Abstract
Rationale:
Standardized dosing of anti-tubercular (TB) drugs leads to variable plasma drug levels, which are associated with adverse drug reactions, delayed treatment response, and relapse. Mutations in genes affecting drug metabolism explain considerable interindividual pharmacokinetic variability; however, pharmacogenomic (PGx) assays that predict metabolism of anti-TB drugs have been lacking.
Objectives:
To develop a Nanopore sequencing panel and validate its performance in active TB patients to personalize treatment dosing.
Measurements and Main Results:
We developed a Nanopore sequencing panel targeting 15 single nucleotide polymorphisms (SNP) in 5 genes affecting the metabolism of isoniazid (INH), rifampin (RIF), linezolid and bedaquiline. For validation, we sequenced DNA samples (n=48) from the 1000 genomes project and compared variant calling accuracy with Illumina genome sequencing. We then sequenced DNA samples from patients with active TB (n=100) from South Africa on a MinION Mk1C and evaluated the relationship between genotypes and pharmacokinetic parameters for INH and RIF.
Results:
The PGx panel achieved 100% concordance with Illumina sequencing in variant identification for the samples from the 1000 Genomes Project. In the clinical cohort, coverage was >100x for 1498/1500 (99.8%) amplicons across the 100 samples. One third (33%) of participants were identified as slow, 47% were intermediate and 20% were rapid isoniazid acetylators. Isoniazid clearance was significantly impacted by acetylator status (p<0.0001) with median (IQR) clearances of 11.2 L/h (9.3–13.4), 27.2 L/h (22.0–31.7), and 45.1 L/h (34.1–51.1) in slow, intermediate, and rapid acetylators. Rifampin clearance was 17.3% (2.50–29.9) lower in individuals with homozygous AADAC rs1803155 G>A substitutions (p=0.0015).
Conclusion:
Targeted sequencing can enable detection of polymorphisms influencing TB drug metabolism on a low-cost, portable instrument to personalize dosing for TB treatment or prevention.
Keywords: isoniazid, Nanopore, NAT2, pharmacogenomics, targeted sequencing, tuberculosis
Summary:
This manuscript describes the development and validation of Nanopore sequencing panel to detect host pharmacogenomic markers to guide personalized drug dosing for treatment or prevention of tuberculosis.
This article has an online data supplement, which is accessible from this issue’s table of content online at www.atsjournals.org
INTRODUCTION
Tuberculosis (TB) continues to be a major cause of morbidity and mortality worldwide. Standardized dosing of anti-tubercular drugs is effective in tuberculosis treatment and prevention, but may result in variable plasma drug levels and risk serious drug-related toxicities1,2. Studies have shown that a substantial proportion of patients treated for active TB experience at least one type of ADR (35%–68%), treatment failure (3%) or relapse (6–10%) within two years3,4,5,6. Liver enzyme elevations and drug-induced liver injury (DILI) are the most common adverse effects, affecting up to 30% of patients undergoing standard therapy7,8,9,10. Interindividual drug pharmacokinetic (PK) variation also affects treatment response. In a clinical cohort in South Africa, individuals who had any plasma drug concentration below target levels had 14-fold increased risk of microbiological failure, death, or relapse11,12,13.
A growing body of literature has identified mutations in genes encoding anti-TB drug-metabolizing enzymes that explain substantial PK variation and predict treatment outcomes and risk of adverse events.14 15. Mutations in N-acetyltransferase-2 (NAT2) and cytochrome P450 2E1 (CYP2E1) genes are known to affect metabolism and clearance of isoniazid16. Polymorphisms in the NAT2 gene explain up to 88% interindividual pharmacokinetic variability of INH17,18. Based on mutations in the NAT2 gene, individuals can be classified into three phenotypes—rapid, intermediate, and slow acetylators. Rapid acetylators typically have the lowest, while slow acetylators have highest plasma INH concentrations19. CYP2E1 gene brings about conversion of acetyl hydrazine to reactive metabolites , which may result in hepatotoxicity20. Patients with CYP2E1 RsaI polymorphism are significantly less likely to experience hepatotoxicity than those with the wild-type (*1A/*1A) genotype21,22. Associations between RIF clearance and mutations in drug transporter gene SLCO1B1 and arylacetamide deacetylase (AADAC) have also been reported23,24. A study in South Africa found that patients with mutation in the SLCO1B1 gene decreased RIF AUC.25 Mutations in cytochrome P450 gene CYP3A5 are associated with faster LZD clearance, putting patients at higher risk of underexposure when treated at standard dose 26,27 In another study on South Africans treated for drug-resistant TB, CYP3A5*3 haplotype was associated with slower BDQ clearance28.
Modification of anti-TB drug doses based on pharmacogenomic data can improve PK target attainment, reduce toxicity risk, and improve treatment outcomes. Observational studies have shown that INH dose modifications enabled rapid and slow acetylators to achieve INH AUC targets comparable to those of intermediate acetylators29. A randomized trial of PGx-guided INH dosing among patients with active TB found that DILI was eliminated in slow acetylators (0% versus 78% in the standard dosing arm) and early treatment failure was reduced in rapid acetylators (15 vs 40% in standard dosing arm).30
Despite this evidence, pharmacogenomic-guided dosing is not widely used in treatment of TB. A major barrier to its implementation is the lack of scalable assays that can be performed quickly in facilities where TB is treated. At present, pharmacogenomic testing for NAT2 and other relevant genes is typically only available in select reference laboratories and is often performed using expensive equipment that is not widely available in clinical laboratories, particularly in low- and middle-income countries where the majority of TB cases occur. In this study, we developed and validated a multiplex targeted sequencing-based panel to detect pharmacogenomic markers for INH, RIF, LZD and BDQ for use on Nanopore MinION sequencers, which are low-cost instruments that are increasingly accessible worldwide. We further validated our panel in a cohort of patients with active TB undergoing treatment, demonstrating utility to identify pharmacogenomic determinants of drug metabolism.
