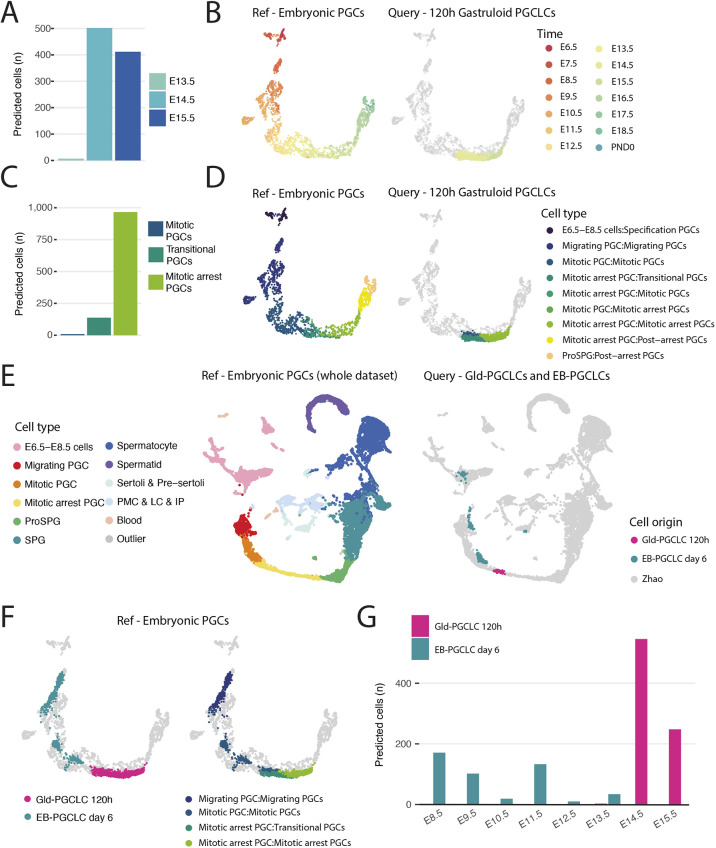
Fig. 5.
Single-cell transcriptomic comparison between expression data from Gld-PGCLCs and an in vivo PGC dataset. (A,C) Quantification of label transfer prediction from Gld-PGCLCs (0.6+ maximum prediction score) in terms of embryonic time (A) and cell stage (C). (B,D) UMAP of PGC-only cell types from Zhao et al. (2021), in terms of time (B) and stage (D) with Gld-PGCLCs embedded. (E) UMAP projection of Gld-PGCLCs (0.9+ maximum prediction score) and published EB-PGCLCs (Ramakrishna et al., 2022) into the in vivo UMAP of the full dataset. (F) Comparison of UMAP projection of Gld-PGCLCs and published EB-PGCLCs onto the in vivo PGC dataset, by origin (left) and cell state (right). (G) Frequency of cell transfer labels from EB-PGCLCs or Gld-PGCLCs (0.6+ maximum prediction score) onto the in vivo PGC dataset, by embryonic time point. IP, interstitial progenitors; LC, Leydig cells; PMC, peritubular myoid cells; PND0, postnatal day 0; ProSPG, pro-spermatogonia; SPG, spermatogonia.