazlauskiene M, Blouin JL, et al. Co-segregation analysis and functional trial in vivo of candidate genes for monogenic diabetes. BMJ Open Diabetes Res Care. 2022;10:e003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li LM, Jiang BG, Sun LL. HNF1A: From Monogenic Diabetes to Type 2 diabetes and gestational diabetes mellitus. Front Endocrinol (Lausanne). 2022;13:829565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang T, Zhao L, Wang S, et al. Common variants in NUS1 and GP2 Genes contributed to the risk of gestational diabetes mellitus. Front Endocrinol (Lausanne). 2021;12:685524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gillery P. HbA1c and biomarkers of diabetes mellitus in clinical chemistry and laboratory medicine: ten years after. Clin Chem Lab Med. 2022;61:861–72. [DOI] [PubMed] [Google Scholar]
- [13].Ghoreishi E, Shahrokhi SZ, Kazerouni F, et al. Circulating miR-148b-3p and miR-27a-3p can be potential biomarkers for diagnosis of pre-diabetes and type 2 diabetes: integrating experimental and in-silico approaches. BMC Endocr Disord. 2022;22:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kubra K, Gaddu GK, Liongue C, et al. Phylogenetic and expression analysis of Fos Transcription Factors in Zebrafish. Int J Mol Sci. 2022;23:10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gupte R, Lin KY, Nandu T, et al. Combinatorial Treatment with PARP-1 inhibitors and cisplatin attenuates cervical cancer growth through Fos-Driven changes in gene expression. Mol Cancer Res. 2022;20:1183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Choi Y, Jeon H, Akin JW, et al. The FOS/AP-1 regulates metabolic changes and cholesterol synthesis in human periovulatory granulosa cells. Endocrinology. 2021;162:bqab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bhosale PB, Kim HH, Abusaliya A, et al. Structural and functional properties of activator protein-1 in cancer and inflammation. Evid Based Complement Alternat Med. 2022;2022:9797929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Na HH, Ryu JM, Kim KC. Knockout of FosB gene changes drug sensitivity and invasion activity via the regulation of Bcl-2, E-cadherin, β-catenin, and vimentin expression. Biochem Biophys Res Commun. 2021;567:131–7. [DOI] [PubMed] [Google Scholar]
- [19].Riedel M, Berthelsen MF, Cai H, et al. In vivo CRISPR inactivation of Fos promotes prostate cancer progression by altering the associated AP-1 subunit Jun. Oncogene. 2021;40:2437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu S, Luan J, Ding Y. miR-144-3p Targets FosB Proto-oncogene, AP-1 Transcription Factor Subunit (FOSB) to suppress proliferation, migration, and invasion of PANC-1 pancreatic cancer cells. Oncol Res. 2018;26:683–90. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Gao S, Gang J, Yu M, et al. Computational analysis for identification of early diagnostic biomarkers and prognostic biomarkers of liver cancer based on GEO and TCGA databases and studies on pathways and biological functions affecting the survival time of liver cancer. BMC Cancer. 2021;21:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pang Z, Xu Y, Zhu Q. Early Growth Response 1 suppresses macrophage phagocytosis by inhibiting NRF2 activation through upregulation of autophagy during pseudomonas aeruginosa infection. Front Cell Infect Microbiol. 2022;11:773665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jha JC, Dai A, Garzarella J, et al. Independent of Renox, NOX5 promotes renal inflammation and fibrosis in diabetes by activating ROS-Sensitive Pathways. Diabetes. 2022;71:1282–98. [DOI] [PubMed] [Google Scholar]
- [24].Leu SY, Kuo LH, Weng WT, et al. Loss of EGR-1 uncouples compensatory responses of pancreatic β cells. Theranostics. 2020;10:4233–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xu P, Zhan H, Zhang R, et al. Early growth response factor 1 upregulates pro-fibrotic genes through activation of TGF-β1/Smad pathway via transcriptional regulation of PAR1 in high-glucose treated HK-2 cells. Mol Cell Endocrinol. 2023;572:111953. [DOI] [PubMed] [Google Scholar]
- [26].Zhao J, Li H, Yuan M. EGR1 promotes stemness and predicts a poor outcome of uterine cervical cancer by inducing SOX9 expression. Genes Genomics. 2021;43:459–70. [DOI] [PubMed] [Google Scholar]
- [27].Xu L, Peng B, Wu H, et al. METTL7B contributes to the malignant progression of glioblastoma by inhibiting EGR1 expression. Metab Brain Dis. 2022;37:1133–43. [DOI] [PubMed] [Google Scholar]