METHODS
Selection of pharmacogenomic markers
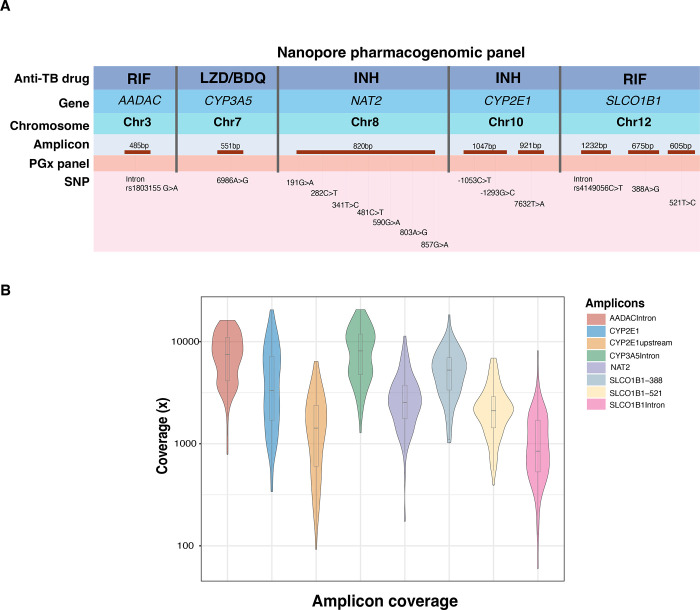
We searched published literature for pharmacogenetic markers of metabolism for drugs that are recommended by the World Health Organization for treatment of TB, including multidrug-resistant or RIF-resistant TB (MDR/RR-TB). We selected 15 well characterized single nucleotide polymorphisms (SNP) for which high quality studies had demonstrated associations with pharmacokinetic parameters, adverse events, or treatment outcomes18–28. The 15 SNPs, which had pharmacogenomic associations with INH, RIF, LZD or BDQ, occurred in five genes: N-acetyltransferase 2 (NAT2), cytochrome P450 family 2 subfamily E member 1 (CYP2E1), solute carrier organic anion transporter family member 1B1 (SLCO1B1), arylacetamide deacetylase (AADAC) and cytochrome P450 family 3 subfamily A member 5 (CYP3A5)31. These variations included 10 single nucleotide polymorphisms (SNP) located in exons, three in introns and two upstream of an exon (Figure 1A).
Figure 1.
(A) Nanopore PGx panel. The top row contains the anti-TB drugs for which pharmacogenomic associations were identified from published studies. Genes and their location on chromosome are listed in rows two and three. Based on the position of targeted SNPs, targets were divided into eight amplicons corresponding to five genes (red). The amplicons are not scaled to their product length. The last row contains information on the positions in pharmacogenes which were included in the PGx panel. (B) Amplicon coverage in PGx panel: Sequencing coverage (log10 scale) per amplicon in 100 samples from the PK cohort, sequenced on a MinION Mk1C sequencer in a single-tube reaction.
Primer design
To perform targeted sequencing, we used a multiplex strategy that relied on anchored primers We developed an 8-plex panel amplifying regions in five genes. Primers were designed to amplify products between 485bp–1232bp using Beacon Designer. (Supplemental methods, Table E1).
DNA samples for panel development and validation
For the Nanopore pharmacogenomic panel development and validation, 48 purified DNA samples from the 1000 Genomes Project for which Illumina whole genome sequencing data was available were procured from the Coriell Institute for Medical Research, USA31. Additionally, we sequenced oral swabs from healthy volunteers (n=20) using custom PGx panel (supplemental methods).
Single-tube multiplex PCR
A single-tube 8-plex PCR reaction was performed for each sample using the LongAmp® Taq DNA Polymerase (NEB). The final reaction was carried out in a 50μl volume containing 1x LongAmp Taq Reaction Buffer (NEB), 2.5M Betaine (Sigma), 5U LongAmp Taq DNA polymerase. Primer efficiency was first evaluated on a single-plex reaction with subsequent addition of primer sets to evaluate the non-specific binding and inhibition of each primer set by the others. The final PCR conditions were optimized to 2 min of DNA denaturation at 94 °C followed by 30 cycles of amplification as follows: 30 sec at 94 °C for denaturing, 60 sec at 60 °C for primer annealing, 1.5min at 65 °C for extension, followed by 10 min at 65 °C for a final extension. The PCR product was purified with PureLink™ PCR Purification Kit (Thermo Fisher Scientific). The purified PCR product was eluted with 50μl nuclease-free water and quantified with Qubit to evaluate reaction yield.
MinION library preparation and sequencing
For panel development and validation, we sequenced a total of 60 Coriell DNA samples (one Coriell DNA sample at 12 dilutions and additional 48 Coriell DNA samples with different genotypes) and 20 oral swabs samples from healthy individuals. The library was prepared using the SQK-LSK110 Ligation Sequencing Kit (Oxford Nanopore Technologies). Samples were barcoded using a Nanopore PCR barcoding expansion (EXP-PBC096 PCR Barcoding Expansion)followed by DNA repair and end-prep using NEBNext FFPE DNA Repair Mix and NEBNext Ultra II End repair / dA-tailing Module reagents in accordance with manufacturer’s instructions. Adaptor ligation was performed using Adapter Mix F (AMX-F) and Quick T4 Ligase. The samples were sequenced on a MinION Mk1C sequencer (Supplemental methods, Table E2).
Sequencing data analysis
Demultiplexing and real-time basecalling was performed on an in-built software MinKNOW (release 22.08.4) using an onboard basecalling software Guppy (Version 3). The run was set on a high accuracy base calling (cutoff >9). Mapping was done aligning reads to a multi-fasta file containing the concatenated sequences of the genome regions included in the panel. Reference fasta genome was uploaded on Epi2ME (version 4.1.3.) cloud and fastq files with passed reads well aligned to the custom genome to generate bam files. To call variants first, the reads were mapped to the reference sequences of target genes included in the gene panel using Minimap2 (v2.26) with default parameters. Aligned reads with a mapping quality score under 60 (MAPQ60) were discarded. Variant calling was performed with Calir3 using default ONT settings. Variants identified were phased using WhatsHap (v1.7).
Samples for clinical validation
Study cohort and ethical approval
Patients with GeneXpert MTB/RIF-confirmed RIF-susceptible pulmonary TB were recruited at the Ubuntu HIV/TB Clinic, Site B, Khayelitsha, South Africa (University of Cape Town Faculty of Health Sciences Human Research Ethics Committee approval 568/2012) as a part of a larger study. Whole blood samples (n=100) from a subcohort of this study who were invited to participate in a nested pharmacokinetic study between July 2013 and April 2014 were used for pharmacogenomic validation32. All patients provided written consent prior to participation. Detailed sociodemographic data, past TB treatment history, and comorbidity data were collected. Weight band-based dosing was used in line with WHO guidelines (Table 1).
Table 1.
Clinical characteristics of tuberculosis pharmacokinetic cohort
| PK cohort (N= 100) | |
|---|---|
|
| |
| Clinical characteristic | |
| Female sex | 43 |
| Age in years, median (IQR) | 33 (29–40) |
| Smear grade at baseline | |
| 3+ | 24 |
| 2+ | 22 |
| 1+ | 20 |
| Scanty/Negative | 34 |
| Baseline time to culture positivity (days), median (IQR) | 10 (7–14) |
| Extensive radiological disease at baseline | 71 |
| Cavities at baseline | 52 |
| Smoking history | |
| Current | 24 |
| Previous | 27 |
| Never | 49 |
| Alcohol consumption | 37 |
| Retreatment | 39 |
| Type 2 diabetes mellitus | 4 |
| BMI at PK study (kg/m2), median (IQR) | 21.5 (20–23) |
| Albumin concentration at PK study (g/liter), median (IQR) | 38 (34–40) |
| Dose (mg/kg), median (range) | |
| Rifampin | 10 (7–11.5) |
| Isoniazid | 5 (3.5–6) |
| Participants reporting side effects of TB treatment | 35 (35) |
Numerical values reflect number of participants with the characteristic unless otherwise specified.
Pharmacokinetic data
A description of the sampling, therapeutic drug monitoring and pharmacokinetic analysis for this cohort has previously published32. Briefly, pharmacokinetic sampling was carried out for RIF and INH 7 to 8 weeks after initiation of anti-TB therapy. Blood samples were obtained immediately before (pre-dose) and 1, 2, 3, 4, 6, and 8 hours after drug ingestion. They were immediately placed on ice, and plasma was separated by centrifugation within 30 minutes before storage at −80°C until analysis. The storage tubes containing the plasma samples were transferred on dry ice to the Clinical Pharmacology Laboratory at the University of Cape Town, where drug concentrations were determined using validated liquid chromatography tandem mass spectrometry (LC-MS/MS) methods35.
Whole blood samples from 100 TB positive patients for which pharmacokinetic data was available were collected in citrate tubes and stored in −80°C until used. DNA was extracted from 100ul whole blood samples using DNeasy Blood & Tissue Kit (Qiagen) and eluted in 50ul nuclease-free water. Approximately 50ng of purified DNA was used for targeted sequencing using custom Nanopore PGx panel as described above.
Haplotype labelling
For INH pharmacogenomic analysis, phased NAT2 haplotypes for PK cohort derived from Nanopore sequencing were labelled based on seven canonical SNPs following an international consensus nomenclature to interpret acetylator phenotype33. The CYP2E allele nomenclature was quoted based on the Human Cytochrome P450 Allele Nomenclature Committee tables34
Statistical analyses
Previously developed population pharmacokinetic models were used to test the effect of polymorphisms or acetylator type on clearance and bioavailability within the models using Monolix (Version 2023R1; Lixoft SAS)35. Stepwise covariate selection was performed using a drop in objective function value of >3.84 as a cutoff for inclusion (corresponding with a p<0.05) and an increase of >6.64 as a cutoff for backward elimination (p<0.01). Pre- and postprocessing of data was done in R (Version 4.3.1).
RESULTS
PGx panel performance and coverage per amplicon
To evaluate the panel’s sensitivity and accuracy, we first performed targeted Nanopore sequencing on six samples in duplicates that were obtained by diluting a Coriell DNA sample with known genotype. At all dilutions, we obtained median coverage above the minimum cutoff (>50x) . Coverage from 500ng to 50ng was 850x, 847x, 972x, 1005x, 1066x and 1076x respectively. The median quality score of diluted samples was 12.8 (std dev=0.1), and the median yield for passed reads was 15Mb (std dev=2.4). (Figure E1). We then validated the PGx panel on 48 purified DNA samples from Coriell Institute. Majority of the Coriell samples selected for panel validation were from sub-Saharan Africa (54.1%) and the Americas (25%) and consisted of 41.3% males. We achieved complete coverage of the targeted regions by aligning eight PCR amplicons in the PGx panel. All amplicons were sequenced with coverage depth above the minimum cutoff. The median sequencing depth across eight amplicons in Coriell samples was ~ 2,281x, with 99.7% amplicons above 100x and 90.2% above 500x. Among the eight amplicons, CYP3A5Intron had the highest coverage (median= 6934x; IQR= 2,522–9,534.7), while SLCO1B1Intron had the lowest coverage (median= 779; IQR= 429.5–1,221.5). We observed 100% concordance between variants identified in Nanopore PGx panel and the reference Illumina whole genome sequencing.
We also sequenced 20 DNA samples extracted from oral swabs collected from healthy volunteers to demonstrate the use of oral swab as an alternate non-invasive sampling method for pharmacogenomic testing. We obtained high quality sequencing data for all oral swab samples with the majority of amplicons above 100x coverage (96.8%).
Clinical validation
For clinical validation, we performed targeted Nanopore sequencing on DNA extracted from whole blood samples from active TB patients enrolled in the INH and RIF PK cohort (Table 1). A majority of the participants were of Xhosa ethnicity 98/100 (98%) and 65% of the participants was living with HIV. The median age was 33 years (range= 29–40), and 43% were women. Median quality score of the samples was 13.8 (IQR=13.5–14.0), We obtained full coverage of the targeted regions for every sample, with a coverage depth that exceeded the minimum cutoff (>50x) for all amplicons. The median sequencing depth across eight amplicons in the PK cohort was ~ 2,963x (IQR 1,512–6,156), with 99.8% amplicons above 100x and 93.6% above 500x. (Figure 1B).
INH and RIF Pharmacogenomic associations
We obtained read depth above 100x for variant alleles at all positions. A total 253 homozygous mutant and 353 heterozygous alleles were detected in 100 samples at 15 genomic positions. The frequency of homozygous wildtype, homozygous alternate and heterozygous variant alleles is shown in Figure 2A and Table 2.
Figure 2.