- [28].Yang L, Li L, Chang P, et al. miR-25 regulates gastric cancer cell growth and apoptosis by targeting EGR2. Front Genet. 2021;12:690196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Geng X, Sun Y, Fu J, et al. MicroRNA-17-5p inhibits thyroid cancer progression by suppressing Early growth response 2 (EGR2). Bioengineered. 2021;12:2713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liang J, Wang J, Zeng J, et al. The Landscape of early growth response family members 1-4 in hepatocellular carcinoma: their biological roles and diagnostic utility. Dis Markers. 2022;2022:3144742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kan HW, Chang CH, Chang YS, et al. Genetic loss-of-function of activating transcription factor 3 but not C-type lectin member 5A prevents diabetic peripheral neuropathy. Lab Invest. 2021;101:1341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tierney JA, Uong CD, Lenert ME, et al. High-fat diet causes mechanical allodynia in the absence of injury or diabetic pathology. Sci Rep. 2022;12:14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li G, Zhang J, Liu D, et al. Identification of Hub Genes and Potential ceRNA networks of diabetic nephropathy by weighted gene co-expression network analysis. Front Genet. 2021;12:767654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Azizi N, Toma J, Martin M, et al. Loss of activating transcription factor 3 prevents KRAS-mediated pancreatic cancer. Oncogene. 2021;40:3118–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xie G, Dong P, Chen H, et al. Decreased expression of ATF3, orchestrated by β-catenin/TCF3, miR-17-5p and HOXA11-AS, promoted gastric cancer progression via increased β-catenin and CEMIP. Exp Mol Med. 2021;53:1706–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gao S, Gao L, Wang S, et al. ATF3 suppresses growth and metastasis of clear cell renal cell carcinoma by deactivating EGFR/AKT/GSK3β/β-Catenin Signaling Pathway. Front Cell Dev Biol. 2021;9:618987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li X, Liu P, Sun X, et al. Analyzing the impact of ATF3 in tumorigenesis and immune cell infiltration of ovarian tumor: a bioinformatics study. Med Oncol. 2021;38:91. [DOI] [PubMed] [Google Scholar]
- [38].Xu M, Cao L, Zhang X, et al. miR-3133 inhibits proliferation and angiogenesis by targeting the JUNB/VEGF pathway in human umbilical vein endothelial cells. Oncol Rep. 2020;44:1699–708. [DOI] [PubMed] [Google Scholar]
- [39].Mieczkowski K, Kitowska K, Braun M, et al. FGF7/FGFR2-JunB signalling counteracts the effect of progesterone in luminal breast cancer. Mol Oncol. 2022;16:2823–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yan P, Zhou B, Ma Y, et al. Tracking the important role of JUNB in hepatocellular carcinoma by single-cell sequencing analysis. Oncol Lett. 2020;19:1478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roumeliotou A, Pantazaka E, Xagara A, et al. phenotypic characterization of circulating tumor cells isolated from non-small and small cell lung cancer patients. Cancers (Basel). 2022;15:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ashraf S, Hegazy YK, Harmancey R. Nuclear receptor subfamily 4 group A member 2 inhibits activation of ERK signaling and cell growth in response to β-adrenergic stimulation in adult rat cardiomyocytes. Am J Physiol Cell Physiol. 2019;317:C513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Odagiu L, May J, Boulet S, et al. Role of the orphan nuclear receptor NR4A Family in T-Cell Biology. Front Endocrinol (Lausanne). 2021;11:624122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Karki K, Li X, Jin UH, et al. Nuclear receptor 4A2 (NR4A2) is a druggable target for glioblastomas. J Neurooncol. 2020;146:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang XZ, Chen MJ, Fan PM, et al. BTG2 serves as a potential prognostic marker and correlates with immune infiltration in lung adenocarcinoma. Int J Gen Med. 2022;15:2727–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang J, Li H, Wang L, et al. Transcriptomic analyses reveal B-Cell Translocation Gene 2 as a Potential Therapeutic Target in Ovarian cancer. Front Oncol. 2021;11:681250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Song P, Xie Z, Chen C, et al. Identification of a novel iron zinc finger protein 36 (ZFP36) for predicting the overall survival of osteosarcoma based on the Gene Expression Omnibus (GEO) database. Ann Transl Med. 2021;9:1552. [DOI] [PMC free article] [PubMed] [Google Scholar]