(A) Distribution of homozygous wildtype (purple), homozygous alternate (blue) and heterozygous alleles (yellow) at 15 polymorphic sites in active TB patients (n=100) from PK cohort sequenced on MinION sequencer. (B) NAT2 haplotypes in red are slow acetylator types, those in green are rapid acetylator haplotypes. Connections in red indicate two slow acetylator haplotypes, those in green indicate two rapid haplotypes, and those in yellow indicate one rapid and one slow haplotype (intermediate acetylation)
Table 2.
Variant calling summary of fifteen PGx panel markers for 100 clinical cohort samples analyzed on a Nanopore MinION sequencer.
| Pharmacogene | SNP identifier | SNP | WT# (%) | MUT* (%) | HET $ (%) | Genotype quality, median | Read depth, median |
|---|---|---|---|---|---|---|---|
|
| |||||||
| NAT2 | rs1801279 | 191G>A | 87 | 2 | 11 | 20.0 | 4801.0 |
| NAT2 | rs1041983 | 282C>T | 41 | 17 | 42 | 22.0 | 5180.0 |
| NAT2 | rs1801280 | 341T>C | 57 | 4 | 39 | 23.0 | 4775.0 |
| NAT2 | rs1799929 | 481C>T | 65 | 3 | 32 | 21 | 4976 |
| NAT2 | rs1799930 | 590G>A | 54 | 4 | 42 | 21.0 | 5339.0 |
| NAT2 | rs1208 | 803A>G | 29 | 21 | 50 | 20.0 | 4715.0 |
| NAT2 | rs1799931 | 857G>A | 99 | 0 | 1 | 23.0 | 6457.0 |
| CYP2E1 | rs3813867 | −1293G>C | 93 | 0 | 7 | 21.0 | 5377.0 |
| CYP2E1 | rs2031920 | −1053C>T | 100 | 0 | 0 | NA | NA |
| CYP2E1 | rs6413432 | 7632T>A | 90 | 0 | 10 | 20.0 | 8377.0 |
| SLCO1B1 | rs4149032 | C>T | 0 | 70 | 30 | 24.0 | 1721.0 |
| SLCO1B1 | rs4149056 | 521T>C | 100 | 0 | 0 | NA | NA |
| SLCO1B1 | rs2306283 | 388A>G | 2 | 66 | 32 | 24.0 | 4979.0 |
| AADAC | rs1803155 | G>A/T | 2 | 65 | 33 | 23.0 | 8047.0 |
| CYP3A5 | rs776746 | 6986A>G | 75 | 1 | 24 | 21.0 | 8053.0 |
Wildtype;
Homozygous alternate;
Heterozygous
Based on the international consensus nomenclature, participants were classified as slow (33/100; 33%), intermediate (47/100; 47%) and rapid (20/100; 20%) acetylators. Demographic and clinical characteristics did not differ by acetylator type (Table 3). The NAT2 haplotype distribution for 100 PK samples is provided in Figure 2B. INH clearance rates were lowest in slow acetylators (median,11.2 L/h; IQR, 9.3–13.4), moderate in intermediate acetylators (27.2 L/h; 22.0–31.7), and highest in fast acetylators (45.1 L/h; 34.1–51.1) (Table 3; Figure 3A). The area under the plasma drug concentration-time curve (AUC) for 0–24h was lowest for rapid acetylators (median 5.80 mg*h/L; IQR, 4.38–9.48), moderate for intermediate acetylators (10.7 mg*h/L; 7.94–14.6) and highest in slow acetylators (23.2 mg*h/L; 18.3–30.9) (Table 3; Figure 3B). NAT2 acetylator status had a significant effect on INH clearance (dOFV=105.5, p<0.0001) (Figure E2). In addition, the effect of HIV, that was previously in the model, was now no longer significant after backward elimination. Individuals who were slow acetylators were more likely to report side effects than intermediate or fast acetylators (52% vs 27%, p=0.027).
Table 3.
Demographic and clinical characteristics, and INH pharmacokinetic parameter estimates, by NAT2 acetylator status
| Slow (n=33) | Intermediate (n=47) | Rapid (n=20) | p-value | |
|---|---|---|---|---|
|
|
||||
| Age (years) | 33.4 (28.9–39.0) | 32.4 (30.3–40.6) | 29.5 (27.1–41.6) | 0.44 |
| Sex, female, n (%) | 16 (48) | 19 (40) | 8 (40) | 0.739 |
| Albumin | 39 (36.0–42.0) | 39.0 (34.0–43.0) | 39.5 (36.3–76.3) | 0.785 |
| Days on TB treatment, median (IQR) | 56.0 (53.0–57.0) | 56.0 (52.0–60.5) | 54.5 (48.8–58.0) | 0.455 |
| HIV infected, n (%) | 8 (24) | 19 (40) | 8 (40) | 0.286 |
| BMI (kg/m2), median (IQR) | 22 (19.3–23.0) | 22.0 (19.3–23.0) | 21.5 (19.8–23.0) | 0.532 |
| Isoniazid PK, median (IQR) | ||||
| Clearance (L/hr) | 11.1 (9.2, 12.9) | 26.2 (20.5, 32.1) | 42.0 (33.5, 52.8) | <0.0001 |
| AUC (mg*hr/L) | 23.1 (18.2, 30.3) | 10.7 (7.9, 14.6) | 5.9 (4.4, 9.5) | <0.0001 |
| Cmax (mg/L) | 4.5 (3.4, 5.5) | 3.3 (2.6, 4.5) | 2.4 (1.7, 3.5) | <0.0001 |
Figure 3.

Predicted phenotype and INH clearance (A) Predicted NAT2 phenotype and INH clearance in active TB patients (n=100) rapid acetylators (purple) retained plasma drug levels for a shorter period than intermediate (blue) and slow (light blue) predicted phenotypes (p-value= <0.0001). (B) The area under the plasma drug concentration-time curve (AUC) for 0–24hrs was lowest for rapid acetylators (5.80 (4.38–9.48) mg*h/L), moderate for intermediate acetylators (10.7 (7.94–14.6) mg*h/L) and highest in slow acetylators (23.2 (18.3–30.9) mg*h/L)
We also evaluated whether polymorphisms in CYP2E1, part of the downstream INH metabolism pathway, could explain pharmacokinetic variability or were associated with adverse events. Eighty-seven participants were *1A/1A (wildtype) genotype, 7 were *5B/*5A and 6 were *1A/*6. After inclusion of acetylator status, 7632T>A (rs6413432) had a significant effect on INH bioavailability. Participants who were heterozygotes (n=10) had a 23% (2.2–50) higher bioavailability than wild type patients (p = 0.0008). We did not observe any significant associations (p=0.28) between CYP2E1 haplotypes and reported side effects.
For RIF pharmacogenomic analysis, we analyzed three SNP sites in SLCO1B1 (rs4149032 C>T, 388A>G and 521T>C) and one in AADAC (rs1803155). At the rs4149032 position, 70/100 patients were homozygous mutant and 30/100 were heterozygous alleles. We identified 2/100 wildtype, 66/100 homozygous alternate 32/100 heterozygous alleles at 388A>G position. All samples were detected as wild type at 521 T>C position in SLCO1B1 gene. We did not observe any significant associations between SLCO1B1 mutations and RIF bioavailability. RIF clearance was 16.5% (1.30–29.3) lower in individuals who were homozygous alternate for AADAC rs1803155 G>A substitutions (p=0.0015; Figure 4, Figure E3).
Figure 4.
(A) AADAC rs1803155 G>T intron mutation and effect on rifampicin clearance: RIF clearance was 16.5% (1.30–29.3) lower in individuals who were homozygous alternate (purple) for AADAC rs1803155 G>A substitutions (p=0.0015) than heterozygous (blue) and wild type (light blue) (B) The area under the plasma drug concentration-time curve (AUC) for 0–24hrs in RIF: Homozygous alternate (purple) for AADAC rs1803155 G>A substitutions heterozygous (blue) and wild type (light blue)
DISCUSSION
While TB is treatable and preventable, a substantial proportion of patients experience drug associated toxicities, treatment failure and relapse under standardized dosing. For preventive therapy, adverse drug events, which are associated with drug metabolism, are a strong predictor of non-completion36. Pharmacogenomic guided dosing has the potential to reduce the risk of these poor outcomes, with observational studies and a randomized trial demonstrating strong premise for feasibility and effectiveness50. However, a major obstacle to using PGx-guided dosing is the lack of access to pharmacogenomic assays in clinical settings where TB is common. To address this gap, we developed a single-tube targeted sequencing panel on Oxford Nanopore MinION platform to detect mutations associated with the metabolism of INH, RIF, LZD and BDQ for which pharmacogenomic associations were previously reported. We achieved high coverage and read depth for all targets in the panel and found that variant identification was 100% concordant in well characterized reference genomes. As proof of principle, we performed the assays on samples from an active TB clinical cohort in Cape Town, confirming that NAT2 acetylator types strongly predicted INH clearance in this population.
Currently available methods used for detection of mutations in pharmacogenes largely rely on qPCR, restriction fragment length polymorphisms (RFLP), SNP array platforms, single gene Sanger sequencing or larger scale (exome or whole genome) sequencing37. While qPCR methods are rapid and easier to perform, they target only limited number of mutations and provide unphased data and in silico haplotype predictions. One consequence of this is that polymorphisms that important in some populations are sometimes neglected. For example, the G191A (R64Q) SNP is common to the NAT2*14 allele cluster, which is frequent in African and African American individuals but is rarely observed in other populations38, leading it to be left out of the popular NAT2 phasing tool nat2pred 39. One study found no correlation between NAT2 genotype and INH metabolism in individuals of Zulu descent, South Africa; however, the study excluded G191A SNP, leading to a population-specific prediction bias 40. SNP array platforms and whole exome/genome sequencing provide data covering more genes and relevant SNP, but typically require expensive laboratory equipment and are not widely available in clinical facilities in resource-constrained settings.
We previously developed a qPCR-based pharmacogenomic assay on the GeneXpert platform to detect polymorphisms in NAT2 gene to guide INH dosing41. This assay predicted INH metabolism with high accuracy based on 5 canonical SNPs; however, there are constraints to including further targets for a single-tube cartridge-based assay. As TB treatment requires at least three drugs and many drugs have several relevant pharmacogenes and multiple important SNP per gene, an optimal panel will require multiple targets. We identified 15 SNP in 5 genes for which there are compelling pharmacogenomic data for important anti-TB drugs. Our search identified many other SNPs for which data were sparse or conflicting; as further studies confirm or reject associations between these polymorphisms, our assay could be easily expanded to include other targets.
In the current study, which covered all 7 canonical SNPs in NAT2, we found that NAT2 haplotypes were strongly predictive of isoniazid clearance and AUC; clearance was nearly 4 times higher, and AUC 4 times lower, in rapid compared with slow acetylators. Prior studies have demonstrated that increasing isoniazid dosing among rapid acetylators (to 7.5–10 mg/kg) and decreasing it among slow acetylators (to 2.5 mg/kg) can achieve PK targets and reduce adverse events29. Given the diversity of NAT2 acetylator types in this population and globally, testing combined with isoniazid dose modification could confer substantial clinical benefits42. We found a modest effect of rs1803155 G>A substitution in the arylacetamide deacetylase gene, AADAC; homozygous individuals had 17.3% lower clearance than heterozygous and wild type alleles (p=0.0015). AADAC appears to be main enzyme in rifampin deacetylation43, and studies have found it affects rifamycin exposure. Chigutsa et al. previously reported that SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations23. In our study, all participants were heterozygous or homozygous alternate for the rs4149032 polymorphism, so we lacked a reference group of wild type individuals for comparison. The high frequency of rs4149032 polymorphisms in this population, together with data showing that low rifampin AUC is predictive of poor outcomes, add to the growing evidence that higher doses of rifampin may be needed14.
We included pharmacogenetic targets associated with LZD and BDQ toxicity, though these are not first line TB drugs. LZD and BDQ are now both included in the primary WHO recommended regimen for treatment of MDR/RR-TB 44. Additionally, a recent trial demonstrated that an 8-week course including LZD and BDQ for RIF-susceptible TB was non-inferior to standard 6-month therapy, opening the door to potential ultra-short-course regimens45. Both drugs are associated with serious adverse events. Peripheral neuropathy and myelosuppression are common with prolonged courses of LZD and can be treatment limiting. QT prolongation, leading to serious arrythmias, can be seen with BDQ, particularly when used in combination with other QT prolonging drugs including moxifloxacin. Recent studies have found that polymorphisms in CYP3A5 may influence LZD and BDQ clearance. The CYP3A5 *1 haplotype was associated with a nearly six-fold risk of LZD underexposure compared with *3/*3, and the *3 haplotype was associated with slower clearance of BDQ, including 30% lower clearance for homozygous individuals (*3/*3)26,27,28. While we analytically validated our assay to correctly identify these polymorphisms, our clinical cohort did not include individuals receiving these drugs, and further studies are needed to confirm the importance of these variants in diverse populations.
Amplicon-based approaches coupled with MinION sequencing offers several advantages over conventional methods. MinION supports real-time base calling that allows users to stop or pause the run when the output is enough for the analysis. This is highly advantageous when used in clinical settings where quick results are needed to decide treatment, improve prognosis, and guide clinical management. A recent study employed a custom variant-prioritization approach with Nanopore sequencing to rapidly diagnose various disorders in critically ill patients46. In the present study, we performed 24 hour and 48-hour runs, resulting in coverage that was several-fold higher than the required cutoff value. Based on these findings, the run time of the PGx assay can be cut down to a few hours. Targeted Nanopore sequencing has been previously used to detect drug resistant strains in MTB from sputum47 and point-of-care diagnosis of viral and bacterial infections48,59. Another advantage of using the Nanopore sequencing approach is availability of a smaller and cheaper Flongle flowcells (~$90), which produce up to 2.8 Gb of output. We found that we could sequence 50 samples on each Flongle run with sufficient coverage. Since targeted sequencing provides information on the entire gene sequence or the targeted amplicons, novel mutations can be identified in these targets which can provide valuable insights into the evolution of these genes in different populations. Our assay requires single tube PCR amplification and library preparation prior to sequencing on Nanopore; instruments to automate these processes are becoming available, which will be important for amplicon sequencing assays to be implemented in clinical laboratories.
The findings of this study are subject to several limitations. To increase the efficiency of the multiplex assay, we split the target genes into two or more amplicons covering a region of the gene instead of the full length. Due to this, we may have missed novel mutations in those regions. Although, we achieved high coverage for all amplicons, there was moderate variability in coverage across the amplicons. Furthermore, while we developed the custom panel for four anti-TB drugs, PK data was available only for INH and RIF to predict pharmacogenomic associations, and there was a lack of genetic diversity at some sites that had been previously identified as important for metabolism of these drugs. Further studies are needed in diverse populations and to assess the impact of CYP3A5 mutations in LZD and BDQ metabolism. Liver enzyme data were not available to assess the effect of CYP2E1 mutations on drug-induced liver injury, which has been previously reported in several studies22.
Amid growing evidence that we can identify individuals at greatest risk of anti-TB drug toxicities and poor treatment outcomes by screening for common genetic variants, there is a need for assays that can be performed near to the point of clinical care in settings where TB is common. We developed and validated a Nanopore amplicon sequencing panel to detect pharmacogenomic markers for key first- and second-line anti-TB drugs. This panel can be further expanded as additional pharmacogenetic markers of TB medications are identified and validated. This assay can be performed on MinION devices, which cost around $1,000 and are increasingly available in public health laboratories in low- and middle-income countries. The movement to optimize TB treatment for each patient will require tools such as this that are scalable for use in settings where TB burden is greatest.
Supplementary Material
At a Glance Commentary.
Scientific Knowledge on the Subject:
Standardized dosing of anti-tubercular (TB) drugs results in variable plasma drug levels, which is associated with adverse drug reactions, poor treatment outcomes, and a risk of relapse. Mutations in genes affecting drug metabolism may explain this pharmacokinetic variability; however, pharmacogenomic (PGx) assays that predict metabolism of anti-TB drugs have been lacking.
What This Study Adds to the Field:
We developed a custom single-tube Nanopore sequencing panel to detect mutations for predicting the metabolism of isoniazid, rifampicin, linezolid, and bedaquiline. Such assays are not currently available in clinical settings to guide drug dosing. We validated our panel on Coriell DNA samples (n=48) and achieved 100% concordance with Illumina whole genome sequencing data. Next, we validated the predicted metabolism of isoniazid and rifampicin based on genotypes derived from the PGx panel in patients with active TB (n=100) undergoing treatment and found strong correlation with INH metabolism. Targeted sequencing on an affordable and portable device can facilitate the identification of polymorphisms that impact TB drug metabolism, allowing for personalized dosing in TB treatment or prevention.
Funding
This study was supported by the National Institutes of Health (R21 AI172182). RJW receives funding from Wellcome (203135) and from the Francis Crick Institute which is supported by Cancer Research UK (FC2112), UK Research and Innovation-Medical Research Council (CC2112) and Wellcome (CC2112). For the purposes of open access, a CC-BY public copyright has been applied to any author-accepted manuscript arising from this submission.
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
Data availability
Data supporting the findings of this manuscript are available in the Supplementary Information files or from the corresponding author upon request.
References:
- 1.Merle CS, Fielding K, Sow OB, Gninafon M, Lo MB, Mthiyane T, et al. OFLOTUB/Gatifloxacin for Tuberculosis Project. A four-month gatifloxacin-containing regimen for treating tuberculosis. N Engl J Med. 2014. Oct 23;371(17):1588–98. [DOI] [PubMed] [Google Scholar]
- 2.Dorman SE, Nahid P, Kurbatova EV, Goldberg SV, Bozeman L, Burman WJ, et al. AIDS Clinical Trials Group and the Tuberculosis Trials Consortium. High-dose rifapentine with or without moxifloxacin for shortening treatment of pulmonary tuberculosis: Study protocol for TBTC study 31/ACTG A5349 phase 3 clinical trial. Contemp Clin Trials. 2020. Mar;90:105938. doi: 10.1016/j.cct.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fei CM, Zainal H, Ali IAH. Evaluation of Adverse Reactions Induced by Anti-Tuberculosis Drugs in Hospital Pulau Pinang. Malays J Med Sci. 2018. Sep;25(5):103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forget EJ, Menzies D. Adverse reactions to first-line antituberculosis drugs. Expert Opin Drug Saf. 2006. Mar;5(2):231–49. [DOI] [PubMed] [Google Scholar]
- 5.Choi H, Park HA, Hyun IG, Kim JH, Hwang YI, Jang SH et al. Incidence and outcomes of adverse drug reactions to first-line anti-tuberculosis drugs and their effects on the quality of life: A multicenter prospective cohort study. Pharmacoepidemiol Drug Saf. 2022. Nov;31(11):1153–1163. [DOI] [PubMed] [Google Scholar]
- 6.Luzze H, Johnson DF, Dickman K, Mayanja-Kizza H, Okwera A, Eisenach K, et al. Tuberculosis Research Unit. Relapse more common than reinfection in recurrent tuberculosis 1–2 years post treatment in urban Uganda. Int J Tuberc Lung Dis. 2013. Mar;17(3):361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell I, Wendon J, Fitt S, Williams R. Anti-tuberculous therapy and acute liver failure. Lancet. 1995. Mar 4;345(8949):555–6. [DOI] [PubMed] [Google Scholar]
- 8.Adverse Drug Reactions Related to Treatment of Drug-Susceptible Tuberculosis in Brazil: A Prospective Cohort Study. 10.3389/fitd.2021.748310 [DOI] [Google Scholar]
- 9.Yee D, Valiquette C, Pelletier M, Parisien I, Rocher I, Menzies D. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am J Respir Crit Care Med. 2003. Jun 1;167(11):1472–7. [DOI] [PubMed] [Google Scholar]
- 10.Shu CC, Lee CH, Lee MC, Wang JY, Yu CJ, Lee LN. Hepatotoxicity due to first-line anti-tuberculosis drugs: a five-year experience in a Taiwan medical centre. Int J Tuberc Lung Dis. 2013. Jul;17(7):934–9. [DOI] [PubMed] [Google Scholar]
- 11.Gumbo T, Louie A, Liu W, Brown D, Ambrose PG, Bhavnani SM, Drusano GL. Isoniazid bactericidal activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics to predict efficacy in different ethnic populations. Antimicrob Agents Chemother. 2007. Jul;51(7):2329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasipanodya JG, McIlleron H, Burger A, Wash PA, Smith P, Gumbo T. Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J Infect Dis. 2013. Nov 1;208(9):1464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasipanodya JG, Srivastava S, Gumbo T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of antituberculosis therapy. Clin Infect Dis. 2012. Jul;55(2):169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahu RK, Singh K, Subodh S. Adverse Drug Reactions to Anti-TB Drugs: Pharmacogenomics Perspective for Identification of Host Genetic Markers. Curr Drug Metab. 2015;16(7):538–52. [DOI] [PubMed] [Google Scholar]
- 15.Metushi I, Uetrecht J, Phillips E. Mechanism of isoniazid-induced hepatotoxicity: then and now. Br J Clin Pharmacol. 2016. Jun;81(6):1030–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinzig-Schippers M, Tomalik-Scharte D, Jetter A, Scheidel B, Jakob V, Rodamer M, et al. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob Agents Chemother. 2005. May;49(5):1733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du H, Chen X, Fang Y, Yan O, Xu H, Li L et al. Huang W. Slow N-acetyltransferase 2 genotype contributes to anti-tuberculosis drug-induced hepatotoxicity: a meta-analysis. Mol Biol Rep. 2013. May;40(5):3591–6. [DOI] [PubMed] [Google Scholar]
- 18.Zabost A, Brzezińska S, Kozińska M, Błachnio M, Jagodziński J, Zwolska Z, et al. Correlation of N-acetyltransferase 2 genotype with isoniazid acetylation in Polish tuberculosis patients. Biomed Res Int. 2013;2013:853602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson SD, Mitchell JR, Timbrell JA, Snodgrass WR, Corcoran GB 3rd. Isoniazid and iproniazid: activation of metabolites to toxic intermediates in man and rat. Science. 1976. Sep 3;193(4256):901–3. [DOI] [PubMed] [Google Scholar]
- 20.Stephens EA, Taylor JA, Kaplan N, Yang CH, Hsieh LL, Lucier GW et al. Ethnic variation in the CYP2E1 gene: polymorphism analysis of 695 African-Americans, European-Americans and Taiwanese. Pharmacogenetics. 1994. Aug;4(4):185–92. [DOI] [PubMed] [Google Scholar]
- 21.Huang YS, Chern HD, Su WJ, Wu JC, Chang SC, Chiang CH et al. Cytochrome P450 2E1 genotype and the susceptibility to antituberculosis drug-induced hepatitis. Hepatology. 2003. Apr;37(4):924–30. [DOI] [PubMed] [Google Scholar]
- 22.Richardson M, Kirkham J, Dwan K, Sloan DJ, Davies G, Jorgensen AL. CYP genetic variants and toxicity related to anti-tubercular agents: a systematic review and meta-analysis. Syst Rev. 2018. Nov 20;7(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chigutsa E, Visser ME, Swart EC, Denti P, Pushpakom S, Egan D et al. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother. 2011. Sep;55(9):4122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiner M, Gelfond J, Johnson-Pais TL, Engle M, Johnson JL, Whitworth WC et al. Pharmacokinetics/Pharmacodynamics Group of Tuberculosis Trials Consortium. Decreased plasma rifapentine concentrations associated with AADAC single nucleotide polymorphism in adults with tuberculosis. J Antimicrob Chemother. 2021. Feb 11;76(3):582–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiner M, Peloquin C, Burman W, Luo CC, Engle M, Prihoda TJ et al. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob Agents Chemother. 2010. Oct;54(10):4192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheli S, Fusi M, De Silvestri A, Bonini I, Clementi E, Cattaneo D et al. In linezolid underexposure, pharmacogenetics matters: The role of CYP3A5. Biomed Pharmacother. 2021. Jul;139:111631. [DOI] [PubMed] [Google Scholar]
- 27.Lau C, Marriott D, Bui J, Figtree M, Gould M, Chubaty et al. LInezolid Monitoring to Minimise Toxicity (LIMMIT1): a multicentre retrospective review of patients receiving linezolid therapy and the impact of therapeutic drug monitoring. Int J Antimicrob Agents. 2023. Mar 13:106783. [DOI] [PubMed] [Google Scholar]
- 28.Haas DW, Abdelwahab MT, van Beek SW, Baker P, Maartens G, Bradford Y, et al. Pharmacogenetics of Between-Individual Variability in Plasma Clearance of Bedaquiline and Clofazimine in South Africa. J Infect Dis. 2022. Aug 12;226(1):147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donald PR, Parkin DP, Seifart HI, Schaaf HS, van Helden PD, Werely CJ, et al. The influence of dose and N-acetyltransferase-2 (NAT2) genotype and phenotype on the pharmacokinetics and pharmacodynamics of isoniazid. Eur J Clin Pharmacol. 2007. Jul;63(7):633–9. [DOI] [PubMed] [Google Scholar]
- 30.Azuma J, Ohno M, Kubota R, Yokota S, Nagai T, Tsuyuguchi K, et al. Pharmacogenetics-based tuberculosis therapy research group. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy. Eur J Clin Pharmacol. 2013. May;69(5):1091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rockwood N, Meintjes G, Chirehwa M, Wiesner L, McIlleron H, Wilkinson RJ, et al. HIV-1 Coinfection Does Not Reduce Exposure to Rifampin, Isoniazid, and Pyrazinamide in South African Tuberculosis Outpatients. Antimicrob Agents Chemother. 2016. Sep 23;60(10):6050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagena E, Fakis G, Boukouvala S. Arylamine N-acetyltransferases in prokaryotic and eukaryotic genomes: a survey of public databases. Curr Drug Metab. 2008. Sep;9(7):628–60. [DOI] [PubMed] [Google Scholar]
- 34.Sim SC, Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum Genomics. 2010. Apr;4(4):278–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravichandran M, Rajaram M, Munusamy M. Pharmacovigilance of Antitubercular Therapy in Tuberculosis. Cureus. 2022. Feb 4;14(2):e21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sant Anna FM, Araújo-Pereira M, Schmaltz CAS, Arriaga MB, Andrade BB, Rolla VC. Impact of adverse drug reactions on the outcomes of tuberculosis treatment. PLoS One. 2023. Feb 7;18(2):e0269765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alwi ZB. The Use of SNPs in Pharmacogenomics Studies. Malays J Med Sci. 2005. Jul;12(2):4–12. [PMC free article] [PubMed] [Google Scholar]
- 38.Sabbagh A, Langaney A, Darlu P, Gérard N, Krishnamoorthy R, Poloni ES. Worldwide distribution of NAT2 diversity: implications for NAT2 evolutionary history. BMC Genet. 2008. Feb 27;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabbagh A, Darlu P, Vidaud M. Evaluating NAT2PRED for inferring the individual acetylation status from unphased genotype data. BMC Med Genet. 2009. Dec 31;10:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mthiyane T, Millard J, Adamson J, Balakrishna Y, Connolly C, Owen A, et al. N-Acetyltransferase 2 Genotypes among Zulu-Speaking South Africans and Isoniazid and N-Acetyl-Isoniazid Pharmacokinetics during Antituberculosis Treatment. Antimicrob Agents Chemother. 2020. Mar 24;64(4):e02376–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma R, Patil S, Zhang N, Moreira FMF, Vitorio MT, Santos ADS, et al. A Rapid Pharmacogenomic Assay to Detect NAT2 Polymorphisms and Guide Isoniazid Dosing for Tuberculosis Treatment. Am J Respir Crit Care Med. 2021. Dec 1;204(11):1317–1326 [DOI] [PubMed] [Google Scholar]
- 42.Nakajima A, Fukami T, Kobayashi Y, Watanabe A, Nakajima M, Yokoi T. Human arylacetamide deacetylase is responsible for deacetylation of rifamycins: rifampicin, rifabutin, and rifapentine. Biochem Pharmacol. 2011. Dec 1;82(11):1747–56. [DOI] [PubMed] [Google Scholar]
- 43.Francis J, Zvada SP, Denti P, Hatherill M, Charalambous S, Mungofa S, , et al. A Population Pharmacokinetic Analysis Shows that Arylacetamide Deacetylase (AADAC) Gene Polymorphism and HIV Infection Affect the Exposure of Rifapentine. Antimicrob Agents Chemother. 2019. Mar 27;63(4):e01964–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.WHO consolidated guidelines on drug-resistant tuberculosis treatment. [Google Scholar]
- 45.Paton NI, Cousins C, Suresh C, Burhan E, Chew KL, Dalay VB, Lu Q, Kusmiati T, Balanag VM, Lee SL, Ruslami R, Pokharkar Y, Djaharuddin I, Sugiri JJR, Veto RS, Sekaggya-Wiltshire C, Avihingsanon A, Sarin R, Papineni P, Nunn AJ, Crook AM; TRUNCATE-TB Trial Team. Treatment Strategy for Rifampin-Susceptible Tuberculosis. N Engl J Med. 2023. Mar 9;388(10):873–887. doi: 10.1056/NEJMoa2212537..Conradie F, Bagdasaryan TR, Borisov S, Howell P, Mikiashvili L, Ngubane N, et al. Bedaquiline-Pretomanid-Linezolid Regimens for Drug-Resistant Tuberculosis. N Engl J Med. 2022 Sep 1;387(9):810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorzynski JE, Goenka SD, Shafin K, Jensen TD, Fisk DG, Grove ME, et al. Ultrarapid Nanopore Genome Sequencing in a Critical Care Setting. N Engl J Med. 2022. Feb 17;386(7):700–702. [DOI] [PubMed] [Google Scholar]
- 47.Mariner-Llicer C, Goig GA, Zaragoza-Infante L, Torres-Puente M, Villamayor L, Navarro D, Borras R, et al. Accuracy of an amplicon-sequencing nanopore approach to identify variants in tuberculosis drug-resistance-associated genes. Microb Genom. 2021. Dec;7(12):000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Lewandowski K, Lumley S, Pullan S, Vipond R, Carroll M, et al. Detection of Viral Pathogens With Multiplex Nanopore MinION Sequencing: Be Careful With Cross-Talk. Front Microbiol. 2018. Sep 19;9:2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017. Jan;72(1):104–114. [DOI] [PubMed] [Google Scholar]
- 50.Rens NE, Uyl-de Groot CA, Goldhaber-Fiebert JD, Croda J, Andrews JR. Cost-effectiveness of a Pharmacogenomic Test for Stratified Isoniazid Dosing in Treatment of Active Tuberculosis. Clin Infect Dis. 2020. Dec 15;71(12):3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available in the Supplementary Information files or from the corresponding author upon request.